Abstract
Formyl peptide receptors (FPR1, FPR2 and FPR3) are innate immune sensors of pathogen and commensal bacteria and have a role in colonic mucosa homeostasis. We identified FPR1 as a tumour suppressor in gastric cancer cells due to its ability to sustain an inflammation resolution response with antiangiogenic potential. Here, we investigate whether FPR1 exerts similar functions in colorectal carcinoma (CRC) cells. Since it has been shown that the commensal bacterium Lactobacillus rhamnosus GG (LGG) can promote intestinal epithelial homeostasis through FPR1, we explored the possibility that it could induce proresolving and antiangiogenic effects in CRC cells. We demonstrated that pharmacologic inhibition or genetic deletion of FPR1 in CRC cells caused a reduction of proresolving mediators and a consequent upregulation of angiogenic factors. The activation of FPR1 mediates opposite effects. Proresolving, antiangiogenic and homeostatic functions were also observed upon treatment of CRC cells with supernatant of LGG culture, but not of other lactic acid or nonprobiotic bacteria (i.e. Bifidobacterium bifidum or Escherichia coli). These activities of LGG are dependent on FPR1 expression and on the subsequent MAPK signalling activation. Thus, the innate immune receptor FPR1 could be a regulator of the balance between microbiota, inflammation and cancer in CRC models.
Keywords: angiogenesis, colon cancer, formyl peptide receptor 1, Lactobacillus rhamnosus GG, LGG, specialized proresolving mediators
The commensal probiotic bacteria Lactobacillus rhamnosus GG (LGG), by activating the innate immune receptor formyl peptide receptor 1 (FPR1), influence normal intestinal epithelial homeostasis and sustain wound restitution. We demonstrated that LGG, but not other lactic acid or nonprobiotic bacteria, activate an inflammation resolution program that dampens the angiogenic response of colorectal carcinoma cells.
Abbreviations
- AA
arachidonic acid
- Abs
antibodies
- Akt
protein kinase B
- ALOX15
arachidonate 15‐lipoxygenase
- ALOX15B
arachidonate 15B‐lipoxygenase
- ALOX5
arachidonate 5‐lipoxygenase
- Ang1
angiopoietin 1
- AnxA1
annexin A1
- B. bifidum
Bifidobacterium bifidum
- BLT‐1
block lipid transport‐1
- cfu
colony‐forming unit
- ChemR23
chemerin receptor 23
- CM
conditioned media
- CRC
colorectal carcinoma
- CsH
cyclosporine
- CXCL1
chemokine (C‐X‐C motif) ligand 1
- DHA
docosahexaenoic acid
- DPP IV
dipeptidyl peptidase‐IV
- E. coli
Escherichia coli
- ECM
extracellular matrix
- ECs
endothelial cells
- EG‐VEGF
human endocrine gland‐derived vascular endothelial growth factor
- EIA
enzyme immunoassay
- ELISA
enzyme‐linked immunosorbent assay
- EPA
eicosapentaenoic acid
- FACS
fluorescence‐activated cell sorting
- fMLF
N‐formyl‐Met‐Leu‐Phe
- FPR
formyl peptide receptor
- FPR1
formyl peptide receptor 1
- FPR2
formyl peptide receptor 2
- GC
gastric cancer
- GDNF
glial cell line‐derived neurotrophic factor
- GPR32
G protein‐coupled receptor 32
- HUVECs
human umbilical vein endothelial cells
- IBD
inflammatory bowel disease
- IGFBP‐1
insulin‐like growth factor‐binding protein 1
- IGFBP‐2
insulin‐like growth factor‐binding protein 2
- LGG
Lactobacillus rhamnosus
- LTB4
leukotriene B4
- LXA4
lipoxin A4
- LXB4
lipoxin B4
- MAPK
mitogen‐activated protein kinase
- MaR
maresins
- MCP‐1
monocyte chemoattractant protein‐1
- MMP8
matrix metallopeptidase 8
- MMP9
matrix metallopeptidase 9
- mRNA
messenger RNA
- MRS
sharpe broth medium
- NSCLC
non‐small‐cell lung cancer
- PCR
polymerase chain reaction
- PD
protectins
- PD‐ECGF
platelet‐derived endothelial cell growth factor
- PDGF‐AA
platelet‐derived growth factor AA
- PDGF‐AB
platelet‐derived growth factor AB
- PDGF‐BB
platelet‐derived growth factor BB
- PGE2
prostaglandin E2
- PIGF
placental growth factor
- PRRs
pattern recognition receptors
- PUFA
polyunsaturated fatty acid
- RNA
ribonucleic acid
- RvD1
resolvin D1
- RvDs
resolvins D
- RvEs
resolvins E
- SD
standard deviation
- shCTR
nontargeting short hairpin
- shFPR1
short hairpin targeting FPR1
- shRNA
short hairpin RNA
- SN
supernatant
- SPMs
specialized proresolving mediators
- STAT3
signal transducer and activator of transcription 3
- TGF‐beta1
transforming growth factor‐beta 1
- TIMP‐1
tissue inhibitor matrix metalloproteinase 1
- TIMP‐4
tissue inhibitor matrix metalloproteinase 4
- TLR7
toll‐like receptor 7
- TS
Tryptic Soy
- uPA
urokinase plasminogen activator plasma
- VEGF‐A
vascular endothelial growth factor A
- VEGF‐B
vascular endothelial growth factor B
- VEGF‐C
vascular endothelial growth factor C
- VEGF‐D
vascular endothelial growth factor D
- ω‐3
omega‐3
- ω‐6
omega‐6
1. Introduction
Chronic inflammation is a risk factor for colorectal carcinoma (CRC) onset [1], as strongly suggested by the increased predisposition to colon carcinogenesis of inflammatory bowel disease (IBD) patients [2].
In recent years, unresolved chronic inflammation has been associated with insufficient production of mediators, which are actively involved in the resolution of inflammation [3, 4]. Molecules diverse in nature are able to actively participate in different moments of the resolution program [3]: (i) lipidic autacoids known as specialized proresolving mediators (SPMs) [4]; (ii) proteic mediators [e.g. annexin A1 (AnxA1), adrenocorticotropic hormone, chemerin peptides, and galectin‐1] [3, 5]; (iii) gaseous mediators (nitric oxide, hydrogen sulfide and carbon monoxide) [3]; (iv) the adenosine, a purine nucleoside generated by the dephosphorylation of adenine [6]; and (v) neuromodulators such as acetylcholine and other neuropeptides produced by vagus nerves [7].
Specialized proresolving mediators are lipidic mediators derived from omega‐6 (ω‐6) arachidonic (AA), or ω‐3 eicosapentaenoic (EPA) and docosahexaenoic (DHA) essential polyunsaturated fatty acids (PUFA) through the activity of lipoxygenases 5 and 15 (ALOX5/15). The best‐characterized SPMs are lipoxins (LXA4, LXB4), E‐ and D‐series resolvins (RvEs and RvDs), protectins (PD) and maresins (MaR) [8]. They exert anti‐inflammatory, antiangiogenic and proresolving effects subsequent to inflammatory conditions [4, 8, 9].
We recently described a novel function of SPMs in gastric cancer (GC) demonstrating that RvD1 and LXB4 suppress angiogenesis, thus inhibiting tumour growth. We also demonstrated that formyl peptide receptor 1 (FPR1), a member of the FPR family, controls SPM production in GC [9, 10], functioning as a tumour suppressor [11, 12]. FPRs are pattern recognition receptors (PRRs) known to balance inflammatory and proresolving responses by sensing host‐derived and bacterial products [13, 14].
Several reports point to a crucial protective role of proresolving pathways also in CRC carcinogenesis [15]. Indeed, it has been demonstrated that CRC is associated with a reduced intake of ω‐3 PUFA [16] and that dietary supplementation of ω‐3 PUFA exerts anticancer effects in CRC [17]. Furthermore, ALOX15 has been described as a tumour suppressor in CRC [18] and specific SPMs (i.e. RvD1, LXA4) have been demonstrated to exert antitumour activity in CRC models [19, 20, 21, 22, 23, 24].
Intestinal inflammatory conditions are strongly influenced and in turn affect microbiota composition [1, 25]. In the last years, several studies in CRC patients and experimental models demonstrated that colon tumorigenesis is associated with significant alterations of intestinal microbial composition termed as dysbiosis [26, 27]: in CRC patients, specific bacterial species are over‐represented compared with those in noncancerous patients and exert protumorigenic function(s), while other species are under‐represented and exert tumour suppressive functions [26, 27, 28].
Lactobacillus rhamnosus GG (LGG) is a commensal bacterium used as probiotic in order to reverse intestinal dysbiosis [29]. Several preclinical studies point to its effects in reducing chronic inflammation linked to CRC development [30]: LGG has been demonstrated to regulate homeostasis and restitution following colonic wounds in mice [31, 32, 33, 34]; in CRC models, LGG activates proapoptotic and antimetastatic responses [35, 36], lowers inflammation and favours adaptive protective immune responses against cancer cells [37]. It has been shown that LGG activity in intestinal epithelial cells depends on the expression of FPR1 [32].
Since the gastrointestinal tract is continuously exposed to external insults, proresolving pathways are particularly important to balance inflammatory responses for its homeostasis [4]. Thus, we investigated the role of FPR1 in CRC cells in order to verify the possibility that it functions as a regulator of inflammation resolution, angiogenesis and cancer. Moreover, we evaluated whether homeostatic and anticancer effects of LGG in CRC models could depend on its ability to activate a proresolving response. In particular, we investigated the possibility that LGG could activate a proresolving and an antiangiogenic response in CRC cells by stimulating FPR1.
Our data confirm this hypothesis and highlight the possibility that FPR1 could be exploited in order to increase the proresolving and inhibit the angiogenic potential of CRC cells, also through the use of probiotics.
2. Materials and methods
2.1. Cell culture
The HCT116 and HT29 cell lines derived from colorectal carcinoma (CRC) were kindly provided by S. Scala (Istituto Nazionale Tumori‐IRCCS‐Fondazione G. Pascale, Napoli, Italy) and grown as elsewhere described [38]. To generate HCT116 cells stably expressing FPR1 shRNA, we used pools of 5 constructs (Qiagen, Valencia, CA, USA) containing 21‐mer short hairpin RNAs (shRNA) directed to various coding regions of the target gene. Transfectants were selected in medium with 500 ng·mL−1 puromycin [10]. Human umbilical vein endothelial cells (HUVECs) from Cell Biologics (Chicago, IL, USA) were grown in human endothelial cell medium with the addition of VEGF, heparin, EGF, FGF, hydrocortisone, L‐glutamine, antibiotic/antimycotic solution and FBS according to the manufacturer's instructions (Cell Biologics) [39].
Cell treatments to verify (a) mRNA changes, (b) enzyme or receptor protein expression, (c) AnxA1 expression, (d) signalling pathway activation and (e) autacoid release were made in serum‐free media and after a 12‐h serum starvation. Instead, cell treatment for VEGF‐A release and to collect cell culture supernatants to be used in capillary formation assay was performed in 1% FBS to improve the stability of VEGF‐A. The same conditions were used when bacterial supernatants (SN) were used to treat CRC cells; the relative control sample of each bacterial strain SN was the same titration of the culture broth.
Treatments of CRC cells were made with fMLF at 1 nm, the concentration at which it binds specifically to FPR1 [13]. The SPMs were used again at 1 nm, the same optimal concentration used to inhibit angiogenesis in the GC model [10].
The functional involvement of GPR32 or MAPK signalling in CRC proresolving responses was evaluated by preincubating cells for 30 min with an anti‐GPR32 neutralizing antibody (Ab) (1 μg·mL−1) (GeneTex, Irvine, CA, USA) or the MAPK signalling inhibitor U0 126 (10 μm) (Cell Signaling, Danvers, MA, USA), respectively, before proceeding with the specific treatment.
2.2. Bacterial culture and supernatant preparation
The bacterial strains used in this study were as follows: Lactobacillus rhamnosus (Hansen) Collins et al. (LGG) (ATCC 53103), obtained from the ATCC (Manassas, VA); Escherichia coli ATCC 13762 (E. coli), used as control of nonprobiotic bacteria; and Bifidobacterium bifidum (B. bifidum), an anaerobic lactic acid bacterium isolated from the ProBiota Bifido (Seeking Health, Bellingham, WA, USA). Bacterial suspensions, at 0.1 optical density (OD) at 600 nm, were inoculated in broth medium and grown in slight motion at 37 °C overnight in aerobic or anaerobic condition, in order to obtain a number of colony‐forming unit (cfu) of ∼ 108/mL, determined by plate counting on medium agar plates. In detail, LGG suspension was inoculated in De Man, Rogosa and Sharpe (MRS) broth and MRS agar medium (Becton Dickinson, Franklin Lakes, NJ, USA) in aerobic condition at 37 °C. E. coli ATCC 13762 was cultured in Tryptic Soy (TS) broth and TS agar (OXOID, Basingstoke, Hampshire, UK) in aerobic condition at 37 °C. B. bifidum was cultured in anaerobic condition at 37 °C, using MRS broth medium supplemented with 0.25% cysteine/HCl (Sigma‐Aldrich, St. Louis, MO, USA). Bacterial supernatant (SN) of each strain was prepared by centrifugation of the overnight cultures in the specific growth medium at 4000 g and 4 °C for 10 min and stored in single‐use aliquots at −80 °C until needed.
2.3. Flow cytometry
Cells were incubated (30 min at 4 °C) with specific or isotype control Abs. ALOX5, ALOX15A and ALOX15B Abs were from Santa Cruz Biotechnology (Dallas, TX, USA), anti‐GPR32 from Acris (Herford, Germany), anti‐BLT1 from LSBio (Seattle, WA, USA), and anti‐ChemR23 and anti‐FPR1 from R&D Systems (Minneapolis, MI, USA). Cells were analysed with a FACSCalibur cytofluorimeter using the cellquest software (BD Biosciences, Lakes, NJ, USA). When necessary, we performed cell membrane permeabilization using the Cytofix/Cytoperm Kit (BD Biosciences). The receptors followed as indicators of resolution responses were the same modulated by FPR1 in the GC model [10]. The concentration used for flow cytometric staining was that indicated by manufacturers. AnxA1 staining was performed using a primary anti‐AnxA1 goat polyclonal Ab (1 : 500) (R&D Systems) followed by the staining with a secondary anti‐goat Ab Alexa Fluor 488 (Invitrogen, Waltham, MA, USA). The secondary antibody alone was used as a negative matched control.
2.4. ELISA and EIA
VEGF‐A contents in culture supernatants were measured in duplicate determinations with a commercially available ELISA (R&D Systems). RvD1, LTB4, PGE2 and LXB4 contents in culture supernatants were measured in triplicate determinations with a commercially available EIA (Cayman Chemical, Ann Arbor, MI, USA) [39]. Cell culture supernatants were assayed, undiluted for autacoid evaluations and diluted 1 : 5 for VEGF‐A release.
2.5. Real‐time PCR
Total RNA was isolated and retrotranscribed according to the manufacturer's instructions (Qiagen) as previously described [40]. Real‐time quantitative PCR was performed on iCycler (Bio‐Rad, Hercules, CA, USA) using the PE SYBR Green PCR kit (Applied Biosystems, Foster City, CA, USA) as reported elsewhere [41]. Normalization was performed using β‐actin mRNA levels. Primer sequences are listed in Table S1.
2.6. Protein studies
Protein extractions and immunoblotting experiments to evaluate signalling pathway activation were performed according to standard procedures [11]. Anti‐phospho‐MAPK, anti‐phospho‐Akt, and anti‐phospho‐STAT3 Abs (1 : 1000) were from Cell Signaling Technology. Antitubulin was from Sigma‐Aldrich (1 : 10 000) (St. Louis, MO, USA), and secondary anti‐mouse and antirabbit Abs coupled to HRP were from Bio‐Rad (1 : 3000).
The expression of angiogenesis‐related proteins in CRC cell culture supernatants was determined using the Human Angiogenesis Array Kit (R&D Systems) according to the manufacturer's instructions. The array allows the detection of 55 angiogenesis‐related proteins by specific capture antibodies spotted onto a nitrocellulose membrane. The data from developed X‐ray film were digitalized and quantified using the ImageJ analysis software [42].
2.7. Tubule formation
The formation of network‐like structures by HUVECs (Cell Biologics) on an extracellular matrix (ECM)‐like 3D gel consisting of Matrigel® (BD Biosciences) was performed as previously described and validated [43]. HUVECs (5 × 104) were seeded on a Matrigel matrix in the presence of cell culture supernatants. After incubation, HUVECs underwent differentiation into capillary‐like tube structures. Tubule formation was defined as a structure exhibiting a length four times its width. Network formation was observed using an inverted‐phase contrast microscope (Zeiss, Oberkochen, Germany). Representative fields were taken [43], and the number of branching points counted in five fields was presented as mean ± SD of three experiments.
2.8. Wound‐healing assay
For wound‐healing assays, confluent monolayers of HCT116 cells were treated with mitomycin (2 μg·mL−1 for 2 h) (Sigma‐Aldrich). The cell monolayers were scraped in three straight lines for each 60‐mm dish with a p10 pipet tip. Cell migration was assessed as previously described [44]. Confluent monolayers were incubated for 30 min with an anti‐GPR32 neutralizing Ab (1 μg·mL−1) (GeneTex) or CsH (800 nm) (Sigma‐Aldrich) and then treated with LGG SN or control broth (1 : 30 titration) for 12 h before assessing cell migration.
2.9. Statistical analysis
Values from groups were compared using the paired Student t test. P < 0.05 was considered statistically significant. Clinic–pathologic parameters in relation to FPR1 or FPR2 expression were plotted using the cBioPortal database. Coexpression data were obtained according to the cBioPortal online instructions: a log‐rank test was provided to identify the significance of Spearman's correlation coefficient between the mRNA expression z‐scores (RNASeq V2 RSEM) [39].
3. Results
3.1. FPR1 pharmacologic stimulation sustains a proresolving response in colorectal carcinoma (CRC) cells
We recently described that FPR1 stimulation induces a proresolving program that relies on the expression of enzymes involved in SPM production (ALOX5, ALOX15A and ALOX15B), the release of specific SPMs (RvD1 and LXB4) and the expression of SPM receptors (GPR32, ChemR23 and BLT1) in gastric cancer (GC) cells [11].
To determine whether FPR1 activates a proresolving program also in colon cancer, we selected two human colorectal carcinoma (CRC) cell lines, HT29 and HCT116 cells. The expression of FPR1 in HCT116 and HT29 cells was assessed by FACS analysis (Fig. S1A). To pharmacologically modulate FPR1, we treated CRC cells with fMLF, an agonist to FPR1 [13], or with cyclosporine H (CsH), an inverse agonist to FPR1 [13], and we analysed the impact of these treatments on SPM pathway.
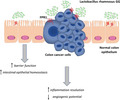
We found that, in HT29 and HCT116 cells, fMLF (10−9 m—3‐h treatment) significantly increased, whereas CsH (800 nm—3‐h treatment) significantly decreased, the mRNA expression of enzymes (ALOX5, ALOX15A and ALOX15B) and receptors (GPR32, ChemR23 and BLT1) involved in SPM synthesis and recognition (Fig. 1A). In accordance with these observations, the protein levels of SPM enzymes (ALOX5, ALOX15A and ALOX15B) and receptors (GPR32, ChemR23 and BLT1) were significantly induced or reduced, compared with relative controls, in HT29 and HCT116 cells treated for 6 h with fMLF or CsH, respectively, as assessed by cytofluorimetric analysis (Fig. 1B,C).
Fig. 1.
Effects of formyl peptide receptor 1 (FPR1) pharmacologic modulation on specialized proresolving mediator (SPM) biosynthetic machinery of colorectal carcinoma (CRC) cells. (A) ALOX5, ALOX15A, ALOX15B, GPR32, ChemR23 and BLT1 mRNA fold change in HT29 and HCT116 cells treated with fMLF (10−9 m) or CsH (800 nm) for 3 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control (dotted line) by Student's t test. (B) ALOX5, ALOX15A, ALOX15B, BLT1 and ChemR23 protein expression levels (mean fluorescence intensity), assessed by cytofluorimetric analysis, in HT29 cells treated with fMLF (10−9 m) or CsH (800 nm) for 6 h. A representative experiment of three independent experiments is shown. (C) ALOX5, ALOX15A, ALOX15B, GPR32, ChemR23 and BLT1 protein expression levels (mean fluorescence intensity), assessed by cytofluorimetric analysis, in HCT116 cells treated with fMLF (10−9 m) or CsH (800 nm) for 6 h. Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the control (NT) by Student's t test. (D) Proresolving and proinflammatory autacoid (RvD1, LXA4, LXB4, PGE2) release in HCT116 and HT29 cells treated or not with fMLF (10−9 m) or CsH (800 nm) for 12 h. Baseline values of each mediator were in HCT116: RvD1 118 ± 18 pg/106 cells, LXB4 34 ± 5 pg/106 cells, LXA4 485 ± 58 pg/106 cells and PGE2 98 ± 12 pg/106 cells. Baseline values of each mediator were in HT29: RvD1 132 ± 24 pg/106 cells, LXB4 32 ± 4 pg/106 cells, LXA4 462 ± 48 pg/106 cells and PGE2 86 ± 11 pg/106 cells. Data are represented as mean ± SD of changes over baseline levels obtained in five independent experiments. *P < 0.05 compared with the control by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
The activation of SPM synthesis during resolution of inflammation counterbalances the production of proinflammatory lipidic mediators (e.g. prostaglandins and leukotrienes) [8, 39]. In CRC, a crucial proinflammatory and protumorigenic role has been described for PGE2 [45]. On the other side, several reports point to an anti‐inflammatory function of LXA4 [23, 46]. Thus, we decided to verify whether FPR1 pharmacologic modulation induces changes in PGE2 and LXA4 release, together with the two SPMs modulated by FPR1 in GC cells (i.e. RvD1 and LXB4) [11].
To this purpose, we treated CRC cells with fMLF (10−9 m) or with CsH (800 nm) for 12 h and looked for RvD1, LXA4, LXB4 and PGE2 release. We found that fMLF significantly increased, while CsH significantly decreased SPM (RvD1, LXA4 and LXB4) release in both HT29 and HCT116 cells (Fig. 1D). Consistently, the treatment of HT29 and HCT116 cells with fMLF (10−9 m—12 h) significantly reduced, whereas CsH (800 nm—12 h) significantly increased, PGE2 release (Fig. 1D).
These data indicate that FPR1 is able to activate in CRC cells an inflammation resolution program, by promoting the induction of SPMs (RvD1, LXA4 and LXB4), SPM enzymes (ALOX5, ALOX15A and ALOX15B) and SPM receptors (GPR32, ChemR23 and BLT1).
3.2. FPR1 pharmacologic modulation controls the angiogenic potential of CRC cells
We previously found that FPR1 ablation/pharmacological inhibition caused a drop in the endogenous levels of SPMs and a concomitant increase in the angiogenic potential of GC cells. We also found that SPMs control the production of angiogenic mediators in GC cells, since the exogenous administration of SPMs (RvD1 or LXB4) to FPR1‐depleted GC cells could suppress their increased angiogenic potential [11].
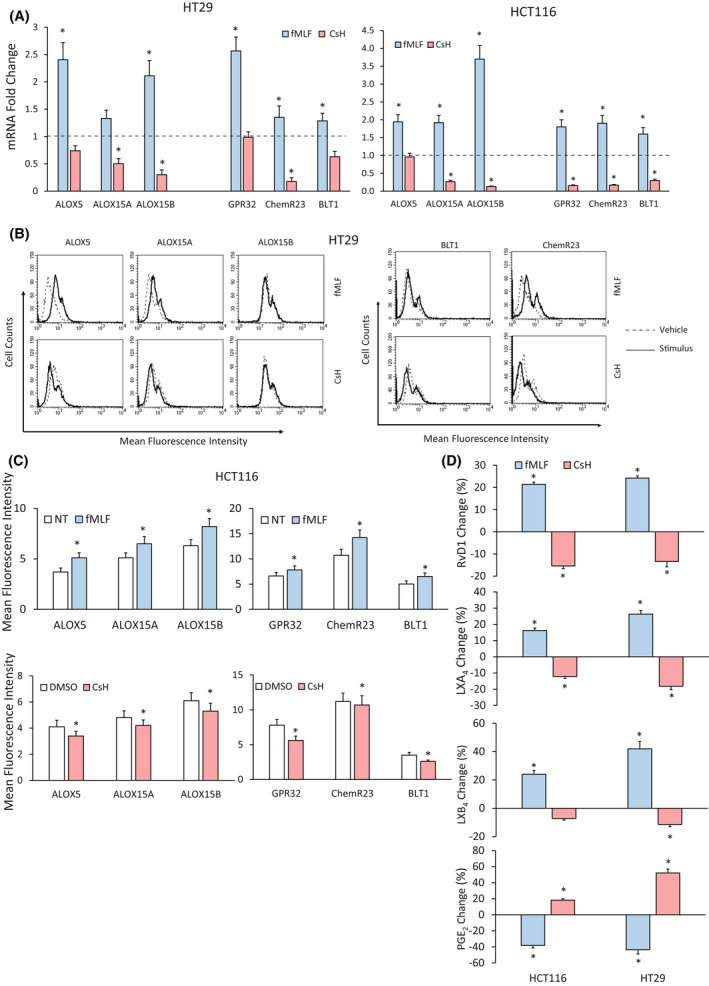
To investigate whether FPR1 acts as a negative modulator of angiogenesis also in CRC cells, we treated HT29 and HCT116 cells with fMLF (10−9 m) or with CsH (800 nm) for 3 h and measured the mRNA expression levels of several proangiogenic mediators (VEGF‐A, VEGF‐B, VEGF‐C, VEGF‐D, Ang1 and CXCL1). We observed that fMLF induced a reduction of mRNA levels for VEGF‐A, VEGF‐C and CXCL1 in HT29 cells and for VEGF‐B, VEGF‐C, Ang1 and CXCL1 in HCT116 cells (Fig. 2A). Consistently, CsH treatment significantly increased mRNA expression of proangiogenic molecules in both HT29 and HCT116 cells (Fig. 2A). Moreover, the release of VEGF‐A was significantly lower in HCT116 treated for 12 h with fMLF (10−9 m) and significantly higher in the same cells treated with CsH (800 nm) compared to relative controls (Fig. 2B). Similar results were obtained in HT29 cells (not shown).
Fig. 2.
Effects of formyl peptide receptor 1 (FPR1) pharmacologic modulation in colorectal carcinoma (CRC) cells on angiogenic response. (A) VEGF‐A, VEGF‐B, VEGF‐C, VEGF‐D, Ang1 and CXCL1 mRNA fold change in HT29 and HCT116 cells treated with fMLF (10−9 m) or CsH (800 nm) for 3 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control (dotted line) by Student's t test. (B) VEGF‐A release in HCT116 cells treated with fMLF (10−9 m) or CsH (800 nm) or the relative controls for 12 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control by Student's t test. (C) Correlation between the mRNA expression levels of the indicated markers in 594 patients affected by colorectal adenocarcinoma. Spearman's factor, Pearson's factor and the relative p are indicated. [Colour figure can be viewed at wileyonlinelibrary.com]
To corroborate our results in CRC cultures, we verified the mRNA coexpression data present in the publicly available cBioPortal for Cancer Genomics database (http://www.cbioportal.org) [47, 48]. Data on 594 colorectal adenocarcinoma revealed that FPR1 mRNA levels significantly and directly correlated with mRNA expression levels of the proresolving factors ALOX5 and ALOX15B (Fig. 2C). Consistently, FPR1 mRNA levels inversely correlated with mRNA levels of two key angiogenic mediators [VEGF‐A and angiopoietin 1 (Ang)] (Fig. 2C).
Finally, in order to search for information on the clinic–pathologic role of FPRs and/or proresolving pathways in CRC, we queried the cBioPortal for Cancer Genomics database (http://www.cbioportal.org): mRNA levels of FPR1 showed a statistic trend of association with the Overall Survival Status (P = 0.11) and a direct and statistically significant association with the disease‐free months (Fig. S2). No statistically significant association with the two parameters was found for FPR2, suggesting that FPR1 in CRC plays a nonredundant role similar to that observed in GC [11].
These data support the hypothesis that FPR1 is responsible for a proresolving and antiangiogenic response in CRC cells.
3.3. FPR1 genetic ablation reduces the proresolving activities and increases the angiogenic potential of CRC cells
To confirm the results presented in 3.1 and 3.2 paragraphs, we generated HCT116 cells stably transfected either with FPR1‐targeting short hairpin RNAs (HCT116 shFPR1) or with nontargeting short hairpin RNAs (HCT116 shCTR). We identified various clones expressing low levels of the receptor (Fig. S1B).
By real‐time PCR, we found that genetic ablation of FPR1 significantly reduced mRNA level of proresolving enzymes (ALOX15A and ALOX15B) and receptors (GPR32, ChemR23) compared with control cells (Fig. 3A). Moreover, HCT116 shFPR1 cells exhibited an increase of mRNA levels for angiogenic mediators compared with controls (Fig. 3A).
Fig. 3.
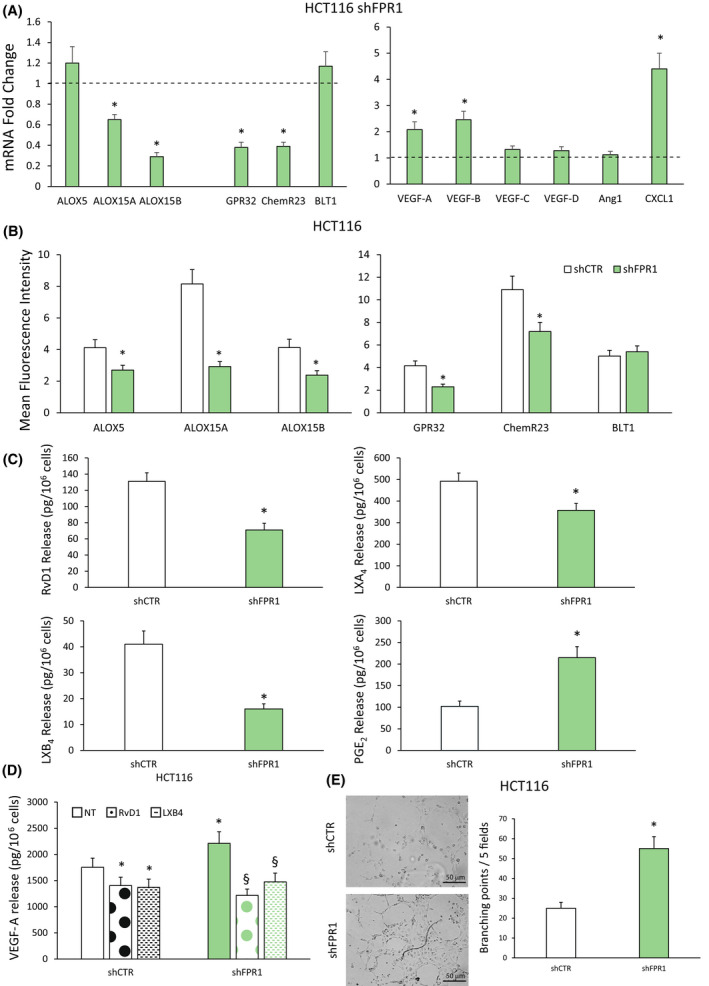
Effects of formyl peptide receptor 1 (FPR1) silencing on specialized proresolving mediator (SPM) biosynthetic machinery and angiogenic potential of colorectal carcinoma (CRC) cells. (A) ALOX5, ALOX15A, ALOX15B, GPR32, ChemR23, BLT1, VEGF‐A, VEGF‐B, VEGF‐C, VEGF‐D, Ang1 and CXCL1 mRNA fold change in HCT116 cells silenced for FPR1 (HCT116 shFPR1, three clones) compared to cells transfected with nontargeting short hairpin RNAs (shCTR cells—dotted line, a mass population). Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control (dotted line) by Student's t test. (B) ALOX5, ALOX15A, ALOX15B, GPR32, ChemR23 and BLT1 expression levels (mean fluorescence intensity), assessed by cytofluorimetric analysis, in HCT116 shFPR1 (three clones) cells and the relative control (shCTR, a mass population). Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the relative control by Student's t test. (C) Proresolving and proinflammatory autacoid (RvD1, LXA4, LXB4, PGE2) release in HCT116 shCTR (a mass population) or shFPR1 (three clones). Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the relative control by Student's t test. (D) VEGF‐A release in HCT116 shCTR cells (a mass population) or shFPR1 cells (three clones) treated or not (NT) with RvD1 (1 nm) or LXB4 (1 nm) for 12 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with shCTR NT by Student's t test. § P < 0.05 compared with the relative control by Student's t test. (E) Human umbilical vein endothelial cells (HUVECs) were cultured in the presence of cell culture conditioned media (CM) from HCT116 shCTR (a mass population) or shFPR1 (three clones) (10× magnifications) in a 24‐well plate. After 12 h, cells were fixed with ice‐cold 70% ethanol, and tubule formation was evaluated. Sample images and a quantification of the angiogenic response are reported. Scale bar 50 μm. Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the control by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
The genetic ablation of FPR1 significantly decreased the protein levels of ALOX5, ALOX15A, ALOX15B, GPR32 and ChemR23, but not BLT1 compared with HCT116 shCTR cells, as assessed by cytofluorimetric analysis (Fig. 3B). Moreover, the amount of RvD1, LXB4 and LXA4 released was significantly reduced in HCT116 shFPR1 compared with that in shCTR cells (Fig. 3C). Furthermore, FPR1 silencing in HCT116 cells caused the release of higher levels of PGE2 compared with that of controls (Fig. 3C). In accordance, these cells displayed increased constitutive release of VEGF‐A (Fig. 3D).
The formation of capillary‐like tube structures in the extracellular matrix by endothelial cells (ECs) is a classic method to measure angiogenesis in vitro [49]. To investigate whether differences in FPR1 expression/activation control functional angiogenic properties of CRC cells, we studied the ability of CRC cell conditioned media (CM) to induce human umbilical vein endothelial cell (HUVEC) network formation on a Matrigel substrate. In particular, we evaluated tubule formation in vitro in response to CM from HCT116 cells silenced or not for FPR1 (shCTR vs shFPR1). As shown in Fig. 3E, HUVECs plated in the presence of CM from HCT116 shCTR cells formed only a few tube structures at 12 h. On the contrary, when the endothelial cells were plated in the presence of CM from HCT116 shFPR1, a significantly higher number of formed tube structures were observed compared with that induced by shCTR CM (Fig. 3E).
To assess whether the increased proangiogenic potential of FPR1‐silenced HCT116 cells could be due to the defective SPM biosynthesis of these cells, we added back LXA4 (1 nm), RvD1 (1 nm) or LXB4 (1 nm) to HCT116 shFPR1 and shCTR cells for 3 h and evaluated their proangiogenic activity. By means of real‐time PCR, we observed that LXA4 had no effects on proangiogenic mediator expression (Fig. S3). Thus, although it has been described that LXA4 exerts a strong anti‐inflammatory potential in CRC [23, 46], our results demonstrate that it has no effect on the modulation of CRC cell angiogenic potential.
RvD1 and LXB4 were able to reduce the expression of proangiogenic mRNAs, in HCT116 shFPR1 and, to a lesser extent, in HCT116 shCTR cells (Fig. S4A). Consistently, as shown in Fig. 3D, we found that RvD1 and LXB4 significantly reduced VEGF‐A protein release in both HCT116 shFPR1 and, to a lesser extent, HCT116 shCTR cells.
Finally, RvD1 and LXB4 displayed the ability to restore the expression of ALOXs and SPM receptor mRNAs in HCT116 shFPR1 cells (Fig. S4B). The RvD1‐ and LXB4‐induced upregulation of SPM enzymes and receptors was also confirmed at the protein level by FACS analysis (Fig. S4C,D).
These results demonstrate that FPR1 exerts an antiangiogenic effect in CRC cells through the modulation of SPM production.
3.4. Lactobacillus rhamnosus GG (LGG) supernatant sustains proresolving and antiangiogenic responses in CRC cells
Previous studies showed that Lactobacillus rhamnosus GG (LGG), a colonic commensal bacterium and one of the most used probiotic strains [30], exerts a homeostatic function in intestinal mucosa [31, 32, 33], decreases levels of procarcinogenic metabolites, reduces chronic inflammation associated with intestinal neoplastic transformation and inhibits the proliferation of malignant cells [30, 35, 36]. Some of these LGG functions were associated with its ability to interact with FPR1 [32]. Thus, we hypothesized that LGG protective effect in normal intestinal mucosa and LGG anticancer activities in CRC cells could be linked to a proresolving and antiangiogenic response activated by the bacteria.
In order to verify our hypothesis, we treated HT29 and HCT116 cells with LGG supernatant (SN) (1 : 30 titration) for 3 h, or with the same dilution of the culture broth, and evaluated, by real‐time PCR, the expression levels of proresolving pathways' components and of proangiogenic markers. The 1 : 30 titration was chosen as optimal one after testing different dilutions (1 : 10–1 : 100) of LGG SN in cell culture media in order to balance pH changes induced by LGG SN and its functional activity (not shown). We found that LGG SN induced, in both CRC cells, a significant increase of ALOX15A, ALOX15B, GPR32, ChemR23 and BLT1 mRNA levels (Fig. S5A) and a statistically significant decrease of proangiogenic mediator mRNA levels (VEGF‐C, VEGF‐D, Ang1 and CXCL1) (Fig. S5B) when compared to control (culture broth).
We confirmed these observations at the protein level: the treatment of HT29 cells with LGG SN for 6 h significantly increased ALOX5, ALOX15A, ALOX15B, BLT1 and ChemR23 protein levels (Fig. 4A). Similar results were obtained in HCT116 cells (not shown). Consistently with the induction of enzymes, LGG SN (12 h treatment) induced a significant increase in SPM release (RvD1, LXB4, LXA4) and a significant decrease in PGE2 levels in both HT29 and HCT116 cells (Fig. 4B).
Fig. 4.
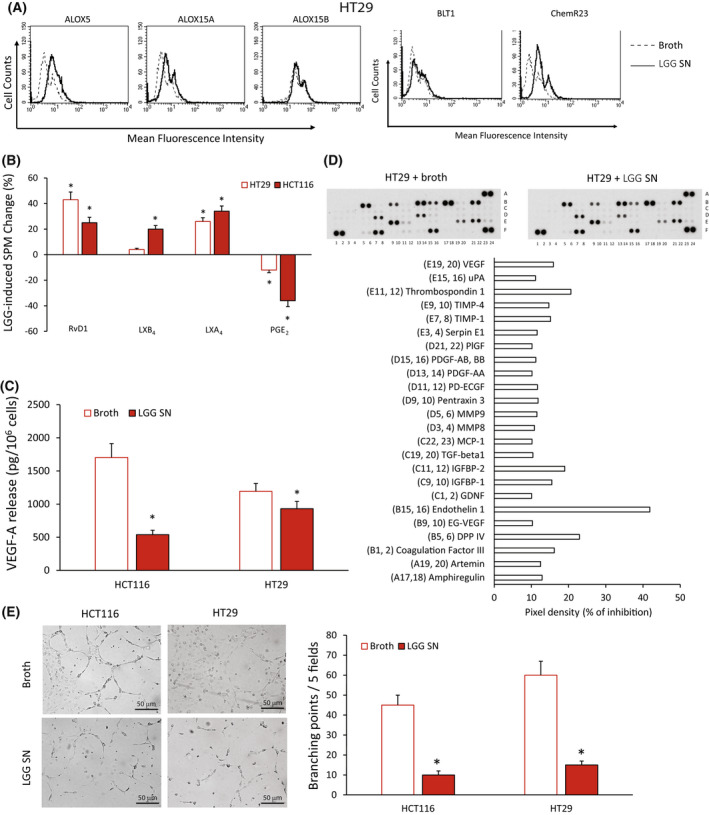
Effects of Lactobacillus rhamnosus GG (LGG) supernatant (SN) on specialized proresolving mediator (SPM) biosynthetic machinery and angiogenic potential of colorectal carcinoma (CRC) cells. (A) ALOX5, ALOX15A, ALOX15B, BLT1, and ChemR23 protein expression levels (mean fluorescence intensity), assessed by cytofluorimetric analysis, in HT29 cells treated with Lactobacillus rhamnosus GG (LGG) supernatant (SN) or its control broth—1 : 30 titration for 3 h. A representative experiment of three independent experiments is shown. (B) Proresolving and proinflammatory autacoid (RvD1, LXB4, LXA4, PGE2) release in HT29 and HCT116 cells treated with LGG SN—1 : 30 titration for 12 h over control (broth). Baseline values of each mediator were in HCT116 cells: RvD1 122 ± 15 pg/106 cells, LXB4 38 ± 4.2 pg/106 cells, LXA4 501 ± 54 pg/106 cells and PGE2 101 ± 11 pg/106 cells. Baseline values of each mediator were in HT29 cells: RvD1 128 ± 27 pg/106 cells, LXB4 36 ± 5 pg/106 cells, LXA4 475 ± 52 pg/106 cells and PGE2 82 ± 9.8 pg/106 cells. Data are represented as mean ± SD of changes over baseline levels obtained in five independent experiments. *P < 0.05 compared with the control by Student's t test. (C) VEGF‐A release in HCT116 and HT29 cells treated with LGG SN—1 : 30 titration or the control culture broth for 12 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control by Student's t test. (D) Analysis of proteins in conditioned media (CM) from HT29 cells treated with LGG SN or its control broth (1 : 30 titration) using angiogenesis‐associated protein antibody arrays. The mean of protein pixel density for each angiogenesis‐related protein, normalized for the reference spots, was calculated and compared with the relative control. The array images and the relative quantitative profiles of protein levels are shown. (E) Human umbilical vein endothelial cells (HUVECs) were cultured in the presence of cell culture conditioned media (CM) from HCT116 or HT29 cells treated with LGG SN or the control culture broth (1 : 30 titration) (10× magnification) in a 24‐well plate. After 12 h, cells were fixed with ice‐cold 70% ethanol, and tubule formation was evaluated. Sample images and a quantification of the angiogenic response are reported. Scale bar 50 μm. Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the control (broth) by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
To confirm the antiangiogenic effect of LGG in CRC cells, we evaluated, by an ELISA assay, VEGF‐A release in HCT116 and HT29 cells treated for 12 h with LGG SN or broth as a control. We found that LGG SN was able to significantly reduce VEGF‐A release in both CRC cells compared with the control broth (Fig. 4C). Furthermore, we evaluated the expression of several other angiogenic proteins using dedicated antibody arrays incubated with CM from HT29 cells treated for 12 h with LGG SN or the relative control (broth) [39]. LGG SN (1 : 30 titration) treatment in HT29 cells downregulated, with changes superior to 10%, the levels of amphiregulin, artemin, coagulation factor III, DPP IV, EG‐VEGF, endothelin 1, GDNF, IGFBP‐1, IGFBP‐2, TGF‐beta1, MCP‐1, MMP8, MMP9, pentraxin 3, PD‐ECGF, PDGF‐AA, PDGF‐AB and BB, PlGF, Serpin E1, TIMP‐1, TIMP‐4, Thrombospondin 1, uPA and VEGF (Fig. 4D).
We then evaluated the ability of LGG SN to modulate CRC cell functional angiogenic potential. HUVECs plated on wells coated with Matrigel with the addition of CM from HCT116 or HT29 cells treated with control culture broth (1 : 30 titration) formed a characteristic capillary‐like network at 12 h. On the contrary, when the cells were plated on Matrigel with the addition of CM from HCT116 or HT29 treated with LGG SN (1 : 30 titration), a significantly lower number of tube structures were observed (Fig. 4E).
Finally, to verify whether the antiangiogenic potential of LGG SN depends on SPM activity, we used a GPR32 neutralizing antibody (Ab) that inhibits the activity of its ligand RvD1. For this purpose, HCT116 cells were stimulated with LGG SN in the presence or absence of the anti‐GPR32 (1 μg·mL−1) Ab, and their angiogenic potential was evaluated. As shown in Fig. S6, at the concentration used for this experiment, the neutralizing anti‐GPR32 Ab was able to significantly reduce the LGG‐mediated inhibition of the angiogenic potential of CRC cells. The anti‐GPR32 effect was partial, as expected, due to its ability to block only RvD1 effects and not that of other SPMs (Fig. S6).
These data suggest that LGG is able to induce a proresolving response and a following antiangiogenic effect in CRC cells.
3.5. The proresolving and antiangiogenic properties of LGG are not common to other commensal bacteria
In order to verify the specificity of action of LGG, we asked whether Escherichia coli (E. coli), as example of a commensal nonprobiotic strain [50], or Bifidobacterium bifidum (B. bifidum), as other lactic acid probiotic strain [51], could sustain a proresolving and antiangiogenic response in CRC cells.
To this aim, we treated HT29 and HCT116 cells with E. coli or B. bifidum SN (1 : 30 titration) and verified their effects on ALOX expression, RvD1 and VEGF‐A release. By flow cytometric analysis, we observed that neither E. coli nor B. bifidum SN were able to increase the level of ALOX5, ALOX15A and ALOX15B proteins in HCT116 cells, while LGG did it (Fig. 5A). Similar results were obtained also in HT29 cells (not shown). Consistently, LGG SN significantly induced RvD1 release, while E. coli and B. bifidum SN did not (Fig. 5B) both in HT29 and in HCT116 CRC cells. Finally, we verified the effects of E. coli or B. bifidum SN on VEGF‐A release. Neither E. coli nor B. bifidum SN were able to reduce VEGF‐A release from HT29 and HCT116 cells (Fig. 5C).
Fig. 5.
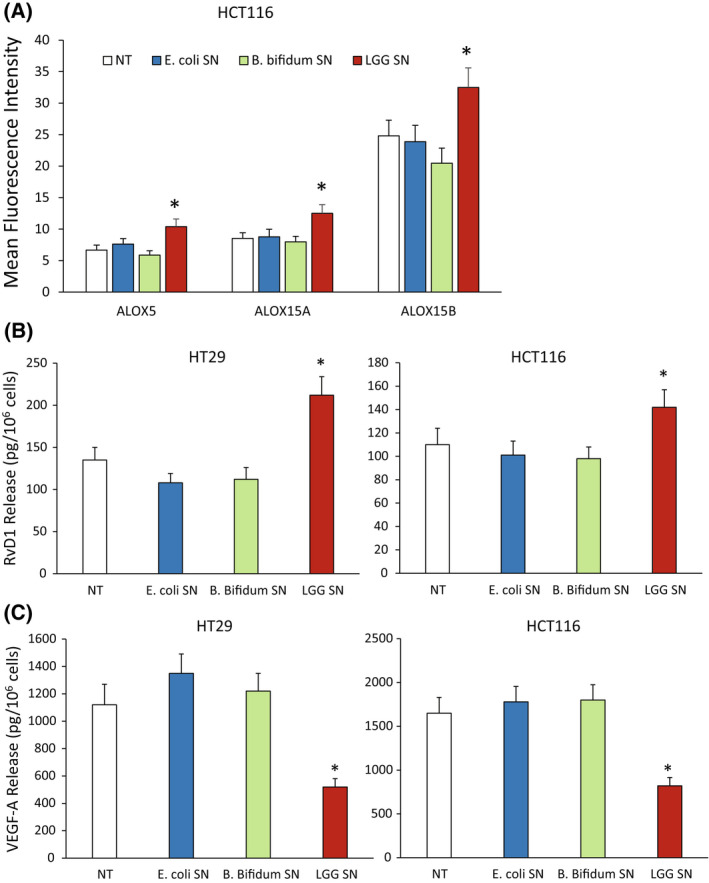
Effects of other commensal bacterial strain on specialized proresolving mediator (SPM) biosynthetic machinery and angiogenic potential in colorectal carcinoma (CRC) cells. (A) ALOX5, ALOX15A and ALOX15B protein expression levels (mean fluorescence intensity), assessed by cytofluorimetric analysis, in HCT116 cells treated or not (NT) with Escherichia coli (E. coli), Bifidobacterium bifidum (B. bifidum) and Lactobacillus rhamnosus GG (LGG) supernatant (SN) (1 : 30 titration) for 6 h. Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the relative control by Student's t test. (B) RvD1 release from HT29 and HCT116 cells treated or not (NT) for 12 h with E. coli, B. bifidum and LGG SN (1 : 30 titration). Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the control by Student's t test. (C) VEGF‐A release from HT29 and HCT116 cells treated or not (NT) for 12 h with E. coli, B. bifidum and LGG SN (1 : 30 titration). Data are represented as mean ± SD of three independent experiments. *P < 0.05 compared with the control by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
Altogether, these data support the evidence that the activation of a proresolving and antiangiogenic response in CRC cells is not general and common to all the commensal or to all the lactic acid bacteria.
3.6. Lactobacillus rhamnosus GG (LGG)‐mediated proresolving response requires FPR1
It has been reported that LGG depends on the expression of FPR1 for its activity in colon cells [32, 52]. To provide evidence that LGG‐mediated proresolving and antiangiogenic responses in CRC cells require FPR1, we treated HCT116 shFPR1 and shCTR with LGG SN or with the control broth and we evaluated the production of proresolving and proangiogenic factors.
LGG SN (1 : 30 titration—3 h) promoted a significant increase of ALOX5, ALOX15A and ALOX15B mRNA levels in shCTR but not in shFPR1 CRC cells (Fig. 6A). Similar results were obtained for the receptors GPR32, ChemR23 and BLT1 (Fig. 6A). Consistently, LGG SN induces the reduction of pro‐angiogenic mediators (VEGF‐A, ‐B, ‐C) only in shCTR but not in shFPR1 CRC cells (Fig. 6A).
Fig. 6.
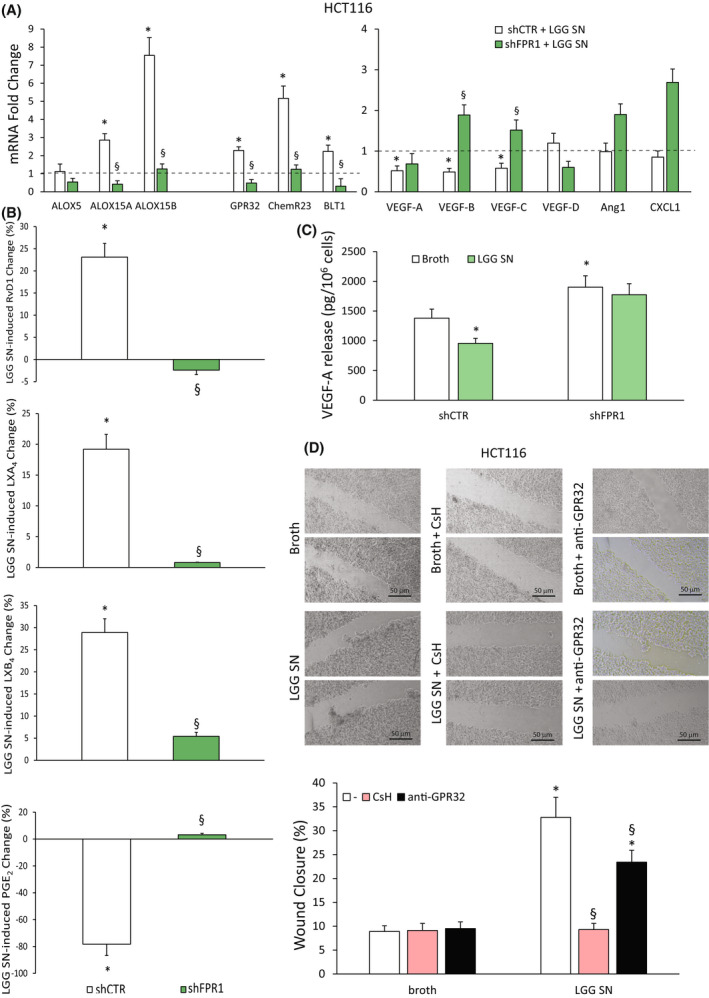
Dependence of Lactobacillus rhamnosus GG (LGG) supernatant (SN) effects on formyl peptide receptor 1 (FPR1) expression in colorectal carcinoma (CRC) cells. (A) ALOX5, ALOX15A, ALOX15B, GPR32, ChemR23, BLT1, VEGF‐A, VEGF‐B, VEGF‐C, VEGF‐D, Ang1 and CXCL1 mRNA fold change in HCT116 cells silenced for FPR1 (HCT116 shFPR1, three clones) or in control cells transfected with nontargeting short hairpin RNAs (shCTR cells, a mass population) upon treatment for 3 h with Lactobacillus rhamnosus GG (LGG) supernatant (SN)—1 : 30 titration. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the control (broth—dotted line) by Student's t test, § P < 0.05 compared with the relative control by Student's t test. (B) Proresolving and proinflammatory autacoid (RvD1, LXA4, LXB4, PGE2) release over control in HCT116 shCTR (a mass population) or shFPR1 (three clones) upon treatment for 12 h with LGG SN—1 : 30 titration. Baseline values of each mediator were in HCT116 shCTR cells: RvD1 128 ± 18 pg/106 cells, LXB4 41 ± 5 pg/106 cells, LXA4 492 ± 51 pg/106 cells and PGE2 98 ± 13 pg/106 cells. Baseline values of each mediator were in HCT116 shFPR1 cells: RvD1 68 ± 7 pg/106 cells, LXB4 18 ± 3 pg/106 cells, LXA4 346 ± 42 pg/106 cells and PGE2 214 ± 28 pg/106 cells. Data are represented as mean ± SD of changes over baseline levels obtained in five independent experiments. *P < 0.05 compared with the broth control by Student's t test, § P < 0.05 compared with the relative control by Student's t test. (C) VEGF‐A release in HCT116 shCTR (a mass population) or shFPR1 (three clones) cells treated with LGG SN—1 : 30 titration or the culture broth for 12 h. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with shCTR broth by Student's t test. (D) Wound‐healing assay of HCT116 cells in the presence of LGG SN (1 : 30 titration) or the same dilution of culture broth for 12 h. Cells were pretreated or not for 30 min with CsH (800 nm) or a neutralizing anti‐GPR32 antibody (1 μg·mL−1). Representative photograms and a quantitative evaluation of the wound closure are shown. Scale bar 50 μm. Values represent the average of triplicate experiments ± SD. *P < 0.05 compared with broth alone by Student's t test. § P < 0.05 compared with the relative control by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
To determine whether LGG depends on FPR1 also for its induction of SPM release, we treated shCTR and shFPR1 HCT116 cells with LGG SN for 12 h. We found that HCT116 shFPR1 treated with LGG SN did not display the increase in RvD1, LXA4 and LXB4 release observed in control cells (Fig. 6B). As expected, LGG SN caused a significant reduction in PGE2 release in shCTR but not in shFPR1 HCT116 cells (Fig. 6B).
To confirm that also the antiangiogenic effect of LGG in CRC cells depends on FPR1, we evaluated the VEGF‐A release in HCT116 shFPR1 and shCTR cells in the presence or absence of LGG SN after 12 h. As shown in Fig. 6C, VEGF‐A release was higher in HCT116 shFPR1 than in controls; LGG SN treatment was able to significantly reduce VEGF‐A release in shCTR but not in shFPR1 HCT116 cells (Fig. 6C).
Taken together, these data demonstrate that the proresolving and antiangiogenic activities of LGG require FPR1.
It has been described that one of the most important homeostatic functions of LGG is mediated by its ability to sustain restitution of injured intestinal epithelial monolayers by interacting with FPR1 [31, 52]. Here, we verified whether the ability of LGG to induce CRC epithelial cell restitution through the activation of FPR1 is dependent, at least in part, on the activation of proresolving pathways. To this aim, we performed a wound‐healing assay on HCT116 in the presence or absence of LGG SN, CsH (800 nm) or a neutralizing anti‐GPR32 Ab (1 μg·mL−1). As shown in Fig. 6D, LGG SN elicits a significant movement of HCT116 cells following injury, which was completely abolished by CsH, confirming the dependence of LGG effects on FPR1 (Fig. 6D). Interestingly, the neutralizing antibody against GPR2 was able to significantly reduce (about 30%) the LGG SN‐induced migration, suggesting that the effects of LGG SN on FPR1 in terms of wound closure imply, at least in part, the production of proresolving mediators (i.e. RvD1).
In other experimental models, the activation of a proresolving response goes through the induction, not only of lipidic SPMs but also of proresolving mediators of a different type (e.g. AnxA1) [53]. In order to verify the possibility that LGG and FPR1 could induce AnxA1 expression in CRC cells, we treated both HT29 and HCT116 cells with LGG SN (1 : 30 titration), fMLF (10−9 m) or the three SPMs [RvD1 (1 nm), LXB4 (1 nm) and LXA4 (1 nm)] for 12 h and verified the expression levels of AnxA1, a potent endogenous proresolving and immunomodulatory protein [5].
FACS analysis shows that both fMLF and LGG SN induced an increase in the protein expression of AnxA1 in CRC cells (Fig. S7). Similarly, the treatment with RvD1, LXB4 and LXA4 increased AnxA1 protein expression levels in CRC cells (Fig. S7). These observations suggest that FPR1 activation sustains a proresolving response that includes AnxA1 and that SPMs could further stimulate AnxA1 expression in a feed‐forward loop [53].
These results confirm previous observations indicating that LGG mediates a homeostatic function in colonic mucosa requiring FPR1 [31, 32]. Moreover, our data indicate that this FPR1 function is maintained also in CRC cells, and is dependent on its ability to sustain a proresolving response.
3.7. FPR1 activation of proresolving program requires MAPK activation
We then explored the signalling pathways, which are involved in the proresolving response of CRC cells to FPR1 activation mediated by LGG or fMLF.
To this aim, we treated HCT116 cells with fMLF (10−9 m) or LGG SN (1 : 30 titration) and the relative controls (not‐treated or broth alone, respectively) and harvested at different time points. fMLF has been demonstrated to classically activate the ERK, PI3K/Akt and STAT3 pathways [54, 55]; thus, we verified the expression levels of phospho‐MAPK, phospho‐Akt, and phospho‐STAT3 in order to evaluate the activated forms of these proteins.
As shown in Fig. 7A, in HCT116 cells a significant activation of MAPK and STAT3 was observed in response to fMLF, while no significant activation levels of Akt were detected. LGG SN, when compared to the broth alone at the same titration (1 : 30), activated MAPK but not STAT3 signalling (Fig. 7A). Thus, the common proresolving and antiangiogenic properties of fMLF and LGG SN could be due to MAPK activation.
Fig. 7.
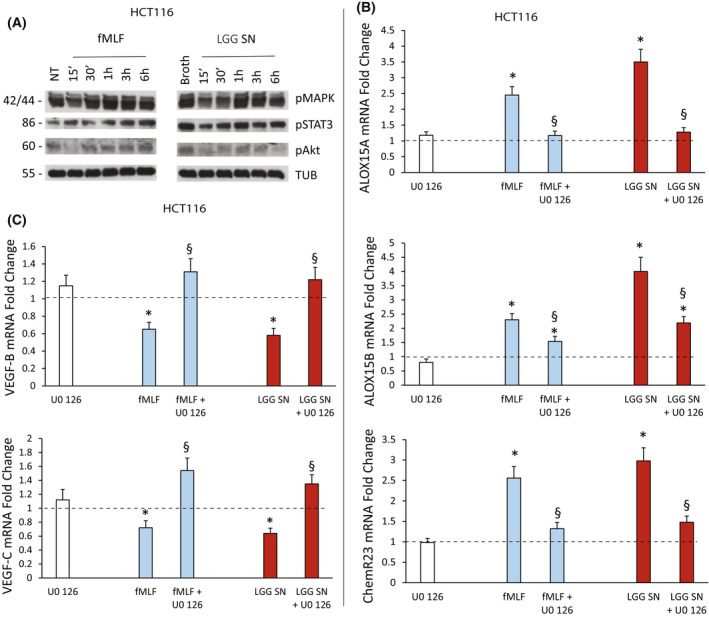
Signaling pathway activation upon fMLF or Lactobacillus rhamnosus GG (LGG) supernatant (SN) treatment of colorectal carcinoma (CRC) cells. (A) Activation of downstream signaling pathways in HCT116 cells induced by fMLF (10−9 m) or Lactobacillus rhamnosus GG (LGG) supernatant (SN) (1 : 30 titration) or the relative controls [not‐treated (NT) or broth alone, respectively]. Total cell lysates were prepared at various time points after stimulation in serum‐free medium. Immunoblots were probed with the indicated phosphospecific Abs. Antitubulin was used for normalization. The figure shows the results of a representative experiment from among three different preparations. (B) ALOX15A, ALOX15B and ChemR23 mRNA fold change in HCT116 cells treated with fMLF (10−9 m) or LGG SN (1 : 30 titration) for 3 h following or not cell preincubation with U0 126 (10 μm) for 30 min. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the negative control (dotted line) by Student's t test; § P < 0.05 compared with the relative control treatment by Student's t test. (C) VEGF‐B and VEGF‐C mRNA fold change in HCT116 cells treated with fMLF (10−9 m) or LGG SN (1 : 30 titration) for 3 h following or not cell preincubation with U0 126 (10 μm) for 30 min. Data are represented as mean ± SD of five independent experiments. *P < 0.05 compared with the negative control (dotted line) by Student's t test; § P < 0.05 compared with the relative control treatment by Student's t test. [Colour figure can be viewed at wileyonlinelibrary.com]
To verify this hypothesis, we treated HCT116 cells with fMLF (10−9 m) or LGG SN (1 : 30 titration) and the relative controls (not‐treated or broth alone, respectively) in the presence or absence of U0 126, a selective inhibitor of mitogen‐activated protein kinase kinase [56]. Figure 7B shows that fMLF and LGG SN significantly upregulated ALOX15A, ALOX15B and ChemR23 expression levels and that the preincubation of cells with U0 126 partially or completely reverted these effects. Consistently, VEGF‐B and VEGF‐C were significantly reduced by fMLF and LGG SN treatments and these effects were not detectable in cells pretreated with U0 126 (Fig. 7C).
These experiments demonstrated that the proresolving and antiangiogenic responses of fMLF and LGG require MAPK signalling activation.
4. Discussion
The homeostasis of intestinal mucosa is tightly regulated by mechanisms able to perceive bacterial species distinguishing between pathogenic and commensal ones, triggering an inflammatory antibacterial and a tolerogenic proresolving response, respectively [1, 57]. Formyl peptide receptors (FPRs), a family of pattern recognition receptors, can recognize several bacterial products and trigger either inflammation or its resolution [13, 14], being optimal candidates to the role of central regulators of intestinal mucosa homeostasis [14, 32, 58].
It has been reported that intestinal epithelial cells sense commensal bacteria using various receptors, including FPRs; as a consequence of this, an increase in barrier function, and improved resolution of epithelial wounds are observed [31, 52, 58, 59]. For instance, it has been demonstrated that the commensal bacteria Lactobacillus rhamnosus GG (LGG), by activating FPR1, influence intestinal epithelial homeostatic signalling and sustain epithelial cell motility enhancing wound restitution [31, 52]. Furthermore, several studies point to an important role of intestinal microbiota, not only in the physiology of intestinal mucosa but also in eliciting a protective antitumour response by both acting directly on cancer cells and modulating the immune response to them [26, 28, 30, 60].
We recently showed that the genetic ablation of FPR1 caused an increase in proinflammatory, angiogenic and tumorigenic potential in gastric cancer (GC) cells [10, 11]. We further showed that these functions of FPR1 are mediated by its ability to sustain the expression and function of proresolving pathways [11]. We also demonstrated that Toll‐like receptor 7 (TLR7) displays similar proresolving and antiangiogenic properties in non‐small‐cell lung cancer cells [39]. Here, we demonstrated, for the first time, that FPR1 exerts a similar function also in the intestinal mucosa. Taken together, our results sustain the hypothesis that specific PRRs could exert homeostatic and proresolving functions in different tissues, which need to balance injuries and inflammatory insults.
More in detail, we showed that FPR1 activation mediated by formyl peptides, which are the main natural ligands to FPR1 [13], or by LGG SN, induces the expression of enzymes and receptors involved in proresolving responses, and the release of a significantly higher amount of SPMs (RvD1, LXB4, LXA4) at the expense of proinflammatory lipid mediators (PGE2). Although we focused our attention specifically on lipidic proresolving autacoids, the proresolving response in CRC cells mediated by FPR1 could be more general, since we also verified that fMLF and LGG are able to induce the expression in CRC cells of AnxA1, a proteic proresolving mediator [53].
The physiology of this type of response is of paramount importance, if one takes into account that SPMs act in vitro at an optimal concentration in the low nanomolar range. Indeed, we observed that the antiangiogenic properties of SPMs are detectable at 0.5–1 nm concentration. By means of EIA, we detected basal concentration of RvD1 released by 106 of two distinct CRC cells around 300 pm; LXB4 was constitutionally released at ∼ 80 pm, while LXA4 was around 1.5 nm. The stimulation of CRC cells with LGG increased RvD1 release of around 40%, LXB4 of 20% and LXA4 of 30%, thus allowing to reach in cell culture media, and presumably in the gut, optimal and active concentrations of each SPM. Furthermore, it should be considered that while we identified 1 nm as the optimal concentration of each SPM alone to inhibit angiogenesis, in the gut the interaction of LGG product(s) with intestinal epithelial cells allows the contemporary production of several SPMs, which could act in synergy.
Although we have still not identified the bacterial products secreted in the LGG SN and responsible for FPR1 stimulation, it is likely that LGG‐derived formylated peptides are the mediators of the observed effects. However, we cannot exclude the presence in LGG SN of other FPR1 agonist(s). Whatever the case, the activity of secreted FPR1 ligands in LGG SN suggests that, in the physiology of the gut, LGG could exert its homeostatic/protective effects on intestinal mucosa by releasing several factors in the extracellular space.
Consistently with previous observations demonstrating that LGG sustains intestinal epithelial restitution via an FPR1‐mediated ERK activation [31, 52], we observed that LGG SN and fMLF shared the ability to activate MAPK signalling in CRC cells. We also verified that the proresolving response activated by LGG in CRC cells is dependent on the activation of this signalling.
In the GC model, the levels of SPMs inversely correlated with the number of proangiogenic mediators produced by cancer cells [11]. Consistently, also in the colorectal carcinoma (CRC) model, SPM production inversely correlated with the angiogenic potential of CRC cells. In particular, we observed that FPR1 activation mediated by fMLF or LGG SN significantly reduced the production of several angiogenic mediators in CRC cells. Furthermore, our experiments also demonstrated that the increased angiogenic potential of CRC cells lacking FPR1 is due to the deficit of RvD1 and/or LXB4 production. On the contrary, LXA4, although modulated by FPR1, did not exert an antiangiogenic response in CRC cells, at least in our experimental models.
The different effects observed between RvD1/LXB4 and LXA4 suggest that probably in dependence of the mediators and on the tissue district, each SPM could exert a typical or at least predominant activity among that ascribed to SPMs (e.g. control of inflammation, limiting tissue damage, promoting resolution, attenuating fibrosis and inhibiting angiogenesis) [61].
In a mouse hepatocarcinoma cell line [62] and in a model of inflammation‐induced pathological neovascularization of the cornea [63], LXA4 has been reported as antiangiogenic. However, in the colon experimental model, it has been described to date that LXA4 could both protect against acute injury [46] and suppress CRC development [23] by specifically regulating intestinal mucosa inflammation: LXA4 inhibits inflammatory mediator expressions [23, 46], and reduced proinflammatory monocyte and neutrophil infiltration in tumours favouring lymphocyte activation [23, 46].
Obviously, these findings deserve a more in‐depth study in order to identify the possible different mechanisms of action (e.g. signalling pathways, receptor expression levels, cell metabolic asset) justifying the antiangiogenic properties of LXA4 in other tumours and not in CRC model and the differences in action compared with RvD1 and LXB4.
We demonstrated that the proresolving and antiangiogenic program in CRC cells could be induced by supernatants obtained by LGG cultures. Our data reinforce the idea that probiotic species contribute to an enhanced repair of mucosal wounds [32, 33] and to a protective antitumour response, which imply not only the already demonstrated antiproliferative effect [35] but also, as here presented, an antiangiogenic response on cancer cells.
These speculations are corroborated by the evidence that the properties observed for LGG are not shared with other commensal nonprobiotic bacteria, as demonstrated by our experiments on CRC cells treated with E. coli and by other evidence in the literature [32, 33, 64]. Furthermore, we did not observe the same effects of LGG on CRC cells neither when we used a different lactic acid probiotic strain, as B. bifidum. We are aware that we did not identify a specific factor/protein produced by LGG and activating FPR1; however, our experiments with E. coli and B. bifidum and previous evidence suggest that the proresolving and homeostatic functions can be ascribed only to some bacterial strains [64].
Several experimental observations in the literature suggest that SPMs or diet supplement of their precursors (ω3/6 PUFA) could integrate CRC treatment because of their ability to counteract intestinal carcinogenesis [15]. In addition, specific commensal bacteria have been identified and described as able to limit colon tumorigenesis by acting on cancer cells or on the protumorigenic inflammatory microenvironment [65]. However, to date no connection between commensal microbiota, a pattern recognition receptor sensing specific probionts and the activation of proresolving pathway has been made. We provide direct evidence of such concept, by showing that LGG can activate FPR1 sustaining the expression and function of proresolving pathways, which, in turn, suppress angiogenesis.
5. Conclusions
Our results consolidate the hypothesis that a correlation exists linking proresolving pathways' deficit and cancer in humans. Our data also highlight the possibility that innate immune receptors, including FPR1, could be the key regulators of the balance between microbiota recognition, inflammation regulation and neoplastic transformation. By defining the molecular mechanisms linking lipid metabolism and inflammation resolution with FPR1 in gastrointestinal (GI) tract, our data will allow the comprehension of the general mechanisms involved in tumour cell growth following their angiogenic switch and will open the possibility to identify new prognostic markers and a novel therapeutic approach for cancers of the GI tract.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
FL participated in the design of the study, carried out experiments, drafted the manuscript, and read and approved this manuscript. MM, DS, CP, VC, GM, ES and PS carried out experiments, and read and approved this manuscript. RMM conceptualized and designed the study, analysed and interpreted the data, wrote the manuscript, and gave final approval of the manuscript and financial support. NP conceptualized and designed the study, carried out experiments, analysed and interpreted the data, wrote the manuscript and gave final approval of the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/1878‐0261.13280.
Supporting information
Fig. S1. Formyl peptide receptor 1 (FPR1) expression levels in colorectal carcinoma (CRC) cells.
Fig. S2. Formyl peptide receptor 1 (FPR1) correlation to colorectal carcinoma (CRC) patients' characteristics.
Fig. S3. Absence of antiangiogenic effect of LXA4 in colorectal carcinoma (CRC) cells.
Fig. S4. Antiangiogenic and proresolving effects of RvD1 and LXB4 in colorectal carcinoma (CRC) cells.
Fig. S5. Effects of Lactobacillus rhamnosus GG (LGG) supernatants LGG (SN) on specialized proresolving mediator (SPM) biosynthetic machinery and angiogenic potential of colorectal carcinoma (CRC) cells.
Fig. S6. Antiangiogenic effects of Lactobacillus rhamnosus GG (LGG) supernatants (SN) in the presence of a neutralizing anti‐GPR32 antibody.
Fig. S7. Annexin A1 (AnxA1) induction upon formyl peptide receptor 1 (FPR1) activation or specialized proresolving mediator (SPM) stimulation of colorectal carcinoma (CRC) cells.
Table S1. List of primers.
Data S1. Supplementary legends.
Acknowledgements
We are grateful to Rosaria Catalano and Mariarosaria Montagna for technical help. This work was supported by PRIN 2017 No. 2017XJ38A4; Istituto Superiore di Oncologia grant (MIUR PON01_02782/12); POR Campania FESR 2014–2020 “SATIN” grant; POR Campania FESR 2014–2020 “RARE.PLAT.NET” grant; “POR Campania FESR 2014‐2020” grant; and PRIN MIUR 2017, grant number 2017SFBFER.
Contributor Information
Rosa Marina Melillo, Email: rosmelil@unina.it.
Nella Prevete, Email: nella.prevete@unina.it.
Data accessibility
All data generated or analysed during this study are included within the article or available from the corresponding author on reasonable request.
References
- 1. Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33:954–64. [DOI] [PubMed] [Google Scholar]
- 2. Jess T, Rungoe C, Peyrin‐Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta‐analysis of population‐based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639–45. [DOI] [PubMed] [Google Scholar]
- 3. Sugimoto MA, Sousa LP, Pinho V, Perretti M, Teixeira MM. Resolution of inflammation: what controls its onset? Front Immunol. 2016;7:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Panigrahy D, Gilligan MM, Serhan CN, Kashfi K. Resolution of inflammation: an organizing principle in biology and medicine. Pharmacol Ther. 2021;227:107879. [DOI] [PubMed] [Google Scholar]
- 5. Perretti M, D'Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat Rev Immunol. 2009;9:62–70. [DOI] [PubMed] [Google Scholar]
- 6. Hasko G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirakaj V, Dalli J, Granja T, Rosenberger P, Serhan CN. Vagus nerve controls resolution and pro‐resolving mediators of inflammation. J Exp Med. 2014;211:1037–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Serhan CN. Pro‐resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prevete N, Liotti F, Amoresano A, Pucci P, de Paulis A, Melillo RM. New perspectives in cancer: modulation of lipid metabolism and inflammation resolution. Pharmacol Res. 2018;128:80–7. [DOI] [PubMed] [Google Scholar]
- 10. Prevete N, Liotti F, Illiano A, Amoresano A, Pucci P, de Paulis A, et al. Formyl peptide receptor 1 suppresses gastric cancer angiogenesis and growth by exploiting inflammation resolution pathways. Onco Targets Ther. 2017;6:e1293213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prevete N, Liotti F, Visciano C, Marone G, Melillo RM, de Paulis A. The formyl peptide receptor 1 exerts a tumor suppressor function in human gastric cancer by inhibiting angiogenesis. Oncogene. 2015;34:3826–38. [DOI] [PubMed] [Google Scholar]
- 12. Shureiqi I, Chen D, Day RS, Zuo X, Hochman FL, Ross WA, et al. Profiling lipoxygenase metabolism in specific steps of colorectal tumorigenesis. Cancer Prev Res (Phila). 2010;3:829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye RD, Boulay F, Wang JM, Dahlgren C, Gerard C, Parmentier M, et al. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol Rev. 2009;61:119–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prevete N, Liotti F, Marone G, Melillo RM, de Paulis A. Formyl peptide receptors at the interface of inflammation, angiogenesis and tumor growth. Pharmacol Res. 2015;102:184–91. [DOI] [PubMed] [Google Scholar]
- 15. Ungaro F, D'Alessio S, Danese S. The role of pro‐resolving lipid mediators in colorectal cancer‐associated inflammation: implications for therapeutic strategies. Cancers (Basel). 2020;12:2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cockbain AJ, Toogood GJ, Hull MA. Omega‐3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut. 2012;61:135–49. [DOI] [PubMed] [Google Scholar]
- 17. Moro K, Nagahashi M, Ramanathan R, Takabe K, Wakai T. Resolvins and omega three polyunsaturated fatty acids: clinical implications in inflammatory diseases and cancer. World J Clin Cases. 2016;4:155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Il Lee S, Zuo X, Shureiqi I. 15‐Lipoxygenase‐1 as a tumor suppressor gene in colon cancer: is the verdict in? Cancer Metastasis Rev. 2011;30:481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sulciner ML, Serhan CN, Gilligan MM, Mudge DK, Chang J, Gartung A, et al. Resolvins suppress tumor growth and enhance cancer therapy. J Exp Med. 2018;215:115–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C, Yu H, Ni X, Shen S, Das UN. Growth inhibitory effect of polyunsaturated fatty acids (PUFAs) on colon cancer cells via their growth inhibitory metabolites and fatty acid composition changes. PLoS One. 2015;10:e0123256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong X, Lee HN, Surh YJ. RvD1 inhibits TNFalpha‐induced c‐Myc expression in normal intestinal epithelial cells and destabilizes hyper‐expressed c‐Myc in colon cancer cells. Biochem Biophys Res Commun. 2018;496:316–23. [DOI] [PubMed] [Google Scholar]
- 22. Wang Z, Cheng Q, Tang K, Sun Y, Zhang K, Zhang Y, et al. Lipid mediator lipoxin A4 inhibits tumor growth by targeting IL‐10‐producing regulatory B (Breg) cells. Cancer Lett. 2015;364:118–24. [DOI] [PubMed] [Google Scholar]
- 23. Liu H, Zeng J, Huang W, Xu Q, Ye D, Sun R, et al. Colorectal cancer is associated with a deficiency of lipoxin A4, an endogenous anti‐inflammatory mediator. J Cancer. 2019;10:4719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong T, Dave P, Yoo E, Ebright B, Ahluwalia K, Zhou E, et al. NAP1051, a lipoxin A4 biomimetic analogue, demonstrates antitumor activity against the tumor microenvironment. Mol Cancer Ther. 2021;20:2384–97. [DOI] [PubMed] [Google Scholar]
- 25. Walsh CJ, Guinane CM, O'Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett. 2014;588:4120–30. [DOI] [PubMed] [Google Scholar]
- 26. Falony G, Joossens M, Vieira‐Silva S, Wang J, Darzi Y, Faust K, et al. Population‐level analysis of gut microbiome variation. Science. 2016;352:560–4. [DOI] [PubMed] [Google Scholar]
- 27. Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23:2061–70. [DOI] [PubMed] [Google Scholar]
- 28. Mori G, Rampelli S, Orena BS, Rengucci C, De Maio G, Barbieri G, et al. Shifts of Faecal microbiota during sporadic colorectal carcinogenesis. Sci Rep. 2018;8:10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Segers ME, Lebeer S. Towards a better understanding of lactobacillus rhamnosus GG ‐ host interactions. Microb Cell Fact. 2014;13(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Banna GL, Torino F, Marletta F, Santagati M, Salemi R, Cannarozzo E, et al. Lactobacillus rhamnosus GG: an overview to explore the rationale of its use in cancer. Front Pharmacol. 2017;8:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Swanson PA 2nd, Kumar A, Samarin S, Vijay‐Kumar M, Kundu K, Murthy N, et al. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species‐mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci USA. 2011;108:8803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alam A, Leoni G, Wentworth CC, Kwal JM, Wu H, Ardita CS, et al. Redox signaling regulates commensal‐mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2014;7:645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, et al. The microenvironment of injured murine gut elicits a local pro‐restitutive microbiota. Nat Microbiol. 2016;1:15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao J, Li Y, Wan Y, Hu T, Liu L, Yang S, et al. A novel postbiotic from lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front Microbiol. 2019;10:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orlando A, Linsalata M, Russo F. Antiproliferative effects on colon adenocarcinoma cells induced by co‐administration of vitamin K1 and lactobacillus rhamnosus GG. Int J Oncol. 2016;48:2629–38. [DOI] [PubMed] [Google Scholar]
- 36. Escamilla J, Lane MA, Maitin V. Cell‐free supernatants from probiotic lactobacillus casei and lactobacillus rhamnosus GG decrease colon cancer cell invasion in vitro. Nutr Cancer. 2012;64:871–8. [DOI] [PubMed] [Google Scholar]
- 37. Cai S, Kandasamy M, Rahmat JN, Tham SM, Bay BH, Lee YK, et al. Lactobacillus rhamnosus GG activation of dendritic cells and neutrophils depends on the dose and time of exposure. J Immunol Res. 2016;2016:7402760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clerici SP, Peppelenbosch M, Fuhler G, Consonni SR, Ferreira‐Halder CV. Colorectal cancer cell‐derived small extracellular vesicles educate human fibroblasts to stimulate migratory capacity. Front Cell Dev Biol. 2021;9:696373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liotti F, Marotta M, Sorriento D, Pone E, Morra F, Melillo RM, et al. Toll‐like receptor 7 mediates inflammation resolution and inhibition of angiogenesis in non‐small cell lung cancer. Cancers (Basel). 2021;13:740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Collina F, La Sala L, Liotti F, Prevete N, La Mantia E, Chiofalo MG, et al. AXL is a novel predictive factor and therapeutic target for radioactive iodine refractory thyroid cancer. Cancers (Basel). 2019;11:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liotti F, Kumar N, Prevete N, Marotta M, Sorriento D, Ierano C, et al. PD‐1 blockade delays tumor growth by inhibiting an intrinsic SHP2/Ras/MAPK signalling in thyroid cancer cells. J Exp Clin Cancer Res. 2021;40:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liotti F, Collina F, Pone E, La Sala L, Franco R, Prevete N, et al. Interleukin‐8, but not the related chemokine CXCL1, sustains an autocrine circuit necessary for the properties and functions of thyroid cancer stem cells. Stem Cells. 2017;35:135–46. [DOI] [PubMed] [Google Scholar]
- 43. Gambardella J, De Rosa M, Sorriento D, Prevete N, Fiordelisi A, Ciccarelli M, et al. Parathyroid hormone causes endothelial dysfunction by inducing mitochondrial ROS and Specific oxidative signal transduction modifications. Oxid Med Cell Longev. 2018;2018:9582319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liotti F, De Pizzol M, Allegretti M, Prevete N, Melillo RM. Multiple anti‐tumor effects of Reparixin on thyroid cancer. Oncotarget. 2017;8:35946–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang D, DuBois RN. An inflammatory mediator, prostaglandin E2, in colorectal cancer. Cancer J. 2013;19:502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gobbetti T, Ducheix S, le Faouder P, Perez T, Riols F, Boue J, et al. Protective effects of n‐6 fatty acids‐enriched diet on intestinal ischaemia/reperfusion injury involve lipoxin A4 and its receptor. Br J Pharmacol. 2015;172:910–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Senger DR, Davis GE. Angiogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramos S, Silva V, Dapkevicius MLE, Canica M, Tejedor‐Junco MT, Igrejas G, et al. Escherichia coli as commensal and pathogenic bacteria among food‐producing animals: health implications of extended Spectrum beta‐lactamase (ESBL) production. Animals (Basel). 2020;10:2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol. 2014;5:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS. Enteric commensal bacteria induce extracellular signal‐regulated kinase pathway signaling via formyl peptide receptor‐dependent redox modulation of dual specific phosphatase 3. J Biol Chem. 2011;286:38448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Borgeson E, Johnson AM, Lee YS, Till A, Syed GH, Ali‐Shah ST, et al. Lipoxin A4 attenuates obesity‐induced adipose inflammation and associated liver and kidney disease. Cell Metab. 2015;22:125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Paulis A, Prevete N, Rossi FW, Rivellese F, Salerno F, Delfino G, et al. Helicobacter pylori hp(2‐20) promotes migration and proliferation of gastric epithelial cells by interacting with formyl peptide receptors in vitro and accelerates gastric mucosal healing in vivo. J Immunol. 2009;183:3761–9. [DOI] [PubMed] [Google Scholar]
- 55. Zhou Y, Bian X, Le Y, Gong W, Hu J, Zhang X, et al. Formylpeptide receptor FPR and the rapid growth of malignant human gliomas. J Natl Cancer Inst. 2005;97:823–35. [DOI] [PubMed] [Google Scholar]
- 56. Smutny T, Bitman M, Urban M, Dubecka M, Vrzal R, Dvorak Z, et al. U0126, a mitogen‐activated protein kinase kinase 1 and 2 (MEK1 and 2) inhibitor, selectively up‐regulates main isoforms of CYP3A subfamily via a pregnane X receptor (PXR) in HepG2 cells. Arch Toxicol. 2014;88:2243–59. [DOI] [PubMed] [Google Scholar]
- 57. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prevete N, de Paulis A, Sgambato D, Melillo RM, D Argenio G, Romano L, et al. Role of formyl peptide receptors in gastrointestinal healing. Curr Pharm Des. 2018;24:1966–71. [DOI] [PubMed] [Google Scholar]
- 59. Lam EK, Yu L, Wong HP, Wu WK, Shin VY, Tai EK, et al. Probiotic Lactobacillus rhamnosus GG enhances gastric ulcer healing in rats. Eur J Pharmacol. 2007;565:171–9. [DOI] [PubMed] [Google Scholar]
- 60. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17:271–85. [DOI] [PubMed] [Google Scholar]
- 61. Basil MC, Levy BD. Specialized pro‐resolving mediators: endogenous regulators of infection and inflammation. Nat Rev Immunol. 2016;16:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Y, Hao H, He S, Cai L, Li Y, Hu S, et al. Lipoxin A4 and its analogue suppress the tumor growth of transplanted H22 in mice: the role of antiangiogenesis. Mol Cancer Ther. 2010;9:2164–74. [DOI] [PubMed] [Google Scholar]
- 63. Leedom AJ, Sullivan AB, Dong B, Lau D, Gronert K. Endogenous LXA4 circuits are determinants of pathological angiogenesis in response to chronic injury. Am J Pathol. 2010;176:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015;81:3655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Drago L. Probiotics and colon cancer. Microorganisms. 2019;7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Formyl peptide receptor 1 (FPR1) expression levels in colorectal carcinoma (CRC) cells.
Fig. S2. Formyl peptide receptor 1 (FPR1) correlation to colorectal carcinoma (CRC) patients' characteristics.
Fig. S3. Absence of antiangiogenic effect of LXA4 in colorectal carcinoma (CRC) cells.
Fig. S4. Antiangiogenic and proresolving effects of RvD1 and LXB4 in colorectal carcinoma (CRC) cells.
Fig. S5. Effects of Lactobacillus rhamnosus GG (LGG) supernatants LGG (SN) on specialized proresolving mediator (SPM) biosynthetic machinery and angiogenic potential of colorectal carcinoma (CRC) cells.
Fig. S6. Antiangiogenic effects of Lactobacillus rhamnosus GG (LGG) supernatants (SN) in the presence of a neutralizing anti‐GPR32 antibody.
Fig. S7. Annexin A1 (AnxA1) induction upon formyl peptide receptor 1 (FPR1) activation or specialized proresolving mediator (SPM) stimulation of colorectal carcinoma (CRC) cells.
Table S1. List of primers.
Data S1. Supplementary legends.
Data Availability Statement
All data generated or analysed during this study are included within the article or available from the corresponding author on reasonable request.


