Abstract
The sequence of a 4.8-kbp DNA fragment adjacent to the right-hand end of the actinorhodin biosynthetic (act) cluster downstream of actVB-orf6 from Streptomyces coelicolor A3(2) reveals six complete open reading frames, named orf7 to orf12. The deduced amino acid sequences from orf7, orf10, and orf11 show significant similarities with the following products in the databases: a putative protein from the S. coelicolor SCP3 plasmid, LysR-type transcriptional regulators, and proteins belonging to the family of short-chain dehydrogenases/reductases, respectively. The deduced product of orf8 reveals low similarities with several methyltransferases from different sources, while orf9 and orf12 products show no similarities with other known proteins. Disruptions of orf10 and orf11 genes in S. coelicolor appear to have no significant effect on the production of actinorhodin. Nevertheless, disruption or deletion of orf10 in Streptomyces lividans causes actinorhodin overproduction. The introduction of extra copies of orf10 and orf11 genes in an S. coelicolor actIII mutant restores the ability to produce actinorhodin. Transcriptional analysis and DNA footprinting indicate that Orf10 represses its own transcription and regulates orf11 transcription, expression of which might require the presence of an unknown inducer. No DNA target for Orf10 protein was found within the act cluster.
Members of the genus Streptomyces have become a major focus of study at the molecular level, in large part because of their ability to undergo both morphological and biochemical differentiation, including the production of bioactive metabolites (9). The activation of antibiotic production, often coupled to morphological development, involves many different pathways in the same organism. Although the multiple and coordinated regulation of secondary metabolism is poorly understood, insight into some of the mechanisms controlling antibiotic biosynthesis is emerging (10).
Streptomyces coelicolor provides an excellent model system for studying the regulation of antibiotic production, because it is genetically well studied and produces at least four quite different antibiotics: actinorhodin (50), undecylprodigiosin (38), methylenomycin (51), and the calcium-dependent peptide antibiotic (28). Their biosynthetic clusters have been isolated, and that for actinorhodin synthesis (act cluster) has been well characterized (7, 8, 16–18). Antibiotic pathway-specific regulatory genes have been found in the biosynthetic clusters for actinorhodin, undecylprodigiosin, and methylenomycin (for reviews, see references 9 and 10). Both ActII-Orf4 and RedD (act- and red-specific regulators) have been proposed to belong to a novel family of Streptomyces antibiotic regulatory proteins (48) that probably have similar mechanisms of transcriptional activation of the genes they regulate. In addition to this type of regulation, several other genes outside the biosynthetic clusters have been shown to pleiotropically affect antibiotic formation. Among them, bld and rel have been implicated in both antibiotic production and morphological differentiation, while a number of genes, including abaA, absA, absB, afsB, afsR, afsS, and afsQ1-afsQ2, have some effect on one or more antibiotic synthesis processes without modifying morphological development.
Streptomyces lividans, a streptomycete closely related to S. coelicolor, has all of the genetic information for actinorhodin biosynthesis but does not produce this antibiotic under usual growth conditions. Because of its ability to show a blue-pigmented phenotype by either the introduction of genes or the generation of mutations, this strain has become a useful tool for understanding the signaling mechanisms involved in the activation of antibiotic biosynthesis. By this procedure, several putative regulatory elements have been isolated. This report describes the characterization of the orf10 gene, encoding a LysR-type transcriptional regulator, disruption or deletion of which induces actinorhodin production in S. lividans.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli strains used for general cloning procedures were JM101 (52) and XL1-Blue (6). E. coli K12ΔH1Δtrp (53) and K38 (39) (containing the helper plasmid pGP-1-2 [45]) were used for the expression of the Orf10 protein. The S. coelicolor A3(2) strains used were J1501 (hisA1 uraA1 strA1 pg1 SCP1− SCP2−) (13) and TK18 (hisA1 uraA1 strA1 actIII141 redE60 SCP1− SCP2−) (37). The S. lividans strain used was TK21 (str-6 SLP2− SPL3−) (25).
Plasmids and bacteriophages.
The E. coli plasmids used were pUC18-19 (52), pIJ2925 (26), pSU19-20-21 (3), pBR329 (14), pT7.7 (45), pAZe3ss (53), and pIJ2333 (32). E. coli M13 derivative phages M13mp18 and M13mp19 (52) were used for DNA sequencing and for in vitro mutagenesis. The Streptomyces vectors and recombinant plasmids used are described in Table 1. The Streptomyces φC31 derivative phage PM1 (32) was used for insertional inactivation.
TABLE 1.
Streptomyces vectors and recombinant plasmids
| Plasmids | Relevant characteristics and constructiona | Reference |
|---|---|---|
| pIJ486 | High-copy-number cloning vector, tsr | 47 |
| pGM9 | pSG5-derived temperature-sensitive replication vector, tsr aphII | 35 |
| pIJ941 | Low-copy-number SCP2* derivative plasmid, tsr hyg | 25 |
| pIJ2314 | BamHI fragment (positions 13–14) cloned in pIJ922, harboring the wild-type actIII gene, tsr | 32 |
| pAM73 | pGM9 derivative obtained from S. lividans TK21::pSCNB06A chromosomal DNA partially digested with Sau3AI, followed by ligation and transformation of S. lividans | This work |
| pAM75 | pGM9 derivative obtained from S. coelicolor J1501::pSCNB06A chromosomal DNA partially digested with Sau3AI, followed by ligation and transformation of S. lividans | This work |
| pSCNB01b | SacII (2274)-XhoI (3465) fragment cloned in pGM9 (EcoRI-HindIII sites); contains the 3′ truncated orf10 gene | This work |
| pSCNB03b | PstI (1881)-XhoI (3465) fragment cloned in pGM9 (EcoRI-HindIII sites); contains orf10 gene | This work |
| pSCNB04b | PstI (1881)-BamHI (4847) fragment cloned in pGM9 (EcoRI-HindIII sites); contains orf10 and orf11 genes | This work |
| pSCNB05c | NcoI (3094)-BamHI (4847) fragment cloned in pGM9 (BglII site); contains orf11 gene | This work |
| pSCNB06Ac | SacII (2274–2674) fragment containing an internal region of orf10 gene cloned in pGM9 (EcoRI-HindIII sites) | This work |
| pSCNB06Bd | pSCNB06A with the internal region of orf10 gene cloned in the opposite orientation | This work |
| pSCNB07c | SmaI (809)-BglII (2389) and NruI (3049)-BamHI (4847) fragments cloned in the same direction in pGM9 (EcoRI-HindIII sites) | This work |
| pSCNB08Bc | SmaI (809)-BamHI (2152) and NcoI (3094)-BamHI (4847) fragments cloned in the same direction in pGM9 (EcoRI-HindIII sites) | This work |
| pSCNB09c | NruI (3049)-BamHI (4847) fragment cloned in pGM9 (EcoRI-HindIII sites); contains orf11 gene | This work |
| pSCNB010d | SacII (2274)-BamHI (4847) fragment cloned in pGM9 (EcoRI-HindIII sites); contains the 3′-truncated orf10 and entire orf11 genes | This work |
| pSCNB013b | PstI (1881)-XhoI (3465) fragment cloned in pIJ941 (EcoRV site) | This work |
| pSCNB014b | PstI (1881)-XhoI (3465) fragment cloned in pIJ486 (EcoRI-HindIII sites) | This work |
tsr, thiostrepton resistance gene; aphII, aminoglycoside phosphotransferase gene from Tn5; hyg, hygromycin resistance gene. Numbers in parentheses after a restriction enzyme indicate nucleotide positions.
Constructed either directly or as blunt-ended fragments through end filling with the Klenow fragment of DNA polymerase I or by T4 DNA polymerase treatment in the intermediate E. coli vector pUC19, rescued, and ligated to Streptomyces vectors as indicated.
Constructed either directly or as blunt-ended fragments through end filling with the Klenow fragment of DNA polymerase I or by T4 DNA polymerase treatment in the intermediate E. coli vector pU2925, then rescued, and ligated to Streptomyces vectors as indicated.
Constructed either directly or as blunt-ended fragments through end filling with the Klenow fragment of DNA polymerase I or by T4 DNA polymerase treatment in an intermediate E. coli vector pSU21, then rescued, and ligated to Streptomyces vectors as indicated.
Media, culture conditions, and microbiological procedures.
E. coli strains were grown on either liquid or solid 2YT medium (40). Appropriate antibiotics were added as required. Streptomyces manipulations were as described previously (25). Thiostrepton (Sigma catalog no. T-8902) was used at concentrations of 50 μg/ml in agar medium and 10 μg/ml in broth cultures. Kanamycin was used at 50 and 15 μg/ml in solid and liquid media, respectively.
DNA sequencing.
DNA sequencing was done by the dideoxy-chain termination method (40); DNA sequence was determined from both strands, using routinely a 7-deaza-dGTP reagent kit from U.S. Biochemical Corp. (catalog no. 70750) as recommended by the manufacturer. Convenient DNA fragments were previously cloned in either M13mp18 or M13mp19 vectors. Identification of DNA sequences in DNase I protection assays were carried out as described above, using a convenient single-stranded DNA as template and the universal 17-mer sequencing primer labeled at its 5′ terminus as primer.
Computer analysis of sequences.
The DNA sequence was analyzed by using the software programs of the University of Wisconsin Genetics Computer Group (version 9.1) (15): analysis for open reading frames (ORFs) was performed with CODONPREFERENCE with a codon usage table made from 100 Streptomyces genes (49); comparisons of sequences were made against the EMBL nucleic acid database (daily updated) and the Swissprot database (daily updated), using FASTA, TFASTA, and BESTFIT. Protein alignments were made with either PILEUP from the same package or CLUSTAL W (version 1.7) (46).
Gene disruption and deletion.
For insertional inactivation of S. coelicolor, internal fragments from either the orf10 or orf11 gene were cloned into the φC31 derivative PM1 vector, and the resulting recombinant phages were used to lysogenize strain J1501 by insert-directed recombination (11). orf10 gene disruptions of S. lividans and S. coelicolor and orf10 deletions of S. lividans were obtained by the procedure of Muth et al. (35), using temperature-sensitive replication pGM9 derivative plasmids. In all cases, the chromosomal arrangements of disruptions and deletions were confirmed by Southern analysis.
DNA and RNA manipulations.
Isolation, cloning, and manipulation of nucleic acids were as previously described for Streptomyces (25) and E. coli (40). Endonuclease restriction sites for further subclonings were generated by using the Sculptor in vitro mutagenesis system (Amersham RPN 1526). Previously, suitable restriction fragments were cloned in M13mp18, and mutagenesis was performed as recommended by the manufacturer with the synthetic oligonucleotides C-079 (5′-CCCGGATCCGTCATCCGGCGTCACGCCCGGTC-3′) and T-051 (5′-CGTCGTCATGCCCATATGTTATCACCGCGGGTCTTGGA-3′). PCRs were carried out with Thermostase according to the manufacturer’s recommendations.
For isolating the complete orf11 and orf12 genes from S. coelicolor and the orf10 and orf11 genes from S. lividans, the chromosomal DNA within this region was obtained by rescuing the pGM9 derivative plasmid that had been used for orf10 disruption. To this end, the chromosomal DNA from both recombinant strains was partially digested with Sau3AI, followed by ligation, transformation of S. lividans, and selection with thiostrepton. Several recombinant plasmids from the resulting transformants were obtained, and those extending beyond the sequenced DNA were selected and named pAM74 or pAM75 for the pGM9 derivative plasmid rescued from either the S. lividans or S. coelicolor chromosome, respectively. This DNA region was subcloned and sequenced as described above. The correct physical arrangement of this region was confirmed by Southern analysis.
RNA was extracted from mycelia grown on the surface of cellophane discs on R5 agar plates as previously described (30).
High-resolution S1 mapping was carried out by the procedure of Hopwood et al. (25). Initially for the orf10 gene, a 316-bp EspI-AvaI fragment (from positions 2983 to 3298) containing the orf10-orf11 intergenic region uniquely labeled at the 5′ end of the EspI site within the orf10 coding region, was used as the probe. When analyzing RNA extracted from S. lividans CNB073, we used as the probe a 525-bp BstEII-XhoI fragment (from positions 2289 to 3465) carrying the orf10 deletion and containing the orf10-orf11 intergenic region uniquely labeled at the 5′ end of the BstEII site within the orf10 coding region. For the orf11 gene, a 471-bp SmaI-SplI fragment (nucleotides 2923 to 3393) that contained the orf10-orf11 intergenic region labeled at the 5′ end of the SplI site within the internal orf11 coding region was used. Nucleotide sequence ladders of the identical fragments were derived as described by Maxam and Gilbert (34). Before assigning a precise RNA initiation site, we subtracted one nucleotide from the length of the protected fragment to account for the difference in 3′ ends resulting from S1 nuclease digestion and the chemical sequencing reactions (24). For actII-orf4 (16), the actII-orf4 promoter region included in a 634-bp fragment (nucleotide positions 4825 to 5458 (16)) was uniquely labeled at the 5′ end of the XhoI (nucleotide 5458) site within the actII-orf4 coding region and used as the probe.
Construction of orf10 expression plasmids.
To express Orf10 from S. coelicolor, a BamHI restriction site at the 3′ end of orf10 was generated by in vitro mutagenesis as described above, using primer C-079 and single-stranded DNA from M13mp18 carrying the PstI (1881)-XhoI(3465) fragment as the template. The BamHI fragment from the resulting M13mp18 derivative (pMCNB018.B) was cloned into BamHI-digested pUC19, yielding plasmid pCNB04. The orf10 gene was cloned under lambda pL promoter control into NcoI-BamHI-digested pAZe3ss, generating plasmid pCNB019. For orf10 expression using the bacteriophage T7 RNA polymerase-promoter system (45), the NcoI site was replaced by an NdeI site by in vitro mutagenesis as described above, using primer T-051. The EcoRI-HindIII fragment from the resulting M13mp18 derivative (pMCNB018.BN) was cloned into EcoRI-HindIII-digested pUC19, generating plasmid pCNB021. The 0.95-kbp NdeI-BamHI fragment from pCNB021 was finally inserted into NdeI-BamHI sites of pT7.7, yielding plasmid pCNB023. To generate the C-terminally truncated Orf10 (amino acid positions 1 to 274), Orf101-274, plasmid pCNB01004 (which contained the internal SacII region of the orf10 gene) was digested with BglII and SmaI; the electroeluted 115-bp fragment was cloned into pCNB04, previously digested with SphI, made blunt ended by T4 DNA polymerase treatment, and then subjected to BglII digestion and alkaline phosphatase treatment. The NcoI-BamHI fragment from the resulting plasmid (pCNB025) was cloned into NcoI-BamHI-digested pAZe3ss, yielding plasmid pCNB031. As a result of this procedure, Orf101-274 had an additional amino acid (Leu) at its C terminus.
To express the Orf10 from S. lividans, the NcoI-BglII fragment from pCNB019 (carrying the orf10 gene from S. coelicolor) was replaced by the homologous DNA from S. lividans, yielding plasmid pCNB086.
Sequencing of suitable fragments confirmed that only the desired mutations had occurred. In all cases, the resulting expressed Orf10 protein incorporated two amino acids (Met and Gly) at its N terminus.
Expression and purification of the Orf10 protein in E. coli.
The Orf10 protein was purified from E. coli K12ΔH1Δtrp carrying pCNB019. The strain was grown overnight at 30°C to stationary phase in 2YT medium; a 3% inoculum was then subcultured on a 0.2-liter scale and grown at 30°C until the optical density at 600 nm reached 0.9. The culture was transferred to 42°C and incubated for an additional 2 h. The culture was harvested by centrifugation, and the resulting cell pellet was washed twice with 50 ml of ice-cold buffer A (50 mM Tris-HCl [pH 8], 100 mM NaCl, 10 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) and resuspended in 10 ml of buffer A. The cells were disrupted by sonication and centrifuged at 4,000 × g for 15 min at 4°C, and the supernatant was again centrifuged. The final supernatant was centrifuged at 25,000 × g for 30 min at 4°C, and the resulting pellet was resuspended in 0.75 ml of buffer A, which was then solubilized by adding 4.25 ml of a denaturing solution containing 7 M urea, 60 mM dithiothreitol, 1.25 mM EDTA, and 62.5 mM Tris-HCl (pH 8). This suspension was incubated on ice for 30 min and then centrifuged at 105,000 × g for 30 min at 4°C. The supernatant, which contained the solubilized Orf10 protein, was dialyzed exhaustively against buffer B (50 mM Tris-HCl [pH 8], 50 mM NaCl, 5 mM dithiothreitol, 10% glycerol) and then centrifuged; the supernatant (approximately 5 ml at 0.2 to 0.4 mg of protein per ml) was loaded onto a 5-ml heparin-agarose column that had been previously equilibrated with buffer B. The column was first washed with 10 volumes of buffer B and then with 5 volumes of buffer B containing 0.25 M NaCl; finally, the Orf10 protein was eluted with 10 volumes of buffer B containing 0.5 M NaCl. Fractions were tested for binding to the orf10-orf11 promoter in gel mobility shift assays and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and the positive fractions were pooled and used for further studies. The Orf10 protein is stable at 4°C for a week and at −20°C for several months. The supernatant obtained at 25,000 × g constituted the crude extract.
Radiolabeled Orf10 protein was obtained from the overexpressed orf10 gene by E. coli K38/pGP-1-2, transformed with plasmid pCNB023. Cell labeling with [35S]methionine was performed as described by Tabor and Richardson (45). The radiolabeled Orf10 protein was purified from the inclusion bodies of 5 ml of culture essentially as described above and used for gel mobility shift assays after the refolding step.
The S. coelicolor truncated Orf101-274 and the complete S. lividans Orf10 proteins were purified as described for the S. coelicolor Orf10 from E. coli K12ΔH1Δtrp carrying pCNB031 and pCNB086, respectively. After the refolding step, the proteins were assayed for the ability to bind to the orf10-orf11 intergenic region.
DNA binding assays.
The standard binding reaction was carried out in a final volume of 20 μl containing the purified Orf10 protein, 1 to 5 ng of labeled DNA fragments, 1 μg of poly(dI-dC) · poly(dI-dC) DNA, 1 μg of bovine serum albumin, 8 mM MgCl2, 10% glycerol, and 1× DNA binding buffer (5 mM Tris-HCl [pH 8], 20 mM NaCl, 1.5 mM 2-mercaptoethanol). Samples were allowed to incubate for 20 min at 4°C and then for 15 min at room temperature. After incubation, the samples were loaded onto a nondenaturing 4% polyacrylamide gel (28:2 acrylamide N,N′-methylenebisacrylamide linkage), with or without the addition of 1× DNA binding buffer containing bromophenol blue. Electrophoresis was performed at 120 V in 0.5× Tris-borate-EDTA buffer until the bromophenol blue had reached the end of the gel, transferred onto Whatman paper, covered with plastic wrap, dried, and exposed to X-ray film. When radiolabeled Orf10 protein was used, the DNA binding assays were performed as described above except that the samples contained unlabeled DNA fragments. The following DNA fragments were tested by gel retardation analysis: the orf10-orf11 intergenic region (EspI-AvaI fragment [nucleotides 2981 to 3297]), the downstream regions from orf10 and orf11 (PstI-SacII [nucleotides 1881 to 2274] and SmaI-HincII [nucleotides 3997 to 4224] fragments, respectively) containing the direct repeats, the actI-actIII intergenic region (MboII fragment containing nucleotides 1 to 240 from Hallam et al. [20]) and 1 to 215 from Fernández-Moreno et al. [17]), the actVI intergenic region (NaeI-SmaI fragment [nucleotides 1914 to 2226] [18]), the actII-orf1-orf2 intergenic region (SphI-SacII fragment [nucleotides 885 to 1171] [16]), and the actII-orf4 promoter region (TaqI fragment [nucleotides 4934 to 5173] [16]).
DNase I protection experiments.
For DNase I footprinting, either the NruI-AvaI (nucleotides 3049 to 3297) or AhaII (nucleotides 2945 to 3151) fragment within the promoter region of orf10-orf11 was cloned as a blunt-ended fragment into HincII-digested pUC19 (pCNB033) or pIJ2921 (pCNB034A), respectively. The inserts of pCNB033 and pCNB034A were used to analyze the protected regions on the upper and lower strands, respectively. The universal 17-mer sequencing primer was labeled with [γ-32P]ATP by treatment with T4 polynucleotide kinase at the 5′ end and used with the M13/pUC 16-mer reverse sequencing primer to amplify by PCR on double-stranded DNA of either pCNB033 or pCNB034A, a 246- or 206-bp DNA fragment, respectively. The binding reaction was carried out as described above. After incubation for 15 min at room temperature, 2 ng (0.09 U) of DNase I was added, and the mixture was incubated for 5 min at 30°C. The reaction was stopped by addition of EDTA up to 10 mM and 20 μl of Sequenase loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). The samples were incubated for 2 min at 90°C and analyzed on a 6% sequencing gel. After electrophoresis, the gels were dried and subjected to autoradiography. To locate the DNase I footprint, sequencing reactions as described above were run concomitantly.
Miscellaneous methods.
Protein was determined as described elsewhere (5), with bovine serum albumin as a standard. SDS-PAGE was carried out with the buffer system described by Laemmli (29) in 10% gels, and protein bands were visualized by staining with Coomassie brilliant blue R-250.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the EMBL-GenBank database under accession no. Y18817 and Y18818.
RESULTS
DNA sequence at the right-hand end of the S. coelicolor act cluster.
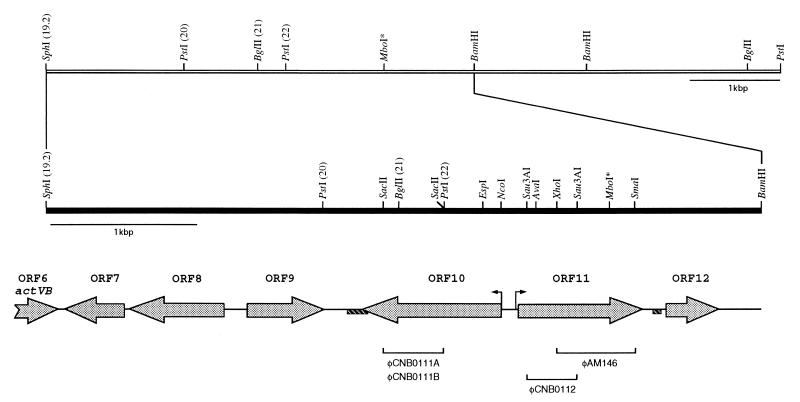
To determine whether there were any other additional regulatory genes involved in the control of actinorhodin biosynthesis, the region next to the right-hand end of the act cluster was explored. Thus, starting at SphI (site 19.2 [17, 32]), a DNA region of 4.8 kbp, adjacent to actVB-orf6 and extending rightward, has been sequenced; the first 31 bp correspond to those already found at the 3′ end by Fernández-Moreno et al. (17). Due to the occurrence of a rearrangement at an MboI site in the original plasmid (pIJ2300) (31) at nucleotide 3818, the entire DNA sequence of this region was reisolated directly from the S. coelicolor J1501 chromosome as described in Materials and Methods (Fig. 1). A scheme of the modified resulting restriction map is shown in Fig. 1.
FIG. 1.
Restriction map of the S. coelicolor A3(2) chromosome next to the right-hand end of the act cluster. The organization of ORFs within this region as deduced by DNA sequencing is given. Only relevant restriction sites are indicated; numbers in parentheses correspond to those reported by Malpartida and Hopwood (32). The MboI restriction site at which DNA rearrangement has occurred is indicated by asterisks. The solid bar shows the extent of the sequenced DNA fragment. The direct repeat sequences at the 3′ ends of orf10 and orf11 are represented by shaded boxes. The DNA fragments used for gene disruptions with the φC31-derived PM1 vector are indicated.
Computer-assisted analysis of the DNA sequence, using CODONPREFERENCE, revealed a set of six putative complete ORFs (Fig. 1), which were named (from left to right) orf7 to orf12. orf9, orf11, and orf12 are transcribed rightward (in the same direction as actVB), whereas orf7, orf8, and orf10 run divergently. The translation start codon for each ORF was tentatively allocated by using the following criteria: (i) the distribution of GC content in the third position (4, 49), (ii) codon usage (49), (iii) the presence of a potential Shine-Dalgarno sequence upstream of the initiation codon (44), and (iv) observed similarities to other putative ORF products from databases. The most relevant features deduced from the DNA sequence are summarized in Table 2.
TABLE 2.
Relevant features of the S. coelicolor 4.8-kbp region deduced from its DNA sequence
| orf | SDa | Start/stop codonsb | No. of amino acids (Mr) | Predicted function |
|---|---|---|---|---|
| 7 | 543GGAGG539 | ATG 529/32 TGA | 165 (17,924) | Minicircle protein |
| 8 | 1221GAAGGAG1215 | ATG 1209/583 TGA | 208 (22,299) | Methyltransferase |
| 9 | None | ATG 1344/1916 TGA | 190 (20,619) | Unknown |
| 10 | None | ATG 3091/2168 TGA | 307 (33,279) | Transcriptional regulator (LysR family) |
| 11 | 3180GAGGGAG3186 | ATG 3192/4022 TGA | 276 (28,780) | SDR family |
| 12 | 4192AGGAG4196 | GTG 4203/4544 TGA | 113 (11,883) | Unknown |
SD, putative Shine-Dalgarno sequence.
Numbers indicate the first and last nucleotides of the start and stop codons, respectively.
The overall GC content of 76% is typical for the genus Streptomyces; a series of direct repeats was found either within the 3′-end region of the orf10 gene (nucleotides 2041 to 3184) or downstream of orf11 (nucleotides 4117 to 4163), their length ranging from 22 to 37 or from 17 to 35 nucleotides, respectively (Fig. 1).
Deduced functions of the sequenced genes.
Searching for similarities of the deduced proteins with others in databases revealed a significant resemblance of the Orf7 protein with a hypothetical 13.3-kDa protein from the S. coelicolor minicircle (22) (although the former is 57 amino acids longer at its N terminus) and with the C terminus of the homologous deduced products, ScI35.38c (EMBL data bank accession no. A1031541) and Sc3c8.21c (EMBL data bank accession no. A1023861), within the IS117-A and IS117-B chromosomal DNA regions (13), respectively.
The Orf8 protein showed low levels of similarity with a number of methyltransferases from different sources, particularly with the Rhodococcus sp. methyltransferase-decarboxylase CobL protein (EMBL database accession no. L21196) (similarity, 44%; identity, 48%).
The sequence of the deduced Orf9 protein showed end-to-end homology to the translated product of Sc6G4.21 (EMBL database accession no. A1031317) from S. coelicolor and the C terminus of the Orf1 protein in the famA-famB DNA region of the Streptomyces purpurascens chromosome (EMBL database accession no. X61931).
The deduced orf10 gene product showed strong resemblance to a number of transcriptional regulators belonging to the LysR family (23, 41). The highest similarity was 46% with BudR, a protein from Klebsiella terrigena believed to be involved in 2,3-butandiol synthesis, and less with other members of the same family (Fig. 2). The similarity or identity levels observed were in the range of 40 to 46% and 32 to 38%, respectively, for the entire amino acid sequence. As a member of the LysR family, the N terminus of Orf10 protein is the most highly conserved region and contains the typical helix-turn-helix (HTH) motif, known to be involved in the binding to the target DNA sequence. Some of the proteins of this family are known to regulate transcription of divergently arranged genes; this might well be the case for Orf10 protein, because a divergently transcribed gene (orf11) is found next to it. Thus, orf10 may encode a protein that is involved in regulating expression of the orf11 gene.
FIG. 2.
Multiple alignment of the N terminus of S. coelicolor Orf10 protein (Orf10_Strco) with those of other LysR-type transcriptional regulators. The HTH motif involved in DNA binding is indicated. Origins of the amino acid sequences and (in parentheses) their Swissprot database accession numbers are as follows: BudR_Klete, Klebsiella terrigena (P52666); AlsR_Bacsu, Bacillus subtilis (Q04778); CbbR_Xanfl, Xanthobacter flavus (P25545); XapR_Ecoli, E. coli (P23841); Ttua_Agrvi, Agrobacterium vitis (P52669); and CynR_Ecoli, E. coli (P27111). Black boxes indicate positions in the alignment where the same amino acid is found in at least four of the seven sequences; gray boxes indicate residues similar to those marked in black.
The Orf11 protein showed significant similarity with proteins of the so-called short-chain dehydrogenases/reductase (SDR) (27) family (Fig. 3). Similarity ranged from 33 to 47% and identity ranged from 25 to 38%, the best score being found with 3-oxoacyl-acyl carrier protein reductase from E. coli. In addition, the actIII gene product (20) was identified as homologous in database searches, as were genes from other Streptomyces species, such as car (36), mon-kr (1), gra-orf5-6 (43), dpsE (19), and jad-orf5 (21), contained in the antibiotic biosynthetic clusters for clavulanic acid, monensin, granaticin, daunorubicin, and jadomicin B, respectively. Most of the enzymes in this family are known to be NAD(H)- or NADP(H)-dependent oxidoreductases. As described for this family of proteins, the putative coenzyme binding domain, represented by three highly conserved glycine residues (Gly-9, Gly-13, and Gly-15), is located in the N-terminal portion of the Orf11 protein (Fig. 3A). Additionally, the conserved Asp-56 may serve as a site for hydrogen bonding to the coenzyme, as in many other dehydrogenases (27). Orf11 protein may contain the second well-conserved domain of these enzymes, corresponding to the active site, represented by the three conserved amino acid residues (Ser-135, Tyr-148, and Lys-152) with a consensus spacing (Fig. 3B). Moreover, as expected, the hydrophilicity pattern within this region of Orf11 exhibited high similarity to that estimated for other oxidoreductases of the same family (data not shown).
FIG. 3.
Alignment of the putative NAD(H)-NADP(H) binding (A) and catalytic (B) sites of S. coelicolor Orf11 protein (Orf11_Strco) with those of other SDRs. Origins of the proteins and (in parentheses) their Swissprot database accession numbers are as follows: Dhb1_human, human (P14061); Fabg_Ecoli, E. coli (P25716); Enta_Ecoli, E. coli (P15047); 2bhd_Strex, Streptomyces exfoliatus (P19992); Ridh_Kleae, Klebsiella aerogenes (P00335); Phbb_Zoora, Zoogloea ramigera (P23238); Act3_Strco, S. coelicolor (P16544); and Dhkr_Strcm, Streptomyces cinnamonensis (P41177). Conserved and similar residues are in black and gray boxes, respectively (plurality, 5). The amino acids reported to play a role in either site are indicated.
A search of databases with the orf12 product gave no significant similarities, and therefore no function could be ascribed for Orf12 protein.
Analysis of orf10 and orf11 function in S. coelicolor.
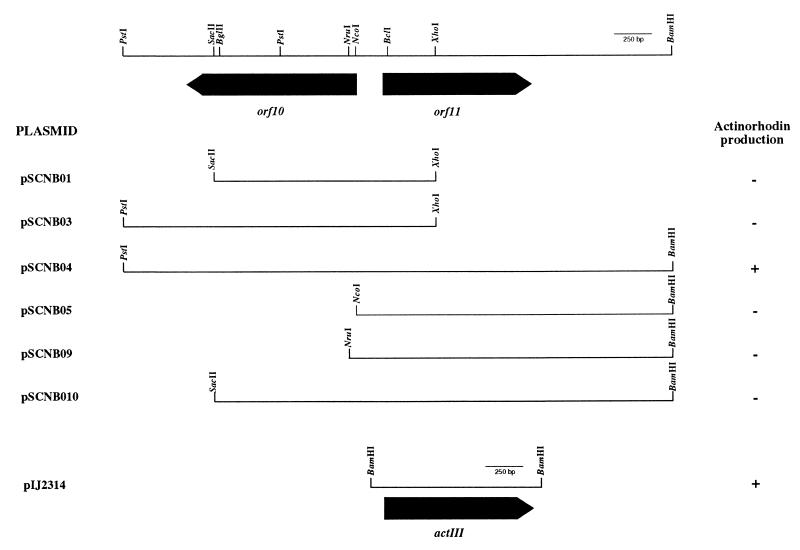
To explore the possible role of orf10 and orf11 gene products in S. coelicolor, mutants were generated by insertional inactivation within both genes, as described in Materials and Methods (Fig. 1). To disrupt orf10, the internal SacII fragment (nucleotides 2274 to 2674) cloned in pIJ2925 (pCNB01004) was recovered by digestion with BglII and ligated to phage PM1, previously digested with BglII. The recombinant phages φCNB0111A and φCNB0111B (Fig. 1), carrying the insert in opposite orientations, were obtained. Similarly, the same fragment was cloned into the pGM9 vector, yielding plasmids pSCNB06A and pSCNB06B for either orientation. For orf11 disruptions, we constructed two different PM1 derivatives, one carrying the 532-bp XhoI-SmaI fragment (nucleotides 3465 to 3997) and the other carrying the 336-bp Sau3AI fragment (nucleotides 3242 to 3578), for φAM146 and φCNB0112, respectively (Fig. 1). All of these constructions were used for gene disruptions in S. coelicolor J1501; that either gene had indeed been interrupted was confirmed by Southern blot analysis. Antibiotic production and auxotrophies were tested in the disruptants, and no obvious phenotypic differences were observed between any of them and the wild-type strain.
Because of the homology between the Orf11 and ActIII proteins, we next explored if the orf11 gene had the ability to complement the actIII mutation in the actinorhodin biosynthetic pathway of S. coelicolor. Therefore, several plasmids were constructed (Table 1; Fig. 4) and used to transform the actIII mutant strain S. coelicolor TK18. S. coelicolor TK18 showed an actinorhodin-producing phenotype only in the presence of both orf10 and orf11 genes (pSCNB04), suggesting that orf10 is required along with the orf11 gene for antibiotic production. It is worth noting that during the cloning steps for orf10 disruptions, a TAG codon was generated four nucleotides downstream of the original SacII restriction site (nucleotide position 2274), thus generating a truncated protein lacking the last 33 amino acids (Orf101-274). This construct was used to generate pSCNB010; interestingly, this plasmid did not allow complementation of the actIII mutation (Fig. 4). All of these results strongly suggest that the Orf10 protein may be involved in the regulation of orf11 expression.
FIG. 4.
Subcloning of the orf10-orf11 DNA region to elucidate the genes required for complementation analysis of actIII mutation. Plasmid construction was as described in Materials and Methods and Table 1. Plasmids pSCNB01 and pSCNB010 contain a TAG stop codon (generated by the cloning procedure) downstream of the 3′ end of the orf10 deletion. Plasmid pIJ2314 contains the actIII gene. Symbols: −, no actinorhodin production; +, actinorhodin production.
Additionally, it is noteworthy that S. coelicolor J1501 showed no obvious change in phenotype when transformed with plasmids (shown in Fig. 4) containing either or both of the orf10 and orf11 genes.
Function of the orf10 gene in S. lividans.
The possible function of the orf10 gene was further investigated in S. lividans, a streptomycete closely related to S. coelicolor. Using the pGM9 derivative plasmids pSCNB06A and pSCNB06B, the orf10 gene was disrupted in S. lividans. Similarly, orf10 deletions in S. lividans were generated by using the pGM9 derivative plasmids pSCNB08B and pSCNB07, yielding strains CNB062 and CNB073, respectively. Unlike the parental strain, both disruption and deletion of the orf10 gene yielded an actinorhodin-producing phenotype, suggesting a function of orf10 in the regulation of actinorhodin production in S. lividans.
To confirm that the resulting phenotype was indeed due to the absence of the orf10 gene, plasmids pSCNB013 and pSCNB014 containing orf10 and its promoter region were constructed and used to transform S. lividans CNB073. When the orf10 gene was introduced in this strain in trans on either a low (pSCNB013)- or high (pSCNB03 or pSCNB014)-copy-number plasmid, S. lividans CNB073 showed no change in phenotype. Nevertheless, when the orf10 gene was introduced in cis into the chromosome by insert-directed recombination through its 3′ end (using the pGM9 derivative pSCNB03), the actinorhodin-producing phenotype of the mutant strain reverted to the wild-type actinorhodin-nonproducing phenotype.
Cloning and characterization of the orf10-orf11 homologous region of S. lividans.
Because all previous constructions were generated by using clones from S. coelicolor, it was of interest to determine if there were within this DNA region relevant sequence differences between this species and S. lividans that could account for the actinorhodin-nonproducing phenotype of S. lividans. Thus, the homologous orf10-orf11 region of S. lividans TK21 was isolated (see Materials and Methods). The 3-kb PstI-BamHI fragment from this region was subcloned and sequenced, and three putative ORFs, orf10, orf11, and orf12, were identified. The DNA sequence was shown to be 99% identical between S. coelicolor and S. lividans. The corresponding products of orf10, orf11, and orf12 were shown to be almost identical between the two species, with the following mismatches: for Orf10 protein, Thr-82, Gln-130, and Ala-192 have been replaced in S. lividans by Ser, Arg, and Val, respectively; for Orf11 protein, Asp-245 has been changed to Ala in S. lividans; and for Orf12 protein, Ser-88 has been replaced by Ala in S. lividans. Such small differences in the Orf10 sequence between the species seem insufficient to explain the actinorhodin-nonproducing phenotype of S. lividans. Moreover, the actinorhodin-producing phenotype of the S. lividans orf10-deleted strain was reverted to the wild-type (actinorhodin-nonproducing) phenotype when the orf10 gene from S. lividans was introduced in cis within the S. lividans orf10 mutant chromosome. Additionally, the orf10 and orf11 genes from S. lividans on a high-copy-number plasmid restored the blue-pigmented phenotype of the S. coelicolor actIII mutant. Thus, orf10 and orf11 appear to function similarly irrespective of their origin.
Transcriptional analysis of the act pathway-specific regulatory gene, actII-orf4.
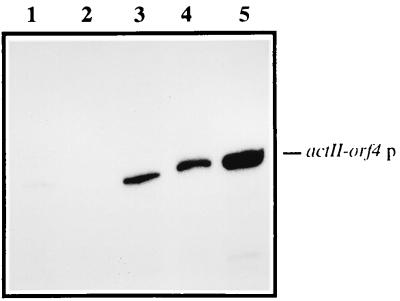
The possible mechanism involved in actinorhodin induction by orf10 disruption and deletion in S. lividans was further examined by analyzing the transcription of the positive regulator of the act genes, the actII-orf4 gene. As shown in Fig. 5, nuclease S1 protection experiments of this gene revealed an increase in its transcription in both S. lividans orf10-disrupted and -deleted strains compared with the basal levels of the parental strain (S. lividans TK21). Thus, the actinorhodin biosynthesis activation caused by orf10 disruption and deletion in S. lividans appears to be mediated by an increase in the transcriptional level of the actII-orf4 gene.
FIG. 5.
Transcription analysis of the actII-orf4 gene. Total RNA was isolated from 3-day-old cultures of S. lividans strains TK21, TK21::pSCNB06A, and CNB073 (lanes 1, 3, and 4, respectively) and of S. coelicolor J1501 (lane 5). E. coli tRNA was used as a control (lane 2). A protected fragment of the expected size (384 nucleotides) was observed. End-labeled HinfI-digested pBR329 was used as a size marker. p, promoter.
Transcriptional analysis of orf10 and orf11 genes.
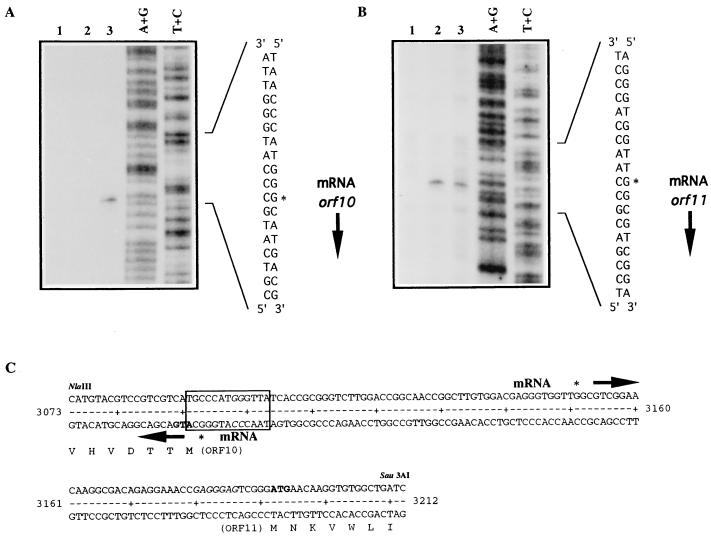
Attempts to determine the transcription start site of orf10 from S. lividans and S. coelicolor from the chromosomal gene as well as the gene carried on a high-copy-number plasmid (pSCNB03, pSCNB04, or pSCNB014) were unsuccessful. Presumably, the apparent low expression of orf10 mRNA could be due to a combination of a weak promoter and tight autoregulation. For that reason, S1 mapping analysis was attempted as described in Materials and Methods, using total RNA extracted from both orf10-disrupted and orf10-deleted mutants of S. lividans. While no protected product was observed with RNA of the former strain, a 152-bp protected fragment was detected (Fig. 6A) with RNA isolated from the orf10-deleted mutant. Thus, the orf10 transcription initiation site was localized one base upstream of its putative start codon, with no room for a ribosome binding site. The lack of a protected fragment in the orf10 disruption mutant suggests that the truncated Orf101-274 might be still functional in regulating its own transcription.
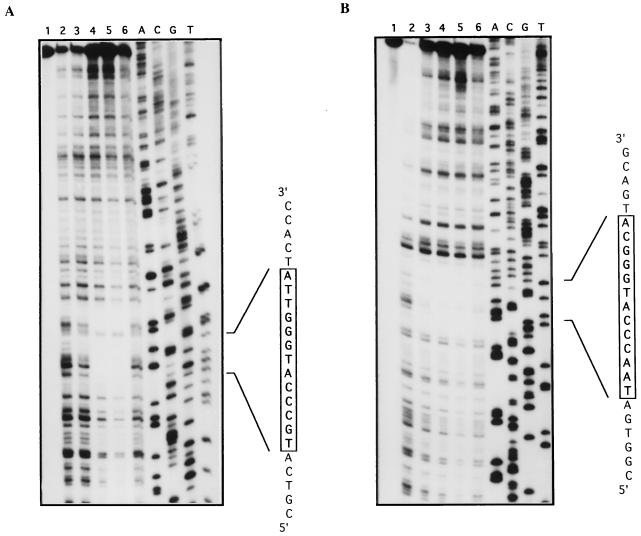
FIG. 6.
Transcriptional analysis of orf10 (A) and orf11 (B) genes. Lanes: 1, E. coli tRNA; 2 and 3, total RNA extracted from 3-day-old cultures of S. lividans TK21 and CNB073, respectively; A+G and T+C, Maxam-and-Gilbert sequence ladders of the corresponding labeled probes. Asterisks indicate the most probable transcription start sites. (C) DNA sequence within the orf10-orf11 intergenic region. orf10 and orf11 mRNAs are initiated at the indicated nucleotides. Arrows indicate direction of transcription. Amino acids are represented in single-letter code below the DNA sequence. The boxed sequence represents the TN11A consensus motif for LysR-type regulators contained in the DNase I-protected region. The GG nucleotides mutated to AT within this region are shown in italics.
The transcription start point of orf11, unlike that of orf10, was easily determined as described in Materials and Methods by using RNA extracted from S. lividans strains (Fig. 6B) and located at nucleotide position 3151. A similar initiation point was obtained with RNA isolated from S. coelicolor J1501 (data not shown). Based on S1 mapping results, neither the orf10 nor the orf11 promoter displays typical −10 and −35 regions (Fig. 6C). Additionally, the proximity of orf10 and orf11 initiation sites (58 bp) indicates that the −35 regions of the two promoters must overlap. As shown in Fig. 6B, we detected no change in orf11 transcript level when analyzing total RNA extracted from S. lividans CNB073, suggesting that Orf10 protein might be required for the activation of orf11 transcription, in agreement with the complementation results of actIII mutation.
Expression and purification of S. coelicolor Orf10 protein.
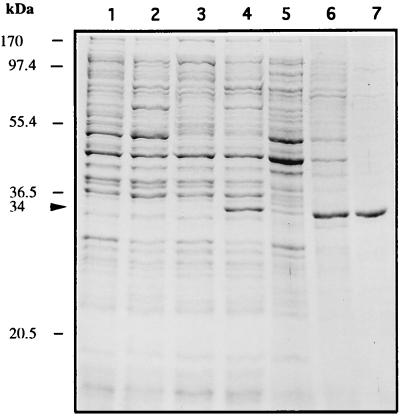
To gain some insight into the biological activity of the orf10 product, the protein was expressed from E. coli and purified (see Materials and Methods). Plasmid pCNB019, in which orf10 is under the control of lambda pL, was constructed and used to transform E. coli K12ΔH1Δtrp. As shown in Fig. 7 (lane 4), a whole-cell extract from an induced culture of this strain harboring pCNB019 showed by SDS-PAGE analysis a 34-kDa protein which corresponds to the predicted apparent molecular mass for the recombinant Orf10 protein. Such a protein was apparently not present in samples from uninduced or induced cultures of E. coli carrying only the plasmid vector (lane 1 or 2, respectively), as well as from uninduced cultures from the same recombinant strain (lane 3). Orf10, mostly in inclusion bodies, was then purified by urea solubilization followed by heparin-agarose chromatography (see Materials and Methods). The Orf10 protein was purified to 98%, as judged by SDS-PAGE analysis (Fig. 7, lane 7).
FIG. 7.
SDS-PAGE analysis of orf10 expression in E. coli and at various stages of its purification. Lanes: 1 and 2, whole-cell extracts from cultures of E. coli K12ΔH1Δtrp carrying the vector plasmid pAZe3ss, grown at 30 and 42°C, respectively; 3 and 4, whole-cell extracts from cultures of the same strain harboring plasmid pCNB019, grown at 30 and 42°C, respectively; 5, supernatant of 25,000 × g centrifugation; 6, pellet of this centrifugation (inclusion bodies) after the refolding step; 7, heparin-agarose chromatography (1 μg of protein). The recombinant purified Orf10 protein is indicated by arrowheads. The positions and molecular masses of marker proteins are shown on the left.
Gel mobility shift assays with the recombinant Orf10 protein.
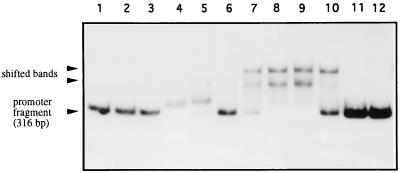
The DNA binding activity of Orf10 protein was tested by band shift analysis. A 316-bp EspI-AvaI fragment containing the intergenic orf10-orf11 region (Fig. 1), uniquely labeled at the 5′ end of the EspI site (nucleotide 2981), was used as the probe. With this fragment, the Orf10 protein showed band shift activity which was dependent on Orf10 protein concentration (Fig. 8). Complete retardation was obtained with 25 ng of the Orf10 protein (Fig. 8, lane 9); the activity was lost partially with use of a fivefold molar excess of the unlabeled fragment (lane 10) and completely when a temperature-denatured Orf10 protein was used (lane 11). No dependence on divalent cations, such as Mg2+, Mn2+, and Ca2+ or K+ (tested up to 200 or 500 mM, respectively), was found for the DNA binding activity of Orf10 (data not shown). It is worth noting that unlike the crude extracts prepared from uninduced and induced cultures of E. coli harboring the vector plasmid (Fig. 8, lanes 2 and 3), those from the respective cultures of E. coli transformed with pCNB019 caused the appearance of a shifted band (Fig. 8, lanes 4 and 5, respectively). This behavior may be due to the presence of either truncated forms of Orf10 or misfolded protein still retaining the DNA binding activity.
FIG. 8.
DNA binding assays. Gel mobility shift analysis with the orf10-orf11 intergenic region was performed as described in Materials and Methods, using the 316-bp EspI-AvaI fragment as the probe. Lanes: 1 and 12, without protein addition; 2 and 3, with crude extracts from cultures of E. coli K12ΔH1Δtrp carrying the control plasmid pAZe3ss, grown at 30 and 42°C, respectively; 4 and 5, with crude extracts from cultures of the same strain harboring plasmid pCNB019, grown at 30 and 42°C, respectively; 6 to 9, with 1.2, 2.5, 12.5 and 25 ng of the purified S. coelicolor Orf10 protein, respectively; 10, with 25 ng of Orf10 and fivefold molar excess of unlabeled orf10-orf11 intergenic region; 11, with 25 ng of Orf10 previously treated for 15 min at 100°C.
To locate the binding region within the orf10 promoter region, the EspI-AvaI (nucleotides 2981 to 3297) probe was shortened by digestion with restriction enzymes Sau3AI, AvaII, and NcoI at nucleotide positions 3242, 3118, and 3094, respectively. Both EspI-Sau3AI and EspI-AvaII fragments still retained the band shift activity, while no band retardation was observed with the EspI-NcoI probe (data not shown). Thus, the interacting region of Orf10 may lie between positions 3094 and 3118 of the orf10-orf11 promoter region.
The truncated Orf101-274, expressed in E. coli and purified as described in Materials and Methods, was also tested for DNA binding activity. A shifted band was detected with the recombinant Orf101-274 (data not shown), in agreement with the previous suggestion (see above). Interestingly, the retarded band migrated similarly to those shown for crude extracts from both uninduced and induced cultures of E. coli carrying the wild-type orf10 gene (Fig. 8, lanes 4 and 5, respectively). Additionally, the purified Orf10 protein from S. lividans showed the same activity as that of S. coelicolor.
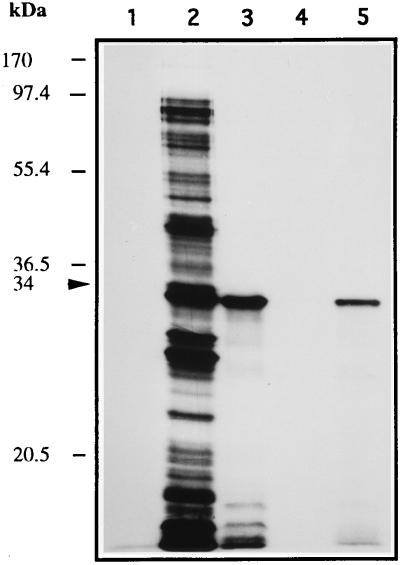
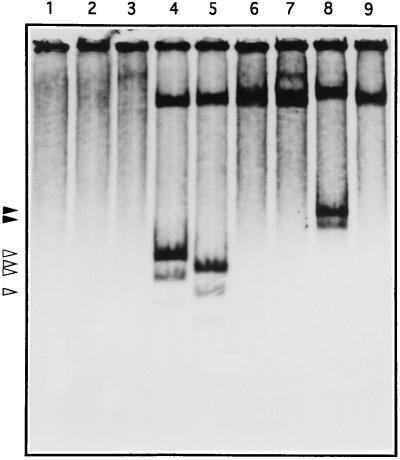
Because of the correlation between actinorhodin activation and expression of the orf10 gene, we next explored possible targets of the Orf10 protein within the promoter regions of some of the act genes. 35S-labeled Orf10 protein was obtained by using a T7 RNA polymerase expression system (see Materials and Methods). SDS-PAGE and autoradiography analysis revealed a high-abundance labeled protein of the size predicted for Orf10 (Fig. 9). This protein was purified from inclusion bodies (Fig. 9, lane 4) and assayed for DNA binding activity (Fig. 10). As expected, either a 246-bp NruI-AvaI or a 206-bp AhaII fragment within the intergenic orf10-orf11 region showed DNA binding activity (Fig. 10, lanes 4 and 5). However, no such activity was observed with any of the DNA fragments tested from the act cluster (data not shown; actIII-actI intergenic region [Fig. 10, lane 9]), as well as those containing the direct repeats at the 3′ end of both orf10 and orf11 (Fig. 10, lanes 6 and 7). Additionally, a 484-bp EspI-XhoI fragment (nucleotides 2981 to 3465) within the orf10 promoter, in which nucleotides GG (at positions 3098 and 3099) had been mutated to AT to generate an NdeI restriction site, retained binding activity (Fig. 10, lane 8).
FIG. 9.
Synthesis of 35S-labeled Orf10 protein. Samples were analyzed by SDS-PAGE and autoradiography as described in Materials and Methods and include whole-cell extract from a culture of E. coli K38/pGP-1-2 carrying plasmid pT7.7 grown at 42°C with rifampin addition (lane 1), whole-cell extract from a culture of E. coli K38/pGP-1-2 carrying plasmid pCNB023 grown at 42°C without and with rifampin addition (lanes 2 and 3, respectively), and supernatant of 25,000 × g centrifugation (lane 4) and its pellet (inclusion bodies) after the refolding step (lane 5). Positions of size markers are indicated on the left. The 34-kDa labeled protein is labeled with an arrow.
FIG. 10.
Analysis of DNA binding activity of 35S-labeled Orf10 purified from inclusion bodies. The binding reaction was carried out as described in Materials and Methods, and products were analyzed by native PAGE and autoradiography. Lanes: 1, in the absence of DNA; 2, with EcoRI-digested pUC19; 3, with EcoRI-digested M13mp18; 4, with the 246-bp NruI-AvaI fragment within the orf10-orf11 intergenic region (pCNB033 insert); 5, with the 206-bp AhaII fragment within the orf10-orf11 intergenic region (pCNB034A insert); 6 and 7, with the 393-bp PstI-SacII and 227-bp SmaI-HincII fragments containing direct repeat sequences at the 3′ ends of orf10 and orf11, respectively; 8, with the 484-bp EspI-XhoI fragment within the orf10-orf11 intergenic region containing the NdeI-engineered restriction site; 9, with the 455-bp MboII fragment within the actIII-actI intergenic region. Arrows indicate mobilities of the complexes between Orf10 protein and its DNA target.
DNase I footprinting.
The interaction between Orf10 and its DNA target within the orf10-orf11 intergenic region was analyzed by DNase I protection assays carried out as described in Materials and Methods. The protected region in either strand (Fig. 11) was shown to expand over a 30-bp region and included the sequence TN11A, described as a general anchor for LysR-type proteins (41) (nucleotide positions 3091 to 3103) (Fig. 6C). The DNA binding site of Orf10 protein was shown to be located within the orf10 transcriptional start point, suggesting a direct competition between this transcriptional regulator and the RNA polymerase as a mechanism for autoregulation. It is interesting that the mutated bases (GG to AT to generate an NdeI restriction site) lie within the TN11A consensus motif (Fig. 6C), and as shown above, these mutations did not seem to affect Orf10 DNA binding activity. Similar DNase I-protected boxes were obtained when either the S. coelicolor Orf101-274 protein or the S. lividans Orf10 protein was used (data not shown).
FIG. 11.
DNase I footprinting analysis of the interactions between Orf10 protein and the orf10-orf11 intergenic region. Upper (A) and lower (B) strands correspond to pCNB033 and pCNB034A inserts. DNase I protection experiments were done as described in Materials and Methods. Lanes: 1, without DNase I; 2, with DNase I but no Orf10; 3 to 5, with 20, 200, and 600 ng of Orf10, respectively; 6, with 600 ng of Orf10 and 10-fold molar excess of unlabeled orf10-orf11 intergenic region. The corresponding sequence reactions (lanes A, C, G, and T) were run in parallel. The boxed regions correspond to the TN11A consensus motif for LysR-type regulators.
DISCUSSION
An additional regulatory gene, orf10, negatively controlling actinorhodin production in S. lividans has been isolated and characterized; this gene is located downstream of the right-hand end of the S. coelicolor actinorhodin biosynthetic cluster. Linked to and divergently transcribed from this new regulator is a gene, orf11, whose deduced product is homologous to the SDR family of proteins (27). Moreover, the ActIII protein (which is implicated in the formation of actinorhodin) as well as other oxidoreductases involved in the synthesis of several other antibiotics belong to this group of proteins.
The nature of Orf10 protein as a transcriptional regulator was inferred from its homology to other proteins belonging to the LysR family. Members of this large family are implicated in the regulation of apparently unrelated metabolic pathways and share several common features (41): (i) a highly conserved HTH motif near the N terminus implicated in DNA binding; (ii) the involvement of a coinducer in regulatory activity; (iii) a divergent promoter structure; and (iv) negative autoregulation. Additionally, a consensus DNA sequence, TN11A, has been proposed as binding site for the LysR-type regulators (41). In this study, we provide evidence that Orf10 may exhibit some (if not all) of the characteristics of the proteins of the LysR family.
Database comparisons indicated similarities of Orf10 with LysR-type regulators, particularly within the N-terminal region, where the HTH motif lies. This might be expected, since the central and C-terminal regions of these proteins have been reported to be implicated in several functions, including the recognition of, and response to, a coinducer (41). By gel retardation and DNase I footprinting analysis, Orf10 was shown to be able to bind to a DNA target (orf10-orf11 intergenic region) which includes a putative TN11A consensus motif. In agreement with this result, a C-terminally truncated Orf10, Orf101-274, retained DNA binding activity.
Regarding the control by Orf10 of the divergently transcribed orf11, two observations suggest that Orf10 can regulate orf11 expression. First, in most cases, the target genes for LysR-type regulators are located next to them and possess a conserved divergent promoter structure with respect to the regulatory gene; such organization may well be found for the orf10-orf11 system. Second, complementation of the actIII mutation requires the presence of both orf10 and orf11, while it was not achieved when orf10 carried a 3′-end deletion. Although potentially expressed, Orf101-274 may be unable to activate orf11 transcription. The presence of detectable levels of orf10 transcript only in orf10-deleted and not in wild-type or orf10-disrupted mutant strains constitutes evidence that the orf10 product negatively regulates its own transcription. However, it should be stressed that additional factors may be involved in the regulation of orf10 expression, as reversion of the orf10 mutant phenotype occurred only by cis complementation.
Mutations (by disruption or deletion) of orf10 induce actinorhodin production in S. lividans. This blue-pigmented phenotype appeared to be caused by an increase in transcription of the pathway-specific regulatory gene actII-orf4, although the mechanism(s) involved in this activation is not known. In this sense, no binding activity of Orf10 to any of the promoter regions of the act cluster was detected; still, the requirement of a coinducer for this activity as well as for in vivo inhibition of actinorhodin biosynthesis cannot be excluded and is currently being investigated. Alternatively, Orf10 may regulate an unknown transcriptional factor that, in turn, negatively controls actII-orf4 gene expression, thus leading to induction of actinorhodin production in orf10-blocked mutants.
Results of complementation experiments using the S. coelicolor actIII mutant revealed that the orf11 gene allowed restitution of a blue-pigmented phenotype (although only in the presence of the entire orf10 gene). It is worth noting that complementation of this strain by gra-orf5 and gra-orf5-orf6 gene products (42), which are also oxidoreductases of the SDR family, has been observed. Furthermore, the actIII product has been shown to function heterologously in an anthracycline system (2). Although the orf11 gene is able to complement the actIII mutation, it does not seem to play a role in the actinorhodin biosynthetic pathway, as orf11 disruptions did not generate a mutant phenotype; the observed complementation might be explained by the small ketoreductase activity of Orf11 protein, which would be increased significantly by extra copies of the gene and/or its activation by the orf10 gene product. It should be noted that orf11 transcription in the actIII mutant strain carrying orf10 and orf11 showed a threefold increase (33). Considering that the orf10-orf11 system appeared to be strictly regulated, these two genes, despite being active, may not be sufficient to overcome the ActIII deficiency when expressed from a single chromosomal copy.
The presence of direct repeat sequences downstream of orf10 and orf11 suggests a role for those sequences in recombination events. Those sequences may have played a role in mediation of chromosomal rearrangement or in duplication of DNA segments.
ACKNOWLEDGMENTS
We are pleased to acknowledge D. Holmes, M. Zalacain, J. Hodgson, and S. Elson for valuable discussions throughout this study and W. Wohlleben for the gift of plasmid pGM9.
This research was supported by grants from the Spanish CICYT (95-0104-OP-02-01 and BIO96-1168-C02-01) and by Smith-Kline Beecham S.A.
REFERENCES
- 1.Arrowsmith T J, Malpartida F, Sherman D H, Birch A, Hopwood D A, Robinson J A. Characterisation of actI-homologous DNA encoding polyketide synthase genes from the monensin producer Streptomyces cinnamonensis. Mol Gen Genet. 1992;234:254–264. doi: 10.1007/BF00283846. [DOI] [PubMed] [Google Scholar]
- 2.Bartel P L, Zhu C B, Lampel J S, Dosch D C, Connors N C, Strohl W R, Beale J J, Floss H G. Biosynthesis of anthraquinones by interspecies cloning of actinorhodin biosynthesis genes in streptomycetes: clarification of actinorhodin gene functions. J Bacteriol. 1990;172:4816–4826. doi: 10.1128/jb.172.9.4816-4826.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bullock W O, Fernández J M, Short J M. XL1 blue: a high efficiency plasmid transforming recA Escherichia coli strain with galactosidase selection. BioTechniques. 1987;5:376. [Google Scholar]
- 7.Caballero J L, Malpartida F, Hopwood D A. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol Gen Genet. 1991;228:372–380. doi: 10.1007/BF00260629. [DOI] [PubMed] [Google Scholar]
- 8.Caballero J L, Martínez E, Malpartida F, Hopwood D A. Organisation and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol Gen Genet. 1991;230:401–412. doi: 10.1007/BF00280297. [DOI] [PubMed] [Google Scholar]
- 9.Champness W C, Chater K F. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp. In: Piggot P, Moran C P J, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C: American Society for Microbiology; 1994. pp. 61–93. [Google Scholar]
- 10.Chater K F, Bibb M J. Regulation of bacterial antibiotic production. In: Kleinkauf H, von Döhren H, editors. Biotechnology. 2nd ed. 7. Products of secondary metabolism. Weinheim, Germany: VCH; 1997. pp. 57–105. [Google Scholar]
- 11.Chater K F, Bruton C J. Mutational cloning in Streptomyces and the isolation of antibiotic production genes. Gene. 1983;26:67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- 12.Chater K F, Bruton C J, King A A, Suarez J E. The expression of Streptomyces and Escherichia coli drug-resistance determinants cloned into the Streptomyces phage φC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 13.Chater K F, Hopwood D A. Streptomyces. In: Sonenshein A L, Hoch J A, Losick L R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 83–89. [Google Scholar]
- 14.Covarrubias L, Bolivar F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene. 1982;17:79–89. doi: 10.1016/0378-1119(82)90103-2. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 17.Fernández-Moreno M A, Martínez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced function of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 18.Fernández-Moreno M A, Martínez E, Caballero J L, Ichinose K, Hopwood D A, Malpartida F. DNA sequence and functions of the actVI region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor A3(2) J Biol Chem. 1994;269:24854–24863. [PubMed] [Google Scholar]
- 19.Grimm A, Madduri K, Ali A, Hutchinson C R. Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene. 1994;151:1–10. doi: 10.1016/0378-1119(94)90625-4. [DOI] [PubMed] [Google Scholar]
- 20.Hallam S E, Malpartida F, Hopwood D A. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene. 1988;74:305–320. doi: 10.1016/0378-1119(88)90165-5. [DOI] [PubMed] [Google Scholar]
- 21.Han L, Yang K Q, Ramalingam E, Mosher R H, Vining L C. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiology. 1994;140:3379–3389. doi: 10.1099/13500872-140-12-3379. [DOI] [PubMed] [Google Scholar]
- 22.Henderson D J, Lydiate D J, Hopwood D A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2) Mol Microbiol. 1989;3:1307–1318. doi: 10.1111/j.1365-2958.1989.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 23.Henikoff S, Haughn G W, Calvo J M, Wallace J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentschel C, Irminger J-C, Bucher P, Birnsteil M L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980;285:147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- 25.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces, a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 26.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 27.Jörnvall H, Persson B, Krook M, Atrian S, González-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 28.Kempter C, Kaiser D, Haag S, Nicholson G, Gnau V, Walk T, Gierling K H, Decker H, Zahner H, Jung G, Metzger J W. CDA: calcium-dependent peptide antibiotics from Streptomyces coelicolor A3(2) containing unusual residues. Angew Chem Int Ed. 1997;36:498–501. [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Leskiw B K, Mah R, Lawlor E J, Chater K F. Accumulation of bldA-specified transfer RNA is temporally regulated in Streptomyces coelicolor A3(2) J Bacteriol. 1993;175:1995–2005. doi: 10.1128/jb.175.7.1995-2005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malpartida F, Hopwood D A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 32.Malpartida F, Hopwood D A. Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2) Mol Gen Genet. 1986;205:66–73. doi: 10.1007/BF02428033. [DOI] [PubMed] [Google Scholar]
- 33.Martínez-Costa, O. H., and F. Malpartida. Unpublished results.
- 34.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 35.Muth G, Nubbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 36.Pérez-Redondo R, Rodríguez-García A, Martín J F, Liras P. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene. 1998;211:311–321. doi: 10.1016/s0378-1119(98)00106-1. [DOI] [PubMed] [Google Scholar]
- 37.Rudd B A, Hopwood D A. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2) J Gen Microbiol. 1979;114:35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- 38.Rudd B A, Hopwood D A. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol. 1980;119:333–340. doi: 10.1099/00221287-119-2-333. [DOI] [PubMed] [Google Scholar]
- 39.Russel M, Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984;159:1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 42.Sherman D H, Kim E S, Bibb M J, Hopwood D A. Functional replacement of genes for individual polyketide synthase components in Streptomyces coelicolor A3(2) by heterologous genes from a different polyketide pathway. J Bacteriol. 1992;174:6184–6190. doi: 10.1128/jb.174.19.6184-6190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman D H, Malpartida F, Bibb M J, Kieser H M, Bibb M J, Hopwood D A. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber TÜ22. EMBO J. 1989;8:2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W, improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward J M, Janssen G R, Kieser T, Bibb M J, Buttner M J, Bibb M J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 48.Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 49.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 50.Wright L F, Hopwood D A. Actinorhodin is a chromosomally determined antibiotic in Streptomyces coelicolor A3(2) J Gen Microbiol. 1976;96:289–297. doi: 10.1099/00221287-96-2-289. [DOI] [PubMed] [Google Scholar]
- 51.Wright L F, Hopwood D A. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2) J Gen Microbiol. 1976;95:96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 cloning vector and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 53.Zaballos A, Salas M, Mellado R P. A set of expression plasmids for the synthesis of fused and unfused polypeptides in Escherichia coli. Gene. 1987;58:67–76. doi: 10.1016/0378-1119(87)90030-8. [DOI] [PubMed] [Google Scholar]