Abstract
Upon infecting populations of susceptible host cells, T-even bacteriophages maximize their yield by switching from lysis at about 25 to 35 min at 37°C after infection by a single phage particle to long-delayed lysis (lysis inhibition) under conditions of sequential infection occurring when free phages outnumber host cells. The timing of lysis depends upon gene t and upon one or more rapid-lysis (r) genes whose inactivation prevents lysis inhibition. t encodes a holin that mediates the movement of the T4 endolysin though the inner cell membrane to its target, the cell wall. The rI protein has been proposed to sense superinfection. Of the five reasonably well characterized r genes, only two, rI and rV, are clearly obligatory for lysis inhibition. We show here that rV mutations are alleles of t that probably render the t protein unable to respond to the lysis inhibition signal. The tr alleles cluster in the 5′ third of t and produce a strong r phenotype, whereas conditional-lethal t alleles produce the classical t phenotype (inability to lyse) and other t alleles produce additional, still poorly understood phenotypes. tr mutations are dominant to t+, a result that suggests specific ways to probe T4 holin function.
The T-even bacteriophages employ a powerful strategy to maximize the yield of virus particles when a population of susceptible Escherichia coli cells is encountered (7, 14). Cells are initially infected by single phage particles and, at 37°C with aeration, lyse 25 to 35 min later, releasing on the order of 102 particles per cell. Eventually, the ratio of released particles to unlysed cells exceeds 1, whereupon cells are infected by one particle and then repeatedly “superinfected” by additional particles. When the interval between infection and superinfection is at least a few minutes, the superinfecting particles trigger lysis inhibition. Lysis is then delayed for up to several hours, and the burst size approaches 103 particles per cell.
Lysis inhibition involves several genes (2, 40). e, a late gene whose protein-encoding DNA sequence resides at kb 66.493 to 66.985 on the standard map (25), encodes an endolysin (gpe in T4 gene product terminology) that accumulates but does not act until released into the periplasmic space; gpe is a lysozyme that degrades the murein layer of the cell envelope (30). Cells singly infected with e mutants cease metabolism after about 30 min at 37°C and synthesize no more phage but neither lyse nor form plaques. t, a late gene that resides at kb 160.219 to 160.873, encodes a 218-residue protein (gpt) (28) that acts as a holin, that is, a (regulated) pore allowing the lysozyme access to the murein layer (27, 32, 40). Cells singly infected with the canonical t amber mutants accumulate endolysin, but rarely lyse, and form tiny plaques or no plaques at all, depending on the plating medium (references 15 and 16 and this report). Unlike e mutants, such t mutants continue to synthesize phage particles long after the normal time of lysis, but “t” refers not to “timing” or “trigger” but to the unfortunate Tithonus, who was granted immortality but not protection against aging (15, 16).
In rapid-lysis (r) mutants, superinfection fails to induce lysis inhibition, resulting in large, sharp-edged plaques instead of the smaller r+ plaques that are surrounded by a dense haze of superinfected, lysis-inhibited cells (14); however, the larger r plaques contain roughly 10-fold-fewer phages than do r+ plaques (10). In the present context, the key r genes are rI and rV. rI is an early gene at kb 59.192 to 59.483, and it has been suggested that gprI senses some primary aspect of superinfection (31). rV was first defined by a single allele mapping to the region between 38 and motA and producing an r phenotype only at higher temperatures (18, 23); additional rV alleles are described in this report. Among the other r genes, rIV and rVI are poorly defined genetically (25, 39) and null mutations in rIIA, rIIB, and rIII produce an r phenotype on some host cells but not on others (2, 31) and thus are not essential for lysis inhibition. These r genes are not further considered here.
Here we show that t and rV mutations are alleles of the same gene. Although rV was discovered in 1965 (23) and t in 1970 (15), we call the gene t because it encodes the holin. We also characterize patterns of lysis and lysis inhibition produced by the two kinds of t alleles and suggest how rI and t alleles may interact.
MATERIALS AND METHODS
T4 strains.
The canonical rV mutant rts64 was obtained from Victor Krylov; it produces the r+ phenotype at 30°C and the r phenotype at 37 to 43°C. r2 and r3 were detected in routine screens for r mutants and were tentatively assigned to rV because of their close linkage to rts64 or their r phenotype on BB cells without a sequence change in rI. The t mutants tsDH634, amtA3, amtB5, hus19, and hus20 were obtained from Dwight Hall, and Rid394 was obtained from Karin Carlson.
E. coli strains.
Bacterial strains were from the laboratory’s E. coli stock collection. The su− strain B was used to score r plaque morphology. Strain B40su+II is a B strain in which amber mutations are suppressed by the insertion of glutamine at positions encoded by UAG, and it was used to grow T4 bearing amber mutations in vital genes. On strain BB, rII mutants display an r+ phenotype whereas rI and rV mutants display an r phenotype. The su− strain KB is a K-12(λ) lysogen that is nonpermissive for rII mutants.
Media and T4 stocks.
Our modified Luria-Bertani (LB) broth, M9 buffer, Drake agars, and general plating methods have been described (6), as have the Hershey (H) media (35). LB broth contains, per liter, 10 g of Bacto Tryptone, 5 g of Bacto Yeast Extract, 5 g of NaCl, and 1 g of glucose. Drake bottom agar contains, per liter, 10 g of Bacto Peptone, 1 g of Bacto Yeast Extract, 5 g of NaCl, 0.2 g of glucose, and 10 g of Bacto Agar. Drake top agar contains 65% of the same components. H broth contains, per liter, 8 g of Bacto Nutrient Broth, 5 g of Bacto Peptone, 5 g of NaCl, and 1 g of glucose. Enriched H top agar contains, per liter, 13 g of Bacto Tryptone, 8 g of NaCl, 2 g of sodium citrate dihydrate, 3 g of glucose, and 6.5 g of Bacto Agar; for enriched H bottom agar, the concentration of glucose was reduced to 1.3 g/liter and that of agar was increased to 10 g/liter.
Stocks were grown by stabbing plaques (with sterile paper strips) into 5-ml volumes of LB broth containing host cells (B cells for ts mutants, 394, and hus mutants and B40su+II cells for t amber mutants; 108 cells/ml) and incubating them on a rotary shaker for several hours at 37°C. Lysis was completed with chloroform. Phages were concentrated by centrifuging the lysate for 10 min at about 3,000 × g to remove debris and then for 1 h at about 50,000 × g to sediment phages; pellets were overlaid for several hours or overnight and then resuspended in storage buffer (1 g of NaCl, 0.2 g of MgCl2 · 6H2O, 1.2 g of Tris buffer, 0.1 g of gelatin, 1,000 ml of distilled water [pH 7]).
PCR and DNA sequencing.
A 3,538-base PCR product was prepared (17) by using the primer sequences starting at kb 163.429 (5′-AAAAAGGCCTGGTGGAGAAATCTGG-3′) from the motA end (kb 163.429 being the standard map starting kilobase) (25) and at kb 159.914 (5′-CCTCGGTCTTCCTGCTCATTCAA-3′) from the 38 end. The PCR products were purified for sequence analysis with QIAquick PCR purification kits (Qiagen). The ABI PRISM dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems) was used to perform fluorescence-based cycle sequencing reactions on PCR products from rV and t mutants. DNA sequencing was conducted on an ABI 377 DNA Sequencer. Each mutation within t was confirmed by sequence analysis in both directions from multiple PCR products. The following primer sequences defined mutations within t (numbers in parentheses designating starting kilobases): 5′-CCTCGGTCTTCCTGCTCATTCAA-3′ (159.914), 5′-GTAGTTTATTTCGGGAGTAGG-3′ (160.730), 5′-CTTTCCTTTTCAATAATTTCA-3′ (160.438), 5′-TGAAATTATTGAAAAGGAAAG-3′ (160.418), and 5′-AGAAAATTACACAGACCAGTT-3′ (161.226).
rts64 backcrosses.
At time zero, rts64 mutant, T4B, and B cells (the last at 5 × 108/ml after mixing) were combined in LB broth at multiplicities of infection (MOIs) of 0.5 for rts64 and 10 for T4B and were incubated on a rotary shaker at 32°C. At 8 min, the complexes were diluted extensively in LB broth and incubation was continued at 32°C. At 40 min, chloroform was added to complete lysis and the progeny were assayed on B cells at 43°C. Four r plaques were picked and grown into stocks, new backcrosses were conducted on all four lines three more times in a linear sequence, and two mutants displaying a temperature-sensitive r phenotype were selected at random from each line at the end. The t regions of the resulting eight r mutants were then sequenced.
One-step growth curves.
Single-infection, single-cycle growth curves were obtained by infecting B cells at MOIs of roughly 0.2. At time zero, 0.1 ml of phage (109 phages/ml) was added to 0.9 ml of B cells at 5 × 108 cells/ml and incubated on a rotary shaker at 37°C. At 4 min, the complexes were diluted to 105 cells/ml and incubation was continued without rotation. Starting at 16 min, samples were taken every 2 min and assayed for PFU. The number of PFU per complex was determined by normalizing all titers to the average of the values at 16 and 20 min.
Lysis inhibition assays.
These were conducted by using MOIs of about 10. At time zero, 1.5 ml of phage (1010 phages/ml) and 1.5 ml of log-phase B cells (109 cells/ml) were mixed and incubated at 37°C on a rotary shaker. At 4 min (by which time typically more than 90% of the phage had adsorbed), 2.8 ml of the culture was added to 18.2 ml of broth and again incubated at 37°C on a rotary shaker. At 15 min, 1.5 ml of phage at (1010 phages/ml) was added to the culture to achieve superinfection. Starting at 20 min, absorbance readings at 600 nm were taken every 10 min with a Beckman DU 640 spectrophotometer. All values were normalized to the value at 20 min.
RESULTS
Locating rV.
By using two-factor crosses, the canonical rV mutation rts64 was previously located roughly 22 map units counterclockwise from rII (24). Later, Dinh Nguyen, of this laboratory, mapped another rV mutant (now called tr2) using three-factor crosses involving widely spaced reference markers together with premature lysis (burst size ≈ 1) (9). Based on such crosses, tr2 was mapped between 38 and motA (data not shown).
Because this region contains several open reading frames that are likely to be T4 genes (25), we focused our initial sequencing on these open reading frames. Using PCR, we amplified and sequenced a 2,565-base fragment from motA to t from three rV mutants (rts64, r2, and r3) but observed no mutations. We therefore extended our search into t. t was not previously sequenced for these mutants because of the marked phenotypic differences between t and rV mutants: on nonsuppressing cells, amber t mutants form either no plaques or small, fuzzy-edged plaques, depending on the medium, while rV mutants form large, sharp-edged plaques. We amplified a 3,538-base fragment that extended from inside motA through part of 38. Using primers focused on t, we then found a mutation in rts64 which consisted of a G:C→A:T substitution at nucleotide 14 causing an arginine→lysine replacement at the position encoded by codon 5.
Because rts64 contained only a conservative missense mutation in t, we sought to confirm that the r phenotype was linked to the t mutation by extensively backcrossing rts64 against T4B (see Materials and Methods). These backcrosses failed to separate the r phenotype from the t mutation. Subsequent sequencing revealed that two other rV mutants (r2 and r3) also contain t mutations. Additional sequencing confirmed that none of the three tr mutants contain mutations in the distant rI gene (whose mutations are phenotypically identical to rV mutations). We next sequenced all of the previously mapped t mutants (11, 13, 26), with the results shown in Fig. 1.
FIG. 1.
The t gene (GenBank accession no. Y00408), with its derived protein sequence and mutations. t is flanked upstream by its late promoter (Pl) and by gene 38 and downstream by asiA and its early promoter (Pe); arrows indicate directions of transcription. The two blocks of sequence in boldface are the putative transmembrane residues (32). Numbers below the sequence identify mutated residues or reference residues. Entries above the sequence identify, in order, amino acid changes (am, amber), then base sequence changes in the complementary (noncoding) strand, and, in italics, names of alleles (alleles producing an r phenotype being shown in boldface).
t is flanked upstream by gene 38, a late gene transcribed in the same direction as t. The first t codon begins 12 bases 3′ to a late promoter, and t is served by no middle or early promoters; the nearest such promoters are about 7 and 10 kb upstream, respectively. Thus, t appears to be an exclusively late gene. Downstream, t is adjacent to the early gene asiA. asiA is transcribed in the opposite direction, with no sign of a transcription terminator that would prevent early asiA transcription from penetrating into t and generating antisense mRNA; however, the function of such a hypothetical antisense RNA remains untested. The three t mutations producing the r phenotype reside in the first third of the gene, and none of the other t missense mutations have an r phenotype. hus mutants grow poorly in the presence of hydroxyurea, while the growth of Rid394 depends on the allelic state of the host rho gene, whose product modulates transcription termination; these traits are considered in Discussion. The locations of most of the t mutations agree with previous mapping studies (11, 13, 38), except that Rid394 does not sequence to the predicted position (26) and the order of amA3 and amB5, while consistent among all later maps, is reversed from that given in the canonical report (15); the amber mutants may have been reversed when originally archived, or the original mapping may have employed too few mutants to secure the order correctly.
Profiles in lysis.
The time of lysis depends on the occurrence and timing of superinfection (1, 2, 7). In the absence of superinfection, at 37°C most lysis occurs between 25 and 35 min following either single or multiple infection. To trigger lysis inhibition, superinfection must occur later than a few minutes after the primary infection but at least shortly before lysis begins. Repeated late superinfection may prolong lysis inhibition, but in most cases the system eventually collapses abruptly (3).
Lysis profiles are measured in two different ways. One method is to infect cells with an MOI of <1, dilute the infected cells to a low density, and periodically measure the number of PFU (which may be either infected cells or released phage particles). To trace the course of superinfection-induced lysis inhibition, a high initial MOI (e.g., 10) is applied, followed later by a second high MOI. However, a few cells may lyse near the minimum time; the released phages can then soon outnumber the unlysed cells and tend to obscure their fate. Therefore, lysis inhibition is best measured at a high cell density by monitoring turbidity or absorbance. Because lysis inhibition is usually measured by using multiple infection for the primary infection, dominance tests can also be performed.
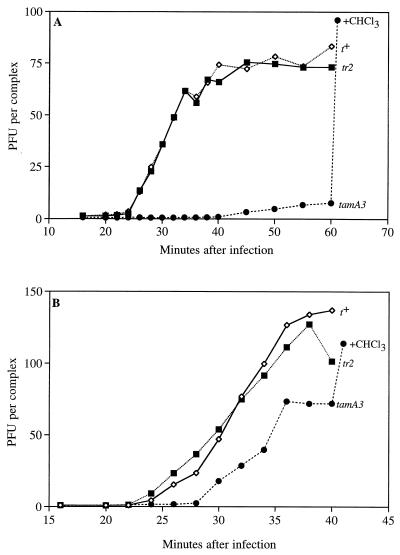
We first investigated whether tr mutants lyse normally or earlier than 25 to 35 min after infection in single-infection experiments with t+, tamA3, and tr2 strains. Typical results with H media are shown in Fig. 2A. As demonstrated originally (15, 16), su− cells infected with tamA3 show very little lysis but accumulate intracellular phages (and continue to do so even after 30 to 35 min) that can be released by artificial lysis with chloroform. The t+ and tr2 lysis profiles are indistinguishable. Therefore, the r phenotype is due not to premature lysis but more probably to an inability to develop lysis inhibition in response to superinfection.
FIG. 2.
One-step growth curves of strains grown in H broth and plated with H agars (A) or grown in LB broth and plated with Drake agars (B).
In contrast to the classical phage release profiles obtained with H media, we were surprised to observe considerable spontaneous release of tamA3 phages with LB broth and Drake agars (Fig. 2B). (The difference in burst sizes shown in Fig. 2A and B is unlikely to be noteworthy because burst sizes often vary among experiments.) Although this dependency on medium has not been explored further, at least three explanations can be entertained, any of which could vary with the medium. First, T4 amber mutants are sometimes leaky (e.g., see reference 34). Second, an additional, inefficient lysis mechanism may operate. Third, tamA3 leaves intact the first 87 amino acids, including the region (discussed below) containing the putative transmembrane residues, and this segment may retain residual holin function.
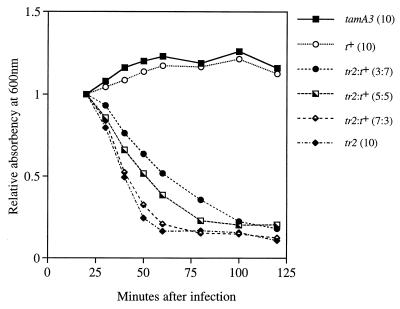
Because models for t function may generate predictions concerning dominance relationships, and in order to confirm the expected phenotype of a tr allele under conditions inducing lysis inhibition, we examined lysis profiles with both single-genotype and two-genotype infections. With H media, cells were rapidly infected with an MOI of 10 phage particles (sometimes including two different genotypes); then, at 15 min, the complexes were superinfected with another 10 particles. In our study, lysis profiles varied somewhat among experiments but retained the topological relations among various infecting genotypes quite robustly. Some results are shown in Fig. 3.
FIG. 3.
Lysis profiles determined after primary infection followed by superinfection with phage particles of various genotypes. Values in parentheses are the MOIs (displayed as a ratio of the MOI of the original infection to the MOI of the superinfection).
Both t+ and tamA3 produced the classical profile of long-delayed lysis, and the same result was produced by mixed infection with t+ plus tamA3 (data not shown). It is characteristic of such curves that absorbency increases after infection, but this behavior has not been explained to date. At later times (not shown in Fig. 3), the cells eventually lyse regardless of the infecting genotype (3).
As expected from its r phenotype and indicated in Fig. 2, tr2 showed no lysis inhibition. Mixed infections with t+ plus tr2 delayed lysis only slightly compared to tr2 alone, and similar results were obtained with trts64 and tr3 (data not shown). Thus, the t+ allele is recessive to tr alleles. (In experiments whose results are not shown here, tamA3 was recessive to tr, an unsurprising result.) The small lysis delay in infections with t+ plus tr2 increased as the t+-to-tr ratio increased from 0.3 through 0.5 to 0.7. Could this result be due to cells that were randomly infected with no tr2 phages but one or more t+ phages? The assortment of phages among cells can be described by the Poisson distribution, which predicts that among cells infected with at least one t+ particle, the fraction infected with no tr2 particles varies from 0.05 to 0.001 as the tr2 MOI ratio varies from 3 to 7. Even 5% of such cells is far too few to produce the observed lysis profiles (e.g., 5% of the t+ value falls well below all of the tr2 curves).
Anomalous r mutants.
Mutations in rII but not rI or rIII were reported to partially suppress tam mutations, and t revertants were often t rII doubles (16, 38). Here, we report several observations in case they may be of interest to other investigators. When stocks of any t mutants grown in LB broth are plated on B cells by using Drake agars, roughly 4% of the PFU produce r plaques. We plated a number of such mutants on KB and BB cells. rI and tr mutants form r plaques on both cells (and on all cells we have tested), while rII mutants from r+ plaques on BB cells but no plaques on K-12(λ) strains such as KB. Of 28 tested r mutants in t stocks, 8 produced r phenotypes on BB cells, 7 behaved like rII mutants, 2 displayed an r phenotype on KB cells but made no plaques on BB cells, and 11 failed to plate on either BB or KB cells. We sequenced both the rI and t regions of the 8 mutants producing r plaques on BB cells and found that while they retained their original t mutation, no additional mutations were found within either t or rI. Putative rI amber mutants producing a suppressible rI phenotype and linked to but not located within rI have also been observed (10, 31). Some of the above-listed mutants presumably reside in additional, less well characterized r loci.
DISCUSSION
Our principal findings are as follows. (i) The bacteriophage T4 genes rV and t are identical. (ii) t mutations producing an r phenotype cluster in the first third of the gene. (iii) In lysis inhibition experiments, these tr mutations are dominant to t+.
Although some of the earliest t mutants were obtained by enriching specifically for the null t phenotype (15, 38), several of the t mutants we sequenced were selected indirectly. First, the tr mutants were selected for their r plaque morphology. Second, tRid394 was selected as a mutant that grows poorly in rho+ host cells but better on strains with nonlethally mutated versions of rho (26). rho encodes an mRNA termination factor important in T4 replication (36), but the reason for the growth advantage of tRid394 on certain rho mutants remains unclear. Third, suppressors of 63 mutations are often t mutations, and ttsDH634 was selected as a temperature-sensitive example of such a suppressor (13). 63 encodes a protein with both RNA ligase and tail fiber attachment activities, but the mechanism of suppression has not been worked out. Fourth, thus19 and thus20 were selected for their sensitivities to growth inhibition in the presence of hydroxyurea (11), which inhibits ribonucleotide reductase. hus mutations map at numerous T4 loci (11, 25), but their appearance in t was surprising. The physiology of the thus mutants is complex (12). They differ from tam mutants in producing few phages when lysis is delayed. A double mutant carrying both thus19 and a ribonucleotide reductase amber mutation (nrd) shows a greater lysis defect than does thus19 alone, while the single nrd mutant shows almost as great a lysis defect as does thus19 itself. In addition, lysozyme levels are reduced in nrd and in thus19 infections (but not in thus20 infections), while nrd also reduces levels of other, unspecified late proteins. Like tamA3, the thus mutants are to various degrees codominant with t+. Unfortunately, this complicated picture does not point very directly at causative mechanisms, although t mutations that delay lysis might simply provide extra time to overcome an incomplete genetically or chemically induced inhibition, as already suggested for Rid394 (40).
One vexing aspect of t is its relation to a gene called stII. st mutants produce small “star” plaques surrounded by numerous sectors or dots of lysis caused by the growth of suppressor mutations. stII mutations map close to t on the asiA side, recombining with trts64 at the opposite end of t at a frequency of 5.5 to 11% (19, 20). In an intensive mapping study of a large set of now-discarded t mutations (38), the two most distal t mutations (one being amB5) recombined at a frequency of roughly 6%. While this value might be an overestimate if the time of lysis was extended by the t mutations, with a resulting increase in recombination (29), 6% is consistent with the estimate that 0.01% recombination corresponds to about 1 bp (29) and thus with the size of t, 654 bp (28). Thus, the mapping studies suggest that stII is outside of but very close to t. asiA is adjacent to t and contains 271 coding base pairs (25). Thus, the mapping data suggest that stII resides beyond asiA but do not exclude the possibility that it is identical either with asiA or with t itself. Both the proximity of t and stII mutations and the facts that stII mutants display a delayed-lysis phenotype and that rII but not rI or rIII mutations partially suppress both t and stII mutations (15, 19) led subsequent authors to conclude with ever-increasing certainty that t and stII were identical (2, 20–22, 38). However, phage is spontaneously released more readily in stII than in t infections (15, 19), although different experimental conditions were used in the two studies and cells infected with t mutants are fragile and can be induced to lyse by apparently gentle manipulations (20, 38). In our studies, t mutants did not produce a star phenotype. Therefore, unless the canonical stII mutations can be recovered or new ones discovered, the relation between stII and t is likely to remain unresolved.
We note in passing that stIII mutations can suppress stII mutations (21, 22), although tests of whether stIII mutations suppress t mutations have not been reported. In addition, E. coli mutants selected as permissive for an e null mutation were sometimes also permissive for tamA3 and tamB5 mutants (37); these were not amber suppressors but might suffer some defect in the cell wall or membrane.
Evidence accumulates that gpt is indeed a holin (40). While our BLAST search revealed no homologies between t and other holin genes except for the nearly identical t gene in phage K3 (33), a BLAST search also revealed no other candidate holins in T4 (32). Although gpt lacks homology to other holins, it localizes in the plasma membrane and can complement a defect in the holin encoded by phage λ S (27, 32). Holins characteristically contain transmembrane regions, and t probably has two (Fig. 1) (27, 32).
The λ S holin and holins in phages P1 and P2 (reviewed in reference 4) provide key insights into holin function. In the S system, both a holin and a holin inhibitor are produced from the same gene (by initiating translation at methionines encoded by either the first or the third codon). The S holin forms oligomers in the plasma membrane and has an intrinsic timing function related to its oligomerization towards an active state. Timing is further modulated by the holin-inhibitor interaction. No such dual-start system is obvious in the T4 t gene. However, lysis inhibition in T4 requires the rI gene, and gprI has been suggested to be the primary sensor of superinfection (31). (Note, however, the possible involvement of yet another rI-like gene [31 and this report].) As is implicit in the recent description of rI (31), we explicitly propose the hypothesis that gprI can exist in two states: only gprI accumulates under conditions of single infection, but gprI is converted to gprI* by superinfection. To achieve lysis inhibition, gprI* would then inhibit gpt holin function. In the λ S system, the holin inhibitor stoichiometrically titrates the pool of holin molecules and even a minority of inhibitor molecules is sufficient to delay lysis (5). Whether sufficient hypothetical T4 gprI* accumulates to act stoichiometrically remains unknown. If few gprI* molecules accumulated, then holin inhibition could still occur if a single gprI* molecule could poison a large gpt oligomer.
In addition to its holin function, gpt is linked both with the timing of lysis and with energy metabolism. In addition to the extreme effects of null and r mutations, other more subtle mutations in t can perturb the time of lysis (32). CN− induces lysis during the last half of the latent period (8), suggesting a balance between lysis and energy metabolism that is perturbed by CN−, and cells infected with tamB5 are largely insusceptible to lysis inducement by CN− (16). Thus, the rate of gpt oligomerization may be perturbed either by gpt structure or by the plasma membrane potential.
All three tr mutations reside in the first third of the gene, two within putative transmembrane domains (Fig. 1). Their amino acid changes are remarkably conservative: R→K (+→+), I→V (two small, nonpolar side chains), and T→I (uncharged polar to nonpolar). (In contrast, two of the other three missense mutations change charges.) It is therefore tempting to anticipate that gprI* interacts precisely with residues in the first third of gpt. It was suggested that gprI is secreted, presumably into the periplasmic space (31). Alternatively, it may have a transmembrane domain: after the initiating methionine residue, 13 of the next 17 residues (or 17 of the next 23) are nonpolar. Thus, gprI might interact with gpt in any of three ways: in the periplasmic space, in the membrane, or in the cytoplasm.
Understanding the dominance of tr alleles begins with the assumption that gptr is unable to interact with the hypothetical gprI*. Assuming that tr mutations do not affect gene expression, even a 7:3 ratio of gpt to gptr results in very little lysis inhibition. If gprI* simply titrated out gpt, then even the remaining minority of gptr molecules could suffice to form oligomers in time for normal lysis. Clearly, these postulated interactions are subject to direct experimental examination.
ACKNOWLEDGMENTS
We thank Victor Krylov for providing us with the canonical rV mutant and Dwight Hall and Karin Carlson for sending us their t mutants. We thank Dwight Hall, Betty Kutter, Erlan Ramanculov, and Ry Young for personal communications of results, some of which are cited herein. Gisela Mosig and Ry Young provided invaluable critical comments during the preparation of this article.
REFERENCES
- 1.Abedon S T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J Bacteriol. 1992;174:8073–8080. doi: 10.1128/jb.174.24.8073-8080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abedon S T. Lysis and the interaction between free phages and infected cells. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 397–405. [Google Scholar]
- 3.Abedon, S. T. Bacteriophage T4 resistance to lysis-inhibition collapse. Genet. Res., in press. [DOI] [PubMed]
- 4.Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang C-Y, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conkling M A, Drake J W. Isolation and characterization of conditional alleles of bacteriophage T4 genes uvsX and uvsY. Genetics. 1984;107:505–523. doi: 10.1093/genetics/107.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doermann A H. Lysis and lysis inhibition with Escherichia coli bacteriophage. J Bacteriol. 1948;55:257–276. doi: 10.1128/jb.55.2.257-276.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doermann A H. The intracellular growth of bacteriophages. I. Liberation of intracellular bacteriophage T4 by premature lysis with another phage or with cyanide. J Gen Physiol. 1952;35:645–656. doi: 10.1085/jgp.35.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake J W. Mapping mutations by phage crosses. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 444–445. [Google Scholar]
- 10.Drake, J. W. Unpublished results.
- 11.Goscin L A, Hall D H. Hydroxyurea-sensitive mutants of bacteriophage T4. Virology. 1972;50:84–94. doi: 10.1016/0042-6822(72)90348-0. [DOI] [PubMed] [Google Scholar]
- 12.Goscin, L. A., G. M. Collins, G. S. Rampe, and D. H. Hall. Unpublished results.
- 13.Hall D H, Sargent R G, Trofatter K F, Russell D L. Suppressors of mutations in the bacteriophage T4 gene coding for both RNA ligase and tail fiber attachment activities. J Virol. 1980;36:103–108. doi: 10.1128/jvi.36.1.103-108.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey A D. Mutation of bacteriophage with respect to type of plaque. Genetics. 1946;31:620–640. doi: 10.1093/genetics/31.6.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josslin R. The lysis mechanism of phage T4: mutants affecting lysis. Virology. 1970;40:719–726. doi: 10.1016/0042-6822(70)90216-3. [DOI] [PubMed] [Google Scholar]
- 16.Josslin R. Physiological studies on the t gene defect in T4-infected Escherichia coli. Virology. 1971;44:101–107. doi: 10.1016/0042-6822(71)90157-7. [DOI] [PubMed] [Google Scholar]
- 17.Jozwik C E, Miller E S. Polymerase chain reaction amplification of DNA from T4 plaques. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 464–465. [Google Scholar]
- 18.Krylov V N. A new r gene of bacteriophage T4B? Microb Genet Bull. 1966;24:4–5. [Google Scholar]
- 19.Krylov V N. Star mutants of the bacteriophage T4B. Genetika. 1971;7(1):112–119. [PubMed] [Google Scholar]
- 20.Krylov V N, Plotnikova T G. Genetic and physiological study of amber mutants in gene stII of T4B phage. Genetika. 1972;8(4):85–95. [PubMed] [Google Scholar]
- 21.Krylov V N, Yankovsky N K. Mutations in the new gene stIII of bacteriophage T4B suppressing the phenotypical defect of mutations in the gene stII in the course of the infection of Escherichia coli K-12 bacteria. Genetika. 1973;9(7):117–124. [Google Scholar]
- 22.Krylov V N, Yankovsky N K. Mutations in the new gene stIII of bacteriophage T4B suppressing the lysis defect of gene stII and gene e mutants. J Virol. 1975;15:22–26. doi: 10.1128/jvi.15.1.22-26.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krylov V N, Zapadnaya A. Bacteriophage T4B r mutations sensitive to temperature (rts) Genetika. 1965;1(1):7–11. [Google Scholar]
- 24.Krylov V N, Alikhanian S I, Morozova A S. The circular genetic map of the phage T4B. Genetika. 1967;3(5):128–132. [Google Scholar]
- 25.Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Rüger W, Mosig G. Genomic map of bacteriophage T4. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 491–519. [Google Scholar]
- 26.Linder C H, Carlson K. Escherichia coli rho factor is involved in lysis of bacteriophage T4-infected cells. Genetics. 1985;111:197–218. doi: 10.1093/genetics/111.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu M-J, Henning U. Lysis protein T of bacteriophage T4. Mol Gen Genet. 1992;235:253–258. doi: 10.1007/BF00279368. [DOI] [PubMed] [Google Scholar]
- 28.Montag D, Degen M, Henning U. Nucleotide sequence of gene t (lysis gene) of the E. coli phage T4. Nucleic Acids Res. 1987;15:6736. doi: 10.1093/nar/15.16.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosig G. Homologous recombination. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 54–82. [Google Scholar]
- 30.Mukai F, Streisinger G, Miller B. The mechanism of lysis in phage T4-infected cells. Virology. 1967;33:398–404. doi: 10.1016/0042-6822(67)90115-8. [DOI] [PubMed] [Google Scholar]
- 31.Paddison P, Abedon S T, Dressman H K, Gailbreath K, Tracy J, Mosser E, Neitzel J, Guttman B, Kutter E M. The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: a new perspective. Genetics. 1998;148:1539–1550. doi: 10.1093/genetics/148.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanculov, E., and R. Young. Unpublished results.
- 33.Riede I. Lysis gene t of T-even bacteriophages: evidence that colicins and bacteriophage genes have common ancestors. J Bacteriol. 1987;169:2956–2961. doi: 10.1128/jb.169.7.2956-2961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosario M O, Drake J W. Frameshift and double-amber mutations in the bacteriophage T4 uvsX gene: analysis of mutant UvsX proteins from infected cells. Mol Gen Genet. 1990;222:112–119. doi: 10.1007/BF00283031. [DOI] [PubMed] [Google Scholar]
- 35.Steinberg C M, Edgar R S. A critical test of a current theory of genetic recombination in bacteriophage. Genetics. 1962;47:187–208. doi: 10.1093/genetics/47.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stitt B, Hinton D. Regulation of middle-mode transcription. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 142–160. [Google Scholar]
- 37.Sundar Raj C V, Wu H C. Escherichia coli mutants permissive for T4 bacteriophage with deletion in e gene (phage lysozyme) J Bacteriol. 1973;114:656–665. doi: 10.1128/jb.114.2.656-665.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel T, Astrachan L. Isolation and mapping of t gene mutants of bacteriophage T4D. J Virol. 1977;24:518–524. doi: 10.1128/jvi.24.2.518-524.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yankovsky N K, Krylov V N. Genetical and physiological study of mutations of phage T4 suppressing the ligase defect of gene stII mutants. Genetika. 1975;11(10):51–60. [PubMed] [Google Scholar]
- 40.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]