Abstract
Background
Endoscopic ultrasonography (EUS) is crucial to diagnose and evaluate gastrointestinal mesenchymal tumors (GIMTs). However, EUS-guided biopsy does not always differentiate gastrointestinal stromal tumors (GISTs) from leiomyomas. We evaluated the ability of a convolutional neural network (CNN) to differentiate GISTs from leiomyomas using EUS images. The conventional EUS features of GISTs were also compared with leiomyomas.
Patients and Methods
Patients who underwent EUS for evaluation of upper GIMTs between 2010 and 2020 were retrospectively reviewed, and 145 patients (73 women and 72 men; mean age 54.8 ± 13.5 years) with GISTs (n = 109) or leiomyomas (n = 36), confirmed by immunohistochemistry, were included. A total of 978 images collected from 100 patients were used to train and test the CNN system, and 384 images from 45 patients were used for validation. EUS images were also evaluated by an EUS expert for comparison with the CNN system.
Results
The sensitivity, specificity, and accuracy of the CNN system for diagnosis of GIST were 92.0%, 64.3%, and 86.98% for the validation dataset, respectively. In contrast, the sensitivity, specificity, and accuracy of the EUS expert interpretations were 60.5%, 74.3%, and 63.0%, respectively. Concerning EUS features, only higher echogenicity was an independent and significant factor for differentiating GISTs from leiomyomas (p < 0.05).
Conclusions
The CNN system could diagnose GIMTs with higher accuracy than an EUS expert and could be helpful in differentiating GISTs from leiomyomas. A higher echogenicity may also aid in differentiation.
Keywords: Artificial intelligence, Convolutional neural network, Deep learning, Endoscopic ultrasonography, Gastrointestinal stromal tumor, Leiomyoma
Introduction
Gastrointestinal mesenchymal tumors are detected usually during upper GI endoscopy. Most are either entirely or partially composed of spindle cells and demonstrate the differentiation of smooth muscles or nerve sheaths. Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors in the gastrointestinal tract, found primarily in the stomach (60%–70%) and small bowel (20%–25%), and are rarely observed in the esophagus [1, 2]. In contrast, leiomyomas are rare in the stomach but most commonly occur in the esophagus [3]. GISTs need to be differentiated from leiomyomas as the former, even if small in size, have a theoretical malignant potential, whereas leiomyomas are almost always benign. A clear differentiation therefore can markedly affect their treatment and prognosis.
Endoscopic ultrasonography (EUS) is currently the standard modality for the evaluation of gastrointestinal mesenchymal tumors [4, 5]. It involves defining the layer of origin, assessment of the lesion size, extent, and sonographic features. A presumptive diagnosis can be made in most cases. However, the differentiation of GISTs from leiomyomas using EUS images alone has been reported inconsistently [6, 7, 8]. GISTs and leiomyomas are seen as relatively homogeneous hypoechoic lesions arising from the fourth hypoechoic layer (muscularis propria) or rarely the second hypoechoic layer (muscularis mucosa) of the gut wall. These cannot be distinguished unless EUS-guided fine-needle aspiration or biopsy (FNAB) followed by immunohistochemical tissue staining is performed. However, in some cases, FNAB may fall short of adequate samples. Therefore, alternative modalities are required to reach a definitive diagnosis to avoid unnecessary surgical resection of incidental leiomyomas.
In recent years, artificial intelligence (AI)-based image processing has gained momentum in gastroenterology and yielded promising results with the emergence of the deep learning algorithm such as convolutional neural networks (CNNs) [9, 10, 11]. In this study, we aimed to evaluate whether CNN-based deep learning can differentiate GISTs from leiomyomas using EUS images as an input. We also assessed the significance of various EUS features in determining the accuracy of diagnosis.
Patients and Methods
Patients
This study was approved by the Institutional Review Board. The medical records of patients referred for EUS to assess subepithelial lesions (SELs) at a single tertiary referral center between October 2010 and June 2020 were retrospectively reviewed. Patients with histologically confirmed GISTs or leiomyomas by surgical resection or EUS-FNAB, whose EUS images were recorded in a digital format, were included in this study. The pathology reports were reassessed to confirm the diagnosis. GIST was confirmed when samples showed descriptive spindle or epithelioid cells together with the expression of c-kit and/or CD34, and a leiomyoma was confirmed when samples were found to be desmin-positive but c-kit-negative.
A total of 145 patients (109 with GISTs and 36 with leiomyomas) were included in this study. The patients were divided into the following 2 groups: 100 patients (74 with GISTs and 26 with leiomyomas) for training and testing of the CNN system and 45 patients (35 with GISTs and 10 with leiomyomas) for validation. The validation cohort was also used to compare the diagnostic performance of the CNN system against a EUS expert. Both training and validation cohorts were used to evaluate and compare the conventional features of GISTs and leiomyomas on EUS.
EUS Images
EUS images were retrieved from the endoscopy database. For training and testing the CNN system, individual images were segmented by offsetting the tumor while retaining only the lesion and the layer of origin using Microsoft Paint (Microsoft, Redmond, WA, USA). Images were also reviewed by a single expert with more than 10 years of experience with EUS procedures (H.S.). The expert was blinded to the final diagnosis as well as to the location of the tumors (esophagus or stomach) by rendering the images such that the expert was unable to guess the location. Diagnostic yield of the CNN system and the EUS expert were compared. For the comparison of the conventional EUS features between GISTs and leiomyomas, the images in both training and validation cohorts were used, and the following EUS features were evaluated for each lesion: the tumor size (in centimeters, cm), border regularity (regular or irregular), shape (oval/round or distorted), echogenicity compared with the surrounding muscular layer (iso/hypoechoic or hyperechoic), homogeneity (homogeneous or heterogeneous), presence of mucosal ulceration, surface lobulation, hyperechogenic foci, anechoic (cystic) spaces, and hypoechoic halo.
Machine Learning Model
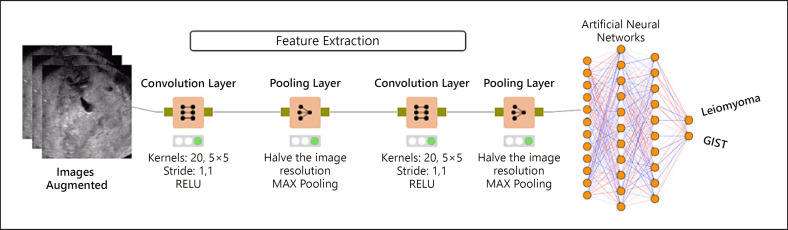
A machine learning model based on a CNN was used in this study. The model was trained, tested, and validated using an external dataset. The CNN is a deep learning algorithm that is designed to analyze information that can be processed as a grid; it is especially well suited for analyzing photographs and other images [12]. The network sees an image as a grid of pixels. It uses a mathematical operation that generates a third function using 2 other functions. The new function defines how the shape of 1 function is modified by the other. The CNN contains multiple layers and hyperparameters that are necessary for training (Fig. 1). First, input images (as pixel values) are sent to the convolution layer as (number) × (width) × (height) × (depth). Here, the user-defined number of kernels produces the feature maps. The kernel is another matrix with a K value of (image width) × (image height) × (image depth). After kernel operations are completed, the produced matrices are sent to pooling layers to rationalize or update the computations. After updating the computations, all productions may be used to feed another convolutional layer, and all operations are repeated in the next layer(s). Finally, the completed model generates a new array to feed a regular artificial neural network (ANN) model. This process is known as flattening. Flattened data are used as inputs for ANNs that have their own fully connected hidden layers and neurons. The optimizer, learning rate, loss function, weight initialization, and dropout rate are very important hyperparameters to be considered. Although these hyperparameters play very important roles in the success of training, there is no thumb rule to determine them beforehand, and these must be adjusted by trial and error. The images taken through EUS were resized by the Lanczos interpolation in a 28 × 28 × 1 format. Two interconnected convolutional layers are built into the model. Twenty CNN kernels were used in the first layer and 50 in the second layer. After each kernel layer, the image resolution was halved. The RELU activation function was used for both layers. MAX pooling was preferred for pooling with 2 × 2 kernel and stride sizes. The learning rate was set to 0.01. The global dropout rate was 0.0. Following these convolutional processes, the featured image information was an input into the ANN model to train the system. After training the CNN system on the training dataset, it was tested to determine the effectiveness of the model.
Fig. 1.
General model for a CNN. CNN, convolutional neural network; GIST, gastrointestinal stromal tumor.
The Training and Validation Dataset
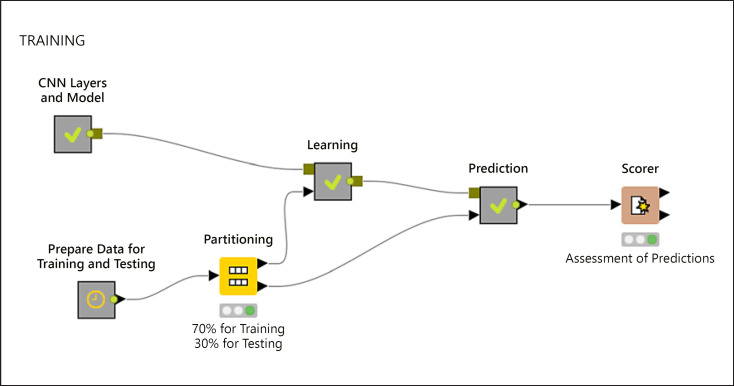
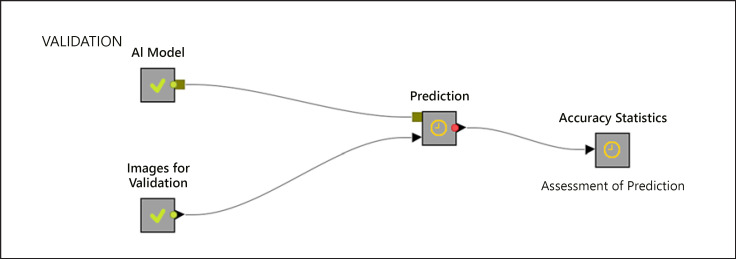
A total of 978 images from 100 patients (733 images from 74 patients with GISTs and 245 images from 26 patients with leiomyomas) were used as the training dataset. As there was an imbalance between the number of patients with GISTs and leiomyomas, the images were augmented with both Python Keras library and synthetic minority over-sampling technique to ensure a balanced class variable and also to reserve more images to be used for training. For the synthetic minority over-sampling technique, k = 3 was used as the nearest neighbor. After the augmentation process, 5,860 EUS images (2,930 images of GIST and 2,930 images of leiomyomas) were used as the final training dataset. This dataset was then partitioned into 2 groups using the stratified sampling method with 70% for training the CNN system and 30% for testing the trained model, as well as to evaluate the overall model (Fig. 2). Ten-fold cross-validation was performed along with stratified sampling when data were divided into 2 groups, that is, training and testing. The images were fed into the system as input parameters, whereas GISTs and leiomyomas that were based on pathological results were used as the target variables. Later, the trained model was validated using 384 images from 45 patients (314 images from 35 patients with GISTs and 70 images from 10 patients with leiomyomas) (Fig. 3). Finally, the trained model was fed with shuffled images, and predictions for each image arrived separately as the model was agnostic to patients' final diagnosis.
Fig. 2.
Representation of a model for training. CNN, convolutional neural network.
Fig. 3.
Representation of a model for validation. AI, artificial intelligence.
Statistical Analysis
The data were presented as mean ± standard deviation or median (min-max) for continuous variables and as the number of cases and percentages (%) for categorical variables. The distributions of continuous variables were evaluated using the Kolmogorov-Smirnov test. The different characteristics were compared between the groups using the χ2 test, Student's t test, or Mann-Whitney U test. Multiple logistic regression analysis was performed to determine the differences in EUS features between the 2 groups. A receiver operating characteristic curve was applied to determine the optimal cutoff value of the tumor size for differentiation. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and the accuracy of tumor size were calculated. The odds ratios (ORs), 95% confidence intervals (CIs), and Wald statistics for each independent variable were also calculated. The statistical significance of the agreement in diagnosis between the gold standard and alternative methods (i.e., human and machine assessments) was evaluated using McNemar's test. The final diagnoses were assessed, and the diagnostic yields of the CNN model and EUS expert were compared. Kappa coefficients were calculated to determine the level of agreement between the assessment methods. All statistical analyses were performed using SPSS Statistics for Windows, version 17.0 (SPSS Inc., Chicago, IL, USA). A p value <0.05 was deemed to be statistically significant.
The KNIME Analytics Platform (University of Konstanz, Zurich, Switzerland) and Python 3.7 (Python Software Foundation, Python Language Reference, version 2.7., available at http://www.phthon.org) were used for the CNN system. The classification was performed on images using the ResNet50 deep-learning network architecture, trained on ImageNet via Keras (TensorFlow).
Study Endpoints
The main objective of this study was to evaluate the diagnostic performance of the CNN system using EUS images from patients having GISTs or leiomyomas to predict the final histopathological diagnosis and to compare its accuracy with that of the EUS expert. The secondary aim was to analyze the conventional EUS features of GISTs in comparison with leiomyomas.
Results
The characteristics of the patients and the tumors are outlined in Table 1. There were 145 patients with pathologically proven mesenchymal tumors (73 women and 72 men, mean age of 54.8 ± 13.8 years [range 22–84 years]). Immunohistochemical (IHC) analyses revealed that 109 and 36 patients had GISTs and leiomyomas, respectively. Off all the GIST cases, 62 (56.9%) patients underwent surgical resection, and 47 (43.1%) were diagnosed by EUS-FNAB. Of 62 surgically resected GISTs, 50 patients had EUS-FNAB, 42 were diagnostic, 6 were nondiagnostic, and 1 showed spindle-cell morphology without IHC staining. In patients with leiomyomas, 14 (38.9%) patients underwent surgical resection, and 22 (61.1%) were diagnosed by EUS-FNAB, respectively. In surgically resected leiomyomas, 13 patients had EUS-FNAB, 5 were diagnostic, 3 were nondiagnostic, and 5 showed spindle-cell morphology without IHC staining. The GISTs were located in the esophagus in 6 patients, cardia in 12, fundus in 10, body in 50, antrum in 23, and duodenum in 8. The leiomyomas were located in the esophagus in 26 patients and cardia in 10. The mean tumor size was 3.3 cm (range 1.4–16.0 cm) in GISTs and 2.5 cm (range 1.0–10.0 cm) in leiomyomas.
Table 1.
Clinicopathological characteristics of patients in the training and validation dataset
| Characteristics | All (n = 145) | Training dataset (n = 100) | Validation dataset (n = 45) |
|---|---|---|---|
| Age (mean±SD), years | 54.8±13.8 | 55.2±14.0 | 55.9±12.9 |
| Sex | |||
| Female | 73 | 51 | 22 |
| Male | 72 | 49 | 23 |
| Histopathology | |||
| GIST | 109 | 74 | 35 |
| Leiomyoma | 36 | 26 | 10 |
| Tumor location | 4 | ||
| Esophagus | 32 | 22 | 10 |
| Stomach | 105 | 72 | 33 |
| Duodenum | 8 | 6 | 2 |
| Tumor size (median, range), cm | |||
| GIST | 3.3 (1.4–16.0) | 3.3 (1.4–16.0) | 2.75 (1.5–14.0) |
| Leiomyoma | 2.5 (1.0–10.0) | 2.5 (1.-10.0) | 2.9 (1.7–6.0) |
GIST, gastrointestinal stromal tumor; SD, standard deviation.
Diagnostic Yields of the CNN and the EUS Expert
The diagnostic yields of the CNN system are summarized in Table 2. The diagnostic sensitivity, specificity, PPV, NPV, and accuracy of the CNN system in the training dataset were 99.5% for each parameter. In the validation cohort, the CNN system yielded a sensitivity of 92.0%, a specificity of 64.3%, a PPV of 92.0%, an NPV of 64.3%, and an accuracy of 86.98%. For the EUS expert, the diagnostic sensitivity, specificity, PPV, NPV, and accuracy were 60.5%, 74.4%, 91.3%, 29.5%, and 63.0%, respectively (Table 3) (Fig. 4a, b). The CNN system fared significantly better than the EUS expert on all assessed parameters (all p < 0.05) except for the specificity (p = 0.281). Kappa coefficient values were 0.563 and 0.219 for the CNN system and the EUS expert, respectively.
Table 2.
Diagnostic performance of the CNN system in differentiating GISTs from leiomyomas in the training and validation dataset
| Diagnosis | Sensitivity, % | Specificity, % | PPV, % | NPV, % | Accuracy, % |
|---|---|---|---|---|---|
| Training dataset | 99.5 (99.4–99.8) | 99.5 (99.4–99.8) | 99.5 (99.4–99.6) | 99.5 (99.4–99.6) | 99.5 (99.4–99.9) |
| Validation dataset | 92.0 (88.5–94.8) | 64.3 (51.9–75.4) | 92.0 (89.4–94.1) | 64.3 (54.3–73.2) | 86.9 (83.2–90.2) |
Values in parentheses are 95% CIs. CNN, convolutional neural network; GIST, gastrointestinal stromal tumor; NPV, negative predictive value; PPV, positive predictive value; CI, confidence interval.
Table 3.
The diagnostic performance of the CNN system and the EUS expert assessment
| EUS expert | CNN system | |
|---|---|---|
| Sensitivity, % | 60.5 (54.9–65.9) | 92.0 (88.5–94.8) |
| Specificity, % | 74.3 (62.4–83.9) | 64.3 (51.9–75.4) |
| PPV, % | 91.3 (87.5–94.1) | 92.0 (89.4–94.1) |
| NPV, % | 29.5 (25.7–33.7) | 64.3 (54.3–73.2) |
| Accuracy, % | 63.0 (57.9–67.9) | 86.9 (83.2–90.2) |
| Kappa | 0.219 | 0.563 |
| p value | <0.001 | >0.999 |
Values in parentheses are 95% CIs. CNN, convolutional neural network; NPV, negative predictive value; PPV, positive predictive value; CI, confidence interval; EUS, endoscopic ultrasonography.
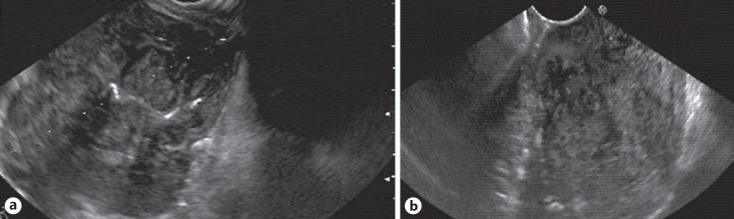
Fig. 4.
a, b Representative cases of leiomyomas. The EUS images of those were diagnosed correctly by the CNN system but not the EUS expert. EUS, endoscopic ultrasonography; CNN, convolutional neural network.
EUS Features Differentiating GISTs from Leiomyomas
Data from 145 patients were included in this analysis (Table 4). The mean age was higher in patients with GISTs than in patients with leiomyomas (58.8 ± 12.1 vs. 43.2 ± 10.3 years, p < 0.001), and there were no sex-specific differences between the 2 types of tumors. The tumor size in patients with GIST was significantly larger than that in patients with leiomyomas (3.3 [1.4–16.0] cm vs. 2.5 [1.0–10.0] cm, p = 0.049). A receiver operating characteristic curve was constructed to identify the discriminating value of tumor size, and the best cutoff value of 2.75 cm yielded a sensitivity of 64.5%, a specificity of 59.4%, a PPV of 82.2%, an NPV of 36.5%, and an accuracy of 63.2%.
Table 4.
Logistic regression analyses of baseline characteristics and EUS features for differentiating GISTs from leiomyomas
| Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|
| GIST (n = 109), n (%) | Leiomyoma (n = 36), n (%) | p value | OR (95% CI) | p value | |
| Age (mean ± SD), years | 58.8±12.1 | 43.2±10.3 | <0.001 | 1.128 (1.067–1.193) | <0.001 |
| Sex | |||||
| Male | 53 (48.4) | 19 (53.1) | 0.797 | ||
| Female | 56 (51.6) | 17 (46.9) | |||
| Size (median, range), cm | 3.3 (1.4–16.0) | 2.5 (1.0–10.0) | 0.049 | 1.922 (0.479–7.706) | 0.357 |
| Location | |||||
| Esophagus | 6 | 26 | |||
| Cardia | 12 | 10 | |||
| Fundus | 10 | <0.001 | |||
| Body | 50 | ||||
| Antrum | 23 | ||||
| Duodenum | 8 | ||||
| Shape | |||||
| Round/oval | 75 (68.8) | 33 (90.6) | 0.028 | 1.239 (0.205–7.477) | 0.815 |
| Distorted | 34 (31.2) | 3 (9.4) | |||
| Surface lobulation | |||||
| No | 69 (63.4) | 30 (84.4) | 0.670 (0.127–3.533) | ||
| Yes | 40 (36.6) | 6 (15.6) | 0.04/ | 0.637 | |
| Border | |||||
| Regular | 43 (39.8) | 25 (68.8) | 0.009 | 2.494 (0.653–9.531) | 0.181 |
| Irregular | 66 (60.2) | 11 (31.2) | |||
| Ulceration | |||||
| No | 84 (77.4) | 36 (100.0) | 0.008 | ||
| Yes | 25 (22.6) | 0 (0.0) | |||
| Echogenicity | |||||
| Hyperechogenic | 48 (44.1) | 2 (6.2) | <0.001 | 6.260 (1.089–35.973) | 0.040 |
| Iso/hypoechogenic | 61 (55.9) | 34 (93.8) | |||
| Homogeneity | |||||
| Homogeneous | 40 (36.6) | 23 (62.5) | 0.019 | 0.677 (0.167–2.743) | 0.585 |
| Heterogeneous | 69 (63.4) | 13 (37.5) | |||
| Anechoic spaces | |||||
| No | 55 (50.5) | 35 (96.9) | <0.001 | ||
| Yes | 54 (49.5) | 1 (3.1) | |||
| Hyperechoic foci | |||||
| No | 69 (63.4) | 32 (87.5) | 0.020 | ||
| Yes | 40 (36.6) | 4 (12.5) | |||
| Hypoechoic halo | |||||
| No | 59 (53.8) | 29 (81.2) | |||
| Yes | 50 (46.2) | 7 (18.8) | 0.011 | 1.225 (0.293–5.112) | 0.781 |
Values in bold indicate statistically significant results. OR, odds ratio; CI, confidence interval; GIST, gastrointestinal stromal tumor; EUS, endoscopic ultrasonography; SD, standard deviation.
Based on univariate analysis, in terms of EUS features, the distorted tumor shape, surface lobulation, border irregularity, higher echogenicity than the surrounding muscle layer, heterogeneity, the presence of mucosal ulceration, anechoic spaces, hyperechogenic foci, and hypoechoic halo were all observed more frequently in GISTs than in leiomyomas (all p < 0.05) (Table 4). However, a multivariate analysis revealed that only higher echogenicity (OR = 6.260, 95% CI: 1.089–35.973, p = 0.040) and age (OR = 1.128, 95% CI: 1.067–1.193, p < 0.001) were significantly differentiating features (Table 4).
Discussion
In the current study, the CNN system was able to differentiate GISTs from leiomyomas with a sensitivity of 99.5%, a specificity of 99.5%, and an accuracy of 99.5% in the training dataset. When the model was tested on a validation dataset, it showed a sensitivity of 92.0%, a specificity of 64.3%, and an accuracy of 86.98%, as against an EUS expert, for whom these values were 60.5%, 74.3%, and 63.0%, respectively. The diagnostic accuracy of the CNN system was significantly higher than that of the EUS expert. Concerning conventional EUS features, only higher echogenicity in comparison with the surrounding muscle echo along with advanced age were significant factors independently associated with the diagnosis of GISTs as compared with leiomyomas.
Integration of AI technology in medical fields has been known to improve the accuracy and speed of diagnosis, aid clinical decision-making, and lead to better health outcomes. AI-guided clinical care has the potential to play an important role in reducing health disparities, particularly in low-resource settings, and the past few years have seen rapid adoption of machine learning-based tools in diagnosis and treatment of patients, especially those with malignancy. We have recently published a report on the ability of a CNN-based machine learning system in predicting the malignant potential of gastric GISTs as well as the mitotic index with high accuracy only using EUS images [11]. None of the conventional EUS features were significantly associated with either the malignant potential or mitotic index of gastric GISTs. A study by Kim et al. [9] revealed the high diagnostic accuracy of the CNN computer-aided diagnosis system for differentiating GISTs from non-GISTs, including leiomyoma and schwannoma, on EUS images. The diagnostic specificity and accuracy of the CNN computer-aided diagnosis system were significantly higher than those of the endoscopists. Another study by Minoda et al. [10] revealed that an EUS diagnostic system with AI (EUS-AI) had high diagnostic yield for differentiating GISTs from non-GISTs of ≥20 mm. The diagnostic yield of EUS-AI for SELs ≥20 mm was higher than that of the endoscopists as previously reported [9]. In the current study, the diagnostic sensitivity and accuracy of the CNN system were higher than those of the EUS expert, but the specificity was similar.
Endosonographically, GISTs generally appear as well-circumscribed, hypoechoic, relatively homogeneous lesions usually arising from the fourth and rarely the second layer of the gut wall. In contrast, leiomyomas are homogenous hypoechoic lesions, more commonly arising from the second layer than GISTs [13]. Some studies have suggested that GISTs have heterogeneity, hyperechoic spots, a marginal hypoechoic halo, and relatively higher echogenicity compared to the muscular echo [14, 15]. Nevertheless, none of these features are specific for differentiating GISTs from leiomyomas since the interpretation of EUS morphological features is highly subjective and depends upon the endoscopist's experience. Moreover, even though the endoscopists were given information about the layer from which the lesion originated as well as the location of the lesion, the sensitivity and specificity for the human diagnosis of GIST were reported to be 75.8% and 85.4%, respectively [16]. As a result, histopathology remains the “gold standard” for making a definitive diagnosis and avoiding unnecessary and invasive treatment for benign mesenchymal lesions. Although combining tissue acquisition by EUS-FNAB of the lesions increases the diagnostic yield, it has been reported to be between 62% and 100%, which is significantly lower for smaller tumors [17, 18]. Minoda et al. [10] reported a higher diagnostic ability of the EUS-AI system for larger tumors (83.3% for SELs <20 mm and 93.3% for SELs ≥20 mm) and recommended it as an alternative to EUS-FNAB for the diagnosis of SELs. In our study, the ability of the CNN did not differ according to tumor size. The diagnostic yield of the CNN system was higher than that of the EUS expert, similar to the results from previous studies [9, 10]. In our study, the endoscopist was provided with only EUS images without informing about the location of the lesions, such as the stomach or esophagus. Furthermore, preparation of EUS images was done to make guessing the location difficult. However, it should be noted that in real-life practice, knowing the location of the lesion and evaluating all lesions in real-time during EUS examination could yield a higher diagnostic performance by EUS experts.
In the present study, higher echogenicity compared with the surrounding muscular echo and age were 2 factors independently associated with GISTs. Hunt et al. showed that a tumor size of 4 cm, the presence of ulceration, or cystic foci were mostly observed in CD-117-positive tumors as compared with CD-117-negative tumors [19]. In addition, the 2 studies that were published before the introduction of GISTs as a different entity suggested that inhomogeneity and hyperechogenic spots were the EUS features that are predictive of malignancy [20, 21]. Another study reported that a marginal halo and relatively higher echogenicity might suggest GISTs [14]. In 2009, Kim et al. [15] compared gastric GISTs with gastric leiomyomas, added inhomogeneity and hyperechoic spots to the foregoing features, and suggested that the presence of at least 2 of the 4 features could predict GISTs with 89.1% sensitivity and 85.7% specificity. Instead, tumor size, the presence of ulceration, and cystic changes were not helpful for differentiating GISTs from leiomyomas. We compared GISTs with only leiomyomas in our study, similar to the previous study [15]. Univariate analysis revealed that larger tumor size, distorted tumor shape, border irregularity, heterogeneity, the presence of surface lobulation, and hypoechoic halo were related to GISTs. However, these factors were not significant in multivariate analysis. Although the presence of mucosal ulceration, anechoic spaces, and hyperechogenic foci was much more related to GISTs, we did not include these features in multivariate analysis because only a few patients with leiomyomas presented these findings (22.6%, 49.5%, 36.6%, and 32.3% vs. 0%, 3.1%, 12.5%, and 1.0%, respectively, all p < 0.05).
The present study showed the efficiency of the CNN-based machine learning model, which was trained on a relatively small dataset, to accurately predict and differentiate GISTs from leiomyoma. While we do not suggest that a machine learning algorithm replaces an expert, the 2 can certainly work in synergy to achieve better. As 1 of its advantages, the CNN system is objective in nature, while the human diagnosis is likely to be interpreted subjectively. A CNN system can provide a rapid diagnosis noninvasively, that is, without the need for tissue sampling using EUS-FNAB. In our study, the diagnostic yield of the CNN was higher for GISTs than for leiomyomas. This could be explained by the fact that fewer cases with leiomyomas than those of GISTs were included in the study. Although an augmentation process was applied to the images in the training dataset, it may be difficult to train a CNN system using EUS images of rare cases as the limited number of available training EUS images may lead to biased or inaccurate detection.
One of the key shortcomings of this study is its retrospective, single-center study design. Second, the study included a small number of patients. However, this was due to the fact that histologically proven GISTs and leiomyoma do not occur frequently. In addition, the number of leiomyomas included in this study was small relative to the number of GISTs. Because only esophageal or cardiac lesions have the possibility of leiomyoma and duodenal or gastric lesions other than cardia lesions were all GISTs in this study, we could not perform a sub-analysis according to the location. In clinical practice, endoscopists make diagnosis based on the location. So taking the location into consideration for diagnosis is crucial.
In conclusion, our study showed that the CNN system is helpful in differentiating GISTs from leiomyomas using EUS images. Even though some EUS features were significant, the diagnostic yield of the EUS expert remained lower than that of the CNN system.
Statement of Ethics
The study was approved by the Institutional Ethics Committee at Bezmialem Vakif University Hospital (IRB No: 2/25). The written informed consent statements have not been obtained for this study as this is not necessary for a retrospective study according to the Institutional Review Board of the Bezmialem University Hospital.
Conflict of Interest Statement
Drs. Seven, Silahtaroglu, Ozluk Seven, and Senturk have no conflicts to report.
Funding Sources
No funding sources have been used for this study.
Author Contributions
Gulseren Seven contributed to the concept and design of the study, acquisition of data, analysis and interpretation of data, drafting and revising articles, writing the manuscript, and final approval of the version to be published. Ozden Ozluk Seven contributed to acquisition of data and drafting and revising articles. Gokhan Silahtaroglu contributed to machine learning and data analysis. Hakan Senturk is the corresponding author and contributed to the concept and design of the study, analysis and interpretation of data, and revising the manuscript. The manuscript has been read and approved by all authors.
Data Availability Statement
Research data are not publicly available on legal and ethical grounds. All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
References
- 1.Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1,765 cases with long-term follow-up. Am J Surg Pathol. 2005 Jan;29((1)):52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- 2.Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006 May;23((2)):70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Miettinen M, Sarlomo-Rikala M, Sobin LH, Lasota J. Esophageal stromal tumors: a clinicopathologic, immunohistochemical, and molecular genetic study of 17 cases and comparison with esophageal leiomyomas and leiomyosarcomas. Am J Surg Pathol. 2000 Feb;24((2)):211–22. doi: 10.1097/00000478-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Cağlar E, Hatemi I, Atasoy D, Sişman G, Sentürk H. Concordance of endoscopic ultrasonography-guided fine needle aspiration diagnosis with the final diagnosis in subepithelial lesions. Clin Endosc. 2013 Jul;46((4)):379–83. doi: 10.5946/ce.2013.46.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baysal B, Masri OA, Eloubeidi MA, Senturk H. The role of EUS and EUS-guided FNA in the management of subepithelial lesions of the esophagus: a large, single-center experience. Endosc Ultrasound. 2017 Sep;6((5)):308–16. doi: 10.4103/2303-9027.155772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DB, Sahai AV, Aabakken L, Penman ID, van Velse A, Webb J, et al. Endoscopic ultrasound guided fine needle aspiration biopsy: a large single centre experience. Gut. 1999 May;44((5)):720–6. doi: 10.1136/gut.44.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritscher-Ravens A, Sriram PV, Schröder S, Topalidis T, Bohnacker S, Soehendra N. Stromal tumor as a pitfall in EUS-guided fine-needle aspiration cytology. Gastrointest Endosc. 2000 Jun;51((6)):746–9. doi: 10.1067/mge.2000.105730. [DOI] [PubMed] [Google Scholar]
- 8.Varadarajulu S, Fraig M, Schmulewitz N, Roberts S, Wildi S, Hawes RH, et al. Comparison of EUS-guided 19-gauge Trucut needle biopsy with EUS-guided fine-needle aspiration. Endoscopy. 2004 May;36((5)):397–401. doi: 10.1055/s-2004-814316. [DOI] [PubMed] [Google Scholar]
- 9.Kim YH, Kim GH, Kim KB, Lee MW, Lee BE, Baek DH, et al. Application of a convolutional neural network in the diagnosis of gastric mesenchymal tumors on endoscopic ultrasonography images. J Clin Med. 2020 Sep 29;9((10)):9. doi: 10.3390/jcm9103162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minoda Y, Ihara E, Komori K, Ogino H, Otsuka Y, Chinen T, et al. Efficacy of endoscopic ultrasound with artificial intelligence for the diagnosis of gastrointestinal stromal tumors. J Gastroenterol. 2020 Dec;55((12)):1119–26. doi: 10.1007/s00535-020-01725-4. [DOI] [PubMed] [Google Scholar]
- 11.Seven G, Silahtaroglu G, Kochan K, Ince AT, Arici DS, Senturk H. Use of artificial intelligence in the prediction of malignant potential of gastric gastrointestinal stromal tumors. Dig Dis Sci. 2021 Feb 6; doi: 10.1007/s10620-021-06830-9. [DOI] [PubMed] [Google Scholar]
- 12.Goodfellow I. Deep Learning. MIT Press; 2016. [Google Scholar]
- 13.Hawes R, Fockens P, Varadarajulu S. Subepithelial lesions Endosonography. Elsevier; 2018. p. 376. [Google Scholar]
- 14.Okai T, Minamoto T, Ohtsubo K, Minato H, Kurumaya H, Oda Y, et al. Endosonographic evaluation of c-kit-positive gastrointestinal stromal tumor. Abdom Imaging. 2003 May;28((3)):301–7. doi: 10.1007/s00261-002-0055-x. [DOI] [PubMed] [Google Scholar]
- 15.Kim GH, Park DY, Kim S, Kim DH, Kim DH, Choi CW, et al. Is it possible to differentiate gastric GISTs from gastric leiomyomas by EUS? World J Gastroenterol. 2009 Jul 21;15((27)):3376–81. doi: 10.3748/wjg.15.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo SW, Hong SJ, Han JP, Choi MH, Song JY, Kim HK, et al. Accuracy of a scoring system for the differential diagnosis of common gastric subepithelial tumors based on endoscopic ultrasonography. J Dig Dis. 2013 Dec;14((12)):647–53. doi: 10.1111/1751-2980.12099. [DOI] [PubMed] [Google Scholar]
- 17.de Moura DTH, McCarty TR, Jirapinyo P, Ribeiro IB, Flumignan VK, Najdawai F, et al. EUS-guided fine-needle biopsy sampling versus FNA in the diagnosis of subepithelial lesions: a large multicenter study. Gastrointest Endosc. 2020 Jul;92((1)):108–e3. doi: 10.1016/j.gie.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minoda Y, Chinen T, Osoegawa T, Itaba S, Haraguchi K, Akiho H, et al. Superiority of mucosal incision-assisted biopsy over ultrasound-guided fine needle aspiration biopsy in diagnosing small gastric subepithelial lesions: a propensity score matching analysis. BMC Gastroenterol. 2020 Jan 21;20((1)):19. doi: 10.1186/s12876-020-1170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt GC, Rader AE, Faigel DO. A comparison of EUS features between CD-117 positive GI stromal tumors and CD-117 negative GI spindle cell tumors. Gastrointest Endosc. 2003 Apr;57((4)):469–74. doi: 10.1067/mge.2003.146. [DOI] [PubMed] [Google Scholar]
- 20.Chak A, Canto MI, Rösch T, Dittler HJ, Hawes RH, Tio TL, et al. Endosonographic differentiation of benign and malignant stromal cell tumors. Gastrointest Endosc. 1997 Jun;45((6)):468–73. doi: 10.1016/s0016-5107(97)70175-5. [DOI] [PubMed] [Google Scholar]
- 21.Palazzo L, Landi B, Cellier C, Cuillerier E, Roseau G, Barbier JP. Endosonographic features predictive of benign and malignant gastrointestinal stromal cell tumours. Gut. 2000 Jan;46((1)):88–92. doi: 10.1136/gut.46.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not publicly available on legal and ethical grounds. All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.