Abstract
In this work, the LysR-type protein XapR has been subjected to a mutational analysis. XapR regulates the expression of xanthosine phosphorylase (XapA), a purine nucleoside phosphorylase in Escherichia coli. In the wild type, full expression of XapA requires both a functional XapR protein and the inducer xanthosine. Here we show that deoxyinosine can also function as an inducer in the wild type, although not to the same extent as xanthosine. We have isolated and characterized in detail the mutants that can be induced by other nucleosides as well as xanthosine. Sequencing of the mutants has revealed that two regions in XapR are important for correct interactions between the inducer and XapR. One region is defined by amino acids 104 and 132, and the other region, containing most of the isolated mutations, is found between amino acids 203 and 210. These regions, when modelled into the three-dimensional structure of CysB from Klebsiella aerogenes, are placed close together and are most probably directly involved in binding the inducer xanthosine.
Escherichia coli possesses the ability to take up purine and pyrimidine nucleosides from the growth medium and use them as sources of nitrogen and carbon. Nucleoside phosphorylases catalyze the phosphorolytic cleavage of the nucleoside, thereby forming the free nucleotide base and (deoxy)ribose-1-phosphate (18, 26). The base can be utilized by the purine or the pyrimidine salvage pathway, and the ribose-1-phosphate and the deoxyribose-1-phosphate can be converted to intermediates of the pentose phosphate shunt and of glycolysis, respectively. Of the four different nucleoside phosphorylases in E. coli, uridine phosphorylase (udp) and thymidine phosphorylase (deoA) are specific for pyrimidine nucleosides (15, 23) whereas purine nucleoside phosphorylase (deoD) and xanthosine phosphorylase (xapA) are specific for purine nucleosides (8, 11, 12).
Purine nucleoside phosphorylase is important for the breakdown of all purine nucleosides and deoxynucleosides except xanthosine (11–13). Xanthosine phosphorylase (XapA), on the other hand, has specificity toward xanthosine and all other purine nucleosides and deoxynucleosides except adenosine and deoxyadenosine (3, 8). Thus, the substrate specificity of XapA resembles the specificity of the mammalian purine nucleoside phosphorylase more than it resembles the specificity of DeoD (10, 16). This is also reflected in the amino acid sequence of XapA which is more similar to the sequence of the mammalian purine nucleoside phosphorylase than to the DeoD sequence (24).
Purine nucleoside phosphorylase (DeoD) is encoded by the last gene of the deoCABD operon. The regulation of these genes is complex and involves two repressors (CytR and DeoR) and an activator (cyclic AMP [cAMP] receptor protein-cAMP complex) (reviewed in reference 7). Despite the action of two repressors, the deo genes are always expressed at a low basal level to ensure a rapid metabolism of purine nucleosides taken up from the medium. In contrast, XapA is expressed only if the inducer xanthosine is present in the growth medium. The xanthosine-induced activation of xapA expression is mediated by the regulatory protein XapR (3, 24). Just downstream of xapA lies another gene, xapB, which encodes a membrane protein, XapB, very similar to the nucleoside transporter NupG; XapB is most probably a nucleoside transporter. Downstream of xapAB is the xapR gene, transcribed constitutively from two promoters. Both XapB and XapR are necessary for full induction of XapA (24).
XapR belongs to the LysR family of regulatory proteins (21). This is a large family of proteins that regulates genes with very diverse functions in prokaryotes. LysR family members have a number of common characteristics, such as a size of about 300 amino acids, a helix-turn-helix DNA-binding domain in the N terminal part, and the requirement for a small molecule to act as an inducer. The homology between LysR family proteins is generally high, with the highest similarity in the N-terminal part. The amino acid sequence of XapR is highly homologous (70%) to that of AlsR from E. coli. Of the 60 N-terminal amino acids in XapR and AlsR, 68% are identical. The work done on LysR family proteins has been hampered by the fact that most members of this family are very insoluble and have a tendency to form inclusion bodies. This is also the case for XapR, and it has not yet been possible to purify the protein for characterization and in vitro work such as footprinting. So far, only one protein, the inducer-binding domain of CysB from Klebsiella aerogenes, has been crystallized and subjected to three-dimensional analysis (25). The isolation of mutants is therefore an important tool for studying LysR family proteins. In some LysR family proteins, point mutations that interfere either with DNA binding (these mutations are located in the N-terminal part of the protein) or with inducer binding have been identified (21). These mutants are mostly null mutants or constitutive mutants that are activated in the absence of any inducer. There are only a few examples (e.g., NahR from Pseudomonas) where mutants that recognize different inducer analogues have been isolated (4). XapR is well suited to examine the inducer-binding regions of LysR family proteins because mutants with a changed inducer specificity can be easily isolated (3). The results described here show that two regions in XapR, and probably also in other LysR family proteins, are important for correct interactions between the inducer and the protein.
MATERIALS AND METHODS
Bacterial strains.
All mutants were isolated in SØ1053 (3). The add mutation was introduced in several steps. First, malI::Tn10 was P1v transduced (17) from ME429 [MC4100 Φ(malK-lacZ) mdoA malQ7 malI::Tn10] (a gift from W. Boos [6]) into SØ430 (SØ003 cdd1 udk add), resulting in GD1030 (SØ003 cdd1 udk add malI::Tn10). Next, P1v grown on GD1030 was used to transduce add malI::Tn10 into SØ1053, SØ1125 (SØ1053 xapR9), SØ1127 (SØ1053 xapR6), SØ1129 (SØ1053 xapR10), and CJM37 (SØ1053 xapR48), resulting in CJ115, CJ116, CJ108, CJ109, and CJ110, respectively.
Plasmids.
pGD111 is a low-copy-number mini-R1 plasmid carrying a xapA-lacZ fusion, and pGD117 is a control plasmid in which the xapA DNA is deleted from pGD111 (24).
DNA techniques.
Methods used for transformation, isolation of DNA fragments, and plasmid isolations are described by Sambrook et al. (20).
Isolation of xapR mutants.
Mutations in the xapR gene were isolated by plating 109 cells of a deoD strain (SØ1053) on plates containing a purine nucleoside other than xanthosine as the carbon source. Colonies were isolated after 4 to 5 days of growth. The isolated strains were tested for temperature sensitivity (growth at 32°C versus growth at 42°C). This provided a test for the presence of the lambda phage inserted in deoD.
Cloning of xapR mutations.
The xapR2, xapR4, xapR6, xapR9, and xapR10 alleles were cloned by isolation of a 2,314-bp PvuII fragment of the E. coli chromosome containing the xapR gene and ligated into the EcoRV site of pBR322, resulting in pCJ10, pCJ3, pCJ8, pCJ2F, and pCJ4, respectively. After transformation into SØ6444 (xapR::Kanr) (24), transformants growing on inosine as the sole carbon source were selected and the presence of the xapR gene was verified by PCR (results not shown).
DNA sequencing.
The xapR gene was amplified by PCR with primers XapR2 (5′-GTATGTCGGATATCTGGTGGTG) and XapR5 (5′-CGGACTACGCGAAGTGAATCG), generating a fragment of 1,022 bp covering the entire xapR gene. Mutated xapR genes were amplified on whole E. coli cells added to the PCR mixture with a toothpick. The PCR product was purified with the QIAquick PCR purification kit purchased from Qiagen. The purified DNA fragment was sequenced by cycle sequencing with dye-labelled terminators and primer XapR2, XapR5, or XapR3 (5′-GGTCGGGGAGAAGCAGGGCGG), using the dye terminator cycle-sequencing ready-reaction kit from Perkin-Elmer. The sequencing reactions were run on a Perkin-Elmer ABI 377 or ABI 310 DNA sequencer.
Growth of cells.
Cells were grown at 32°C in AB minimal medium containing 0.2% fructose as the carbon source and complemented with thiamine, biotin, and Casamino Acids (5). Purine nucleosides were supplemented to a concentration of 1 mg/ml. Strains containing pGD111 or pGD117 were grown in the presence of 25 μg of ampicillin per ml, while strains containing plasmid derivatives of pBR322 were grown in the presence of 100 μg of ampicillin per ml. When cell extracts were prepared for use in enzyme assays, cells were first grown overnight in a minimal-medium culture with the inducer present to ensure full induction of xapA. The next day, the culture was diluted and grown for at least four doublings to a final optical density at 436 nm of 0.8. The cells were harvested by centrifugation, washed with AB minimal medium, and resuspended in 100 mM Tris-HCl–2 mM EDTA (pH 7.0) to a cell density of approximately 2 × 109 bacteria per ml. The harvested cells were sonicated twice for 30 s each, and cell debris was removed by centrifugation.
Enzyme assays.
Xanthosine phosphorylase activity was determined at 37°C as described by Hammer-Jespersen et al. (8). β-Galactosidase activity was assayed at 28°C as described by Miller (17). The amount of protein was determined with the bicinchoninic acid protein assay kit from Pierce with bovine serum albumin as the standard. The amount of enzyme that catalyzes the conversion of 1 nmol of substrate per min defines 1 enzyme unit.
RESULTS
Isolation of XapR mutants.
Spontaneous XapR mutants were isolated by plating 109 cells of a deoD strain (SØ1053) on minimal medium containing a purine nucleoside other than xanthosine as the carbon source, as previously described (3). Colonies appearing on the plates can utilize the purine nucleoside as carbon source. Five of the mutants isolated by Buxton et al. (3) were still viable and were tested for growth on different nucleosides together with new mutants (see Table 1). Originally, Buxton et al. selected the mutants on inosine or adenosine as the carbon source. To isolate new classes, we selected for mutants that can grow on adenosine, deoxyadenosine, inosine, or guanosine as the carbon source. A total of 43 candidates were isolated, and 18 of these were discarded as deoD revertants because they had become temperature resistant and probably had lost the lambda phage inserted into deoD. Four different classes were identified based on growth pattern and were compared with a deoD revertant (CJM25), as shown in Table 1. All four classes grew equally well on glucose and the normal inducer xanthosine. Likewise, they all grew fairly well on uridine, indicating that the genes for pyrimidine metabolism are intact. Class I mutants have gained the ability to grow on all purine nucleosides and deoxynucleosides tested. Class II mutants grew well on adenosine, deoxyadenosine, inosine, and deoxyinosine but did not grow on guanosine or deoxyguanosine. Class III mutants, on the other hand, grew well on all purine nucleosides except adenosine, whereas class IV mutants grew only on xanthosine, deoxyadenosine, and deoxyinosine. Of the 25 new XapR mutants, 17 were isolated on adenosine, 4 were isolated on deoxyadenosine, and 4 were isolated on inosine. No mutants appeared on plates containing guanosine as the carbon source. A total of 20 mutants belonged to the three previously defined classes (classes I, II, and III). The most common class of mutants is class II (11 of 17 mutants isolated on adenosine). On adenosine, mutants appeared with a frequency of 3.0 × 10−8, including the deoD revertants. This corresponds to the frequency obtained by Buxton et al. of 3.3 × 10−8 (3). On deoxyadenosine and inosine, the frequencies of mutation were too low to be determined accurately but were lower than 10−8.
TABLE 1.
Growth pattern of xapR mutants derived from SØ1053 on nucleosides and deoxynucleosidesa
| xapR allele | Selected onb: | Growth onb:
|
Classc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glu | XR | AR | AdR | IR | IdR | GR | GdR | UR | |||
| xapR+ | **** | **** | − | − | − | − | − | − | ** | WT | |
| xapR9 | IR | **** | **** | **** | **** | **** | **** | **** | **** | ** | I |
| xapR6 | AR | **** | **** | *** | **** | **** | **** | − | − | ** | II |
| xapR10 | AR | **** | **** | − | **** | *** | **** | *** | **** | ** | III |
| xapR48 | AR | **** | **** | − | **** | − | **** | − | − | ** | IV |
| xapR36 | AR | **** | *** | *** | *** | *** | NDd | *** | ND | ** | Rev |
The bacteria were streaked out on minimal plates containing the indicated carbon sources. (Deoxy)nucleosides were present at 1 mg/ml, and glucose was present at 2 mg/ml.
−, no growth; ****, good growth after 4 to 5 days of incubation at 32°C. Abbreviations: AdR, deoxyadenosine; AR, adenosine; GdR, deoxyguanosine; Glu, glucose; GR, guanosine; IdR, deoxyinosine; IR, inosine; UR, uridine; XR, xanthosine.
WT, wild type; Rev, revertant.
ND, not determined.
Sequencing of XapR mutants.
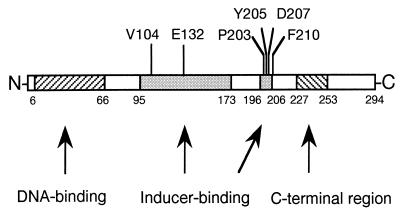
Of the 30 isolated strains carrying xapR mutations, including some of the mutants isolated by Buxton et al. (3), 21 were selected for sequencing of the xapR gene. In most cases the xapR gene was PCR amplified and sequenced directly on the PCR product, whereas a few mutants were cloned into pBR322 and sequenced. All the sequenced xapR genes contained 1-bp substitutions, resulting in single-amino-acid replacements in the protein. The identified mutations are shown in Table 2. At some positions, more than one mutant had the same substitution. The mutations fall in two regions in XapR (Fig. 1). Fifteen mutations were located in a region between amino acids 203 and 210. All but 1 of the 12 class II mutants were found to have mutations here, with changes of amino acids 203 and 207. Nine strains had substitutions leading to alteration of the aspartic acid at position 207 (it was changed to an asparagine in six strains, to a glycine in one, to a tyrosine in one, and to a glutamic acid in one); two strains had identical substitutions leading to a change of a proline at 203 to an arginine. The last class II mutant contained a mutation at position 132, another region in XapR. Apparently, mutations at positions separated by more than 70 amino acids result in similar phenotypes. Two class IV mutations were found at the extremities of the region between 203 and 210 (Fig. 1). Also, one of the class I mutations was found in this region at amino acid 205. The second region of mutations in XapR is defined by mutations found at positions 104 and 132. Mutation at valine 104 occurred in all but one of the constitutive mutants (it was changed to a glutamic acid in two strains, to a leucine in one, and to a methionine in one). The two class III mutants had a mutation that was also found in another class. In SØ1129, aspartic acid 207 was changed to glutamic acid as in CJM28 (class II), and in CJM36, valine 104 was changed to glutamic acid, as in CJM30 (class I). In each of these duplicates, one of the mutants probably contains an additional mutation outside xapR that interferes with the expression of xapA. These mutations are presently unknown, but they are closely linked to xapR, since P1 transduction of xapR+ restores the wild-type phenotypes (data not shown) and since all the mutants isolated by Buxton et al. transduce as a single locus (3).
TABLE 2.
SØ1053 strains carrying xapR mutations
| Strain | xapR allele | Selected ona: | Codon change | Amino acid change | XapR classa |
|---|---|---|---|---|---|
| SØ1053 | xapR+ | WT | |||
| SØ1180 | xapR2 | AR | TAC→GAC | Tyr205Asp | I |
| SØ1126 | xapR4 | AR | GAA→GAC | Glu132Asp | II |
| SØ1127 | xapR6 | AR | GAC→AAC | Asp207Asn | II |
| SØ1125 | xapR9 | IR | GTG→GAG | Val104Glu | I |
| SØ1129 | xapR10 | AR | GAC→GAA | Asp207Glu | III |
| CJM2 | xapR13 | AR | GTG→TTG | Val104Leu | I |
| CJM8 | xapR19 | AR | GAC→AAC | Asp207Asn | II |
| CJM13 | xapR24 | AR | CCC→CGC | Pro203Arg | II |
| CJM15 | xapR26 | AR | GAC→TAC | Asp207Tyr | II |
| CJM16 | xapR27 | AR | GTG→ATG | Val104Met | I |
| CJM17 | xapR28 | AR | GAC→AAC | Asp207Asn | II |
| CJM18 | xapR29 | AR | GAC→AAC | Asp207Asn | II |
| CJM21 | xapR32 | AR | GAC→AAC | Asp207Asn | II |
| CJM23 | xapR34 | AR | GAC→AAC | Asp207Asn | II |
| CJM26 | xapR37 | AR | GAC→GGC | Asp207Gly | II |
| CJM28 | xapR39 | AR | GAC→GAA | Asp207Glu | II |
| CJM30 | xapR41 | AR | GTG→GAG | Val104Glu | I |
| CJM31 | xapR42 | AR | CCC→CGC | Pro203Arg | II |
| CJM36 | xapR47 | AR | CTG→GAG | Val104Glu | III |
| CJM37 | xapR48 | AR | CCC→ACC | Pro203Thr | IV |
| CJM49 | xapR60 | AdR | TTT→TAT | Phe210Tyr | IV |
AR, adenosine; IR, inosine; AdR, deoxyadenosine; WT, wild type.
FIG. 1.
Inducer-binding regions in XapR as defined by the isolated point mutations (shown above the figure). The regions proposed by Schell (21) are shown as grey boxes. Also shown is the proposed DNA-binding region at the N-terminal part of the protein.
XapA induction by xanthosine in XapR mutants.
The activity of xanthosine phosphorylase was determined in selected mutants from each class and compared to the activity in the wild type (Table 3). Table 3 shows that xapA expression in SØ1053 is dependent on the presence of xanthosine in the growth medium, with an induction ratio of more than 300-fold, thus confirming previously published results (3). In the xapR9 mutant (V104E), the expression appears to be constitutive, since the XapA level was high in the absence as well as the presence of xanthosine. For the five tested class II mutants, a different induction of XapA by xanthosine was seen, indicating that these mutants are not identical. For most of the class II mutants, the induction ratio with xanthosine was lower than for the wild type. In xapR4 (E132D) and xapR24 (P203R), this is due to a slightly higher uninduced level of XapA. In xapR26 (D207Y) and xapR37 (D207G), the induced level was lower than in the other mutants and the wild type, giving induction ratios of 44 and 150, respectively. In the xapR6 mutant (E132D), the induction by xanthosine was similar to the induction seen in the wild type. In xapR10 (D207E), a class III mutant, xapA expression was constitutive, although the uninduced level was not as high as the induced level. This is the opposite of the situation for the class I mutant (xapR9). The induction of xapA by xanthosine in xapR48 (P203T) was similar to the induction in the wild type, although the induced level was slightly lower.
TABLE 3.
Xanthosine phosphorylase activity in the presence or absence of xanthosine
| Strain/plasmid | xapR allele | Class | XapA activity (U/mg of protein)a
|
Induction ratiob | |
|---|---|---|---|---|---|
| Without xanthosine | With xanthosine | ||||
| SØ1053/pGD111 | xapR+ | WTc | 5 | 1,540 | 308 |
| SØ1125/pGD111 | xapR9 | I | 1,120 | 820 | 0.8 |
| SØ1126/pGD111 | xapR4 | II | 9 | 1,450 | 161 |
| SØ1127/pGD111 | xapR6 | II | 5 | 1,490 | 298 |
| CJM13/pGD111 | xapR24 | II | 12 | 1,260 | 105 |
| CJM15/pGD111 | xapR26 | II | 5 | 220 | 44 |
| CJM26/pGD111 | xapR37 | II | 4 | 600 | 150 |
| SØ1129/pGD111 | xapR10 | III | 80 | 1,550 | 14 |
| CJM37/pGD111 | xapR48 | IV | <5 | 1,100 | >220 |
The data are averages of two to four independent experiments.
The induction ratio is induced level (with xanthosine)/uninduced level (without xanthosine).
WT, wild type.
Deoxyadenosine and deoxyinosine can act as inducers in the wild type.
SØ1053 and the mutants were transformed with the low-copy-number plasmid pGD111, which carries a translational xapA-lacZ fusion (24). This allows both the chromosomal xanthosine phosphorylase (XapA) activity and the β-galactosidase activity to be measured. When XapA and β-galactosidase activities were compared in cultures grown with or without xanthosine, similar results were obtained (Tables 3 and 4). However, the induction ratio was higher for the β-galactosidase activity (1,700-fold) than for the chromosomal xapA activity (300-fold). When other purine nucleosides were used a surprising result was obtained. Deoxyadenosine and deoxyinosine induced xapA from an uninduced level of 20 U/mg of protein to 835 and 1,285 U/mg of protein, respectively. This is unexpected since a deoD strain cannot grow on plates when either of these compounds is supplied as the carbon source (Table 1). The other tested purine nucleosides could not function as inducers of xapA in the wild type.
TABLE 4.
β-Galactosidase activity in xapR mutants grown with different purine nucleosides
| Strain/plasmid | xapR allele | Class | β-Galactosidase activity (U/mg of protein)a on:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| NI | XR | GR | AR | IR | AdR | IdR | |||
| SØ1053/pGD117 | xapR+ | WTb | 2 | 1 | 1 | 1 | 2 | 5 | 3 |
| SØ1053/pGD111 | xapR+ | WT | 20 | 34,500 | 32 | 26 | 24 | 840 | 1,280 |
| SØ1125/pGD111 | xapR9 | I | 52,900 | 42,200 | 19,300 | 15,500 | 18,900 | 20,100 | NDc |
| SØ1127/pGD111 | xapR6 | II | 45 | 29,900 | 54 | 12,700 | 14,100 | 21,700 | ND |
| SØ1129/pGD111 | xapR10 | III | 6,200 | 34,900 | 2,700 | 3,900 | 5,000 | 31,800 | ND |
| CJM37/pGD111 | xapR48 | IV | 30 | 39,000 | 21 | 640 | 520 | 9,100 | 11,300 |
The data are averages of three to five independent experiments. Abbreviations: NI, no inducer; XR, xanthosine; GR, guanosine; AR, adenosine; IR, inosine; AdR, deoxyadenosine; IdR, deoxyinosine.
WT, wild type.
ND, not determined.
Induction of a xapA-lacZ fusion by purine nucleosides in XapR mutants.
To further characterize the isolated XapR mutants, the β-galactosidase activity was measured in strains carrying pGD111 (Table 4). The activity of the xapA-lacZ fusion in mutants from each class was also determined in the presence of purine nucleosides other than xanthosine (Table 4). In xapR9 (V104E), xapA was clearly expressed constitutively, since high enzyme levels could be measured on all tested purine nucleosides. The levels obtained with guanosine, adenosine, inosine, and deoxyadenosine in the medium were nearly the same as each other but were lower than the uninduced level and the level obtained with xanthosine. For xapR6 (D207N) (class II), XapA was not induced by guanosine but was induced by the other purine nucleosides tested, correlating well with the growth pattern of class II mutants. The enzyme level for the xapR6 mutant with guanosine was comparable to the uninduced level. In the xapR10 mutant, transcription of xapA was partly constitutive. The enzyme level in this class III mutant in the presence of guanosine, adenosine, or inosine was lower than without inducer, similar to what can be seen for a class I mutant. With deoxyadenosine, the induction was comparable to that with xanthosine. On plates, adenosine was the only purine nucleoside on which this mutant could not grow. This is difficult to explain, since adenosine induces XapA to a higher level than does guanosine (3,940 U/mg of protein with adenosine compared to 2,700 U/mg of protein with guanosine), which is enough to allow growth on this nucleoside. Since the xapR10 mutant can grow on deoxyadenosine, the add gene must be intact. This indicates that in this xapR strain an additional mutation probably has occurred that prevents the growth of this mutant on adenosine. In the class IV mutant, guanosine could not function as an inducer for XapA, as found for class II. Adenosine and inosine induced XapA more than 15-fold, but this was not enough to allow growth on plates. The two deoxynucleosides, on the other hand, induced XapA to a level of about 10,000 U/mg of protein. This is not an induction like that seen for xanthosine, but it is sufficient for growth on plates. XapA is therefore induced by the same purine nucleosides as the wild type.
XapA induction in add strains.
In E. coli, adenosine and deoxyadenosine can either be cleaved phosphorylytically by DeoD or be deaminated by adenosine deaminase (add) to form inosine or deoxyinosine. To find whether (deoxy)adenosine is the actual inducer in the XapR mutants or whether the deaminated product (deoxy)inosine is the inducer, we introduced an add mutation in the XapR mutants. The resulting strains were grown in the presence of purine nucleosides, and β-galactosidase activity from the xapA-lacZ fusion on pGD111 was determined (Table 5). Table 5 shows that adenosine cannot function as an inducer in class II or class IV mutants. The levels of induction by adenosine are lower than the uninduced levels in these two classes. The level of induction by adenosine was higher in the class III mutant than in the class II and IV mutants. This is in agreement with the previous results showing that class III mutants are partly constitutive. However, the level of induction by adenosine was 25 times lower than the uninduced level.
TABLE 5.
β-Galactosidase activity in add strains grown in the presence of different purine nucleosides
| Strain/plasmid | xapR allele | Class | β-Galactosidase activity (U/mg of protein)a on:
|
||||
|---|---|---|---|---|---|---|---|
| NI | XR | IR | AR | AdR | |||
| CJ115/pGD111 | xapR+ | WTb | 28 | 42,100 | 28 | 11 | 17 |
| CJ116/pGD111 | xapR9 | I | 43,600 | 33,800 | 25,000 | NDc | 15,100 |
| CJ108/pGD111 | xapR6 | II | 16 | 14,200 | 5,600 | 5 | ND |
| CJ109/pGD111 | xapR10 | III | 8,200 | 28,200 | 3,000 | 325 | ND |
| CJ110/pGD111 | xapR48 | IV | 16 | 35,100 | 940 | 8 | ND |
The data are averages of two or three independent experiments. Abbreviations: NI, no inducer; XR, xanthosine; IR, inosine; AR, adenosine; AdR, deoxyadenosine.
WT, wild type.
ND, not determined due to heavy growth inhibition.
DISCUSSION
A number of XapR mutants with an altered induction pattern of xapA compared to the wild type have been isolated in the past (3). These mutants have now been sequenced and analyzed in further detail. Additionally, more mutants have been isolated and characterized. The isolated mutants all contain single-amino-acid substitutions in XapR. The altered residues fall in two domains and suggest that these regions are directly involved in binding of the inducer xanthosine. One region, between residue 203 and 210, includes 15 of the 21 found alterations. The other region includes five substitutions at position 104 leading to constitutive expression of xapA and one substitution at position 132 (Fig. 1). The isolated mutants have been divided into four classes depending on their growth on various purine nucleosides. Three of these classes were defined previously, and one is new. The expression of xapA in representatives from each class with different purine nucleosides present in the growth medium was determined by using a xapA-lacZ fusion. This has led to a more detailed description of the XapR mutants.
In the wild type (i.e., SØ1053), xapA and the xapA-lacZ fusion are induced by xanthosine by a factor of more than 300- and 1,700-fold, respectively (Tables 3 and 4). Deoxyinosine is an inducer as well, inducing the xapA-lacZ fusion by a factor of 65. A strain containing a xapR9 mutation expresses XapA constitutively. With no inducer present, the activity is 1.5 times higher than in the wild type induced with xanthosine. With xanthosine present, the xapR9 mutation leads to an activity that is reduced to about 80% of that in the absence of inducer. This effect is even more pronounced with other (deoxy)nucleosides present in the growth medium. Typically, they reduce the activity by a factor of 3. A possible explanation for this is that these nucleosides cause a weak catabolite repression that changes the metabolic state of the cell. It is also possible that the inducers bind to the mutated XapR protein, changing the structure of the protein and making it less active.
XapR mutants belonging to class II are the most common mutants. They have retained the ability to respond to inducers. The activity from the xapA-lacZ fusion with no inducer present is only slightly above the uninduced level for the wild type (Table 4). These mutants most probably contain a mutation that broadens the specificity of XapR for binding (deoxy)nucleosides. This binding resembles the binding of xanthosine to the wild type, thus causing activation at the xap promoter. XapA activity can be measured with all the tested nucleosides except guanosine. Since class II mutants are unable to grow on guanosine or deoxyguanosine on plates, deoxyguanosine would not be expected to function as an inducer either. Experiments with SØ1127, with an add mutation, showed that adenosine has to be converted to inosine before XapA activity can be detected (Table 5). The same probably applies for deoxyadenosine. This means that class II mutants are altered in a way that allows them to respond to inosine. Like the wild type, they also induce xapA expression in the presence of xanthosine and deoxyinosine.
Class III mutants are partly constitutive with respect to xapA expression. Only two class III mutants have been isolated, and in both cases another mutant with the same mutation has been found that belongs to a different class (compare xapR10 with xapR39 and xapR47 with xapR41). Buxton et al. (3) reported that class III mutants (exemplified by SØ1129) were very sensitive to the metabolic state of the cell and that XapR perhaps mutated to recognize a normal metabolite as an inducer since the enzyme levels varied from experiment to experiment. The finding of identical mutations in different classes indicates that some of these mutants contain additional mutations outside xapR. The add gene is intact in SØ1129, because the activity with adenosine is lowered 10-fold when an add mutation is introduced (Tables 4 and 5). Still, this mutant class cannot grow on adenosine (Table 1). Therefore, it is likely that the class III mutants contain additional mutations that interfere with the growth on purine nucleosides. To test if the class III mutants and their class I and class II analogues contained an unlinked mutation, they were transduced with a P1 lysate grown on an orf254::Kanr xapABR+ strain, selecting for kanamycin resistance. Since more than 90% became xapABR+, the additional unknown mutation(s) is somewhere in the region near xap (data not shown), in agreement with the P1 transduction experiments by Buxton et al., who found that all of their mutants transduced as a single locus (3).
Class IV mutants respond to xanthosine in the same way as the wild type does. With guanosine, there is no induction of xapA at all, while inosine causes an induction of about 20-fold and deoxyinosine causes an induction of more than 300-fold. In class IV mutants, xapA is therefore induced by the same compounds as in the wild type, although the effect of deoxyinosine is stronger in the presence of xapR48.
It is possible that the activities seen for the different mutants are affected by an altered protein stability of the mutants with respect to the wild type. The finding that induction by xanthosine in all four mutant classes gives similar activities of XapA or β-galactosidase (Tables 3 and 4) indicates that all mutants have similar levels of XapR protein. One mutant (containing xapR26) gives a sevenfold-lower xanthosine phosphorylase activity compared to the wild type when induced by xanthosine. We cannot exclude the possibility that differences in XapR protein stability account for this lower activity.
None of the isolated XapR mutants have gained the ability to induce xapA in the presence of guanosine. On the other hand, mutations leading to induction with inosine present are very common. This suggests that the presence of an amino group at position 2 in the nucleotide base prevents binding to XapR. For all the isolated mutants except the constitutive ones, adenosine must be converted to inosine before induction can take place. This indicates that the oxo-group at position 6, which is also present in xanthosine, is important for binding to XapR. The oxo-group at position 2 in xanthosine is less important for binding to XapR, since mutations leading to the recognition of inosine are common. The ability of class IV mutants to grow on deoxynucleosides suggests that the hydroxy-group at position 2 in the ribose part of the purine nucleosides is also involved in binding to the protein. It is possible that it is not the binding per se but, rather, a conformational change caused by the bound nucleoside that is necessary for activation to take place. This suggests that XapR can bind all purine nucleosides but that only a few of these are capable of acting as an activator. Binding studies with purified XapR protein will most probably clarify this.
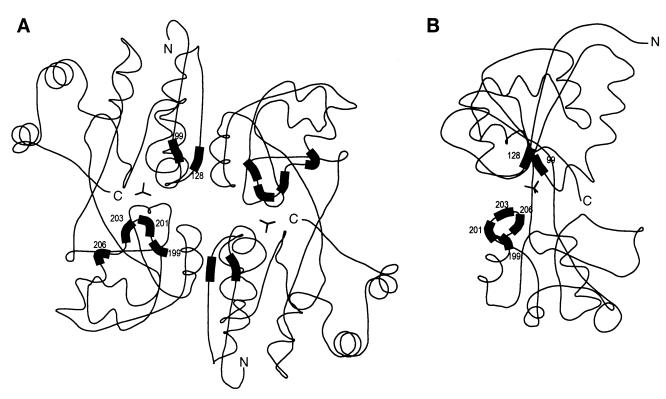
The three-dimensional structure of a C-terminal fragment of the LysR family protein CysB from K. aerogenes has been solved (25). The close similarity between the amino acid sequences of LysR family proteins and common physical characteristics such as insolubility could suggest that all LysR family proteins have very similar structures. In a computer alignment between CysB and XapR, all the identified mutations at positions 104, 132, and 203 to 210 correspond to positions in CysB that are placed close together in the three-dimensional structure (Fig. 2). These residues are also close to the predicted inducer-binding domain in CysB, where a sulfate ion was found. That these residues are in fact part of an inducer-binding site is underscored by the presence of point mutations in other LysR family proteins. The most extensively examined region is from amino acids 196 to 206 (21). Examples of mutations in this area are C199S in OxyR (14), H195Y and L204F in NodD (2), T201A in AlsR (19), and A201V, G203D, T204I, and H206Y in NahR (9, 22). The mutations found at position 104 in XapR can be compared to mutations found in AmpR from Citrobacter freundii (1). A stretch of five identical amino acids with the sequence GVVGT is found at position 102 in XapR and at position 99 in AmpR. The mutation G102E was isolated as a spontaneous mutation in AmpR, resulting in a constitutive phenotype. Site-directed mutagenesis of this residue resulted in a similar phenotype. These mutations lie in an area predicted to be between an α-helix and a β-strand. Most of the mutants described for other LysR-type mutants are null or constitutive mutants; however, Pseudomonas putida mutants with mutations in NahR have been isolated that bind various salicylate analogues (4). Four mutations were identified; three of these were positioned in the proposed inducer-binding domain (M116, R132 and N169), while the fourth (R248) mapped in the C-terminal domain, which is proposed to be important for multimerization.
FIG. 2.
Structure of the C-terminal fragment of CysB from K. aerogenes (25) modelled with XapR mutations. The positions in CysB (amino acids 99, 128, 199, 201, 203, and 206 [highlighted]) that correspond to the mutations in XapR were found by ClustalW sequence alignment of the two amino acid sequences (accession no. P45600 [CysB] and P23841 [XapR]). The sulfate ions believed to be positioned in the inducer-binding domain are shown. (A) Dimeric form of CysB(88–324). (B) The molecule is turned 90° relative to that in panel A, and only the left subunit is shown.
The isolated E132D mutation in XapR lies in an area predicted to be involved in interactions between monomers (Fig. 2). In NahR, a mutation at exactly the same position (R132C) also changed the inducer specificity (4). Most other mutations isolated in this area in LysR family proteins are associated with different phenotypes, perhaps because of differences in interactions between monomers.
ACKNOWLEDGMENTS
We thank Koen H. G. Verschueren for the coordinates of the dimeric form of CysB, Winfried Boos for the malI::Tn10 strain, and Kaj Frank Jensen for reading and commenting on the manuscript.
REFERENCES
- 1.Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol. 1991;5:1715–1725. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 2.Burn J E, Hamilton W D, Wootton J C, Johnston A W B. Single and multiple mutations affecting properties of the regulatory gene nodD of Rhizobium. Mol Microbiol. 1989;3:1567–1577. doi: 10.1111/j.1365-2958.1989.tb00142.x. [DOI] [PubMed] [Google Scholar]
- 3.Buxton R S, Hammer-Jespersen K H, Valentin-Hansen P. A second purine nucleoside phosphorylase in Escherichia coli K-12. I. Xanthosine phosphorylase regulatory mutants isolated as secondary-site revertants of a deoD mutant. Mol Gen Genet. 1980;179:331–340. doi: 10.1007/BF00425461. [DOI] [PubMed] [Google Scholar]
- 4.Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 5.Clark D J, Maaløe O. DNA replication and the division cycle of Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 6.Ehrmann M, Boos W. Identification of endogenous inducers of the mal regulon in Escherichia coli. J Bacteriol. 1987;169:3539–3545. doi: 10.1128/jb.169.8.3539-3545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammer-Jespersen K. Nucleoside catabolism. In: Munch-Petersen A, editor. Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. London, United Kingdom: Academic Press, Ltd.; 1983. pp. 203–258. [Google Scholar]
- 8.Hammer-Jespersen K, Buxton R S, Hansen T D H. A second purine nucleoside phosphorylase in Escherichia coli K-12. II. Properties of xanthosine phosphorylase and its induction by xanthosine. Mol Gen Genet. 1980;179:341–348. doi: 10.1007/BF00425462. [DOI] [PubMed] [Google Scholar]
- 9.Huang J Z, Schell M A. In vivo interactions of the NahR transcriptional activator with its target sequences. Inducer-mediated changes resulting in transcription activation. J Biol Chem. 1991;266:10830–10838. [PubMed] [Google Scholar]
- 10.Ikezawa Z, Nishino T, Murakami K, Tsushima K. Purine nucleoside phosphorylase from bovine liver. Comp Biochem Physiol Ser B. 1978;60:111–116. doi: 10.1016/0305-0491(78)90113-x. [DOI] [PubMed] [Google Scholar]
- 11.Jensen K F. Purine nucleoside phosphorylase from Salmonella typhimurium and Escherichia coli. Initial velocity kinetics, ligand binding, and reaction mechanism. Eur J Biochem. 1976;61:377–386. doi: 10.1111/j.1432-1033.1976.tb10031.x. [DOI] [PubMed] [Google Scholar]
- 12.Jensen K F, Nygaard P. Purine nucleoside phosphorylase from Escherichia coli and Salmonella typhimurium. Purification and some properties. Eur J Biochem. 1975;51:253–265. doi: 10.1111/j.1432-1033.1975.tb03925.x. [DOI] [PubMed] [Google Scholar]
- 13.Kocharyan S M, Smirnov Y V. Escherichia coli K-12 mutants capable of catabolizing purine nucleosides without the participation of purine nucleoside phosphorylase. Sov Genet. 1977;13:962–968. [PubMed] [Google Scholar]
- 14.Kullik I, Tolodano M B, Targalia L A, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leer J C, Hammer-Jespersen K, Schwartz M. Uridine phosphorylase from Escherichia coli: physical and chemical characterization. Eur J Biochem. 1977;75:217–234. doi: 10.1111/j.1432-1033.1977.tb11520.x. [DOI] [PubMed] [Google Scholar]
- 16.Lewis A S, Lowy B A. Human erythrocyte purine nucleoside phosphorylase: molecular weight and physical properties. A Theorell-Chance catalytic mechanism. J Biol Chem. 1979;254:9927–9932. [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Neuhard J, Kelln R A. Biosynthesis and conversions of pyrimidines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 580–599. [Google Scholar]
- 19.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 22.Schell M A, Brown P H, Raju S. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcriptional activator. J Biol Chem. 1990;265:3844–3850. [PubMed] [Google Scholar]
- 23.Schwartz M. Thymidine phosphorylase from Escherichia coli. Methods Enzymol. 1978;51:442–453. doi: 10.1016/s0076-6879(78)51061-6. [DOI] [PubMed] [Google Scholar]
- 24.Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–5516. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyrrell R, Verschueren K H G, Dodson E J, Murshudov G N, Addy C, Wilkinson A J. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure. 1997;5:1017–1032. doi: 10.1016/s0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]
- 26.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 561–579. [Google Scholar]