Abstract
New Delhi metallo-β-lactamase-13 (NDM-13) is an NDM variant that was first identified in 2015 and has not been detected in Salmonella species prior to this study. Here we describe the first identification of a Salmonella Rissen strain SR33 carrying bla NDM-13. The aim of this study was to molecularly characterize SR33’s antimicrobial resistance and virulence features as well as investigate the genetic environment of bla NDM-13. The Salmonella Rissen SR33 strain was isolated from a patient with fever and diarrhea. SR33 belonged to ST469, and it was found to be multidrug-resistant (MDR) and to carry many virulence genes. Phylogenetic analysis showed that SR33 shared a close relationship with most of the Chinese S. Rissen ST469 strains. bla NDM-13 was located in a transmissible IncI1 plasmid pNDM13-SR33. Sequence analysis of bla NDM-13-positive genomes downloaded from GenBank revealed that a genetic context (ΔISAba125-bla NDM-13-ble MBL-trpF) and a hybrid promoter (consisting of −35 sequences provided by ISAba125 and −10 sequences) were conserved. ISAba125 was truncated by IS1294 in three plasmids carrying bla NDM-13, including pNDM13-SR33. To our knowledge, this is the first report of bla NDM-13 carried by Salmonella. The emergence of bla NDM-13 in a clinical MDR S. Rissen ST469 strain highlights the critical need for monitoring and controlling the dissemination of bla NDM-13. bla NDM-13 carried by a transmissible IncI1 plasmid may result in an increased risk of bla NDM-13 transmission. IS1294 may be involved in the movement of bla NDM-13.
Keywords: bla NDM-13 , Salmonella Rissen, ST469, ISAba125 , IS1294
Introduction
Carbapenems have been used for decades to treat severe gram-negative bacterial infections, particularly in resistant and multidrug-resistant (MDR) infections (Hansen, 2021). According to the World Health Organization’s Global Priority List, carbapenem-resistant Enterobacteriaceae (CRE) pose a growing threat to public health worldwide (Tacconelli et al., 2018). New Delhi metallo-β-lactamase (NDM) is a subclass B1 metallo-β-lactamase that is capable of hydrolyzing almost all β-lactams including carbapenems (Yong et al., 2009; Nordmann et al., 2011). Worse still, clinically available β-lactamase inhibitors are ineffective in preventing carbapenem hydrolysis by NDM enzymes (Wu et al., 2019). NDM-positive strains are usually resistant to most of antimicrobial agents, due to coexistence of other resistance mechanisms (Nordmann et al., 2011), leading to a variety of infections that are associated with high mortality (Guducuoglu et al., 2017). Since NDM-1 was first identified in clinical isolates in India in 2008 (Yong et al., 2009), 31 variants have been reported worldwide, representing a significant challenge for public health and clinical management (Moellering, 2010; Dortet et al., 2014; Li et al., 2021). Of these, NDM-13 is a variant that has two amino acid substitutions (D95N and M154L) compared with NDM-1, resulting in the increased hydrolytic activity against cefotaxime (Shrestha et al., 2015). NDM-13 has been detected in five Escherichia coli strains obtained from Nepal (n = 1) (Shrestha et al., 2015), China (n = 3) (Lv et al., 2016), and Korea (n = 1) (Kim et al., 2019). Here we aim to characterize a bla NDM-13-positive Salmonella Rissen strain SR33 isolated in China. To our knowledge, this is the first report of bla NDM-13 detected in Salmonella.
Materials and methods
Bacterial strain
Strain SR33 was isolated from a fecal sample of an old patient. This patient was hospitalized due to occasional fever and diarrhea. During hospitalization, cefixime was ineffective against this infection, but it improved after treatment with levofloxacin. SR33 was identified by the VITEK-2 COMPACT automatic microbial identification system (bioMérieux, Marcy-l’Étoile, France), and its serotype was confirmed by slide agglutination technique (Kauffmann-White-Le Minor scheme) (Grimont and Weill, 2007)
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) for imipenem, ertapenem, ceftazidime, ceftriaxone, cefepime, amoxicillin/clavulanic acid, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, levofloxacin, ampicillin, tetracycline, ciprofloxacin, chloramphenicol, and azithromycin were determined by broth microdilution following the CLSI guidelines, and MIC results were interpreted according to the CLSI breakpoints (Wayne, 2021).
Whole-genome sequencing and bioinformatics analysis
The genomic DNA of SR33 was extracted by the bacterial genomic DNA extraction kit (Tiangen, Beijing, China) and sequenced on an Oxford Nanopore platform (Novogene, Tianjin, China). Sequence reads were assembled by Unicycler 0.4.8 (Wick et al., 2017) and annotated by Prokka 1.14.5 (Seemann,, 2014). The serotype was further confirmed by SISTR 1.1.1 (Yoshida et al., 2016), and the sequence type (ST) was determined using MLST 2.18.0 (Larsen et al., 2012). The distance matrix based on the core-genome single-nucleotide polymorphism (SNP) profiles of 37 Chinese S. Rissen ST469 isolates was generated using Parsnp and HarvestTools (Treangen et al., 2014). The phylogenetic tree was constructed by MEGA X (Kumar et al., 2018). Resistance genes and plasmid replicons were identified using Abricate (https://github.com/tseemann/abricate) with the ResFinder (Zankari et al., 2012) and PlasmidFinder (Carattoli et al., 2014) databases, respectively. The filtering criteria of antimicrobial resistance genes were >90% identity and >90% coverage. The virulence genes were analyzed by the database of Virulence Factors of Pathogenic Bacteria (VFDB) using BLASTn with a threshold of >70% identity and >70% coverage (Chen et al., 2016). The presence of Salmonella pathogenicity islands (SPIs) was explored by SPIFinder (https://cge.cbs.dtu.dk/services/SPIFinder/). Circular maps of plasmids were generated using the BLAST Ring Image Generator (BRIG) tool (Alikhan et al., 2011). Transposon and insertion sequence (IS) elements were scanned using the ISFinder database (Siguier et al., 2006). BLASTn (Altschul et al., 1990) was used to determine the identity of the genetic environment between NDM-13-positive sequences. The genetic environment was visualized by EasyFig (Sullivan et al., 2011).
Plasmid conjugation experiments
Transferability of plasmid harboring bla NDM-13 was assessed by the conjugation experiment, using rifampin-resistant E. coli C600 as the recipient strain. Transconjugants were selected on Luria-Bertani agar plates containing rifampin (100 µg/ml) and imipenem (2 µg/ml). Transconjugants containing the bla NDM-13 gene were verified by PCR sequencing (forward primer sequence: ATGGAATTGCCCAATATTATGCAC and reverse primer sequence: TCAGCGCAGCTTGTCGGC). The antimicrobial susceptibility of the transconjugant was confirmed by the broth microdilution method.
Nucleotide sequence accession number
The whole-genome sequence of SR33 has been submitted to the GenBank database with accession numbers CP092911–CP092914. The nucleotide sequence of plasmid pNDM13-SR33 has been deposited under accession number CP092912.
Results
Antimicrobial susceptibility testing and antimicrobial resistance genes
As shown in Table 1, SR33 was multidrug resistant to all tested β-lactams, trimethoprim/sulfamethoxazole, and tetracycline and was susceptible to quinolones (levofloxacin and ciprofloxacin), azithromycin, and chloramphenicol. In addition to bla NDM-13, SR33 carried genes that mediate resistance to β-lactams (bla TEM-1), bleomycin (ble MBL), streptomycin (aadA1, aadA2), chloramphenicol (cmlA1), trimethoprim (dfrA12), sulfonamide (sul3), and tetracycline [tet(A)]. The information of resistance genes detected in SR33 is listed in Supplementary Table S1.
Table 1.
MIC values of antimicrobials for SR33 and its transconjugant.
| SR33 | C600 | SR33-C600 | ||||
|---|---|---|---|---|---|---|
| Antimicrobials | MIC values (µg/mL) | Interpretation | MIC values (µg/mL) | Interpretation | MIC values (µg/mL) | Interpretation |
| Imipenem | ≥16 | R | ≤1 | S | ≥16 | R |
| Ertapenem | ≥8 | R | ≤0.5 | S | ≥8 | R |
| Ceftazidime | ≥64 | R | ≤4 | S | ≥64 | R |
| Ceftriaxone | ≥64 | R | ≤1 | S | ≥64 | R |
| Cefepime | 16 | R | ≤2 | S | 16 | R |
| Amoxicillin/clavulanic acid | ≥32 | R | ≤4 | S | ≥32 | R |
| Piperacillin/tazobactam | ≥128 | R | ≤16 | S | ≥128 | R |
| Trimethoprim/sulfamethoxazole | ≥320 | R | ≤20 | S | ≤20 | S |
| Levofloxacin | ≤0.12 | S | ≤0.5 | S | 0.5 | S |
| Ampicillin | ≥32 | R | ≤8 | S | ≥32 | R |
| Tetracycline | ≥16 | R | ≤4 | S | ≤4 | S |
| Ciprofloxacin | ≤0.06 | S | ≤0.25 | S | ≤0.25 | S |
| Chloramphenicol | ≤8 | S | ≤8 | S | ≤8 | S |
| Azithromycin | ≤16 | S | ≤16 | S | ≤16 | S |
MIC, minimum inhibitory concentration; R, resistant; I, intermediate; S, sensitive.
Whole-genome sequencing (WGS) showed that bla NDM-13 and ble MBL were located on an IncI1 plasmid designated as pNDM13-SR33, which is 88,258 bp in length with an average GC content of 50.37%. The other resistance genes were found on the chromosome. pNDM13-SR33 was successfully self-transferred into C600, and the transconjugant SR33-C600 was resistant to all tested β-lactams (Table 1).
Characterization of the SR33 strain and phylogenetic analysis of Chinese S. Rissen ST469 isolates
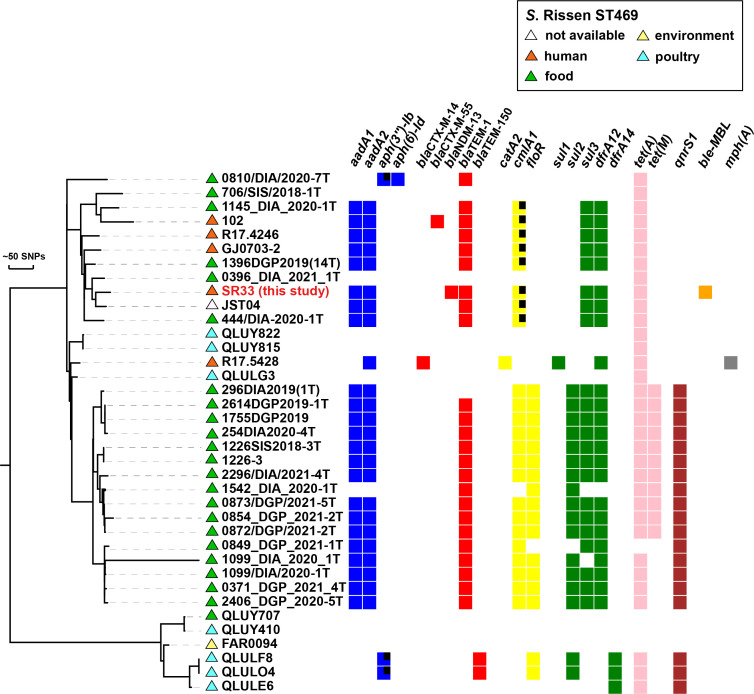
The serotype and sequence type of SR33 were determined to be serovar Rissen and ST469. Phylogenetic analysis of SR33 with other 36 Chinese S. Rissen ST469 isolates (retrieved and downloaded from EnteroBase in February 2022, https://enterobase.warwick.ac.uk/species/index/senterica) revealed that SR33 differed from the other isolates by 41–418 SNPs (Figure 1). The information of these strains is listed in Supplementary Table S2. Besides, these strains were mainly isolated from food, poultry, and humans. Meanwhile, the majority of Chinese S. Rissen ST469 strains were MDR. The drug resistance profiles of these MDR strains were similar, and common drug resistance genes include aadA1, aadA2, bla TEM-1, cmlA1, sul3, dfrA12, and tet(A). Since the common drug resistance genes in SR33 were located on chromosomes, and 29/37 Chinese S. Rissen ST469 isolates did not carry resistance plasmids, we speculated that the antimicrobial resistance genes were mainly located on the chromosomes of these closely related MDR strains.
Figure 1.
Phylogenetic distribution of antimicrobial resistance genes of SR33 of this study and other Chinese S. Rissen ST469 isolates. Antimicrobial resistant genes are represented by colored squares, genes with partial deletions are marked with black squares. The source of each isolate is shown with colored triangles. Bars represent unit distance of 50 SNPs.
Salmonella pathogenicity islands and virulence-associated genes
According to SPIFinder, SR33 contained SPI-1 to SPI-5, SPI-8, and SPI-9. All VFDB-annotated genes are listed in Table 2. Based on the annotation of the VFDB database, SR33 harbored 124 virulence genes. The virulence genes are involved in adhesion systems, iron uptake, magnesium uptake, macrophage, flagella, type III secretion systems (T3SS), and serum resistance.
Table 2.
Virulence-associated genes in SR33.
| VF classes | Virulence factors | Genes |
|---|---|---|
| Fimbrial adherence determinants | Agf (thin aggregative fimbriae/curli) | csgABCDEFG, steABC |
| Lpf (long polar fimbriae) | lpfABCE | |
| Type 1 fimbriae | fimCDFHI | |
| Non-fimbrial adherence determinants | MisL | misL |
| SinH | sinH | |
| Iron uptake | Enterobactin | entABCES, fepABCDG |
| Salmochelin | iroBCN | |
| Magnesium uptake | Magnesium uptake/transporter | mgtBC |
| Macrophage inducible gene | Mig-14 | mig-14 |
| Motility | Flagella | cheWY, flgGH, fliAGMP |
| Secretion system | T3SS (SPI-1 encoded) | invABCEFGHIJ, orgABC, prgHIJK, sicAP, sipD, spaOPQRS |
| T3SS-1 translocated effectors | avrA, sipABC/sspABC, sopABDE2, sptP, slrP | |
| T3SS (SPI-2 encoded) | ssaCDEGHIJKLMNOPQRSTUV, sscAB, sseABCDE | |
| T3SS-2 translocated effectors | pipBB2, sifABH, sopD2, sseFGJK1K2L, spiC/ssaB | |
| Serum resistance | OmpA (Outer membrane protein A) | ompA |
| Others | Lipooligosaccharide | gmhA/lpcA |
| Lipopolysaccharide | gtrAB |
VF, virulence factors.
Plasmid analysis of bla NDM-13-positive isolates
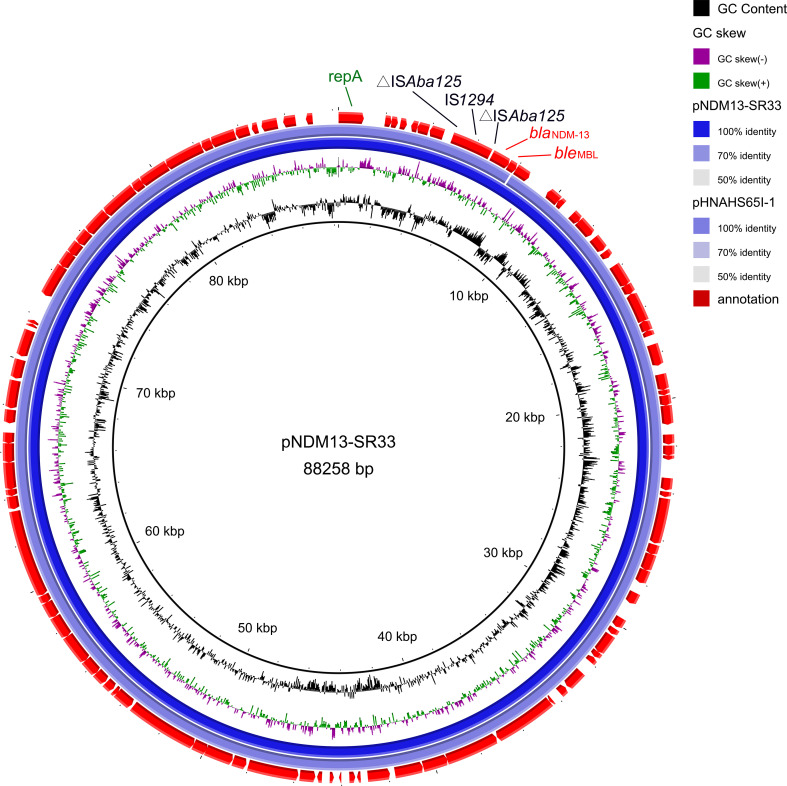
NDM-13 has been identified in plasmids of three E. coli stains, including an IncX3 plasmid pNDM13-DC33(accession no. KX094555), an IncFIB plasmid pSECR18-0956 (accession no. MK157018), and an IncI1 plasmid pHNAHS65I-1 (accession no. MN219406). Of note, pNDM13-SR33 shared 99% coverage and 100% identity with an IncI1-bla NDM-13 plasmid pHNAHS65I (accession no. MN219406) of E. coli discovered in 2020 (Figure 2), which has a truncated ble MBL.
Figure 2.
Genetic map of pNDM13-SR33 (no. CP092912) and pHNAHS65I-1 (no. MN219406). The repA, IS elements, resistance genes are annotated by green, black, red fonts, respectively.
Comparative analysis of the genetic environment of bla NDM-13
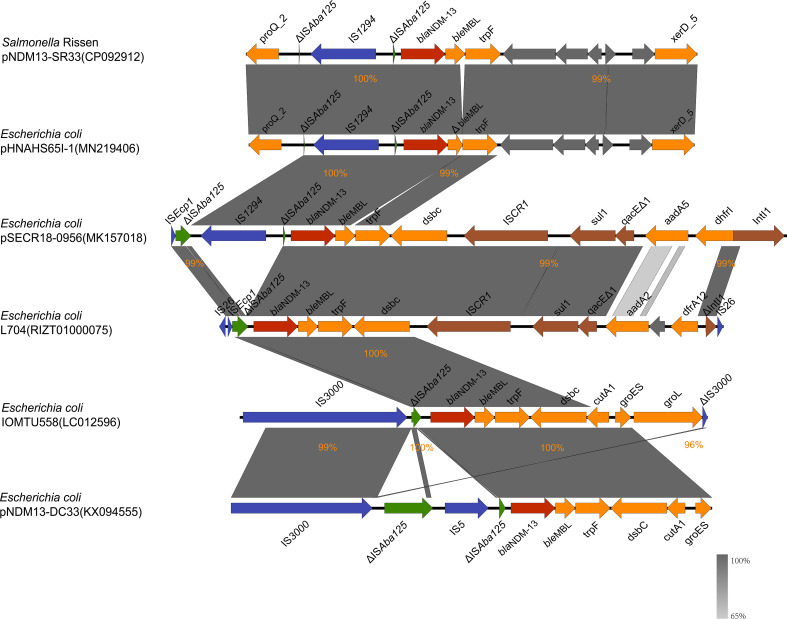
As shown in Figure 3, the bla NDM-13-producing strains shared a conserved genetic structure (ΔISAba125-bla NDM-13-ble MBL-trpF). The conserved region was found involved in various genetic contexts with different insertion sequences. The genetic context of bla NDM-13 in SR33 was highly similar to pHNAHS65I-1 (no. MN219406) with ΔISAba125 truncated by the insertion of an IS1294 upstream, which was also detected in pSECR18-0956 (no. MK157018). In L704 (no. RIZT01000075) and pSECR18-0956 (no. MK157018), the bla NDM-13 region was adjacent to an ISCR1 complex class 1 integron (ISCR1-sul1-qacEΔ1-IntI1). The sequences of L704 and IOMTU558 (accession no. LC012596) were flanked by IS26 and IS3000, respectively. In addition, a cluster (IS3000-ΔISAba125-IS5-ΔISAba125) was found upstream of bla NDM-13 in pNDM13-DC33 (no. KX094555). Moreover, a hybrid promoter (consisting of −35 sequences within the inverted repeat left of ISAba125 and −10 sequences) located upstream of bla NDM-13 was conservative in bla NDM-13-producing strains.
Figure 3.
bla NDM-13 flanking sequence of pNDM13-SR33 (no. CP092912), pHNAHS65I-1 (no. MN219406), L704 (no. RIZT01000075), pSECR18-0956 (no. MK157018), IOMTU558 (no. LC012596), and pNDM13-DC33 (no. KX094555). The bla NDM-13 gene, ISAba125, other insertion sequences, integron elements, and genes encoding hypothetical proteins are shown in red, green, blue, brown, and gray, respectively. The rest of genes are colored in orange. Δ; indicates a truncated gene or mobile element. The percentages signify the genetic identity between these sequences.
Discussion
To date, New Delhi metallo-β-lactamase-13 (NDM-13) has been detected in five E. coli stains with different genetic backgrounds. Here, we report the emergence of an NDM-13-positive Salmonella strain SR33. The serotype of SR33 was determined to be serovar Rissen, which is regarded as one of the 20 most common serovars to cause human salmonellosis (European Food Safety Authority, E.C.f.D.P.a.C, 2017). SR33 was assigned to ST469, an MDR clone that has been reported in multiple countries (Campos et al., 2019).
SR33 was found to be MDR and to harbor nine resistance genes. These resistance genes were consistent with the phenotypes except for cmlA1. SR33 remained sensitive to chloramphenicol, which might be due to the fact that the cmlA1 gene had a sequence deletion of 96 bp. Since SR33 was resistant to all β-lactams and susceptible to quinolones, it explains well why cefixime was ineffective against this infection and levofloxacin was effective.
Based on phylogenetic analysis, SR33 was closely related to the majority of the Chinese S. Rissen ST469 strains downloaded from EnteroBase. Since the available 37 Chinese S. Rissen ST469 isolates were mostly isolated from food, poultry, and humans, it is in agreement with the idea that S. Rissen infection occurs in humans as a zoonosis through food chain transmission (Xu et al., 2020). Therefore, it is possible that this patient had a foodborne infection. Another important finding is that most Chinese S. Rissen ST469 strains were MDR and shared similar drug resistance profiles. Since the antimicrobial resistance genes were mainly located on chromosomes, we should pay close attention to the vertical transmission of MDR S. Rissen ST469 strains. These observations emphasize the necessity of the surveillance of S. Rissen ST469 pathogens.
SPIs are gene clusters located on chromosomes and encode various virulence components (Foley et al., 2008). SR33 contained five important SPIs (SPI-1 to SPI-5) that are correlated with the pathogenesis of Salmonella (Cui et al., 2021) and additional two SPIs (SPI-8, SPI-9). Based on the annotation of the VFDB database, most of the virulence genes carried by SR33 are associated with flagella, type III secretion systems (T3SS), and adhesion systems, which have been demonstrated to play a variety of roles in the pathogenesis of Salmonella (Jajere, 2019). Of these, T3SS is regarded as the most important virulence factor of Salmonella (Lou et al., 2019). In general, MDR strain SR33 possessed important pathogenicity islands and many virulence-associated genes, which highlights the pathogenesis of SR33.
NDM-13 was first identified on the chromosome of E. coli IOMUT558 (ST101) from Nepal (Shrestha et al., 2015), and it was subsequently detected in four E. coli stains, namely, an IncFIB plasmid pSECR18-0956 of SECR18-0956 (ST8499) from Korea (Kim et al., 2019), an IncX3 plasmid pNDM13-DC33 carried by DC33 (ST5138) (Lv et al., 2016), an IncI1 plasmid pHNAHS65I-1 of AHS8C65RI, and L704 strain (the location of bla NDM-13 is unclear) from China. In our study, bla NDM-13 was found in a transmissible IncI1 plasmid pNDM13-SR33 of S. Rissen (ST469). The high coverage and identity between pNDM13-SR33 and pHNAHS65I-1 suggest that cross-species dissemination of bla NDM-13 plasmids had occurred. The bla NDM-carrying plasmids mostly belong to IncX3, IncFII, and IncC replicon types (Wu et al., 2019), indicating that the vector of NDM-13 may be different from the other variants. Previous studies showed that the IncI1 plasmids are often associated with clinically relevant strains (García-Fernández et al., 2008) and it is the major vehicle of extended spectrum β-lactamase (Carattoli et al., 2021). Thus, bla NDM-13 in SR33 carried by an IncI1 transmissible plasmid may result in an increased risk of bla NDM-13 transmission.
Comparative analysis of the bla NDM-13 genetic contents revealed that bla NDM-13 was bracketed by multi-insertional sequences. Of these, ISAba125 was conservative in bla NDM-13-positive isolates. It is consistent with the finding that ISAba125 (intact or truncated) upstream of bla NDM is common in bla NDM genetic contexts (Ahmad et al., 2018; Pérez-Vázquez et al., 2019; Das et al., 2019; Wu et al., 2019), implying a role in the transmission of bla NDM. IS3000, IS26, and IS5 have also been reported to be associated with dissemination of NDM-encoding genes, while the role of IS1294 is still unclear (Zhao et al., 2021; Acman et al., 2022). IS1294 belongs to the IS91 family, and previous reports demonstrated that the disruption of the ISEcp1 element by IS1294 was linked to the promotion of bla CMY-2 (Sidjabat et al., 2014; Tagg et al., 2014) and bla CTX-55 (Pan et al., 2013; Hu et al., 2018) gene dissemination. In this study, ΔISAba125 truncated by IS1294 was found in three bla NDM-13-harboring plasmids including pNDM13-SR33. We thus suspected that IS1294 may be involved in the mobilization and dissemination of bla NDM-13.
Expression of the bla NDM-1 gene is under the control of a hybrid promoter (consisting of −35 sequences within the inverted repeat left of ISAba125 and −10 sequences) located upstream of bla NDM-1 (Poirel et al., 2011). BLASTn analysis revealed that this hybrid promoter was also conservative in bla NDM-13-producing strains. This finding further supports that bla NDM-13 is derived from bla NDM-1 (Lv et al., 2016; Wu et al., 2019).
Conclusion
To the best of our knowledge, this study first reports an NDM-13-producing Salmonella isolate. The emergence of bla NDM-13 in a clinical MDR Salmonella Rissen ST469 strain poses a significant threat to public health. Most of the S. Rissen ST469 strains isolated from China were MDR, which highlights the importance of the surveillance for S. Rissen ST469. The bla NDM-13 carried by a transmissible IncI1 plasmid may cause an increased risk of bla NDM-13 transmission. IS1294 may be involved in the mobilization and dissemination of bla NDM-13.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP092911-CP092914.
Ethics statement
The studies involving human participant were reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Xiamen University. The participant provided his written informed consent to participate in this study.
Author contributions
HX and XL contributed to the conception and design of the study. HX and XM provided this strain. YH and SZ performed laboratory experiments. YH, XM, SZ, and LF analyzed the data. YH wrote the manuscript. XL revised the manuscript. All authors have read and approved the manuscript.
Funding
This study was funded by the Youth Foundation of the National Natural Science Foundation of China (81902104), Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515220153), Basic and Applied Basic Research Foundation of Guangdong Province Natural Science Foundation (2022A1515012481), and Joint Research Projects of Health and Education Commission of Fujian Province (2019-WJ-42).
Acknowledgments
We thank Dr. Kai Zhou (Shenzhen Institute of Respiratory Diseases, The First Affiliated Hospital (Shenzhen People’s Hospital), Southern University of Science and Technology, Shenzhen, China) for his revision of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.936649/full#supplementary-material
References
- Acman M., Wang R., van Dorp L., Shaw L. P., Wang Q., Luhmann N., et al. (2022). Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene bla(NDM). Nat. Commun. 13 (1), 1131. doi: 10.1038/s41467-022-28819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N., Ali S.M., Khan A.U.. (2018). Detection of New Delhi Metallo-β-Lactamase Variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes Isolated from a Neonatal Intensive Care Unit of a North India Hospital: A First Report. Microb. Drug Resist. 24 (2), 161–165. doi: 10.1089/mdr.2017.0038 [DOI] [PubMed] [Google Scholar]
- Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A.. (2011). BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/s0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Campos J., Mourão J., Peixe L., Antunes P. (2019). Non-typhoidal salmonella in the pig production chain: a comprehensive analysis of its impact on human health. Pathogens 8 (1), 19. doi: 10.3390/pathogens8010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Fortini D., García-Fernández A. (2021). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in enterobacteriaceae. Plasmid 118, 102392. doi: 10.1016/j.plasmid.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 (7), 3895–3903. doi: 10.1128/aac.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zheng D., Liu B., Yang J., Jin Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44 (D1), D694–D697. doi: 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Wang X., Zhao Y., Peng Z., Gao P., Cao Z., et al. (2021). Virulence comparison of salmonella enterica subsp. enterica isolates from chicken and whole genome analysis of the high virulent strain s. enteritidis 211. Microorganisms 9 (11), 2239. doi: 10.3390/microorganisms9112239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U. N., Singh A. S., Lekshmi M., Nayak B. B., Kumar S. (2019). Characterization of bla(NDM)-harboring, multidrug-resistant Enterobacteriaceae isolated from seafood. Environ Sci Pollut Res Int 26 (3), 2455–2463. doi: 10.3390/microorganisms9112239 [DOI] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Nordmann P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res. Int. 2014, 249856. doi: 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority, E.C.f.D.P.a.C (2017). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. Efsa J. 15 (12), e05077. doi: 10.2903/j.efsa.2017.5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley S. L., Lynne A. M., Nayak R. (2008). Salmonella challenges: prevalence swine poultry potential pathogenicity such isolates. J. Anim. Sci. 86 (14 Suppl), E149–E162. doi: 10.2527/jas.2007-0464 [DOI] [PubMed] [Google Scholar]
- García-Fernández A., Chiaretto G., Bertini A., Villa L., Fortini D., Ricci A., et al. (2008). Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in escherichia coli and salmonella of human and animal origin. J. Antimicrob. Chemother. 61 (6), 1229–1233. doi: 10.1093/jac/dkn131 [DOI] [PubMed] [Google Scholar]
- Grimont P. A., Weill F. X.. Antigenic formulae of the Salmonella serovars. WHO collaborating centre for reference and research on Salmonella 9 1–166. Available online at: https://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%20Serovars%202007%209th%20edition.pdf. [Google Scholar]
- Hansen G. T. (2021). Continuous evolution: perspective on the epidemiology of carbapenemase resistance among enterobacterales and other gram-negative bacteria. Infect. Dis. Ther. 10 (1), 75–92. doi: 10.1007/s40121-020-00395-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Gou J., Guo X., Cao Z., Li Y., Jiao H., et al. (2018). Genetic contexts related to the diffusion of plasmid-mediated CTX-M-55 extended-spectrum beta-lactamase isolated from enterobacteriaceae in China. Ann. Clin. Microbiol. Antimicrob. 17 (1), 12. doi: 10.1186/s12941-018-0265-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guducuoglu H., Gursoy N.C., Yakupogullari Y., Parlak M., Karasin G., Sunnetcioglu M., et al. (2017). Hospital outbreak of a colistin-resistant, NDM-1- and OXA-48-Producing klebsiella pneumoniae: High mortality from pandrug resistance. Microbial Drug resistance (Larchmont N.Y.) 24 (7), 966–972. doi: 10.1089/mdr.2017.0173 [DOI] [PubMed] [Google Scholar]
- Jajere S. M. (2019). A review of salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 12 (4), 504–521. doi: 10.14202/vetworld.2019.504-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Jin Y. H., Park S. H., Han S., Kim H. S., Park J. H., et al. (2019). Emergence of a multidrug-resistant clinical isolate of escherichia coli st8499 strain producing ndm-13 carbapenemase in the republic of Korea. Diagn. Microbiol. Infect. Dis. 94 (4), 410–412. doi: 10.1016/j.diagmicrobio.2019.02.013 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 (4), 1355–1361. doi: 10.1128/jcm.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhao D., Li W., Sun J., Zhang X. (2021). Enzyme inhibitors: The best strategy to tackle superbug NDM-1 and its variants. Int. J. Mol. Sci. 23 (1) , 197. doi: 10.3390/ijms23010197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L., Zhang P., Piao R., Wang Y. (2019). Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell Infect. Microbiol. 9, 270. doi: 10.3389/fcimb.2019.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Qi X., Zhang D., Zheng Z., Chen Y., Guo Y., et al. (2016). First report of complete sequence of a bla(ndm-13)-harboring plasmid from an escherichia coli st5138 clinical isolate. Front. Cell Infect. Microbiol. 6, 130. doi: 10.3389/fcimb.2016.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R. C., Jr. (2010). NDM-1–a cause for worldwide concern. N Engl. J. Med. 363 (25), 2377–2379. doi: 10.1056/NEJMp1011715 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Walsh T. R., Livermore D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19 (12), 588–595. doi: 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Pan Y. S., Liu J. H., Hu H., Zhao J. F., Yuan L., Wu H., et al. (2013). Novel arrangement of the blaCTX-M-55 gene in an escherichia coli isolate coproducing 16S rRNA methylase. J. Basic Microbiol. 53 (11), 928–933. doi: 10.1002/jobm.201200318 [DOI] [PubMed] [Google Scholar]
- Pérez-Vázquez M., Sola Campoy P.J., Ortega A., Bautista V., Monzón S., Ruiz-Carrascoso G., et al. (2019). Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: Phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J Antimicrob Chemother 74 (12), 3489–3496. doi: 10.1093/jac/dkz366 [DOI] [PubMed] [Google Scholar]
- Poirel L., Bonnin R.A., Nordmann P.. (2011). Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother 45 (9), 4224–4229. doi: 10.1128/aac.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B., Tada T., Miyoshi-Akiyama T., Shimada K., Ohara H., Kirikae T., et al. (2015). Identification of a novel NDM variant, NDM-13, from a multidrug-resistant escherichia coli clinical isolate in Nepal. Antimicrob. Agents Chemother. 59 (9), 5847–5850. doi: 10.1128/aac.00332-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjabat H. E., Seah K. Y., Coleman L., Sartor A., Derrington P., Heney C., et al. (2014). Expansive spread of IncI1 plasmids carrying blaCMY-2 amongst escherichia coli. Int. J. Antimicrob. Agents 44 (3), 203–208. doi: 10.1016/j.ijantimicag.2014.04.016 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahilon J., Chandler M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–36. doi: 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: A genome comparison visualizer. Bioinf. (Oxford England) 27 (7), 1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18 (3), 318–327. doi: 10.1016/s1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tagg K. A., Iredell J. R., Partridge S. R. (2014). Complete sequencing of IncI1 sequence type 2 plasmid pJIE512b indicates mobilization of blaCMY-2 from an IncA/C plasmid. Antimicrob. Agents Chemother. 58 (8), 4949–4952. doi: 10.1128/aac.02773-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinf. (Oxford England) 30 (14) 2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Treangen T. J., Ondov B. D., Koren S., Phillippy A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15 (11), 524. doi: 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne P. (2021). Performance Standards for Antimicrobial Susceptibility Testing. 31st ed CLSI supplement M100, (USA: Clinical and Laboratory Standards Institute; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13 (6), e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z., et al. (2019). NDM metallo-β-Lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32 (2), e00115–18. doi: 10.1128/cmr.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Biswas S., Gu G., Elbediwi M., Li Y., Yue M. (2020). Characterization of multidrug resistance patterns of emerging salmonella enterica serovar rissen along the food chain in China. Antibiotics (Basel) 9 (10), 660. doi: 10.3390/antibiotics9100660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 (12), 5046–5054. doi: 10.1128/aac.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C. E., Peter K., Laing C. R., Lingohr E. J., Gannon V. P., Nash J. H. E., et al. (2016). The salmonella in silico typing resource (sistr): An open web-accessible tool for rapidly typing and subtyping draft salmonella genome assemblies. PloS One 11 (1), e0147101. doi: 10.1371/journal.pone.0147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 (11), 2640–2644. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q. Y., Zhu J. H., Cai R. M., Zheng X. R., Zhang L. J., Chang M. X., et al. (2021). IS26 is responsible for the evolution and transmission of bla(NDM)-harboring plasmids in escherichia coli of poultry origin in China. mSystems 6 (4), e0064621. doi: 10.1128/mSystems.00646-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP092911-CP092914.