Abstract
The definition of bacterial species is traditionally a taxonomic issue while bacterial populations are identified by population genetics. These assignments are species specific, and depend on the practitioner. Legacy multilocus sequence typing is commonly used to identify sequence types (STs) and clusters (ST Complexes). However, these approaches are not adequate for the millions of genomic sequences from bacterial pathogens that have been generated since 2012. EnteroBase (http://enterobase.warwick.ac.uk) automatically clusters core genome MLST allelic profiles into hierarchical clusters (HierCC) after assembling annotated draft genomes from short-read sequences. HierCC clusters span core sequence diversity from the species level down to individual transmission chains. Here we evaluate HierCC's ability to correctly assign 100 000s of genomes to the species/subspecies and population levels for Salmonella, Escherichia, Clostridoides, Yersinia, Vibrio and Streptococcus. HierCC assignments were more consistent with maximum-likelihood super-trees of core SNPs or presence/absence of accessory genes than classical taxonomic assignments or 95% ANI. However, neither HierCC nor ANI were uniformly consistent with classical taxonomy of Streptococcus. HierCC was also consistent with legacy eBGs/ST Complexes in Salmonella or Escherichia and with O serogroups in Salmonella. Thus, EnteroBase HierCC supports the automated identification of and assignment to species/subspecies and populations for multiple genera.
This article is part of a discussion meeting issue ‘Genomic population structures of microbial pathogens’.
Keywords: big data, hierarchical clustering, genomic databases, EnteroBase, cgMLST, accessory genome
1. Introduction
Microbiologists need large databases to identify and communicate about clusters of related bacteria, … Such databases should contain the reconstructed genomes of bacterial isolates, … together with metadata describing their sources and phenotypic properties … and we are currently developing EnteroBase, a genome-based successor to the E. coli and S. enterica MLST databases.
Achtman and Zhou, 2014 [1].
The Linnean system of genus and species designations was applied to bacterial nomenclature in the late ninteenth century, soon after their ability to cause infectious diseases had been recognized. These taxonomic labels initially reflected disease specificity and phenotypic similarities. Over the following decades, phenotypic criteria for distinguishing bacterial taxa were extended to include serologically distinct groupings and even differential sensitivity to bacteriophages. A primary goal for taxonomic designations for bacterial pathogens was that they be useful for epidemiology and clinical diagnoses. As a result, serovars of Salmonella enterica, which differ by dominant epitopes on lipopolysaccharide and flagella, were each assigned a distinct species designation, often referring to the disease syndrome and host, e.g. Salmonella typhimurium [2]. Similarly, even though Yersinia pestis is a clone of Yersinia pseudotuberculosis [3,4], it was designated as a distinct species because Y. pestis causes plague whereas Y. pseudotuberculosis causes gastroenteritis. The use of species designations for Salmonella serovars is now disparaged, although it is still in common use [5], but Y. pestis has retained its species designation.
Percentage DNA–DNA hybridization levels became the ‘Gold Standard’ metric for new taxonomic designations after 1987 [6]. DNA-DNA hybridization reflects genomic relationships but distinctive phenotypic differences have remained a requirement for the definition of a novel species until the present. Indeed, the international committee which controls taxonomic designations continues to reject the validity of taxonomic designations based solely on DNA similarities [7,8].
(a) . Average nucleotide identity and multilocus sequence typing
An alternative genomic approach for taxonomic designations was proposed in 2005 by Konstantinidis [9], namely the definition of species on the basis of average nucleotide identity (ANI). Pairs of genomes with 95% ANI usually belong to the same species whereas pairs with lower ANI values belong to different species. Computational methods based on k-mer searches, such as FastANI [10], can rapidly perform ANI-like calculations on large numbers of genomes, and these calculations are now routinely used by bioinformaticians for taxonomic assignments. An approach based on ANI is enticing because it could provide defined criteria for the definition of a species across all Bacteria, and allow species assignment based exclusively on genome sequences. However, the use of ANI for species assignments has been criticized because it does not completely correlate with DNA-DNA hybridization [11,12]. Furthermore, multiple taxa that are designated as single species each contain multiple 95% ANI groups [13,14]. For example, strains of Streptococcus mitis colonize the human oropharyngeal tract, have similar phenotypes and are considered to represent a single species. However, S. mitis encompasses myriad, genetically distinct strains [15–17], and encompasses at least 44 distinct 95% ANI clusters [17]. Other problematic genera include Pseudomonas [12,18] and Aeromonas [19]. Furthermore, large databases including 100 000s of genomes per genus would struggle to implement methods such as pairwise clustering by ANI because each new entry would require testing against all genomes. There is also no consensus on ANI criteria for recognizing lower taxonomic entities, such as subspecies and populations. Indeed, there is not even a consensus on the definitive properties of what constitutes a bacterial species [20].
Bottom-up population genetic approaches such as multilocus sequence typing (MLST) can provide an alternative to top-down taxonomy, and deal efficiently with large numbers of bacterial strains. Legacy MLST based on the sequence differences of several housekeeping genes was introduced in 1998 [21], and has now been applied to more than 100 bacterial species [22]. MLST defines sequence types (STs), consisting of unique integer designations for each unique sequence (allele) of each of the MLST loci. Some STs mark individual clones with special pathogenic properties and which seem to have arisen fairly recently, such as Escherichia coli ST131, which is a globally prominent cause of urinary tract infection (UTI) and invasive disease [23]. Similarly, Salmonella enterica subsp. enterica serovar Typhimurium ST313 is a common cause of extra-intestinal, invasive salmonellosis in Africa [24]. Higher order clusters of related STs are also well known. Such clusters can be recognized by eBurst analyses [25], and are referred to as ST Complexes in E. coli [26] and eBGs (eBurst groups) in S. enterica [27]. ST Complexes and eBGs seem to reflect natural populations, but their broad properties are still not well understood [28].
The principles of legacy MLST were extended to rMLST, which uses sequences of 53 universal bacterial genes encoding ribosomal proteins and provides a universal MLST scheme for all Bacteria [29]. For both Escherichia and Salmonella, rMLST offers a slight improvement in resolution over legacy MLST, and the identification of Salmonella eBGs is reasonably consistent across both approaches [30]. rMLST has been used to identify tractable sets of representative genomes from large collections for calculating phylogenetic trees [28] and pan-genomes [17].
MLST has also been extended to cgMLST, which encompasses all the genes in a soft core genome [29–31], and cgMLST nomenclatures have been implemented for multiple bacterial genera [28]. The large number of loci encompassed by cgMLST schemes results in enormous numbers of cgMLST STs (cgSTs), but these can be clustered into groups of bacterial genomes at multiple levels of genomic diversity (hierarchical clustering) [32]. cgMLST provides considerably higher resolution than legacy MLST or rMLST, and initial analyses indicated that it is ideal for investigating transmission chains within single source outbreaks or for identifying population structures up to the genus level [28]. Here we focus on the automated assignments of genome assemblies from six genera of important bacterial pathogens to taxonomic and population structures by hierarchical clustering of cgSTs with the EnteroBase HierCC pipeline (table 1) [32]. For five of those genera, HierCC is a full solution for taxonomic designations of species.
Table 1.
Legacy data, newly assembled genomes and hierarchical cgST clusters in EnteroBase (09/2021). NOTE: no. genomic loci is numbers of coding sequences whose alleles are automatically called for the wgMLST and cgMLST schemes; HierCC level: maximum numbers of allelic differences within minimal spanning trees that define HC clusters of cgSTs. EnteroBase automatically calculates existing Legacy MLST STs according to legacy schemes for Escherichia/Shigella (Wirth et al. [26]), Salmonella (Achtman et al. [27]), and Clostridioides (Griffiths et al. [33]), but does not assign new STs nor does it maintain a database of legacy data from ABI sequencing. Additional public databases are presented by EnteroBase for Helicobacter (>3500 genomes) and Moraxella (>2350 genomes), but these lack a cgMLST scheme. EnteroBase also has a database for >80 000 Mycobacterium genomes, but this currently (March, 2022) lacks a cgMLST scheme.
| genus | legacy MLST (no. strains) | cgMLST (no. genomes) | no. genomic loci |
no. HierCC clusters (HierCC level) |
|||
|---|---|---|---|---|---|---|---|
| wgMLST | cgMLST | ST complexes | Lineages | species/subspecies | |||
| Salmonella | 4930 | 312 196 | 21 065 | 3002 | 3648 (900) | 2185 (2000) | 15 (2850) |
| Escherichia/Shigella | 9525 | 175 256 | 25 002 | 2513 | 1379 (1100) | 343 (2000) | 16 (2350) |
| Streptocococcus | 76 718 | 33 887 | 372 | 3681 (100) | 132 (363) | ||
| Clostridioides | 23 197 | 11 490 | 2556 | 299 (950) | 12 (2500) | ||
| Vibrio | 12 267 | 152 249 | 1128 | 2000 (800) | 155 (1090) | ||
| Yersinia | 4286 | 4023 | 19 591 | 1553 | 451 (600) | 34 (1490) | |
(b) . EnteroBase and HierCC
EnteroBase (http://enterobase.warwick.ac.uk) includes tools for automatic downloading of short-read sequences and their metadata from the public domain, assembly into draft genomes and population genetic analyses of the core and accessory genomes (table 2) [28]. Draft genomes are annotated according to genus-wide pan-genome schemes created with PEPPAN [17]. In September 2021, EnteroBase contained over 600 000 draft genomes from the six genera in table 1, and likely provides a nearly comprehensive overview of their global diversity. Many of these samples reflect a focus on food-borne disease in the US and United Kingdom, but this bias is increasingly being reduced by the global sources of many genomes (see electronic supplementary material, text, Sample Bias).
Table 2.
EnteroBase-related software tools that support the analysis of large numbers of bacterial genomes. NOTE: PEPPAN is a stand-alone program that was used to generate the wgMLST schemes used in EnteroBase. SPARSE is a second stand-alone program that has been used to extract taxon-specific reads from metagenomes of ancient DNA that were then used with EToKi to define pseudo-MAGs (metagenomic assembled genomes) that were uploaded to EnteroBase for phylogenetic comparisons. HierCC was used as a stand-alone program in development mode to define an initial set of clusters from a representative set of genomes. Subsequent assignments to existing clusters or to novel clusters were performed automatically within EnteroBase using pHierCC in production mode. BlastFrost is used by EnteroBase to determine the presence/absence of toxins and other pathovar characteristics of gastrointestinal pathogens within E. coli, and to assign gastrointestinal pathovar designations.
| name | purpose | citation | URL for stand-alone version |
|---|---|---|---|
| GrapeTree | GUI for depicting and analysing minimal spanning and NJ trees of character data | [34] | https://github.com/achtman-lab/GrapeTree |
| PEPPAN | pan-genome calculation including pseudo-genes from numerous representatives of an entire genus | [17] | https://github.com/zheminzhou/PEPPAN |
| SPARSE | assignment of metagenomic reads to individual taxa | [35,36] | https://github.com/zheminzhou/SPARSE |
| EToKi | EnteroBase toolkit of useful functions and pipelines | [28] | https://github.com/zheminzhou/EToKi |
| BlastFrost | efficient k-mer based search for DNA sequences in large genomic datasets | [37] | https://github.com/nluhmann/BlastFrost |
| HierCC | automated hierarchical clustering of cgSTs to existing or novel clusters | [32] | https://github.com/zheminzhou/pHierCC |
One of the primary goals for EnteroBase was a hierarchical overview of the population structure of the genera in table 1. We therefore developed HierCC [32] based on cgMLST assignments to support the rapid recognition and detailed investigation of differing levels of population structure. EnteroBase reports cluster assignments and designations at 10–13 levels of allelic differences for all six genera [28] (table 1). HC5–HC10 clusters with maximal internal pair-wise distances within minimal spanning trees of 5 or 10 alleles, respectively, have been used to identify short-term, single source outbreaks of S. enterica and E. coli/Shigella that extended to multiple European countries [38–42]. Pathogen species can also include higher level clusters, which can correspond to somewhat more distantly related bacterial populations that cause endemic or epidemic disease over longer time periods in one or more countries [43–46]. EnteroBase HierCC has even been used to classify all Shigella [47], which correspond to discrete lineages of E. coli [26,48,49].
Classical taxonomic approaches for assigning individual strains and genomes to species depend on human expertise and are not suitable for automated pipelines in real-time databases such as EnteroBase. This problem is acute because many short read sequences in the European Nucleotide Archive (ENA) have incorrect taxonomic assignments, or none at all, and phenotypic distinctions are inappropriate for databases containing 100 000s of genomes. Zhou et al. compared taxonomic designations with peak normalized mutual information and silhouette scores for HierCC clusters and identified HierCC levels that corresponded with well-defined species/subspecies in each genus [32]. For example, for Salmonella with a total of 3002 loci in the cgMLST scheme, HC2850 (94.9% of all cgMLST alleles) was chosen as the optimal HC level for identifying species and subspecies. The optimal HC levels for the other five genera with cgMLST schemes ranged from HC363 to HC2500 (93.6–97.8%) (table 1). Here we compare the consistency of those HierCC assignments with assignments based on classical taxonomic approaches and 95% ANI. The results illustrate problems with classical approaches and with 95% ANI, and we conclude that HierCC is preferable for automated assignment of genomes to species/subspecies for those genera. However, neither ANI nor HierCC is universally satisfactory for species/subspecies assignments within Streptococcus. This manuscript also provides an initial overview of the abilities of HierCC to assign genomes to populations and Lineages [50,51] within Salmonella and Escherichia/Shigella, and compares those assignments with the distributions of O antigens within lipopolysaccharide.
2. Results
(a) . Species and subspecies
In order to test the efficacy of HierCC at identifying species and subspecies, we extracted collections of representative genomes from all six genera in EnteroBase for which HierCC clusters had been implemented (table 3). We wished to calculate maximum-likelihood (ML) phylogenetic topologies of these genomes based on their core genome single nucleotide polymorphisms (SNPs) or the presence/absence of accessory genes from the pan-genome. Such ML trees are very slow to calculate with large datasets, but disjointed tree merging (DTM) within a divide-and-conquer approach allows large phylogenetic trees to be calculated in a reasonable time [52]. We therefore developed cgMLSA (see electronic supplementary material, Methods in Supplementary Text), a novel DTM approach based on ASTRID [53] and ASTRAL [54] that enabled the calculations of ML super-trees from up to 10 000 representative genomes within hours to days, and applied it to each of the datasets. The resulting ML super-trees were annotated with taxonomic designations and cluster designations according to a 95% ANI cutoff calculated with FastANI [10] (95% ANI clusters). We then compared those annotations with cluster assignments based on the species-specific HierCC levels in table 1.
Table 3.
Parameters of datasets used for calculating ML super-trees. Each dataset consists of genomes representing the entire diversity of a genus as described in Methods (see electronic supplementary material, Supplemental Text).
| genus | no. type strains | total no. genomes | no. strict core genes | no. accessory genes | no. soft core genes (cgMLST) | no. SNPs |
|---|---|---|---|---|---|---|
| Salmonella | 7 | 10 002 | 1409 | 19 486 | 3002 | 1 410 331 |
| Escherichia (EcoRPlus) | 8 | 9479 | 134 | 20 294 | 2513 | 1 172 200 |
| Escherichia reps | 2 | 967 | 192 | 17 983 | 2513 | 971 516 |
| Streptocococcus | 84 | 5937 | 90 | 31 630 | 372 | 263 080 |
| Clostridioides | 1 | 6725 | 515 | 10 975 | 2556 | 725 240 |
| Vibrio | 119 | 5032 | 124 | 146 041 | 1128 | 776 439 |
| Yersinia | 23 | 1847 | 744 | 18 799 | 1553 | 656 143 |
(i) . Salmonella
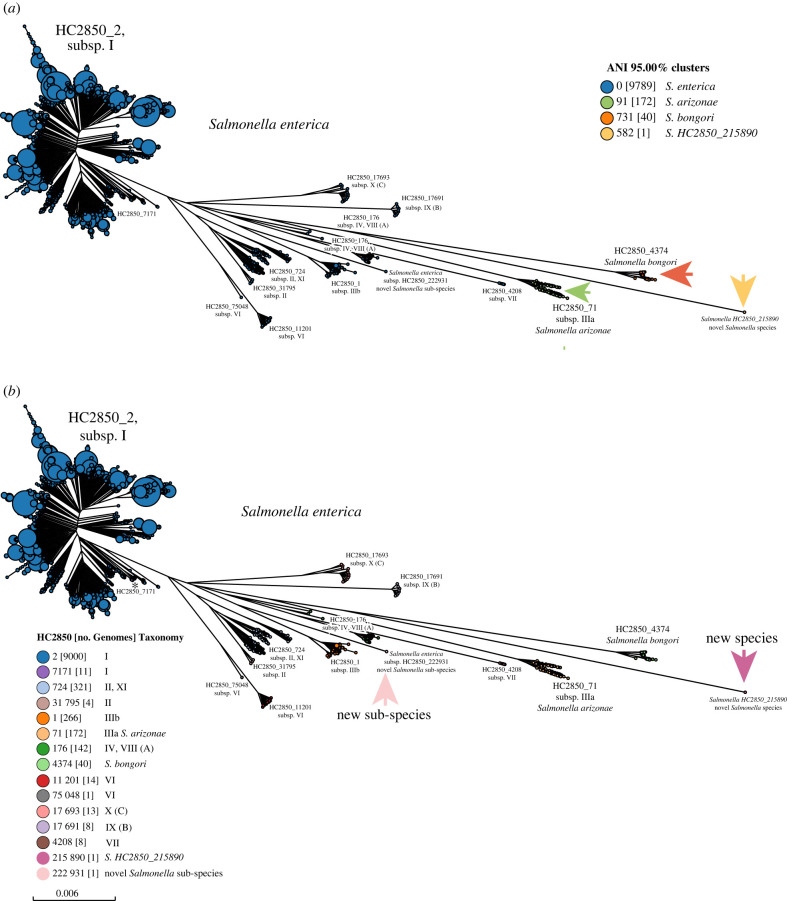
The topologies of ML super-trees based on 1 410 331 core SNPs from 10 002 representative genomes were in large part concordant with traditional taxonomic assignments, and with clustering according to 95% ANI (figure 1a) or HC2850 (figure 1b). The three sets of assignments were also largely concordant with the topology of an ML super-tree based on the presence or absence of accessory genes (https://enterobase.warwick.ac.uk/ms_tree/53258). HC2850 and 95% ANI distinguished S. enterica from S. bongori, and from former subspecies IIIa, which was recently designated S. arizonae by Pearce et al. [55]. HC2850 also identified another new Salmonella species, cluster HC2850_215890, and that identification was confirmed by 95% ANI. HC2850_215890 consists of five strains that have been isolated from humans since 2018 in the UK, and a gene for gene comparison indicated that roughly half of their core genes were more similar to S. enterica and the other half to S. bongori.
Figure 1.
A comparison of species and subspecies assignments within Salmonella with HierCC and ANI. The figure shows an ML super-tree of 1 410 331 SNPs among 3002 core genes from 10 002 representative genomes of Salmonella (table 3). Former subspecies IIIa is designated S. arizonae in accordance with Pearce et al. 2021 [55]. (a) Partitions differentiated by ANI 95% clusters (legend) correspond to species S. enterica, S. bongori, S. arizonae and a new species, S. HC2850_215890 (five strains from the UK, 2018–2020), as indicated by arrows, and subspecies are not differentiated. (b) Partitions coloured by HC2850 clusters (legend). Arrows indicate HC2850_215890, and a new subspecies, HC2850_222931 (one strain from France, 2018). All other HC2850 clusters correspond to species (S. bongori and S. arizonae) or subspecies, except for HC2850_7171 (starred), which is subsp. enterica (I) according to the ML tree. An interactive version of this GrapeTree rendition can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53257. The corresponding presence/absence tree can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53258.
Ninety-five percent ANI did not detect any subspecies structure within S. enterica whereas HC2850 differentiated a total of 12 clusters that corresponded to distinct phylogenetic branches in the ML super-tree (figure 1b). Most of these have been designated as subspecies by traditional taxonomy [30,55], except for a singleton genome in HC2850_222931, which likely represents a novel subspecies. Despite the correct identification of so many subspecies, HC2850 did not support Pearce's definition of subspecies XI and did not distinguish those genomes from subsp. II. Similarly, HC2850 did not differentiate Pearce's subsp. VIII from subsp. IV. The prior definition of these subspecies was largely dependent on branch topologies within phylogenetic trees based on rMLST loci, and was also contradicted by population statistics [55].
HierCC also differentiated HC2850_7171, which consists of a short phylogenetic branch within subsp. I that would not have been assigned to a subspecies according to its ML topology. Genomic comparisons of multiple single genes from genomes within HC2850_7171 suggest that HC2850_7171 may be a hybrid between subsp. I and II because some gene sequences were most similar to the former and others were most similar to the latter. With this sole possible exception, HierCC seems to be suitable for the automated detection of new Salmonella species and subspecies, and for routinely assigning novel genomes to the appropriate taxa.
(ii) . Escherichia
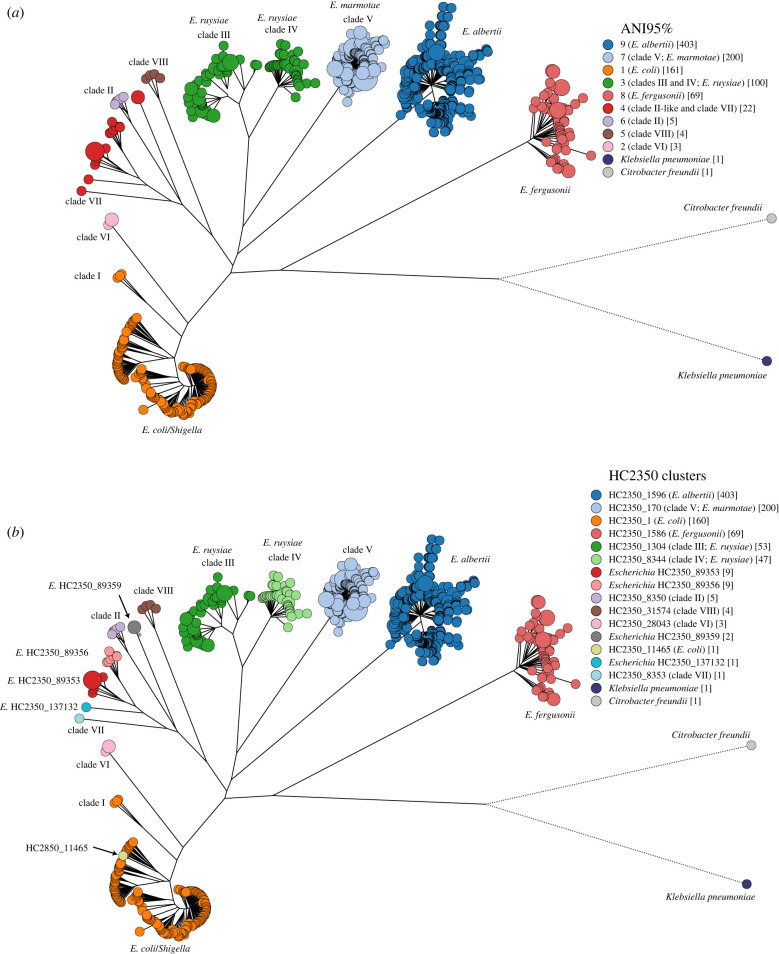
In addition to Escherichia coli, Escherichia includes the named species albertii [56–58], fergusonii [59,60], marmotae [61,62] and ruysiae [63]. And despite their apparently distinct genus and species names, Shigella boydii, Shigella dysenteriae, Shigella flexneri and Shigella sonnei, all common causes of dysentery, correspond to phylogenetic lineages within E. coli [48] rather than to discrete taxonomic units. Still other, unusual Escherichia strains from lake and ocean water are associated with long phylogenetic branches, and were designated as ‘cryptic clades’ I–VIII by population geneticists [64–69]. The branch leading to clade I is simply a long phylogenetic branch within E. coli [64]. However, clade V encompasses E. marmotae [61,62] and E. ruysiae consists of the union of clades III and IV [63]. Initial analyses with the EcoRPlus collection of 9479 genomes [28] (table 3) yielded results that were compatible with these interpretations. Almost all E. coli or Shigella genomes were within HierCC cluster HC2350_1, and genomes with other taxonomic or clade designations belonged to other HC2350 clusters. However, soon after the definition of the EcoRPlus collection, additional genomes of Escherichia which were related to clade II were described from inter-tidal marine and fresh-water sediments near Hong Kong [66,70]. Furthermore, due to its numerical predominance, E. coli overshadows other Escherichia species and subspecies within EcoRPlus. We therefore, created Escherichia reps in Jan 2021, a novel set of 967 representative genomes which included one genome from each of the 160 most common HC1100 clusters in HC2350_1 and all 807 genomes from other HC2350 clusters which existed in EnteroBase at that time.
Figure 2 shows that E. fergusonii and E. albertii form discrete 95% ANI clusters as do E. marmotae and other clade V genomes. As previously reported [63], E. ruysiae consists of the distinct clusters of clades III and IV. Three distinct 95% ANI clusters were found within clades II, VI and VIII while the Hong Kong genomes represented multiple, related phylogenetic branches, one of which has previously been designated clade VII. All E. coli and Shigella genomes are in a common 95% ANI group, as is clade I. Comparable clustering results were also found in an ML tree of presence/absence of accessory genes, albeit with different topology of the deepest branches resulting in E. fergusonii and clade VIII being most closely related to E. coli (https://enterobase.warwick.ac.uk/ms_tree?tree_id=71125). These observations indicate that the taxonomy and population genetic designations for Escherichia are incomplete, and also partially inconsistent.
Figure 2.
Maximum-likelihood core SNP tree of 967 Escherichia genomes consisting of one genome from each of 161 HC1100 clusters containing E. coli or Shigella as well as all 806 other Escherichia genomes in EnteroBase as of November 2020. The tree is coloured by (a) pairwise FastANI values clustered at the 95% level and (b) HC2350 cluster designations. The key legends indicate taxonomic designations in the literature which best match the cluster groupings. In (b), HC2350 cluster designations were used to mark novel taxonomic groupings in HC2350 clusters 89353, 89356, 89359 and 137132. An interactive version of this GrapeTree rendition can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=52101. The corresponding presence/absence tree can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=71125.
HierCC clustering of the same data provided a consistent and uniform nomenclature. Each discrete phylogenetic cluster received its own HC2850 designation, including clades III and IV and four phylogenetic clusters among the Hong Kong isolates which were designated according to their HC cluster number: e.g. Escherichia HC2350_89353. E. coli, Shigella and clade I all clustered together in HC2350_1, and the only discordance between topological clustering and HierCC clustering among the 967 genomes in Escherichia reps was a single genome which belonged to E. coli by phylogenetic topology but was assigned to HC2350_11465 by HierCC. These results were so convincing that HierCC groupings were used in late 2021 to curate and update all species designations for Escherichia entries within EnteroBase.
(iii) . Clostridioides
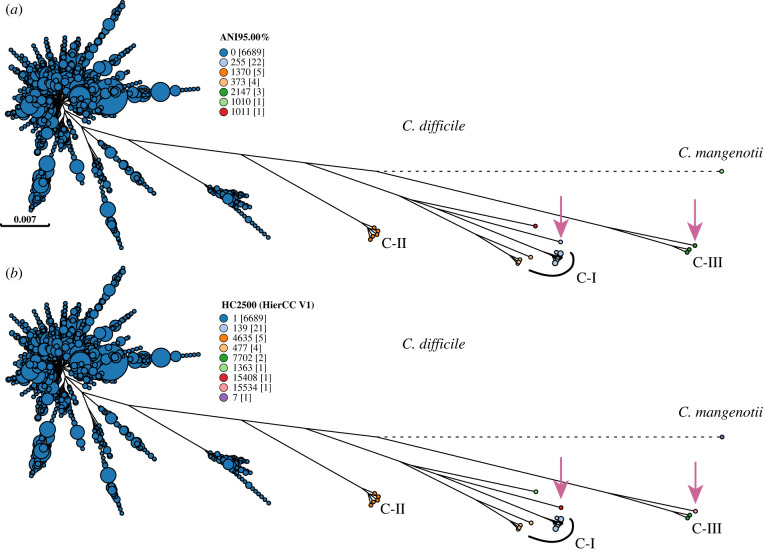
Clostridium difficile clustered distinctly from C. mangenotii according to 95% ANI, and ANI also distinguished five other clusters among genomes that were designated C. difficile. These clusters seem to be novel species according to their phylogenetic topology in the ML SNP super-tree (figure 3). HC2500 identified the same clusters, and also separated out two additional distinct clusters which resemble novel subspecies (arrows: HC2500_15334, HC2500_15408). A subset of the genomes in these clusters (figure 3) were assigned to cryptic clades C-I, CII and C-III by a recent publication [71], which also concluded that they represent novel species. Thus, HierCC is also suitable for the automated detection of new Clostridioides species and subspecies, and for routinely assigning novel genomes to the appropriate taxa.
Figure 3.
Species and subspecies assignments within Clostridioides according to 95% ANI (a) and HierCC (b). ML super-tree of 725 240 SNPs among 2556 core genes from 6724 representative genomes of Clostridioides difficile and one genome of Clostridioides mangenotii (table 3). (a) ANI 95% clusters differentiate C. mangenotii (cluster 1010) and five other clusters (clusters 255, 1370, 373, 2147, 1011) from C. difficile (cluster 0). Four of the 95% ANI clusters correspond to cryptic clades C-I (clusters 255, 373), C-II (cluster 1370) and C-III (cluster 2147) in the designations by Knight et al. [71]. Arrows indicate two additional clusters that were distinguished by HierCC in (b). (b) Partitions coloured by HierCC assign the same genomes to HC2500 clusters as ANI, except that HierCC assigns HC2500_15334 and HC2500_15408 designations to one genome each. An interactive version of this GrapeTree rendition can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53253 and a presence/absence tree can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53254.
(iv) . Yersinia and Vibrio
Detailed results with Yersinia and Vibrio can be found in the electronic supplementary material, text. The general take-home lessons from those analyses resemble those summarized above: 95% ANI clustering is largely congruent with existing taxonomic designations, and HierCC identifies both species and subspecies with better resolution than 95% ANI. However, multiple, unanticipated taxonomic problems were evident in both genera. Firstly, ENA uses the strain designation as a species designation when a species is not specified during the submission process. The resulting public metadata is bloated with inaccurate species names. Secondly, multiple distinct phylogenetic branches were found without any species identifier. Thirdly, in multiple cases the species identifier did not correspond with ANI and/or HierCC, and comparisons with both the SNP and presence/absence ML topologies confirmed that the species designations in ENA were incorrect. We addressed these problems within EnteroBase by performing radical manual curation to the metadata to ensure that the taxonomic species designations correspond to the phylogenies and population structures. We also encountered multiple, problematical taxonomic designations which are described in the next paragraphs.
(v) . Yersinia
Ninety-five percent ANI supports the taxonomic convention that Y. enterocolitica represents a single species, and all genomes designated as Y. enterocolitica were in a single 95% ANI cluster. However, in accord with the conclusions by Reuter et al. [72], HierCC clustering assigned the non-pathogenic biotype 1A genomes to HC1490_73 and HC1490_764, the highly pathogenic biotype 1B to HC1490_2, and pathogenic biotypes 2–5 to HC1490_10. We therefore renamed these four groups by adding the HierCC cluster to the species name, e.g. Y. enterocolitica HC1490_2.
In contrast to Y. enterocolitica, the Yersinia pseudotuberculosis Complex corresponds to a single phylogenetic cluster according to both HC1490 and 95% ANI (electronic supplementary material, figure S1). Taxonomists have split these bacteria into Y. pseudotuberculosis, Y. pestis, Y. similis and Y. wautersii [73,74]. DNA–DNA hybridization [75] and gene sequences of several housekeeping genes [4] previously demonstrated that Y. pestis is a clade of Y. pseudotuberculosis, and the new observations demonstrate that a distinct species status is not consistent with the genomic data for Y. similis and Y. wautersii. We have therefore downgraded all three taxa within EnteroBase to the category of subspecies of Y. pseudotuberculosis, and extended the designation of Y. pseudotuberculosis to Y. pseudotuberculosis sensu stricto. These assignments are not reflected by distinct HC1490 clusters, and HC1490 clustering can only be used to automatically assign new genomes to the Y. pseudotuberculosis Complex.
We also defined eight other novel species/subspecies designations within Yersinia and assigned unique designations based on HierCC clusters: e.g. Yersinia HC1490_419. These clusters were previously unnamed, or had been incorrectly designated with the names of other species which formed distinct ANI and HC1490 clusters (electronic supplementary material, text, figure S1).
(vi) . Vibrio
Vibrio encompassed 152 HC1090 clusters (electronic supplementary material, figure S2, text), which corresponds to much greater taxonomic diversity than for the other genera dealt with above. The concordance between HC1090 and 95% ANI clusters was absolute for most clusters, including a large numbers of genomes from V. cholerae, V. parahaemolyticus, V. vulnificans and V. anguillarum (electronic supplementary material, table S1), and did not support the existence of additional subspecies. However, eleven HC1090 clusters each encompassed between two and five ANI clusters (electronic supplementary material, table S2) and three ANI clusters each encompassed 2–3 HC1090 clusters (electronic supplementary material, table S3). In order to support the automated assignment of genomes to named species, we implemented taxonomic assignments according to HC1090 and renamed the species of all genomes that were contradictory to this principle. The resulting dataset contains 109 HC1090 clusters with a unique, classical species designation as well as 43 other species level clusters designated as Vibrio HC1090_xxxx. Seventeen species names were eliminated because they were contradictory to the phylogenetic topologies or were incoherently applied (electronic supplementary material, text, table S4). These taxonomic changes now permit future automated assignment of novel genomes to species designations, and the recognition of novel species, and have provided a clean and consistent basic taxonomy that can be progressively expanded.
Prior work has assigned many Vibrio species into so-called higher order clades of species on the basis of MLSA (multilocus sequence analysis) [76]. These clades are also apparent in the ML super-tree of core SNPs, and electronic supplementary material, figure S2 indicates the three largest (Cholerae, Harveyii, Splendidus).
(vii) . Streptococcus
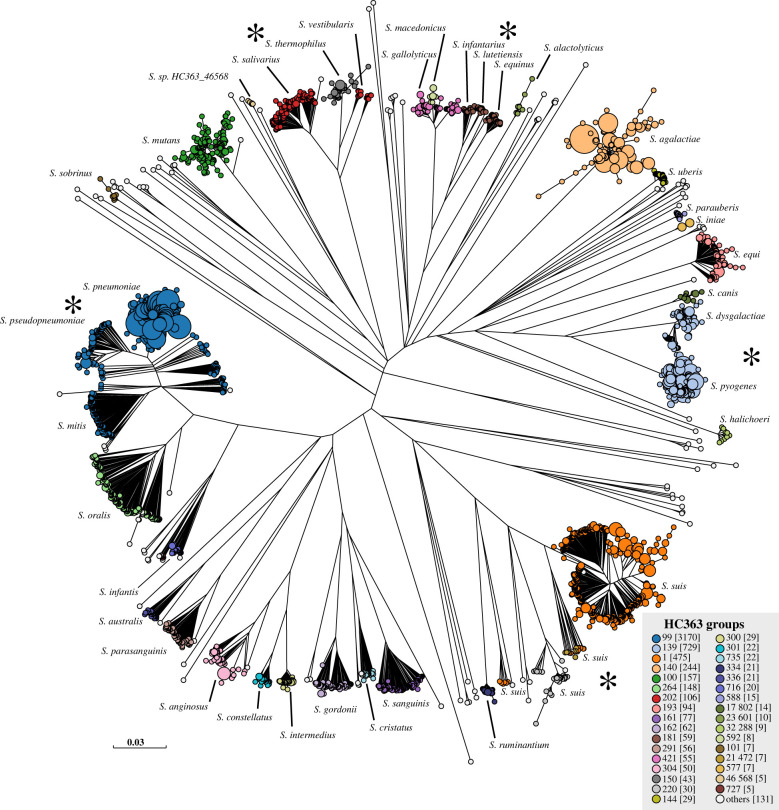
For all five genera summarized above, clustering according to HierCC was largely concordant with the ML super-trees based on core SNPs or presence/absence of accessory genes. Ninety-five percent ANI and taxonomic designations were also largely concordant with the phylogenetic trees, although to a lesser degree. Similar concordances with the trees based on core SNPs (figure 4) and presence/absence of accessory genes (electronic supplementary material, figure S3) were also found for a majority of the named species within Streptococcus. One hundred and two HC363 clusters were concordant with 95% ANI clusters, and each of those HierCC clusters was specific for a single species after eliminating S. milleri, S. periodonticum and S. ursoris (electronic supplementary material, table S5). These results also demonstrated strong agreement between the two methods with classical taxonomy. However, genomes from a large number of other species within Streptococcus were not clustered satisfactorily by either method.
Figure 4.
Comparison of HC363 clusters with taxonomic designations in Streptococcus. ML super-tree of 263 080 SNPs among 372 core genes from 5937 representative Streptococcus genomes (table 3). Species names are indicated next to the phylogenetic clusters according to the locations of genomes from type strains and public metadata. Nodes were coloured by HC363 clusters, and exceptional assignments are indicated by asterisks next to S. pneumoniae and S. pseudopneumoniae, which were both HC363_99; multiple phylogenetic and HierCC clusters within S. suis; S. salivarius and S. vestibularis, which were both HC353_202; S. lutetiensis and S. equinus, which were both HC363_181; and S. dysgalactiae and S. pyogenes, which were both HC363_139. An interactive version of the GrapeTree rendition of the SNP tree can be found at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53261 and the presence/absence tree at https://enterobase.warwick.ac.uk/ms_tree?tree_id=53262.
Earlier publications [15,16,77,78], had demonstrated that Streptococcus mitis and oralis each represents multiple related sequence clusters rather than two single species, and similar results were obtained with 95% ANI clustering [17]. These observations were confirmed by the current data. Of the 102 concordant HC363 and 95% ANI clusters, 23 clusters from S. cristatus, S. infantis, S. mitis, S. oralis, S. sanguinis or S. suis each corresponded to only a sub-set of the topological diversity within their species. Furthermore, single HC363 clusters from 18 species or groups of species were subdivided extensively by 95% ANI for a total of 155 discrepancies between the two methods (electronic supplementary material, table S6). These discrepant clusters were within the species S. suis, S. criceti, S. sanguinis, S. oralis, S. hyointestinalis, S. parasanguinis, S. australis, S. infantis and S. cristatus. Ninety-five percent ANI was not able to differentiate S. pneumoniae from S. pseudopneumoniae, S. pyogenes from S. dysgalactiae, S. salivarius from S. vestibularis, or between S. equinus, S. infantarius and S. lutentiensis. Contrariwise, three other 95% ANI clusters each encompassed two HC363 clusters (electronic supplementary material, table S7). HC363 could not distinguish S. pneumoniae or S. pseudopneumoniae from 57 95% ANI clusters within S. mitis. In addition to these problems, eight additional species should not have been given a species name because their type strains belonged to one of these chaotic clusters. We conclude that numerous species have been defined on taxonomic grounds that cannot be correctly identified by either 95% ANI or HierCC, and that multiple discrepancies exist between the two approaches within Streptococcus.
(b) . Populations, eBurst groups and Lineages
HierCC supports the identification of populations at multiple hierarchical levels ranging from the species/subspecies down to individual transmission chains. We also expected that intermediate HierCC cluster levels could reliably detect the natural populations defined by legacy MLST. Here we focus on examples of such populations from Salmonella, E. coli/Shigella and S. pneumonia, taxa where they have been examined in greatest detail.
(i) . Salmonella
eBurstGroups (eBGs) defined by legacy MLST in Salmonella generally correspond to HierCC HC900 clusters [32]. In some cases, genetically related groups of eBGs correspond to HC2000 clusters [50]; we refer to those as Lineages. By contrast, none of the HierCC levels corresponded consistently with other, prior intra-species subdivisions of S. enterica subsp. enterica into lineages [79], clades [80–83] or branches [84]. This failure may reflect a high frequency of extensive homologous recombination at these deeper branches, which can obscure topological relationships [51,84], whereas HC900 (and HC2000 clusters) diverged more recently, and have not yet undergone extensive recombination [51].
Traditional subgrouping nomenclature of Salmonella below the subspecies level predates DNA sequencing and is predominantly based on serovar designations. Serovar designations consist of common names that have been assigned to a total of more than 2500 unique antigenic formulae based on epitopes within lipopolysaccharide (O antigen) and two alternately expressed flagellar subunits (H1, H2). These antigenic formulae are written in the form O epitopes: H1 epitopes: H2 epitopes in the revised Kauffmann-White scheme [85], and O epitopes are summarized as O serogroups with distinct numeric designations [86] , e.g. O:4 for the O group of serovar Typhimurium [87]. Detailed metabolic maps for lipopolysaccharide (LPS) synthesis have been elucidated for 47 O serogroups [87,88]. Serogroups from natural isolates can be determined serologically by agglutination reactions with specific antisera or in silico from genomic sequences with the programs SeqSero [89] or SISTR [90]. In practice, each of these methods has an error rate of a few percent [27,91].
Several hundred legacy eBGs within subsp. enterica were relatively uniform for serovar [27], with multiple exceptions, and many serovars were associated with multiple, apparently unrelated eBGs, likely due to the extensive exchange of genes encoding LPS and flagellar epitopes between eBGs early in their evolutionary history. We proposed that legacy MLST was a better metric for identifying the natural populations than serotyping, and could completely replace this traditional method [27]. Our new data indicate that HC900 clusters are even more reliable than legacy eBGs for recognizing natural populations within Salmonella.
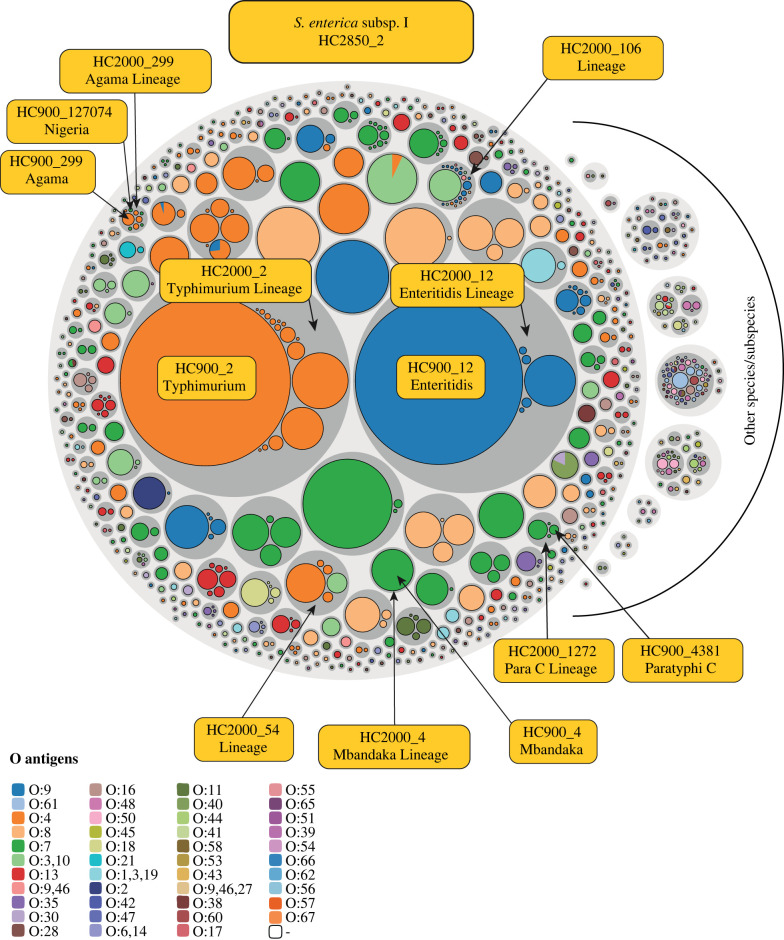
Several phylogenetic comparisons indicated that HC900 clusters based on cgMLST are concordant with eBGs in Salmonella [32,50]. Here we test this concordance quantitatively over a total of 319 490 genomes (table 4). The concordance between eBG and HC900 clusters was 0.985 according to the Adjusted Mutual Information Score (AMI) (table 4), a metric that is suitable for samples with a heterogeneous size distribution [94], and almost as high with the Adjusted Rand Index (ARI: 0.982), which is less suitable for heterogeneous data. Comparisons of eBG or HC900 with HC2000 clustering yielded slightly lower AMI scores (table 4). We also tested whether HC900 clusters are uniform for serovar. In late 2019, we corrected false serological data in the metadata Serovar field in EnteroBase by manual curation of 790 HC900 clusters that contained at least five entries. Ninety-seven percent (770/790) of those HC900 clusters were uniform (greater than or equal to 95%) for serovar (electronic supplementary material, table S8), as were most HC2000 clusters (437/473 clusters; 92%). The data also indicated that the predominant O groups were uniform over the multiple HC900 clusters within 95% (382/403) of HC2000 clusters within S. enterica subsp. enterica and 79% (58/70) of HC2000 clusters in other Salmonella species and subspecies (figure 5; electronic supplementary material, table S9).
Table 4.
Quantitative concordance between eBG and HierCC clustering. NOTE: Salmonella: Calculations were performed on 319 490 entries in EnteroBase which had been assigned to eBGs, HC900 and HC2000 clusters by December, 2021. The dataset only represented 411 eBGs, 690 HC900 clusters and 312 HC2000 clusters, and consists of a subset of all genomes because new eBGs have not been created in recent years. Escherichia/Shigella: HC1100 and HC2000 assignments were from 143 520 genomes which had been assigned to ST Complexes as well as to HC1100 and HC2350 clusters in December, 2021. At that time 186 852 genomes had been assigned to HC1100 and HC2000 clusters, with AMI of 0.59 and ARI of 0.2. Streptococcus pneumoniae consisted of GSPC assignments for 18 147 genomes from electronic supplementary material, table S2, and CC assignments for 13 396 genomes from electronic supplementary material, table S1 by Gladstone et al. [92] which were imported into User defined fields in EnteroBase as described [28]. ARI and AMI were calculated with the functions adjusted_rand_score() and adjusted_mutual_info_score() in the Python 3 library sklearn.metrics v. 1.0.1 [93].
| Salmonella | |||
| Test statistic | eBG-HC900 | eBG-HC2000 | HC900-HC2000 |
| Adjusted Mutual Information Score (AMI) | 0.985 | 0.948 | 0.942 |
| Adjusted Rand Index (ARI) | 0.982 | 0.856 | 0.866 |
| Escherichia/Shigella | |||
| Test statistic | ST Cplx-HC1100 | ST Cplx-HC2000 | HC1100-HC2000 |
| Adjusted Mutual Information Score (AMI) | 0.940 | 0.688 | 0.692 |
| Adjusted Rand Index (ARI) | 0.871 | 0.299 | 0.337 |
| Streptococcus pneumoniae | |||
| Test statistic | CC-HC100 | GSPC-HC100 | GSPC-HC160 |
| Adjusted Mutual Information Score (AMI) | 0.959 | 0.956 | 0.980 |
| Adjusted Rand Index (ARI) | 0.987 | 0.906 | 0.949 |
Figure 5.
Hierarchical population structure of O serogroups in Salmonella. Hierarchical bubble plot of 310 901 Salmonella genomes in 790 HC900 clusters for which a consensus O serogroup could be deduced by metadata, or bioinformatic analyses with SeqSero V2 [89] or SISTR 1.1.1 [90]. Taxonomic level (HC level; colours): species/subspecies (HC2850; light grey circles), Lineages (HC2000; dark grey circles) and eBurst groups (HC900; O:group specific colours). Additional information is indicated by yellow text for selected HC2000 and HC900 circles which are specifically mentioned in the text. The diameters of HC900 circles are proportional to the numbers of genomes. An interactive version of this figure can be found at https://observablehq.com/@laurabaxter/salmonella-serovar-piechart from which the representation, raw data and d3 Java code [95] for generating the plot can be downloaded.
HC2000 Lineages with uniform O group included the Para C Lineage (HC2000_1272; serovars Paratyphi C, Typhisuis, Choleraesuis and Lomita [51]) which emerged about 3500 years ago (electronic supplementary information, Supplemental Text) [51,84]. The Para C Lineage is uniformly serogroup O:7, which is not particularly surprising because all its serovars have identical antigenic formulaes, and are only distinguished by biotype. The Enteritidis Lineage (HC2000_12) consists of serovars Enteritidis (1,9,12:g,m:-), Gallinarum and its variant Pullorum (1,9,12:-:-), and Dublin (1,9,12:g,p:-:-) [50] (figure 5), and is uniform for O:9. The Typhimurium Lineage (HC2000_2) includes serovars Typhimurium, Heidelberg, Reading, Saintpaul, Haifa, Stanleyville and others [50]. This Lineage is also uniform for O group because all these serovars are O:4. The Mbandaka Lineage (HC2000_4) includes serovars Mbandaka (6,7,14: z10:e,n,z15) and Lubbock (6,7:g,m,s:e,n,z15), and is uniformly O:7. However, three Lineages were exceptional, and did contain more than one O serogroup. HC2000_299 is a mixture of O:4 (HC900_299; serovar Agama [28]) and O:7 (HC900_127074; serovar Nigeria). HC900 clusters within HC2000_54 are O:4 (HC900_54: Bredeney, Schwarzengrund; HC24937: Kimuenza) or O:9,46 (HC900_57: Give). Similarly, HC900 clusters within HC2000_106 are O:3,10; O:9; O:9,46; O:4: or O:8. These observations confirm and extend the prior conclusions about concordance between natural populations and serogroup within Salmonella.
(ii) . Escherichia coli
Patterns of multilocus isoenzyme electrophoresis were used in the 1980s to provide an overview of the genetic diversity of E. coli (see overview by Chaudhuri and Henderson [96]). Those analyses yielded a representative collection of 72 isolates [97], the EcoR collection, whose deep phylogenetic branches were designated haplogroups A, B1, B2, C, D and E [98]. Several haplogroups have since been added [99,100]. The presence or absence of several accessory genes can be used for the assignment of genomes to haplogroups with the Clermont scheme [101,102], and the haplogroup can also be predicted in silico from genomic assemblies with ClermontTyping [103] or EZClermont [104], both of which are implemented within EnteroBase. However, the Clermont scheme ignores Shigella, which consists of E. coli clades despite its differing genus designation [26,47,48], and makes multiple discrepant assignments according to phylogenetic trees [28]. The Clermont scheme also does not properly handle the entire diversity of species and environmental clades/subspecies within the genus Escherichia [28,63,64,66].
EnteroBase HierCC automatically assigns genomes within the Escherichia/Shigella database to the cgMLST equivalents of ST Complexes (HC1100 clusters) and Lineages (HC2000 clusters) (table 1). It also perpetuates the ST Complexes that were initially defined for legacy MLST by Wirth et al. [26]. However, the numbers and composition of legacy ST Complexes have not been updated since 2009 because additional sequencing data defined intermediate, recombinant genotypes that were equidistant to multiple ST Complexes, and threatened to merge existing ST Complexes. By contrast, HC1100 clusters do not merge: intermediate genotypes are rare because cgMLST involves 2512 loci while legacy MLST was based on only seven. Secondly, new genotypes which are similar to and equidistant to multiple clusters do not trigger merging because HierCC arbitrarily assigns them to the oldest of the existing alternatives [32]. Finally, unlike ST Complex designations in legacy MLST, which have not been actively updated in Escherichia, HierCC clusters are automatically created as necessary for new genotypes.
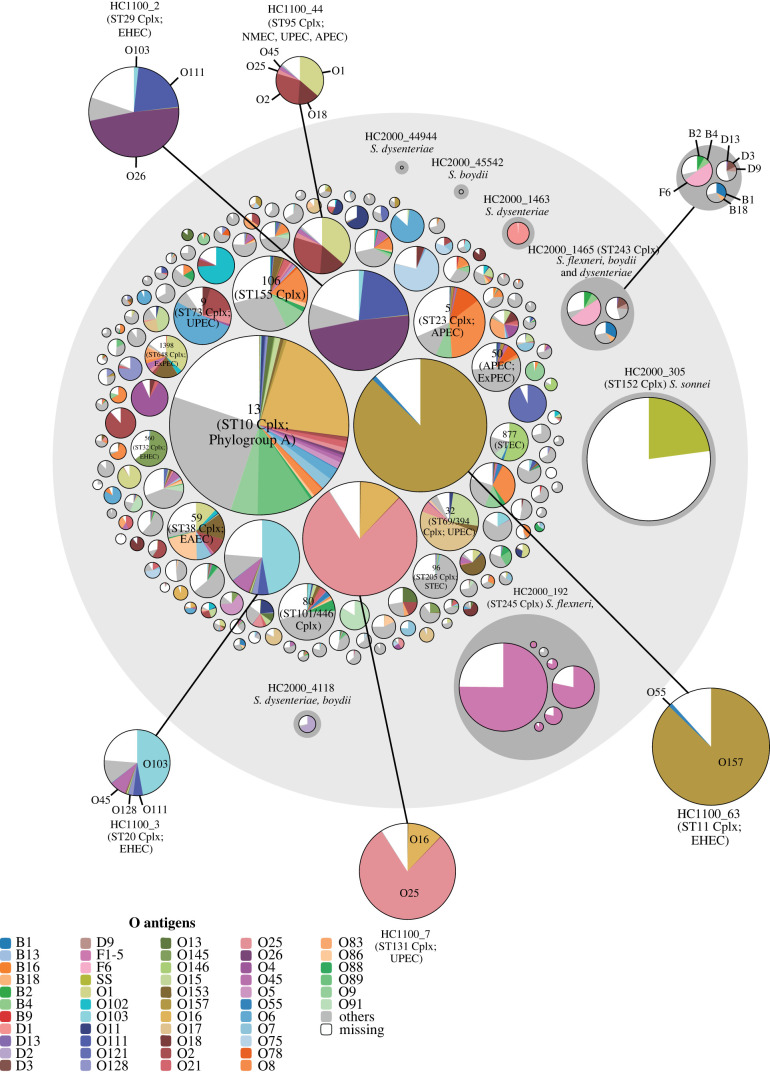
Frequent recombination in E. coli results in poor bootstrap support for the deep branches in phylogenetic trees of concatenated genes [26], and we were unable to identify a unique HierCC level which was largely concordant with haplogroups according to the Clermont scheme. However, similar to Salmonella, HC1100 clusters identify clear population groups, which are highly concordant with legacy ST Complexes (AMI = 0.94) (table 4). Unlike Salmonella, HC2000 clusters did not in general mark recognizable additional population structure beyond that which was provided by ST Complexes, and HC2000 clusters are only moderately concordant with either legacy ST Complexes (AMI = 0.69) or their cgMLST equivalent, HC1100 clusters (AMI = 0.69) (table 4). Multiple Shigella species are exceptions to this rule and their legacy ST Complexes [26] equate to cgMLST HC2000 clusters rather than HC1100 clusters according to phylogenetic trees. HC2000_305 replaces ST152 Cplx, and encompasses all S. sonnei (figure 6). HC2000_192 replaces ST245 Cplx, and contains many S. flexneri. However, S. flexneri O group F6 is in an HC1100 cluster within HC2000_1465 (ST243 Cplx), as are S. boydii B2 and B4. HC2000_1465 also includes a second HC1100 cluster with S. boydii O groups B1 and B18, and a third with S. dysenteriae D3, D9 and D13 (figure 6). HC2000_4118 replaces the combination of ST250 Complex and ST149 Complex, which have merged, and contains both S. dysenteriae as well as S. boydii.
Figure 6.
Hierarchical population structure of O serogroups in Escherichia coli/Shigella. Hierarchical bubble plot for 167 312 genomes of Escherichia coli or Shigella in HC2350_1 (large light grey circle) that were available in EnteroBase in April, 2021. Seven HC2000 Lineages encompassing 15 HC1100/ST Complexes of Shigella are shown at the right. HC2350_1 also includes 15 other E. coli HC2000 Lineages that each contains at least 50 E. coli genomes and encompass 144 other HC1100 clusters. The remainder of the figure shows those HC1100 clusters and not the corresponding HC2000 clusters. Numbers of genomes assigned to individual O serogroups (legend) are indicated by pie chart wedges within the HC1100 circles. Selected HC1100 clusters are also depicted with indications of phenotype and nomenclature at a larger scale outside the main circle, connected to the original circles by lines. An interactive version of this figure can be found at https://observablehq.com/@laurabaxter/escherichia-serovar-piechart, from which the representation, raw data and d3 Java code [95] for generating the plot can be downloaded.
Unlike Salmonella, O serogroups are remarkably variable within HC1100 clusters of E. coli (figure 6). HC1100_63 (ST11 Complex) combines classical EHEC strains of serovar O157:H7 as well as O55:H7 [105], and other, atypical ancestral EPEC isolates of serovar O55:H7 [106]. Multiple other HC1100 clusters that contain EHEC strains also encompass multiple O groups, including HC1100_2 (ST29 Cplx; O26, O103, O111) [107] and HC1100_3 (ST20 Cplx; O45, O103, O111, O128). Similarly, multiple HC1100 clusters of E. coli that cause extra-intestinal diseases also contain a variety of O groups, including HC1100_7 (ST131 Cplx; O16, O25) [23,108,109] and HC1100_44 (K1-encapsulated bacteria of the ST95 Cplx; O1, O2, O18, O25, O45) [26,110,111]. Indeed, the primary impression from figure 6 is that O group variation within an HC1100 cluster is nearly universal throughout E. coli, confirming prior conclusions about the high frequency of homologous recombination in this species [26,99].
(iii) . Other genera
Correlations between intra-specific HierCC clusters and population groupings at multiple levels, including ST Complexes and ribotypes have been described in Clostridioides difficile [112]. It remains to be tested whether HierCC can identify populations within Yersinia because bacterial populations in Y. pseudotuberculosis are largely obscured by recombination [113]. Similarly, we are not aware of extensive efforts to identify bacterial populations which could be compared with HierCC within Vibrio. However, recent work has described the assignment of large numbers of genomes of Streptococcus pneumoniae to legacy Clonal Complexes as well as GSPC (Global Pneumococcal Sequence Clusters) [92], initially with PopPunk [114] and more recently with Mandrake [115]. Most of those genomes have been assembled within EnteroBase, and assigned to HierCC clusters. We therefore compared the HierCC and GSPC assignments for 18 147 genomes, and also compared them to Clonal Complexes (CCs) based on legacy MLST (13 396 genomes). HC160 clusters yielded the highest AMI and ARI scores with GSPC clusters while CC was most concordant with HC100 clusters (table 4). These conclusions were also supported by visual comparisons of the colour-coding of nodes within a Neighbor-Joining tree based on cgMLST distances according to the various clustering criteria (electronic supplementary material, figure S4). Thus, HC100 clusters seem to be concordant with Clonal Complexes within S. pneumonia and Mandrake/PopPunk clustering equates roughly to HC160 clusters.
3. Discussion
In 2013, we anticipated that EnteroBase might eventually contain as many as 10 000 genomes of Salmonella and Escherichia. By the end of 2021 it hosted greater than 600 000 assembled genomes of Salmonella, Escherichia/Shigella, Streptococcus, Vibrio, Clostridioides and Yersinia, and has become one of the primary global sources of their genomic data. An analyses of their genomic properties is facilitated by software tools (table 2) that support indexing of genetic diversity, searching for genomes with specific metadata or genetic relations and investigating stable population structures at multiple levels. Here we focused on the automated identification and designation of species/subspecies and populations on the basis of HierCC of core genome MLST genotypes. The data demonstrate that HierCC can completely replace classical taxonomic methods for genomes of Salmonella, Escherichia/Shigella, Vibrio, Clostridioides and Yersinia, with only few minor exceptions. HierCC is also a complete replacement for legacy MLST regarding the assignments to populations within Salmonella, Escherichia/Shigella and Streptococcus pneumoniae.
(a) . Taxonomic assignments
The paired ML super-trees based on core SNPs and presence/absence of accessory genes from each genus were generally concordant in their clustering patterns, indicating that those genera contain well-defined and reproducible species/subspecies independent of the genetic criteria. Those phylogenetic clusterings were then used to resolve the most appropriate designations for genomes which were assigned contradictory designations by their taxonomic metadata versus clusters based on 95% ANI or HierCC. The frequency of agreement with current taxonomy was comparable for HierCC and 95% ANI at the species level within Salmonella, Escherichia/Shigella, Vibrio, Clostridioides and Yersinia but 95% ANI was generally unable to resolve subspecies whereas HierCC did not distinguish between subspecies and species clusters. Classical taxonomical designations exhibited multiple, glaring problems in each genus and we replaced those problematical designations in EnteroBase with labels based on the automated, HierCC-based species/subspecies taxonomy. We also defined novel species/subspecies, and labelled them with HierCC-based designations.
Why were there so many obvious discrepancies between the different approaches? One obvious reason is that we have discarded the medical tradition of retaining discrete species names for pathogens that cause particular diseases, and recommend designating Y. pestis as a subspecies of Y. pseudotuberculosis. Other discrepancies may reflect technical errors in manipulating data or uploading information to public databases, or simple strain mix-ups [46,50]. Possibly the discrepancies were particularly obvious because our analyses were performed at an unprecedented scale over multiple species. Many insights can be attributed to the facile ability to investigate large databases within a graphic framework that is provided by EnteroBase, and to our optimization of HierCC levels for each genus. However, we failed in a similar attempt to optimize ANI levels for recognizing both species and subspecies.
Our taxonomic changes in Yersinia are likely to be highly controversial. According to both ML trees and HierCC, the current designation ‘Y. enterocolitica’ encompasses four, previously unnamed species/subspecies. These taxa are not distinguished by 95% ANI and had not previously been assigned distinct names. We maintain continuity with the traditional designation of Y. enterocolitica, and simply add HierCC affiliations to the species name, e.g. Y. enterocolitica HC1490_2. Alternatively, these clusters of strains could be considered to represent subspecies and their name modified slightly, e.g. Y. enterocolitica subspecies HC1490_2, similar to our downgrading of the named species Y. pestis, Y. similis [74] and Y. wautersii [73] on phylogenetic grounds to subspecies of Y. pseudotuberculosis (electronic supplementary material, figure S1). Finally, we defined eight new species/subspecies within Yersinia without submitting traditional evidence for simple phenotypic differences, and named them after their HC cluster, e.g. Yersinia HC1490_4399. We also followed comparable strategies for the other genera analysed here.
In Salmonella, HierCC and 95% ANI identified a new species, Salmonella HC2850_215890, and a new subspecies, S. enterica subsp. HC2850_222931 (figure 1). Five new species were identified within C. difficile by HierCC as well as ANI, and HierCC identified two additional new subspecies. In Escherichia we provide HierCC designations for multiple clusters of bacteria from environmental sources near Hong Kong that were previously unnamed.
The ML trees presented here (electronic supplementary material, figure S2) support prior definitions of high order clades containing multiple species in Vibrio on the basis of MLSA [76]. We also observed a general concordance between ANI and HierCC for 109 named Vibrio species. However, 43 HierCC clusters of species rank had not previously been properly classified with species designations, and 17 other species names were superfluous. These changes are fully described in the electronic supplementary material (text), allowing closer scrutiny by the Vibrio taxonomic community for consistency with other properties.
HierCC was so effective in clarifying the taxonomies of these five species that we had hoped that it could also facilitate the taxonomic classification of Streptococcus. One hundred and two HC363 clusters were indeed concordant with 95% ANI and taxonomic designations, with only minor exceptions. However, extensive discrepancies existed between 95% ANI and HierCC for numerous other clusters, and both approaches showed multiple discrepancies with classical taxonomic designations. The existence of multiple 95% ANI clusters within S. mitis and S. oralis and other species of Streptococcus has previously been commented on by Kilian and his colleagues [15,16,77,78]. Our observations extend these taxonomic problems to multiple additional species in which HC363 does not distinguish between pairs of named species (figure 4). Neither 95% ANI nor cgMLST HierCC seems to provide a suitable general strategy for elucidating the taxonomy of all of Streptococcus, and we failed to find a general solution to this problem.
(b) . HierCC versus classical taxonomy
Classical microbiological species taxonomy involves identifying a type strain whose phenotype can be distinguished from all other type strains, identifying additional isolates with similar phenotypes, demonstrating distinct clustering from other known species in phylogenetic trees based on DNA or amino acid sequences, and publishing a report in one of a very limited number of acceptable journals. Such species definitions are then considered tentative until an international committee has approved them. Other forms of identifying species that include sole reliance on DNA sequence differences are not acceptable [7,8]. The metagenomics community and scientists working with uncultivated organisms from the environment have largely liberated themselves from such regulations, and tend to use operational taxonomic units (OTUs) as taxonomic entities. However, the taxonomic species structure of many microbes from environmental sources remains fuzzy, or does not clearly correspond to classical taxonomy (e.g. Prochlorococcus [116] or Synechococcus [117]). Furthermore, we are not aware of any other method that is able to assign 1000s of genomes per day to existing taxa and also reliably identify new taxa automatically as they appear. HierCC performs this task with bravura for the genera in table 1, with the notable exception of Streptococcus.
We have taken the liberty of dropping all attempts to reconcile HierCC species/subspecies clusters with classical prescriptions for how to define a species. Instead we have adopted the practice of using HierCC cluster designations within EnteroBase for the nomenclatures for species/subspecies groupings that had not yet been identified by others and eliminated from EnteroBase multiple species designations of type strains in Vibrio and Streptococcus which did not match the ML tree topologies and HierCC clusters. These actions provide a uniform base for the future additions of additional species and ensured that future genomes can be correctly assigned to uniform clusters of related strains. Scientists wishing to use these schemes to identify the species of their bacterial isolates can upload their sequenced genomes to EnteroBase. The HierCC assignments will be available within hours. We also welcome additional curators of these databases who are willing to test and improve the current taxonomic assignments we have implemented. But we reject the concept of designating our assignments as tentative until they are confirmed in several years by an international committee.
(c) . HierCC and populations
The first bacterial taxonomic designations were assigned over 100 years ago. Bacterial population genetics is much younger and has many fewer practitioners. Initial population genetic analyses in the early 1980s subdivided multiple bacterial species into intra-specific lineages based on multilocus enzyme electrophoresis [97]. This methodology was replaced in the late 1990s by legacy MLST based on several housekeeping genes [21], which is currently being replaced by cgMLST based on all genes in the soft core genome [22,29]. EnteroBase calculates genotypes for both types of MLST, as well as for rMLST [28]. Legacy STs differ from cgSTs due to different levels of resolution. However, the boundaries of ST Complexes/eBGs are highly concordant with HierCC clusters, with AMI indices of 0.985 for eBGs versus HC900 in Salmonella and 0.94 for ST Complexes versus HC1100 clusters in Escherichia/Shigella (table 4). We recommended previously that serovars should be replaced by eBGs in S. enterica [27] and the data presented here show that HC900 clusters are an even better replacement. Our data also indicate that Lineages in S. enterica correspond to HC2000 clusters, and that these tend to be highly uniform for O group. The data presented here also indicate that HC1100 groups are a good replacement for detecting populations within Escherichia as are HC100 clusters for CCs in S. pneumoniae [92] (table 4).
We interpret these consistencies between legacy MLST and cgMLST as reflecting the existence of natural populations. We previously claimed that legacy ST Complexes in E. coli were unstable due to frequent recombination [26], and were therefore not surprised at their tendency to merge as additional isolates were sequenced. However, legacy MLST is based on only seven genes. The high resolution of cgMLST and the decision to assign new genotypes that are equidistant from multiple HierCC clusters to the oldest cluster have largely negated these problems, and E. coli HC1100 clusters correspond to bacterial groupings that have been independently identified by multiple phenotypic patterns. In the early 1980s, MA began his academic research on bacterial pathogens with E. coli that expressed the K1 polysaccharide capsule [110]. He observed unusually uniform patterns of electrophoretic mobility of major outer membrane proteins across multiple isolates, and interpreted those bacteria as representing ‘clones’. These ‘clones’ correspond to HC1100 clusters, including HC1100_44 (ST95 Complex) which continues to cause invasive disease in humans and animals around the globe [111]. The 1980s analyses showed that these K1 bacteria were variably O1, O2 or O18, and that the serotype variants differed both in their invasiveness and in the hosts which they infected [118]. As indicated in figure 6, HC1100_44 includes O groups O25 and O45 in addition to O1, O2 and O18. Similarly, HC1100_7 (ST131) represents another major cause of extra-intestinal disease in humans [23,108,109]. EHEC bacteria are notorious for their association with hemolytic uremic syndrome (HUS). Many of them belong to distinctive HC1100 clusters, including O157:H7/O55:H7 (HC1100_63, ST11 Cplx [106,119]) and O26, O103 and O111 EHEC bacteria (HC1100_2, ST29 Cplx [107]) (figure 5).
By contrast to these correlations with epidemiological groupings, we were unable to identify any other HierCC levels that were consistently concordant with other intra-specific phylogenetic subdivisions based on SNP trees, including haplogroups [98] or Clermont typing [101,102] in E. coli [28] and clades [80–83], Lineages [79] or branches [84] in S. enterica. Instead, we found that a subdivision we refer to as Lineages is marked by some HC2000 clusters in Salmonella and Escherichia/Shigella.
(d) . Lineages
In E. coli, HC2000 Lineages were particularly appropriate for defining the clustering level of seven groups of Shigella genomes (figure 6). Lineage designations did not add additional insights into other populations because they were almost entirely encompassed by HC1100 clusters or occasionally even lower level clusters, such as HC400. In S. enterica subspecies enterica, multiple HC2000 lineages corresponded to genetically related combinations of multiple HC900 clusters, in some cases with distinctive serovar designations. However, all but three of the major Lineages were predominantly uniform in O group. The three exceptional HC2000 Lineages (HC2000_54, HC2000_106, HC2000_299) might be worth exploring in greater detail to reconstruct the recombination that has resulted in their differing LPS epitopes.
HC160 clusters, the Lineage equivalent in S. pneumonia, were strongly concordant with PopPunk GSPC clustering. Otherwise, little is yet known about the general properties of Lineages in other genera, except that their properties are likely to vary with species or genus. For example, S. enterica HC2000 Lineages were uniform for O group whereas O serogroups were already heterogeneous within HC1100 clusters/ST Complexes within E. coli (figure 6).
(e) . Future prospects
EnteroBase was conceived to satisfy a need that MA and ZZ perceived in 2014 [1]. We contend that it is now fit for purpose for investigations of multiple bacterial genera by scientists ranging from beginners through to experts in the areas of microbial epidemiology and population genetics. Its original creators have now all left this project, but EnteroBase is being maintained as a service for the global community by the University of Warwick. Maintenance of its technical functions and databases are thereby assured for the near future. Further functional developments will, however, depend on increased participation and perception of ownership by its users. We perceive a general trend to focus on insular solutions that can satisfy the demands of individual bioinformaticians and regional diagnostic laboratories. Such approaches can yield relatively rapid progress in solving short-term needs. However, a global overview of genomic diversity needs central databases to ensure definitive terminology. Pooling efforts on a central endeavour at the scale represented by EnteroBase would ensure that it continues to function over decades, is representative over all continents, and serves the global community even better. We therefore welcome additional curators and scientific experts as well as bioinformatics collaborations to help improve EnteroBase even more.
4. Methods (see supplemental text)
Acknowledgements
We are very grateful to François-Xavier Weill for permission to include electronic supplementary material, table S8 for public access in Supplemental Material. We are very grateful to François-Xavier Weill and Tandy Warnow for their comments on an earlier draft of this manuscript.
Contributor Information
Mark Achtman, Email: m.achtman@warwick.ac.uk.
Zhemin Zhou, Email: zheminzhou@qq.com.
Data accessibility
cgMLSA is available for downloading at https://github.com/zheminzhou/cgMLSA. All trees in figures and genomic data are freely available within EnteroBase for interactive examination. Other code is described in electronic supplementary material, text. Datasets used for the analyses presented here have been deposited for permanent public access at http://wrap.warwick.ac.uk/162247/.
The data are provided in electronic supplementary material [120].
Authors' contributions
M.A.: conceptualization, data curation, formal analysis, funding acquisition, investigation, project administration, supervision, visualization, writing—original draft, writing—review and editing; Z.Z.: conceptualization, data curation, formal analysis, investigation, methodology, software, visualization, writing—review and editing; J.C.: data curation, formal analysis, investigation, methodology, project administration, validation, writing—review and editing; L.B.: data curation, formal analysis, investigation, methodology, visualization, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
M.A. was supported by grant no. 202792/Z/16/Z from the Wellcome Trust (https://wellcome.org/).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Achtman M, Zhou Z. 2014. Distinct genealogies for plasmids and chromosome. PLoS Genet. 10, e1004874. ( 10.1371/journal.pgen.1004874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kauffmann F. 1961. Die Bakteriologie der Salmonella-Species. Copenhagen, Denmark: Munksgaard. [Google Scholar]
- 3.Morelli G, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nature Genet. 42, 1140-1143. ( 10.1038/ng.705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA 96, 14 043-14 048. ( 10.1073/pnas.96.24.14043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulford CV, et al. 2021. Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa. Nat. Microbiol. 6, 327-338. ( 10.1038/s41564-020-00836-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne LG, et al. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463-464. ( 10.1099/00207713-37-4-463) [DOI] [Google Scholar]
- 7.Murray AE, et al. 2020. Roadmap for naming uncultivated Archaea and Bacteria. Nat. Microbiol. 5, 987-994. ( 10.1038/s41564-020-0733-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutcliffe IC, Dijkshoorn L, Whitman WB. and Executive Board. 2020. Minutes of the International Committee on Systematics of Prokaryotes online discussion on the proposed use of gene sequences as type for naming of prokaryotes, and outcome of vote. Int. J. Syst. Evol. Microbiol. 70, 4416-4417. ( 10.1099/ijsem.0.004303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl Acad. Sci. USA 102, 2567-2572. ( 10.1073/pnas.0409727102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain C, Rodriguez R, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. ( 10.1038/s41467-018-07641-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auch AF, von Jan M, Klenk HP, Göker M. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2, 117-134. ( 10.4056/sigs.531120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrido-Sanz D, Meier-Kolthoff JP, Goker M, Martin M, Rivilla R, Redondo-Nieto M. 2016. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 11, e0150183. ( 10.1371/journal.pone.0150183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter M, Rosselló-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl Acad. Sci. USA 106, 19 126-19 131. ( 10.1073/pnas.0906412106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maderankova D, Jugas R, Sedlar K, Vitek M, Skutkova H. 2019. Rapid bacterial species delineation based on parameters derived from genome numerical representations. Comput. Struct. Biotechnol. J. 17, 118-126. ( 10.1016/j.csbj.2018.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen A, Scholz CF, Kilian M. 2016. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int. J. Syst. Evol. Microbiol. 66, 4803-4820. ( 10.1099/ijsem.0.001433) [DOI] [PubMed] [Google Scholar]
- 16.Kilian M, Poulsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, Tettelin H, Sorensen UB. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS ONE 3, e2683. ( 10.1371/journal.pone.0002683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z, Charlesworth J, Achtman M. 2020. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 30, 1667-1679. ( 10.1011/gr.260828.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomila M, Pena A, Mulet M, Lalucat J, Garcia-Valdes E. 2015. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 6, 214. ( 10.3389/fmicb.2015.00214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaz-Hidalgo R, Hossain MJ, Liles MR, Figueras MJ. 2015. Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for Aeromonas genomes in the GenBank database. PLoS ONE 10, e0115813. ( 10.1371/journal.pone.0115813) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Achtman M, Wagner M. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6, 431-440. ( 10.1038/nrmicro1872) [DOI] [PubMed] [Google Scholar]
- 21.Maiden MCJ, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA 95, 3140-3145. ( 10.1073/pnas.95.6.3140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolley KA, Bray JE, Maiden MC. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124. ( 10.12688/wellcomeopenres.14826.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoesser N, et al. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 7, e02162. ( 10.1128/mBio.02162-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingsley RA, et al. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279-2287. ( 10.1101/gr.091017.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from Multilocus Sequence Typing data. J. Bacteriol. 186, 1518-1530. ( 10.1128/JB.186.5.1518-1530.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136-1151. ( 10.1111/j.1365-2958.2006.05172.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achtman M, et al. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8, e1002776. ( 10.1371/journal.ppat.1002776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. 2020. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 30, 138-152. ( 10.1101/gr.251678.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiden MC, van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728-736. ( 10.1038/nrmicro3093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet. 14, e1007261. ( 10.1371/journal.pgen.1007261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moura A, et al. 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 16185. ( 10.1038/nmicrobiol.2016.185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Charlesworth J, Achtman M. 2021. HierCC: a multi-level clustering scheme for population assignments based on core genome MLST. Bioinformatics 37, 3645-3646. ( 10.1093/bioinformatics/btab234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths D, et al. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48, 770-778. ( 10.1128/JCM.01796-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carrico JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 28, 1395-1404. ( 10.1101/gr.232397.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Achtman M, Zhou Z. 2020. Metagenomics of the modern and historical human oral microbiome with phylogenetic studies on Streptococcus mutans and Streptococcus sobrinus. Phil. Trans. R. Soc. B 375, 20190573. ( 10.1098/rstb.2019.0573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Luhmann N, Alikhan N-F, Quince C, Achtman M. 2018. Accurate reconstruction of microbial strains from metagenomic sequencing using representative reference genomes. In RECOMB 2018, pp. 225-240. Cham, Switzerland: Springer. [Google Scholar]
- 37.Luhmann N, Holley G, Achtman M. 2021. BlastFrost: fast querying of 100,000s of bacterial genomes in Bifrost graphs. Genome Biol. 22, 30. ( 10.1186/s13059-020-02237-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones G, et al. 2019. Outbreak of Salmonella enterica serotype Poona in infants linked to persistent Salmonella contamination in an infant formula manufacturing facility, France, August 2018 to February 2019. Euro Surveill. 24, 13. ( 10.2807/1560-7917.ES.2019.24.13.1900161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones G, et al. 2019. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O26 paediatric haemolytic uraemic syndrome (HUS) cases associated with the consumption of soft raw cow's milk cheeses, France, March to May 2019. Euro Surveill. 24, 1900305. ( 10.2807/1560-7917.ES.2019.24.22.1900305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Bossche A, Ceyssens PJ, Denayer S, Hammami N, van den Beld M, Dallman TJ, Mattheus W. 2021. Outbreak of Central American born Shigella sonnei in two youth camps in Belgium in the summer of 2019. Eur. J. Clin. Microbiol. Infect. Dis. 40, 1573-1577. ( 10.1007/s10096-021-04164-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.European Centre for Disease Prevention and Control, EFSA. 2020. Multi-country outbreak of Salmonella Typhimurium and S. Anatum infections linked to Brazil nuts - 21 October 2020. EFSA Supporting Publications 17, 1944E. ( 10.2903/sp.efsa.2020.EN-1944) [DOI] [Google Scholar]
- 42.European Food Safety Authority European Centre for Disease Prevention and Control. 2022. Multi-country outbreak of monophasic Salmonella typhimurium sequence type 34 linked to chocolate products: first update, 18 May 2022. EFSA Supporting Publications 19 (6): 7352E.
- 43.Hawkey J, et al. 2021. Global population structure and genotyping framework for genomic surveillance of the major dysentery pathogen, Shigella sonnei. Nat. Commun. 12, 2684. ( 10.1038/s41467-021-22700-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SE, et al. 2018. The phylogeography and incidence of multi-drug resistant typhoid fever in sub-Saharan Africa. Nat. Commun. 9, 5094. ( 10.1038/s41467-018-07370-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weill FX, et al. 2017. Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785-789. ( 10.1126/science.aad5901) [DOI] [PubMed] [Google Scholar]
- 46.Achtman M, van den Broeck F, Cooper KK, Lemey P, Parker CT, Zhou Z, and the ATCC14028s Study Group. 2021. Genomic population structure associated with repeated escape of Salmonella enterica ATCC14028s from the laboratory into nature. PLoS Genet. 17, e1009820. ( 10.1371/journal.pgen.1009820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yassine I, et al. 2022. Population structure analysis and laboratory monitoring of Shigella by core-genome multilocus sequence typing. Nat. Commun. 13, 551. ( 10.1038/s41467-022-28121-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl Acad. Sci. USA 97, 10 567-10 572. ( 10.1073/pnas.180094797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Payne M, Nguyen T, Kaur S, Lan R. 2021. Cluster-specific gene markers enhance Shigella and enteroinvasive Escherichia coli in silico serotyping. Microb. Genom. 7, 000704. ( 10.1099/mgen.0.000704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achtman M, et al. 2020. Genomic diversity of Salmonella enterica—the UoWUCC 10K genomes project. Wellcome Open Res. 5, 223. ( 10.12688/wellcomeopenres.16291.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, et al. 2018. Pan-genome analysis of ancient and modern Salmonella enterica demonstrates genomic stability of the invasive Para C lineage for millennia. Curr. Biol. 28, 2420-2428. ( 10.1016/j.cub.2018.05.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaharias P, Warnow T. 2022. Recent progress on methods for estimating and updating large phylogenies. Phil. Trans. R. Soc. B 377, 2021100258. ( 10.20944/preprints202110.0258.v1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vachaspati P, Warnow T. 2015. ASTRID: Accurate Species TRees from Internode Distances. BMC Genom. 16(Suppl. 10), S3. (doi:1471-2164-16-S10-S3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sayyari E, Mirarab S. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 33, 1654-1668. ( 10.1093/molbev/msw079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearce ME, Langridge GC, Lauer AC, Grant K, Maiden MCJ, Chattaway MA. 2021. An evaluation of the species and subspecies of the genus Salmonella with whole genome sequence data: proposal of type strains and epithets for novel S. enterica subspecies VII, VIII, IX, X and XI. Genomics 113, 3152-3162. ( 10.1016/j.ygeno.2021.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huys G, Cnockaert M, Janda JM, Swings J. 2003. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 53, 807-810. ( 10.1099/ijs.0.02475-0) [DOI] [PubMed] [Google Scholar]
- 57.Hyma KE, Lacher DW, Nelson AM, Bumbaugh AC, Janda JM, Strockbine NA, Young VB, Whittam TS. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187, 619-628. ( 10.1128/JB.187.2.619-628.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ooka T, et al. 2015. Defining the genome features of Escherichia albertii, an emerging enteropathogen closely related to Escherichia coli. Genome Biol. Evol. 7, 3170-3179. ( 10.1093/gbe/evv211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farmer JJ III, Fanning GR, Davis BR, O'Hara CM, Riddle C, Hickman-Brenner FW, Asbury MA, Lowery VA III, Brenner DJ. 1985. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21, 77-81. ( 10.1128/jcm.21.1.77-81.1985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaastra W, Kusters JG, Van Duijkeren E, Lipman LJ. 2014. Escherichia fergusonii. Vet. Microbiol. 172, 7-12. ( 10.1016/j.vetmic.2014.04.016) [DOI] [PubMed] [Google Scholar]
- 61.Liu S, Jin D, Lan R, Wang Y, Meng Q, Dai H, Lu S, Hu S, Xu J. 2015. Escherichia marmotae sp. nov., isolated from faeces of Marmota himalayana. Int. J. Syst. Evol. Microbiol. 65, 2130-2134. ( 10.1099/ijs.0.000228) [DOI] [PubMed] [Google Scholar]
- 62.Liu S, et al. 2019. Genomic and molecular characterisation of Escherichia marmotae from wild rodents in Qinghai-Tibet plateau as a potential pathogen. Sci. Rep. 9, 10619. ( 10.1038/s41598-019-46831-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Putten BCL, Matamoros S, COMBAT Consortium, and Schultsz C. 2021. Escherichia ruysiae sp. nov., a novel Gram-stain-negative bacterium, isolated from a faecal sample of an international traveller. Int. J. Syst. Evol. Microbiol. 71, 004609. ( 10.1099/ijsem.0.004609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walk ST. 2015. The ‘cryptic’ Escherichia. EcoSal Plus 6, 0002. ( 10.1128/ecosalplus.ESP-0002-2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vignaroli C, Di SL, Magi G, Luna GM, Di CA, Pasquaroli S, Facinelli B, Biavasco F. 2015. Adhesion of marine cryptic Escherichia isolates to human intestinal epithelial cells. ISME J. 9, 508-515. ( 10.1038/ismej.2014.164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen Z, Koh XP, Yu Y, Woo CF, Tong Y, Lau SCK. 2019. Draft genome sequences of 16 strains of Escherichia cryptic clade II isolated from intertidal sediment in Hong Kong. Microbiol. Resour. Announc. 8, e00415-19. ( 10.1128/MRA.00416-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl Acad. Sci. USA 108, 7200-7205. ( 10.1073/pnas.1015622108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75, 6534-6544. ( 10.1128/AEM.01262-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9, 2274-2288. ( 10.1111/j.1462-2920.2007.01341.x) [DOI] [PubMed] [Google Scholar]
- 70.Shen ZY, Koh XP, Yu YP, Lau SCK. 2020. Genetic variation and preliminary indications of divergent niche adaptation in cryptic clade II of Escherichia. Microorganisms 8, 1713. ( 10.3390/microorganisms8111713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knight DR, et al. 2021. Major genetic discontinuity and novel toxigenic species in Clostridioides difficile taxonomy. Elife 10, e64325. ( 10.7554/eLife.64325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reuter S, et al. 2014. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl Acad. Sci. USA 111, 6768-6773. ( 10.1073/pnas.1317161111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savin C, et al. 2014. The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii. Int. J. Med. Microbiol. 304, 452-463. ( 10.1016/j.ijmm.2014.02.002) [DOI] [PubMed] [Google Scholar]
- 74.Sprague LD, Scholz HC, Amann S, Busse HJ, Neubauer H. 2008. Yersinia similis sp. nov. Int. J. Syst. Evol. Microbiol. 58, 952-958. ( 10.1099/ijs.0.65417-0) [DOI] [PubMed] [Google Scholar]
- 75.Bercovier H, Mollaret HH, Alonso JM, Brault J, Fanning GR, Steigerwalt AG, Brenner DJ. 1980. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis. Curr. Microbiol. 4, 225-229. ( 10.1007/BF02605861) [DOI] [Google Scholar]
- 76.Gomez-Gil B, Thompson CC, Matsumura Y, Sawabe T, Iida T, Christen R, Thompson F, Sawabe T. 2014. The family Vibrionaceae. In The Prokaryotes: Gammaproteobacteria (eds Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F), pp. 660-747. Berlin, Germany: Springer. [Google Scholar]
- 77.Kilian M, Riley DR, Jensen A, Bruggemann H, Tettelin H. 2014. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio 5, e01490-14. ( 10.1128/mBio.01490-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7, 3. ( 10.1186/1741-7007-7-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Didelot X, et al. 2011. Recombination and population structure in Salmonella enterica. PLoS Pathog. 7, e1002191. ( 10.1371/journal.ppat.1002191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.den Bakker HC, et al. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genom. 12, 425. ( 10.1186/1471-2164-12-425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Timme RE, et al. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol. Evol. 5, 2109-2123. ( 10.1093/gbe/evt159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons SK, Bull CM, Gordon DM. 2011. Substructure within Salmonella enterica subspecies enterica isolated from Australian wildlife. Appl. Environ. Microbiol. 77, 3151-3153. ( 10.1128/AEM.02764-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Worley J, Meng J, Allard MW, Brown EW, Timme RE. 2018. Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. MBio 9, e02303-18. ( 10.1128/mBio.02303-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Key FM, et al. 2020. Emergence of human-specific Salmonella enterica is linked to the Neolithization process. Nature Ecol. Evol. 4, 324-333. ( 10.1038/s41559-020-1106-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill F-X. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161, 26-29. ( 10.1016/j.resmic.2009.10.002) [DOI] [PubMed] [Google Scholar]
- 86.Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, De PE, Nair S, Fields PI, Weill F-X. 2014. Supplement 2008–2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 165, 526-530. ( 10.1016/j.resmic.2014.07.004) [DOI] [PubMed] [Google Scholar]
- 87.Grimont PA, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th edn. Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella. [Google Scholar]
- 88.Seif Y, Monk JM, Machado H, Kavvas E, Palsson BO. 2019. Systems biology and pangenome of Salmonella O-Antigens. MBio 10, e01247-19. ( 10.1128/mBio.01247-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, Lauer AC, Fields PI, Deng X. 2019. SeqSero2: rapid and improved Salmonella serotype determination using whole genome sequencing data. Appl. Environ. Microbiol. 85, e01746-19. ( 10.1128/AEM.01746-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robertson J, Yoshida C, Kruczkiewicz P, Nadon C, Nichani A, Taboada EN, Nash JHE. 2018. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb. Genom. 4, 1-11. ( 10.1099/mgen.0.000151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uelze L, Borowiak M, Deneke C, Szabo I, Fischer J, Tausch SH, Malorny B. 2020. Performance and accuracy of four open-source tools for in silico serotyping of Salmonella spp. based on whole-genome short-read sequencing data. Appl. Environ. Microbiol. 86, e02265-19. ( 10.1128/AEM.02265-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gladstone RA, et al. 2019. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMed. 43, 338-346. ( 10.1016/j.ebiom.2019.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pedregosa F, et al. 2011. Scikit-learn: machine learning in Python. J. Machine Learn. Res. 12, 2825-2830. [Google Scholar]
- 94.Romano S, Vinh NX, Bailey J, Verspoor K. 2016. Adjusting for chance clustering comparison measures. J. Machine Learn. Res. 17, 1-32. [Google Scholar]
- 95.Bostock M, Ogievetsky V, Heer J. 2011. D3: Data-Driven Documents. IEEE Trans. Vis. Comput. Graph 17, 2301-2309. ( 10.1109/TVCG.2011.185) [DOI] [PubMed] [Google Scholar]
- 96.Chaudhuri RR, Henderson IR. 2012. The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 12, 214-226. ( 10.1016/j.meegid.2012.01.005) [DOI] [PubMed] [Google Scholar]
- 97.Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690-693. ( 10.1128/jb.157.2.690-693.1984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Selander RK, Caugant DA, Whittam TS. 1987. Genetic structure and variation in natural populations of Escherichia coli. In Escherichia coli and Salmonella typhimurium cellular and molecular biology, vol. II (eds Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE), pp. 1625-1648. Washington, DC: American Society for Microbiology. [Google Scholar]
- 99.Touchon M, Perrin A, de Sousa JAM, Vangchhia B, Burn S, O'Brien CL, Denamur E, Gordon D, Rocha EP. 2020. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 16, e1008866. ( 10.1371/journal.pgen.1008866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Denamur E, Clermont O, Bonacorsi S, Gordon D. 2021. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 19, 37-54. ( 10.1038/s41579-020-0416-x) [DOI] [PubMed] [Google Scholar]
- 101.Gordon DM, Clermont O, Tolley H, Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10, 2484-2496. ( 10.1111/j.1462-2920.2008.01669.x) [DOI] [PubMed] [Google Scholar]
- 102.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58-65. ( 10.1111/1758-2229.12019) [DOI] [PubMed] [Google Scholar]
- 103.Beghain J, Bridier-Nahmias A, Le NH, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4, e000192. ( 10.1099/mgen.0.000192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waters NR, Abram F, Brennan F, Holmes A, Pritchard L. 2020. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2, 1-5. ( 10.1099/acmi.0.000143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sawyer C, et al. 2021. Epidemiological investigation of recurrent outbreaks of haemolytic uraemic syndrome caused by Shiga toxin-producing Escherichia coli serotype O55:H7 in England, 2014–2018. Epidemiol. Infect. 149, e108. ( 10.1017/S0950268821000844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187, 1783-1791. ( 10.1128/JB.187.5.1783-1791.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eichhorn I, Semmler T, Mellmann A, Pickard D, Anjum MF, Fruth A, Karch H, Wieler LH. 2018. Microevolution of epidemiological highly relevant non-O157 enterohemorrhagic Escherichia coli of serogroups O26 and O111. Int. J. Med. Microbiol. 308, 1085-1095. ( 10.1016/j.ijmm.2018.08.003) [DOI] [PubMed] [Google Scholar]
- 108.Liu CM, et al. 2018. Escherichia coli ST131-H22 as a foodborne uropathogen. MBio 9, e00470-18. ( 10.1128/mBio.00470-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Price LB, et al. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 4, e00377-13. ( 10.1128/mBio.00377-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39, 315-335. ( 10.1128/iai.39.1.315-335.1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gordon DM, et al. 2017. Fine-scale structure analysis shows epidemic patterns of Clonal Complex 95, a cosmopolitan Escherichia coli lineage responsible for extraintestinal infection. mSphere 2, e00168-17. ( 10.1128/mSphere.00168-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frentrup M, et al. 2020. A publicly accessible database for Clostridioides difficile genome sequences supports tracing of transmission chains and epidemics. Microbial Genom. 6, mgen.0.000410. ( 10.1099/mgen.0.000410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laukkanen-Ninios R, et al. 2011. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ. Microbiol. 13, 3114-3127. ( 10.1111/j.1462-2920.2011.02588.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, Weiser JN, Corander J, Bentley SD, Croucher NJ. 2019. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29, 304-316. ( 10.1101/gr.241455.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lees JA, Tonkin-Hill G, Yang Z, Corander J. 2022. Mandrake: visualising microbial population structure by embedding millions of genomes into a low-dimensional representation. Phil. Trans. R. Soc. B 377, 20210237. ( 10.1098/rstb.2021.0237) [DOI] [PMC free article] [PubMed]
- 116.Tschoeke D, Salazar VW, Vidal L, Campeao M, Swings J, Thompson F, Thompson C. 2020. Unlocking the genomic taxonomy of the Prochlorococcus collective. Microb. Ecol. 80, 546-558. ( 10.1007/s00248-020-01526-5) [DOI] [PubMed] [Google Scholar]
- 117.Salazar VW, Tschoeke DA, Swings J, Cosenza CA, Mattoso M, Thompson CC, Thompson FL. 2020. A new genomic taxonomy system for the Synechococcus collective. Environ. Microbiol. 22, 4557-4570. ( 10.1111/1462-2920.15173) [DOI] [PubMed] [Google Scholar]
- 118.Achtman M, Pluschke G. 1986. Clonal analysis of descent and virulence among selected Escherichia coli. Annu. Rev. Microbiol. 40, 185-210. ( 10.1146/annurev.mi.40.100186.001153) [DOI] [PubMed] [Google Scholar]
- 119.Carroll KJ, Jenkins C, Harvey-Vince L, Mohan K, Balasegaram S. 2021. Shiga toxin-producing Escherichia coli diagnosed by Stx PCR: assessing the public health risk of non-O157 strains. Eur. J. Public Health 31, 576-582. ( 10.1093/eurpub/ckaa232) [DOI] [PubMed] [Google Scholar]
- 120.Achtman M, Zhou Z, Charlesworth J, Baxter L. 2022. EnteroBase: hierarchical clustering of 100 000s of bacterial genomes into species/subspecies and populations. Figshare. ( 10.6084/m9.figshare.c.6097222) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Achtman M, Zhou Z, Charlesworth J, Baxter L. 2022. EnteroBase: hierarchical clustering of 100 000s of bacterial genomes into species/subspecies and populations. Figshare. ( 10.6084/m9.figshare.c.6097222) [DOI] [PMC free article] [PubMed]
Data Availability Statement
cgMLSA is available for downloading at https://github.com/zheminzhou/cgMLSA. All trees in figures and genomic data are freely available within EnteroBase for interactive examination. Other code is described in electronic supplementary material, text. Datasets used for the analyses presented here have been deposited for permanent public access at http://wrap.warwick.ac.uk/162247/.
The data are provided in electronic supplementary material [120].