Abstract
Highly multiplexed protein and RNA in situ detection on a single tissue section concurrently is highly desirable for both basic and applied biomedical research. CO-detection by inDEXing (CODEX) is a new and powerful platform to visualize up to 60 protein biomarkers in situ, and RNAscope in situ hybridization (RNAscope) is a novel RNA detection system with high sensitivity and unprecedent specificity at a single-cell level. Nevertheless, to our knowledge, the combination of CODEX and RNAscope remained unreported until this study. Here, we report a simple and reproducible combination of CODEX and RNAscope. We also determined the cross-reactivities of CODEX anti-human antibodies to rhesus macaques, a widely used animal model of human disease.
Keywords: CODEX, RNAscope ISH, spatial information
Introduction
Highly multiplexed protein in situ detection (HMPISD) on a single tissue section at subcellular resolution is a powerful new technology to visualize and quantify proteins in their native tissue microenvironment with spatial context.1 –3 The HMPISD technologies have been wildly used in basic and applied biological research. The HMPISD has also been used in cancer biology to explore the complex immune landscapes and tumor microenvironment1,3 –7 and in the study of pathogenesis of autoimmune disease. 8 Several strategies and platforms of HMPISD have been developed and utilized including (1) Time-of-flight Mass Spectrometry based platforms of Imagine Mass Cytometry (IMC)9,10 and Multiplexed Ion Beam Imaging (MIBI).11,12 Both methods combined metal-labeled antibody immunostaining, ultraviolet (UV) laser ablation in IMC or ion beam gun ablation in MIBI, and CyTOF mass cytometry; (2) iterative fluorescent-conjugated antibody staining, imaging, and removing antibody or inactivating fluorophores, for example, MultiOmyx13 –15 and the tissue-based CyClic Immunofluorescence16,17; and (3) DNA-barcoded antibody platforms, for example, the Immunostaining with Signal Amplification by Exchange Reaction (Immuno-SABER)18,19 and CO-detection by inDEXing (CODEX). In Immuno-SABER, tissue target proteins are first bound with a panel of DNA-conjugated antibodies, followed by multiple rounds of hybridizing with corresponding concatemer DNA and fluorescent-DNA, imaging, and removing the fluorophores. CODEX is a new and powerful platform, of which multiple epitopes are first bound with a panel of barcoded-oligonucleotide-conjugated antibodies followed by multiple cycles of adding corresponding fluorescently labeled–oligonucleotide probes, imaging, and removing fluorescently labeled–oligonucleotide probes to visualize up to 60 protein biomarkers in situ.6,8,20 While the prototype CODEX using dNTP analogs plus DNA polymerase primer extension to amplify signal of DNA barcode conjugated to antibody, 8 the current CODEX uses chaotropic solvents to facilitate sequential room temperature annealing and stripping process without polymerase reaction. 6 Compared with other HMPISD methods, CODEX enlarges multiplexing ability,3,20 increases detection resolution to subcellular level, 1 preserves tissue architecture, and reduces experiment duration through single staining procedure. 1 Simultaneous visualization of spatially resolved and highly multiplexed proteins and RNA in situ is highly desirable for both basic and applied biomedical research.21,22 RNAscope in situ hybridization (RNAscope) from the Advanced Cell Diagnostics (ACD) is a novel RNA detection system with high sensitivity and unprecedent specificity at a single-cell level.19,23 Nevertheless, RNAscope and CODEX involve complex chemical and physical procedures. To our knowledge, the combination of CODEX and RNAscope (Comb-CODEX-RNAscope) remained unknown until this study.
In this study, we tested various methods of combination and found a simple and reproducible approach of CODEX first and RNAscope second (Comb-CODEX-RNAscope) to visualize highly multiplexed proteins and RNA simultaneously in a single tissue section. Moreover, current HMPISD platforms including CODEX are designed for human sample detection using anti-human antibodies. Non-human primates (NHP) evolutionally, anatomically, and physiologically are the closest species to humans and are regarded as one of the best animal models of many human diseases. We therefore determined the cross-reactivities of CODEX anti-human antibodies to rhesus macaques, which are the most widely used NHP to model human diseases including HIV.24,25
Materials and Methods
Rhesus Macaque Tissues
Archived fixed lymph node and rectum tissues from adult rhesus macaques infected with SIVmac251 from our previous reported studies, which were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Nebraska–Lincoln, were used.26,27 Rhesus macaques were intra-rectally inoculated with SIVmac251 (3.1 × 104 TCID50) and were euthanized at different days post-inoculation, and rhesus macaques without virus inoculation served as uninfected controls. Lymph node and rectal tissues were fixed with SafeFix II (cat. no. 23-042600, Fisher Scientific, Waltham, MA, USA), 4% paraformaldehyde or neutral-buffered formalin and embedded with paraffin.
Antibody-oligonucleotide Conjugation
The major materials and reagents used in these experiments are listed in Supplemental Table 1. To find a rhesus macaque reactive CD3 antibody for CODEX, an anti-human CD3 antibody in a carrier-free phosphate-buffered saline (PBS) solution (clone no. SP162, cat. no. ab245731, Abcam, Boston, MA, USA) was conjugated with the barcode-oligonucleotide (cat. no. 5350002, Akoya Biosciences, Menlo Park, CA, USA) using CODEX conjugation kit (cat. no. 7000009, Akoya Biosciences, Menlo Park, CA, USA) following CODEX conjugation manual.
CODEX Immunostaining
CODEX immunostaining was performed according to CODEX user manual and previously published method. 20 Six-μm tissue sections were cut using Leica RM2235 rotary microtome and mounted to poly-lysine-coated coverslips. The following steps were performed in six-well tissue culture plate. Tissue-coverslips were deparaffinized by sequentially heating on a hotplate at 60C for 1 hr and immersed in xylene for 5 min twice and rehydrated with gradually decreased concentration of ethanol and diethylpyrocarbonate (DEPC) water. The CODEX pretreament was performed in a high-pressure cooker (Decloaking Chamber, Biocare Medical, Pacheco, CA, USA) for 20 min at high-pressure or hotplate (Cat# SP88857104, Thermo Fisher, Waltham, MA, USA) boiling for 20 min, of which the tissue-coverslips in the coverslip staining rack (PN no. 72240, Electron Microscopy Science, Hatfield, PA, USA) were immersed in citrate buffer (pH 6, Millipore 21545, Sigma-Aldrich, Burlington, MA, USA) in 50 ml glass beaker. After the pretreatment, the tissue-coverslips were stained with a cocktail of DNA-barcoded antibodies for 3 hr at room temperature. After washing in PBS 3 times, 2 min each to remove unbound antibodies, the tissue-coverslips were post-fixed with 1.6% paraformaldehyde (PFA) for 10 min at room temperature, washed, post-fixed with ice-cold methanal for 5 min and washed. The tissue-coverslips then could be stored in the storage buffer for 5 days at 4C or immediately loaded into CODEX instrument (Akoyo Biosciences, Menlo Park, CA, USA) for multiple cycle immunostaining. The CODEX fluidics and Keyence microscope was set up using CODEX instrument manager software and Keyence software according to the manufacturer’s protocol. The master mix of fluorescently labeled–oligonucleotide probes (fluorescent-reporters) that corresponded to DNA-barcoded antibodies for multiple cycle immunostaining was prepared in a 96-well plate according to CODEX manual. Each cycle of CODEX immunostaining consisted of three steps of adding nuclear staining 4′,6-diamidino-2-phenylindole (DAPI); Atto550-, Cy5-, and Cy7-fluorescent-reporters; imaging; and removing fluorescent-reporters. Two blank cycles (only DAPI nuclear stain without any fluorescent-reporters) were ran for evaluating the level of autofluorescence and subtracting background using CODEX processor software.
RNAscope
RNAscope was performed according to the manufacturer’s protocol and our previously published method28,29 with slight modifications. All the procedures including deparaffinization, hydration, and pretreatment were conducted to minimize RNase contamination. The tissue-coverslips were deparaffinized and rehydrated as described in the CODEX section above. The RNAscope pretreament included (1) incubation of tissue-coverslips in 3% hydrogen peroxide for 10 min at room temperature, (2) after washing in mili-water (2700PRD, Aqua Solution, Deer Park, TX, USA), the tissue-coverslips were subjected to antigen retrieval by boiling for 15 min in RNAscope Target Retrieval Reagent solution (cat. no. 322000, ACD, Farmington, UT, USA) in a 50 ml glass beaker on hotplate to undo the cross-linking, and (3) treatment with RNAscope Protease Plus (cat. no. 322330, ACD, Farmington, UT, USA) at 40°C for 20 min. After the pretreatment, tissue-coverslips were hybridized with RNAscope Probe-SIVmac239 (antisense, cat. no. 312811, ACD, Farmington, UT, USA) at 40°C for 2 hr. The signals were amplified and detected with RNAscope 2.5 HD assay-RED kit (cat. no. 322360, ACD, Farmington, UT, USA), where the fast red can be visualized both in regular light and in far-red fluorescent channel in CODEX instrument. The RNAscope negative control probe-DapB (cat. no. 310043, ACD, Farmington, UT, USA) was used as negative control.
Combination of CODEX Immunostaining With RNAscope
Two different combination methods, RNAscope and subsequent CODEX as well as CODEX and subsequent RNAscope, were tested. For RNAscope and subsequent CODEX approach, tissue deparaffinization, hydration, RNAscope pretreatment, probe hybridization, signal amplification, and development were conducted by following the single RNAscope protocol as described above. After completion of the RNAscope, tissue-coverslip underwent CODEX pretreatment as described above by following single CODEX protocol. For the CODEX and subsequent RNAscope approach, two protocols were tested. For the first protocol, CODEX was conducted by following the single CODEX protocol as described above. After the completion of last cycle of CODEX, the tissue-coverslip was removed from the CODEX instrument and continued with the single RNAscope procedure from 3% hydrogen peroxide incubation step on (Supplemental Fig. 2). For the second protocol, we combined the pretreatments of CODEX and RNAscope together at the beginning of the experiment (Fig. 3). The tissue-coverslips were deparaffinized, hydrated, and pretreated with 3% hydrogen peroxide for 10 min at room temperature, either citrate buffer (pH 6, ipore 21545, Sigma-Aldrich, Burlington, MA, USA) or RNAscope Target Retrieval Reagent solution (cat. no. 322000, ACD, Farmington, Utah, USA) in a 50 ml glass beaker on hotplate to boil for 15 min, and RNAscope Protease Plus (cat. no. 322330, ACD, Farmington, UT, USA) at 40C for 20 min. After the pretreament, the tissue-coverslips were stained with a cocktail of DNA-barcoded antibodies by following the single CODEX protocol as described above. After the completion of the CODEX, tissue-coverslip underwent RNAscope procedure as described above by following single RNAscope protocol without RNAscope pretreament. After the completion of RNAscope procedure, the tissue-coverslip was stained with DAPI (1:2000) for 5 min at room temperature, washed, and loaded into the CODEX instrument for capturing RNAscope fluorescent image. To integrate RNAscope image to CODEX images, Keyence microscope was set using Keyence software as described above and a new blank cycle was added, where Cy5-channel was assigned to RNAscope. After capturing the RNAscope fluorescent image, image data were transferred into the CODEX file location, where raw data of CODEX plus RNAscope were processed by CODEX processor software.
Figure 3.
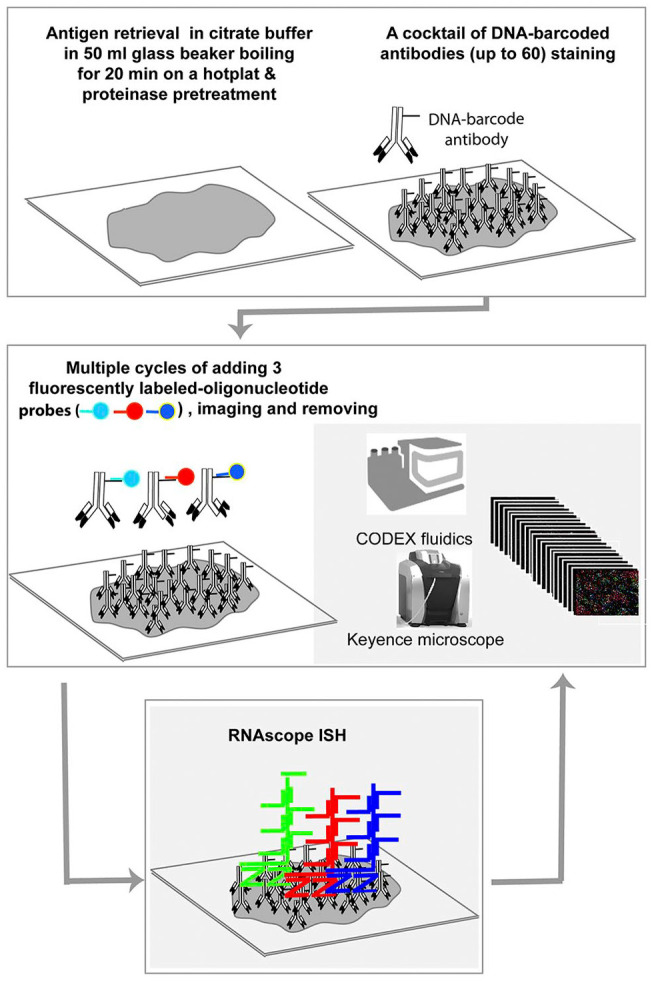
The simplified workflow of CODEX and subsequent RNAscope ISH combination (Comb-CODEX-RNAscope). All pretreatments of CODEX and RNAscope were combined together at the beginning of the experiment. Abbreviations: CODEX, CO-detection by indexing; ISH, in situ hybridization.
Results
Determine the Cross-reactivities of CODEX Anti-human Antibodies to Rhesus Macaques
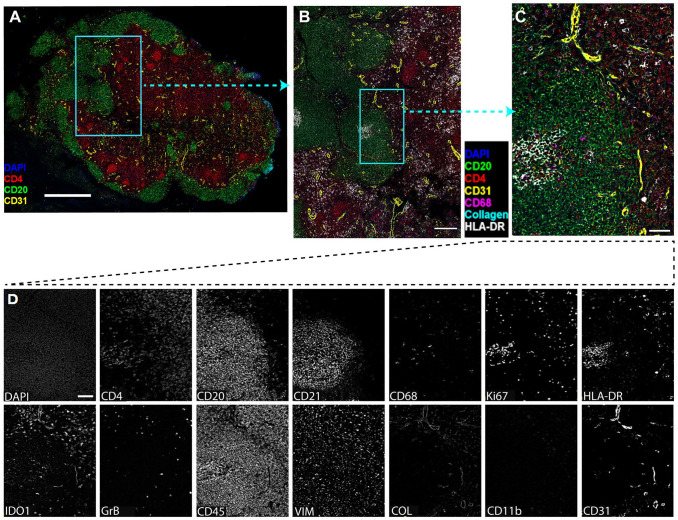
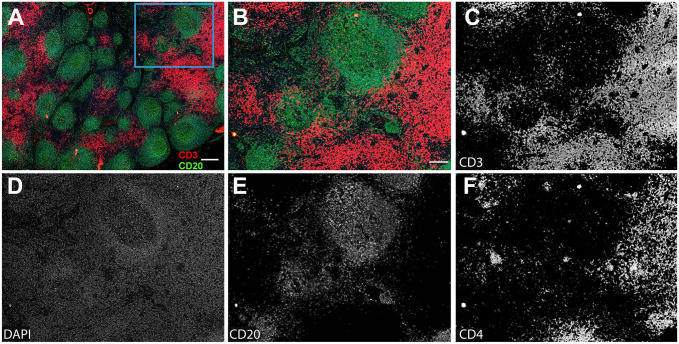
To determine the cross-reactivities of CODEX anti-human antibodies to rhesus macaques, a panel of DNA-barcoded anti-human antibodies from the Akoya Biosciences was tested in various fixative-fixed (SafeFixII, 4% Paraformaldehyde, neutral-buffered formalin) and paraffin-embedded lymph node tissues of rhesus macaques who were infected with simian immunodeficiency virus (SIV). We found 13 out of 24 CODEX anti-human antibodies from Akoya were reactive specifically with rhesus macaques (Table 1 and Fig. 1). The DNA-barcoded anti-human CD3 antibody from Akoya (CD3e, clone no. EP449E) did not react with rhesus macaque tissues. We therefore conjugated another anti-human CD3 antibody (SP162, cat. no. ab245731, Abcam, Boston, MA, USA) with a DNA-barcoded oligonucleotide (cat. no. 5350002, Akoya) by following the conjugating kit manual (cat. no. 7000009, Akoya). After verifying the size of antibody after conjugation was correct through protein gel electrophoresis, the conjugated CD3 antibody was tested using CODEX. It worked well for rhesus macaque tissues, where CD3 signals were clearly separated from CD20 signals and partially colocalized with CD4 signals and primarily distributed in T-cell zone of lymph node tissues (Fig. 2). The specificity of antibody detection was confirmed with isotypes control antibodies, where there were no detectable signals (data not shown).
Table 1.
The Cross-reactivities of CODEX Anti-human
DNA-barcoded-antibodies From Akoya to Rhesus Macaques.
| Antibody | Clone no. | Cross-reactivity to Rhesus Macaque |
|---|---|---|
| CD107a | H4A3 | + |
| CD11c | A118/A5 | − |
| CD14 | EPR3653 | − |
| CD20 | L26 | + |
| CD21 | EP3093 | + |
| CD31 | EP3095 | + |
| CD3e | EP449E | − |
| CD4 | EPR6855 | + |
| CD44 | 156-3C11 | − |
| CD45 | D9M81 | + |
| CD45R0 | UCHL1 | − |
| CD11b | EP1345Y | + |
| CD68 | KP1 | + |
| CD8 | C8/144B | − |
| CollagenIV | EPR20966 | + |
| E-Cadherin | 4A2C7 | − |
| Granzyme B | D6E9W | + |
| HLA-DR | EPR3692 | + |
| Pan-Cytokeratin | AE1/AE3 | − |
| IDO-1 | V1NC3IDO | + |
| Vimentin | O91D3 | + |
| CD57 | HNK-1 | − |
| CD34 | QBEND/10 | − |
| Ki67 | B56 | + |
Abbreviation: CODEX, CO-detection by inDEXing.
Figure 1.
The cross-reactivities of 24 DNA-barcoded anti-human antibodies from Akoya to rhesus macaques using CODEX. (A) Overview image of lymph node tissue from a rhesus macaque infected with SIVmac251 (Rh4979, 10 dpi), showing CD4 (red), CD20 (green), DAPI (blue), and CD31 (yellow). Scale bar equals 1000 microns. (B) The boxed area in A was zoomed to show DAPI (blue), CD4 (red), CD20 (green), CD68 (magenta), Collagen (cyan), HLA-DR (white), and CD31 (yellow). Scale bar equals 200 microns. (C) The boxed area in B was zoomed. Scale bar equals 50 microns. (D) Individual channel image showing 13 cross-reactive anti-human antibodies to rhesus macaque. Scale bar equals 50 microns. Abbreviations: CODEX, CO-detection by indexing; DAPI, 4′,6-diamidino-2-phenylindole.
Figure 2.
The validation of a home conjugated anti-human CD3 antibody (SP162, cat. no. ab245731, Abcam) with a DNA barcode using CODEX. (A) Overview image of lymph node tissue from a rhesus macaque infected with SIVmac251 (Rh4979, 10 dpi). Scale bar equals 200 microns. (B) The boxed area in A was zoomed. Scale bar equals 50 microns. (C–F) Individual channel image of antibodies, showing CD3, CD20, CD4, and DAPI. Scale bar equals 50 microns. Abbreviations: CODEX, CO-detection by indexing; DAPI, 4′,6-diamidino-2-phenylindole.
The Combination of CODEX Immunostaining With RNAscope
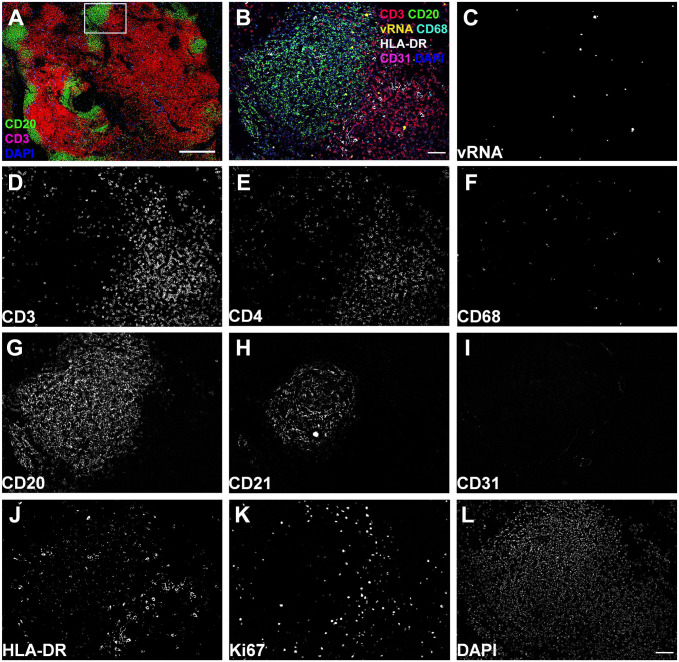
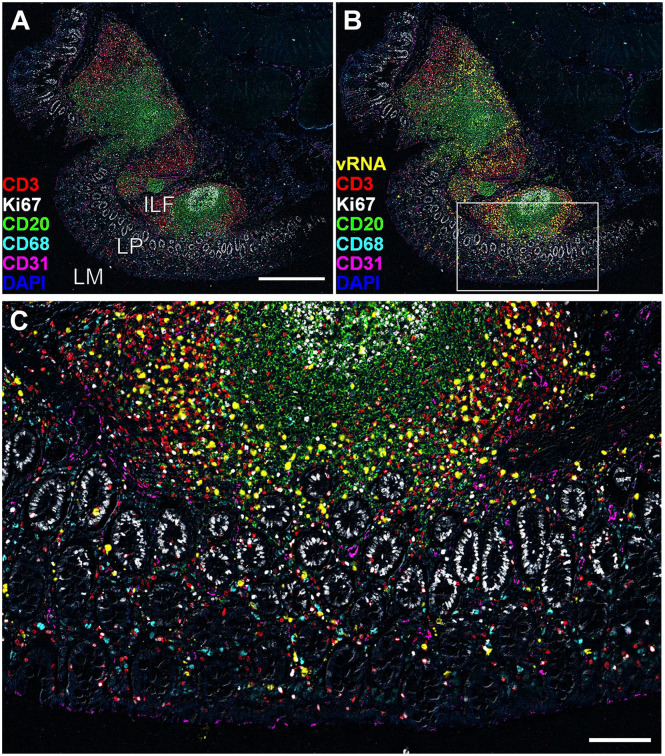
After identifying rhesus macaque compatible antibodies, we selected a subset of them that are important for SIV immunopathogenesis to test various methods of combining CODEX with RNAscope. Two different orders of combination, RNAscope and subsequent CODEX as well as CODEX and subsequent RNAscope, were tested. All the procedures including deparaffinization, tissue hydration, and antigen retrieval were conducted to minimize RNase contamination. RNAscope and subsequent CODEX method significantly induced nonspecific signals and reduced the specific signals of CODEX (Supplemental Fig. 1). This combination led to unaccepted low signal-noise-ratio (SNR) of CODEX as compared with a single CODEX, although RNAscope signals maintained before and after CODEX (data not shown). We reasoned that complexed process of RNAscope procedure may damage some epitopes of CODEX target proteins. Furthermore, RNAscope fluorescent signal is hard to be removed during the CODEX cyclic fluorescent detection, which overlaps and interferes with the subsequent fluorescent channel in CODEX. As RNAscope and subsequent CODEX approach could not get satisfying results, next efforts were focused on the approach of CODEX and subsequent RNAscope. When following the protocol of single CODEX and single RNAscope sequentially, RNAscope signals could not be consistently detected, although CODEX signal detection worked well. It was plausible that some steps of CODEX including the antigen-retrieval step might impair RNAscope signal detection. It was tested and excluded the impact of number of cycles of CODEX on the RNAscope signal detection by comparing single cycle versus multiple cycles of CODEX immunostaining. The impact of RNase contamination of CODEX fluidics instrument was further excluded by adding CODEX fluidics washing step into a single RNAscope procedure. The washing with CODEX fluidics did not affect RNAscope signal detection. It was narrowed down that the source of inconsistent RNAscope signal detection was in the antigen-retrieval step using a high-pressure cooker (Decloaking Chamber, Biocare Medical). An alternative method for antigen retrieval using 50 ml glass beaker on a hotplate (SP88857104, Thermo Fisher, Waltham, MA, USA) was tested, where the coverslips with tissue sections were placed in the coverslip staining rack (PN no. 72240, Electron Microscopy Science, Hatfield, PA, USA) and immersed in boiling citrate buffer (pH 6, Millipore 21545, Sigma-Aldrich, Burlington, MA, USA) for 20 min. This method is simple and easy to eliminate potential RNase contamination by pre-baking the glass beaker and coverslip staining rack covered with aluminum foil at 300°C for 6 hr. The developed CODEX and subsequent RNAscope approach with hotplate antigen retrieval is simple and reproducible for both highly multiplexed protein and RNA detections for SafeFixII, 4% PFA, and neutral-buffered formalin fixed and paraffin-embedded tissues (Supplemental Fig. 2). SIV viral RNA (vRNA) signals from RNAscope (Supplemental Fig. 3B and C) in addition to CD3, CD4 CD68, CD20, CD21, CD31, HLA-DR, Ki67, and DAPI from CODEX (Supplemental Fig. 3D to L) are specific and intense enabling downstream spatial analyses. To simplify the CODEX and subsequent RNAscope protocol described above, we tested if it is possible to combine all the pretreatments for CODEX and RNAscope together at the beginning of the experiment (Comb-CODEX-RNAscope; Fig. 3). It worked very well and all the signals from CODEX and RNAscope were as clear as separate pretreatments (Fig. 4 and Supplemental Fig. 4). We quantitatively compared the RNAscope signals in the Comb-CODEX-RNAscope with RNAscope along in adjacent lymph node (LN) tissue sections (Supplemental Fig. 4). The ratio of vRNA signal pixel area out of total tissue pixel area in Comb-CODEX-RNAscope versus RNAscope alone from obturate and axillary LN tissues from different monkeys were quantified using a positive pixel count algorithm in Aperio’s Spectrum Plus analysis program (version 9.1; Aperio ePathology Solutions, Leica Biosystems, Deer Park, IL, USA), as we described previously. 30 There is no statistical difference in the Comb-CODEX-RNAscope versus RNAscope along (two-way analysis of variance, p<0.0001; Supplemental Fig. 4E). To determine the applicability of the Comb-CODEX-RNAscope to non-lymphoid tissue, we successfully detected multiplexed proteins and SIV vRNA in rectal tissues of rhesus macaques infected with SIV (Fig. 5 and Supplemental Fig. 5)
Figure 4.
Representative images of the Comb-CODEX-RNAscope where all pretreatments of CODEX and RNAscope were combined together at the beginning of the experiment. (A) Overview image of lymph node tissues from a rhesus macaque infected with SIVmac251 (Rh4979, 10 dpi), showing CD3 (red), CD20 (green), and DAPI (blue). Scale bar equals 500 microns. (B) The boxed area in A was zoomed to show SIV viral RNA detected by RNAscope and other seven protein markers by CODEX. Scale bar equals 50 microns. (C–L) Individual channel image showing vRNA and protein biomarkers. Scale bar equals 50 microns. Abbreviations: CODEX, CO-detection by indexing; DAPI, 4′,6-diamidino-2-phenylindole; SIV, simian immunodeficiency virus.
Figure 5.
Representative non-lymphatic tissue images detected using the Comb-CODEX-RNAscope where all pretreatments of CODEX and RNAscope were combined at the begin of the experiment. (A) The merged image of rectal tissue of a rhesus macaque infected with SIVmac251 (Rh4978, 10 dpi), showing CD3 (red), Ki67 (white), CD20 (green), CD68 (cyan), CD31 (magenta), and DAPI (blue). Scale bar equals 500 microns. (B) The image of A adds SIV vRNA (yellow). (C) The boxed area in image B was zoomed in, scale bar equals 100 microns. Abbreviations: CODEX, CO-detection by inDEXing; DAPI, 4′,6-diamidino-2-phenylindole; ILF, isolated lymphoid follicle; LP, lamina propria; LM, lumen; SIV, simian immunodeficiency virus.
Discussion
Simultaneous detection of highly multiplexed proteins and RNA in situ is important for better understanding health and disease. The spatially resolved relationships of different cells populations, protein, and RNA and DNA molecules in their native tissue architecture bears crucial information for disease diagnosis, pathogenesis, and treatment. CODEX represents a powerful new platform for highly multiplexed proteins (up to 60) in situ detection.6,8,20 RNAscope represents another power multiplex RNA/DNA detection system (up to 12) with high sensitivity and specificity,19,23 where the unique double-Z-probes and corresponding amplification probes enable concurrent signal amplification and background noise reduction. The combination of RNAscope with regular immunohistochemical or immunofluorescent staining to concurrently detect RNA and protein within the same tissue section has been reported by us 29 and others.31,32 However, to our knowledge, there is no reported study that combined CODEX and RNAscope together. To that end, we tested different combination methods and found CODEX and subsequent RNAscope worked reliably (Figs. 3 and 4 and Supplemental Figs. 2 and 3) while RNAscope and subsequent CODEX did not work well for CODEX signal detection (Supplemental Fig. 1). The following two factors in the RNAscope procedure may damage some epitopes for CODEX detection. First, RNAscope uses protease to digest the tissue to facilitate probes access to target sequences. The protease treatment has been demonstrated to be harsh for many antigenic sites, although some epitopes may survive this treatment. 33 By assessing the combined pretreatments before CODEX cycles, we found protease is not the main factor affecting CODEX staining. Another factor is the dehydration step in the RNAscope method. In a typical protocol, after antigen retrieval, the slide will be dipped in ethanol and air dried before adding probes for hybridization. However, preservation immune-recognizable epitopes in tissues requires maintaining a certain amount of water within tissues after antigen retrieval.34,35 Possible solution for this negative effect of dehydration can be overcome using disaccharides to protect tissue 35 or carefully avoiding tissue drying after antigen retrieval. Moreover, to process RNAscope and multiple cycles of CODEX data, we need to capture and then eliminate fluorescent signals from RNAscope, which is difficult if we conduct RNAscope first. For the CODEX and subsequent RNAscope approach, we refined antigen-retrieval procedure after finding it was crucial for RNAscope signal detection by using the hotplate antigen-retrieval method, which is easier to eliminate RNase contamination by baking the glass beaker and coverslip holder. Both protocols of the CODEX and subsequent RNAscope described in this article worked well; nevertheless, the combination of pretreatments of CODEX and RNAscope together at the beginning of the experiment (Comb-CODEX-RNAscope) is simpler and timesaving, which works for both lymphatic and non-lymphatic tissues (Fig. 5 and Supplemental Fig. 5). The Comb-CODEX-RNAscope described in this article can be further enhanced with multiplex RNAscope published recently36,37 to maximize the detection of interested RNA signals in addition to highly multiplexed proteins.
The cross-reactivities of 24 CODEX anti-human antibodies to rhesus macaques were also evaluated and 13 were compatible with rhesus macaques. The anti-human CD3 antibody from Akoya did not work for rhesus macaques. Another anti-human CD3 antibody (clone no. SP162, cat. no. ab245731, Abcam, Boston, MA, USA) was conjugated with a barcode-oligonucleotide (cat. no. 5350002, Akoya Biosciences, Menlo Park, CA, USA), which works for rhesus macaque tissues in CODEX. Of note, after the submission of this manuscript and during the revision, one study reported the cross-reactivity of human antibodies to rhesus macaque tissues, 38 which reenforced our results.
In summary, a simple and reproducible CODEX and subsequent RNAscope protocol (Comb-CODEX-RNAscope) reported here enables spatially resolved highly multiplexed protein and RNA in situ detection, which will facilitate better understanding of the health and disease at single-cell level in tissue spatial context.
Supplemental Material
Supplemental material, sj-docx-1-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-2-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-3-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-4-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-5-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-6-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: QL and YC designed the experiments. YC conducted CODEX, RNAscope, and combination experiments. RKB conducted some RNAscope experiments. YC and QL prepared the manuscript, and all authors provided manuscript editing. The authors would like to thank Oliver Braubach, Najiba Mammadova, and Kristin Schmidt from Akoya for their CODEX technical support. The authors would like to thank the current and previous laboratory members of QL, especially Subhra Mandal and Saroj Candra Lohani, for their contribution to rhesus macaque experiments.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by NIH R01 AI136756 (Y.L., Q.L.), R01 DK087625-01 (L.Q.), R21 AI143405 (Q.L.).
ORCID iD: Qingsheng Li  https://orcid.org/0000-0003-4101-4073
https://orcid.org/0000-0003-4101-4073
Contributor Information
Yilun Cheng, Nebraska Center for Virology, School of Biological Sciences, University of Nebraska–Lincoln, Lincoln, Nebraska.
Rachel K. Burrack, Nebraska Center for Virology, School of Biological Sciences, University of Nebraska–Lincoln, Lincoln, Nebraska
Qingsheng Li, Nebraska Center for Virology, School of Biological Sciences, University of Nebraska–Lincoln, Lincoln, Nebraska.
Literature Cited
- 1. McGinnis LM, Ibarra-Lopez V, Rost S, Ziai J. Clinical and research applications of multiplexed immunohistochemistry and in situ hybridization. J Pathol. 2021;254:405–17. [DOI] [PubMed] [Google Scholar]
- 2. Parra ER, Francisco-Cruz A, Wistuba II. State-of-the-art of profiling immune contexture in the era of multiplexed staining and digital analysis to study paraffin tumor tissues. Cancers (Basel). 2019;11:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis SM, Asselin-Labat M-L, Nguyen Q, Berthelet J, Tan X, Wimmer VC, Merino D, Rogers KL, Naik SH. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18:997–1012. [DOI] [PubMed] [Google Scholar]
- 4. Parra ER, Zhai J, Tamegnon A, Zhou N, Pandurengan RK, Barreto C, Jiang M, Rice DC, Creasy C, Vaporciyan AA, Hofstetter WL, Tsao AS, Wistuba II, Sepesi B, Haymaker C. Identification of distinct immune landscapes using an automated nine-color multiplex immunofluorescence staining panel and image analysis in paraffin tumor tissues. Sci Rep. 2021;11:4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, Balter A, Kawashima R, Choe G, Sauer D, El Rassi E, Clayburgh DR, Kulesz-Martin MF, Lutz ER, Zheng L, Jaffee EM, Leyshock P, Margolin AA, Mori M, Gray JW, Flint PW, Coussens LM. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19:203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kennedy-Darling J, Bhate SS, Hickey JW, Black S, Barlow GL, Vazquez G, Venkataraaman VG, Samusik N, Goltsev Y, Schürch CM, Nolan GP. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur J Immunol. 2021;51:1262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schürch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, Chu P, Black S, Demeter J, McIlwain DR, Kinoshita S, Samusik N, Goltsev Y, Nolan GP. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive. Front Cell. 2020;182:1341–59.e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, Nolan GP. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–81.e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Günther D, Bodenmiller B. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–22. [DOI] [PubMed] [Google Scholar]
- 10. Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, Moch H, Muenst S, Varga Z, Weber WP, Bodenmiller B. The single-cell pathology landscape of breast cancer. Nature. 2020;578:615–20. [DOI] [PubMed] [Google Scholar]
- 11. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Multiplexed ion beam imaging of human breast tumors. Nat Methods. 2014;20:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang SR, Kurian A, Van Valen D, West R, Bendall SC, Angelo M. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174:1373–87.e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hollman-Hewgley D, Lazare M, Bordwell A, Zebadua E, Tripathi P, Ross AS, Fisher D, Adams A, Bouman D, O’Malley DP, Weiss LM. A single slide multiplex assay for the evaluation of classical Hodgkin lymphoma. Am J Surg Pathol. 2014;38:1193–202. [DOI] [PubMed] [Google Scholar]
- 15. Xu-Monette ZY, Xiao M, Au Q, Padmanabhan R, Xu B, Hoe N, Rodríguez-Perales S, Torres-Ruiz R, Manyam GC, Visco C, Miao Y, Tan X, Zhang H, Tzankov A, Wang J, Dybkær K, Tam W, You H, Bhagat G, Hsi ED, Ponzoni M, Ferreri AJM, Møller MB, Piris MA, van Krieken JH, Winter JN, Westin JR, Pham LV, Medeiros LJ, Rassidakis GZ, Li Y, Freeman GJ, Young KH. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol Res. 2019;7:644–57. [DOI] [PubMed] [Google Scholar]
- 16. Lin J-R, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 2015;6:8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S, Sorger PK. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, Kirli K, Yapp C, Cicconet M, Beliveau BJ, Lapan SW, Yin S, Lin M, Boyden ES, Kaeser PS, Pihan G, Church GM, Yin P. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol. 2019;37:1080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Black S, Phillips D, Hickey JW, Kennedy-Darling J, Venkataraaman VG, Samusik N, Goltsev Y, Schurch CM, Nolan GP. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16:3802–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes—next generation tools for tissue exploration. Bioessays. 2020;42:1900221. [DOI] [PubMed] [Google Scholar]
- 22. Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun X-K, Jackson HW, Bodenmiller B. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6:25–36.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Atout S, Shurrab S, Loveridge C. Evaluation of the suitability of RNAscope as a technique to measure gene expression in clinical diagnostics: a systematic review. Mol Diagn Ther. 2022;26:19–37. doi: 10.1007/s40291-021-00570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hessell AJ, Haigwood NL. Animal models in HIV-1 protection and therapy. Curr Opin HIV AIDS. 2015;10:170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang B, Li H, Li L, Omange RW, Hai Y, Luo M. Current advances in HIV vaccine preclinical studies using Macaque models. Vaccine. 2019;37:3388–99. [DOI] [PubMed] [Google Scholar]
- 26. Lu W, Demers AJ, Ma F, Kang G, Yuan Z, Wan Y, Li Y, Xu J, Lewis M, Li Q. Next-generation mRNA sequencing reveals pyroptosis-induced CD4+ T cell death in early simian immunodeficiency virus-infected lymphoid tissues. J Virol. 2015;90:1080–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Demers A, Kang G, Ma F, Lu W, Yuan Z, Li Y, Lewis M, Kraiselburd EN, Montaner L, Li Q. The mucosal expression pattern of interferon-epsilon in rhesus macaques. J Leukoc Biol. 2014;96:1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan Z, Wang N, Kang G, Niu W, Li Q, Guo J. Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth Biol. 2017;6:721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ko A, Kang G, Hattler JB, Galadima HI, Zhang J, Li Q, Kim WK. Macrophages but not astrocytes harbor HIV DNA in the brains of HIV-1-infected aviremic individuals on suppressive antiretroviral therapy. J Neuroimmune Pharmacol. 2019;14:110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L-X, Kang G, Kumar P, Lu W, Li Y, Zhou Y, Li Q, Wood C. Humanized-BLT mouse model of Kaposi’s sarcoma-associated herpesvirus infection. Proc Natl Acad Sci U S A. 2014;111:3146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Annese T, Tamma R, De Giorgis M, Ruggieri S, Maiorano E, Specchia G, Ribatti D. RNAscope dual ISH-IHC technology to study angiogenesis in diffuse large B-cell lymphomas. Histochem Cell Biol. 2020;153:185–92. [DOI] [PubMed] [Google Scholar]
- 32. Kersigo J, Pan N, Lederman JD, Chatterjee S, Abel T, Pavlinkova G, Silos-Santiago I, Fritzsch B. A RNAscope whole mount approach that can be combined with immunofluorescence to quantify differential distribution of mRNA. Cell Tissue Res. 2018;374:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duncan JS, Elliott KL, Kersigo J, Gray B, Fritzsch B. Combining whole-mount in situ hybridization with neuronal tracing and immunohistochemistry. In: In situ hybridization methods. 2015. p. 339–52. doi: 10.1007/978-1-4939-2303-8_17. [DOI] [Google Scholar]
- 34. Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, Cattoretti G. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 2017;65:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boi G, Scalia CR, Gendusa R, Ronchi S, Cattoretti G. Disaccharides protect antigens from drying-induced damage in routinely processed tissue sections. J Histochem Cytochem. 2016;64:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan S, Filezac de, L’Etang A, Rangell L, Caplazi P, Lowe JB, Romeo V. A method for manual and automated multiplex RNAscope in situ hybridization and immunocytochemistry on cytospin samples. PLoS ONE. 2018;13:e0207619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao L, Labaer J, Guo J. Highly sensitive and multiplexed in situ RNA profiling with cleavable fluorescent tyramide. Cells. 2021;10:1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang S, Mukherjee N, Bennett RS, Chen H, Logue J, Dighero-Kemp B, Kurtz JR, Adams R, Phillips D, Schürch CM, Goltsev Y, Hickey JW, McCaffrey EF, Delmastro A, Chu P, Reader JR, Keesler RI, Galván JA, Zlobec I, Van Rompay KKA, Liu DX, Hensley LE, Nolan GP, McIlwain DR. Rhesus macaque CODEX Multiplexed immunohistochemistry panel for studying immune responses during Ebola infection. Front Immunol. 2021;12:729845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-2-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-3-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-4-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-5-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-tif-6-jhc-10.1369_00221554221114174 for Spatially Resolved and Highly Multiplexed Protein and RNA In Situ Detection by Combining CODEX With RNAscope In Situ Hybridization by Yilun Cheng, Rachel K. Burrack and Qingsheng Li in Journal of Histochemistry & Cytochemistry