The polyphenol castalagin acts as a prebiotic and exerts antitumor activity as well as potentiates anti–PD-1 immune efficacy through a beneficial shift in microbiome composition.
Abstract
Several approaches to manipulate the gut microbiome for improving the activity of cancer immune-checkpoint inhibitors (ICI) are currently under evaluation. Here, we show that oral supplementation with the polyphenol-rich berry camu-camu (CC; Myrciaria dubia) in mice shifted gut microbial composition, which translated into antitumor activity and a stronger anti–PD-1 response. We identified castalagin, an ellagitannin, as the active compound in CC. Oral administration of castalagin enriched for bacteria associated with efficient immunotherapeutic responses (Ruminococcaceae and Alistipes) and improved the CD8+/FOXP3+CD4+ ratio within the tumor microenvironment. Moreover, castalagin induced metabolic changes, resulting in an increase in taurine-conjugated bile acids. Oral supplementation of castalagin following fecal microbiota transplantation from ICI-refractory patients into mice supported anti–PD-1 activity. Finally, we found that castalagin binds to Ruminococcus bromii and promoted an anticancer response. Altogether, our results identify castalagin as a polyphenol that acts as a prebiotic to circumvent anti–PD-1 resistance.
Significance:
The polyphenol castalagin isolated from a berry has an antitumor effect through direct interactions with commensal bacteria, thus reprogramming the tumor microenvironment. In addition, in preclinical ICI-resistant models, castalagin reestablishes the efficacy of anti–PD-1. Together, these results provide a strong biological rationale to test castalagin as part of a clinical trial.
This article is highlighted in the In This Issue feature, p. 873
Introduction
The advent of immune-checkpoint inhibitors (ICI) has improved overall survival in multiple malignancies (1–3). However, primary therapeutic resistance remains a major hurdle. Defining mechanisms of resistance implicates several factors, including PD-L1 expression, tumor microenvironment contexture, and the tumor mutational burden determining the cancer-immune set point or threshold (4, 5). Recent metagenomic sequencing studies coupled with experiments in germ-free (GF) mice have revealed that the gut microbiome serves as another key mediator of this cancer-immune set point (5, 6). Preclinical results suggest that modulation of the gut microbiome by antibiotics (ATB) is associated with ICI resistance, and multiple reports in more than 11,000 patients have demonstrated the deleterious impact of ATB on ICI efficacy (7–9). In addition, experiments in GF or ATB-treated mice revealed that fecal microbiota transplantation (FMT) using fecal samples of patients with cancer reproduced the patients’ clinical phenotype in terms of response or nonresponse upon tumor implantation and treatment with ICI (10).
Gut microbiome profiling in patients amenable to ICI revealed that high gut bacterial diversity and the presence of immunogenic bacteria, such as Ruminococcus, Akkermansia muciniphila, and Bifidobacterium, were associated with a stronger CD8+ T-cell and CD4+ Th1-dependent antitumor response and resulted in favorable clinical outcomes (10–13). FMT from ICI-responsive patients or oral supplementation with A. muciniphila in mice initially colonized with FMT from nonresponder (NR) patients was shown to enhance the efficacy of ICI (10, 13). Furthermore, a consortium of 11 beneficial gut bacteria induced a stronger CD8+ T-cell and IFNγ immune response in the colon of mice and improved ICI activity (14). Evidence has also shown that ATB use prior to ICI initiation decreased gut microbiome diversity and decreased the presence of Ruminococcaceae in patients with advanced non–small cell lung cancer (NSCLC) and led to unfavorable outcomes (15). Taken together, these observations have initiated efforts to harness the gut microbiome as a therapeutic adjuvant to ICI (16). Recently, the first proof-of-principle FMT clinical trials in melanoma were reported, and several others evaluating FMT, probiotics, or targeted dietary modifications in combination with ICI are ongoing (17, 18). However, the use of prebiotics—compounds that favorably modify the host gut microbiome—has not yet been investigated as a microbiome-based intervention to improve ICI efficacy. Camu-camu (CC), also known as Myrciaria dubia, is an Amazonian berry containing several phytochemicals and has been shown to exert protective prebiotic effects against obesity and related metabolic disorders in mice through increasing the abundance of A. muciniphila and Bifidobacterium in the gut (19). Here, we evaluated whether the prebiotic action of CC could also be harnessed to shift the gut microbiome and improve antitumor activity.
Results
Antitumor Activity of CC Is Microbiome-Dependent and Circumvents Anti–PD-1 Resistance
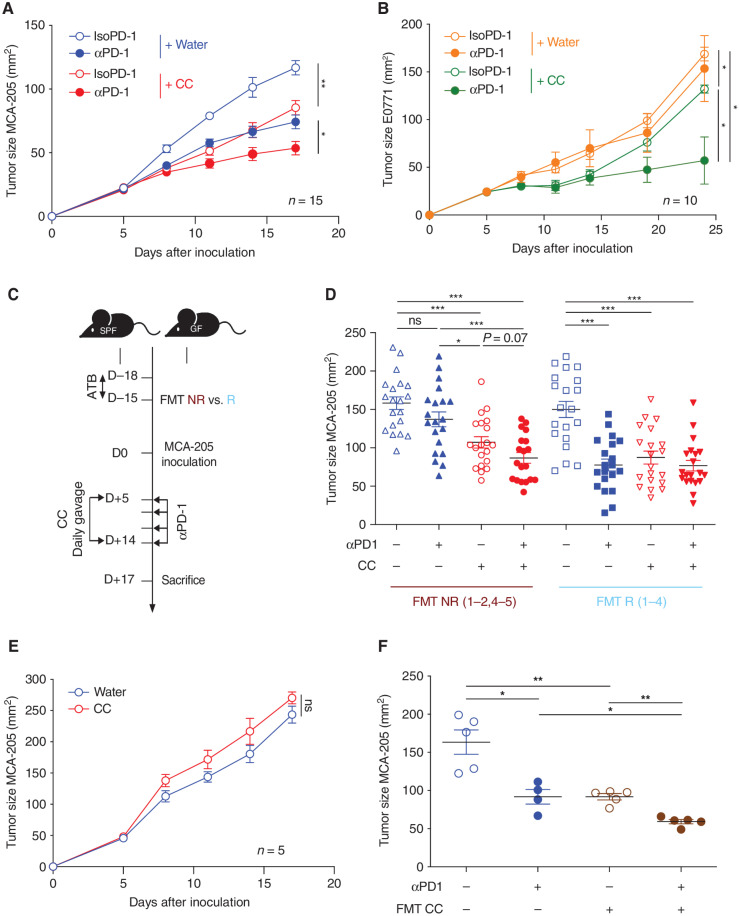
To assess the antitumor effect of CC oral supplementation, specific pathogen-free (SPF) mice received daily oral gavage of CC after MCA-205 sarcoma (ICI-sensitive) tumor inoculation (10) and were treated intraperitoneally with either anti–PD-1 (αPD-1) or an isotype control for αPD-1 (IsoPD-1) every three days. We observed a similar therapeutic efficacy of CC/IsoPD-1 and water/αPD-1 compared with water/IsoPD-1, whereas the combination of CC and αPD-1 led to further inhibition of tumor growth compared with each individual treatment (Fig. 1A). We then tested CC in an αPD-1–resistant E0771 breast cancer tumor mouse model (20) and confirmed that the αPD-1 alone did not exhibit any antitumor activity in this model (Fig. 1B). CC/IsoPD-1 demonstrated minimal antitumor activity, whereas the combination of CC/αPD-1 increased αPD-1 activity. These drug combinations were well tolerated by the animals, with no weight loss or diarrhea observed (Supplementary Fig. S1A). To further explore the therapeutic potential of CC alone or in combination with αPD-1, we recolonized ATB-treated mice by performing FMT from four responder (R) patients and four NR patients with advanced NSCLC amenable to αPD-1 (Supplementary Table S1). MCA-205 tumors were inoculated in these “avatar” mouse models, and mice were treated with CC or water, with or without αPD-1 (Fig. 1C). As previously published, FMT from NR patients conferred resistance to αPD-1, whereas FMT from R patients supported the αPD-1 antitumor effect (Fig. 1D; ref. 10). In the FMT NR avatar mice, oral supplementation of CC/IsoPD-1 induced an antitumor effect. Moreover, the combination of CC/αPD-1 further improved tumor response compared with water/αPD-1 or CC/IsoPD-1. Conversely, in FMT R avatar mice, CC alone or in combination with αPD-1 did not further enhance the antitumor response compared with water/αPD-1 (Fig. 1D).
Figure 1.
CC supplementation is associated with antitumor activity that is microbiome dependent and circumvents the αPD-1 resistance in mice conferred by FMT from NR patients with NSCLC. A and B, Tumor growth kinetics in SPF C57BL/6 mice after sequential injections of αPD-1 or IsoPD-1 and daily oral gavage with CC or water are depicted for (A) MCA-205 sarcoma (n = 15 mice/group) and (B) E0771 breast cancer (n = 10 mice/group). C, Experimental design of avatar mice experiments. FMT from feces samples from NR and R patients with NSCLC were individually performed in GF C57BL/6 mice or after 3 days of ATB in SPF C57BL/6 mice. Two weeks later, MCA-205 tumors were inoculated, and daily gavage with CC or water was performed in combination with sequential injections of αPD-1 or IsoPD-1. D, Pooled analysis of the mean tumor size ± SEM at sacrifice post-FMT from four NR (NR1–2, 4–5) and four R (R1–4) groups for each CC and water group. Each symbol represents one mouse. E, MCA-205 tumor kinetics in mice reared in GF conditions and receiving daily oral gavage with CC or water (n = 5 mice/group). F, Tumor size at sacrifice of mice bearing MCA-205 treated with αPD-1 or IsoPD-1 in combination with daily FMT with mouse feces previously supplemented with CC. Each circle represents one mouse. Means ± SEM are represented in all experiments. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test our hypothesis that the antitumor effect of CC was dependent on the gut microbiota, we used three different strategies. We first demonstrated that in mice reared in GF conditions, CC oral supplementation had no antitumor activity (Fig. 1E). Second, treatment of mice with broad-spectrum ATB during CC treatment compromised the antitumor activity conferred by CC (Supplementary Fig. S1B). Third, we used fecal samples from mice treated with or without CC to perform daily FMT in SPF mice over the course of 14 days after MCA-205 inoculation (10, 21). Only FMT from CC-treated mice further improved αPD-1 activity (Fig. 1F). These experiments demonstrate that the antitumor activity of CC is microbiota-dependent.
CC Increases Microbiome Diversity and Beneficially Shifts the Composition of the Microbiome
We first sought to define the differences in microbiome composition in our avatar murine models between those receiving FMT from R or NR prior to any CC ± αPD-1 treatment. Microbiome profiling based on 16S rRNA revealed that FMT from R, which translated into αPD-1 activity, was associated with greater alpha diversity and different objective clusters when compared with mice that received FMT from NR (resistant to αPD-1; Supplementary Fig. S2A and S2B). Engraftment of Ruminococcaceae UBA1819, Christensenellaceae R-7 group, Paraprevotella, Bifidobacterium, and Oscillospiraceae UCG-003 was enriched in mice that underwent FMT from R and had tumors that were sensitive to water/αPD-1, whereas Lachnospiraceae UCG-006, NK4B4, and FCS020 were more abundant in FMT from NR (Supplementary Fig. S2C). Altogether, these results confirm the importance of specific bacteria associated with the patient-level αPD-1 response in our murine experiments.
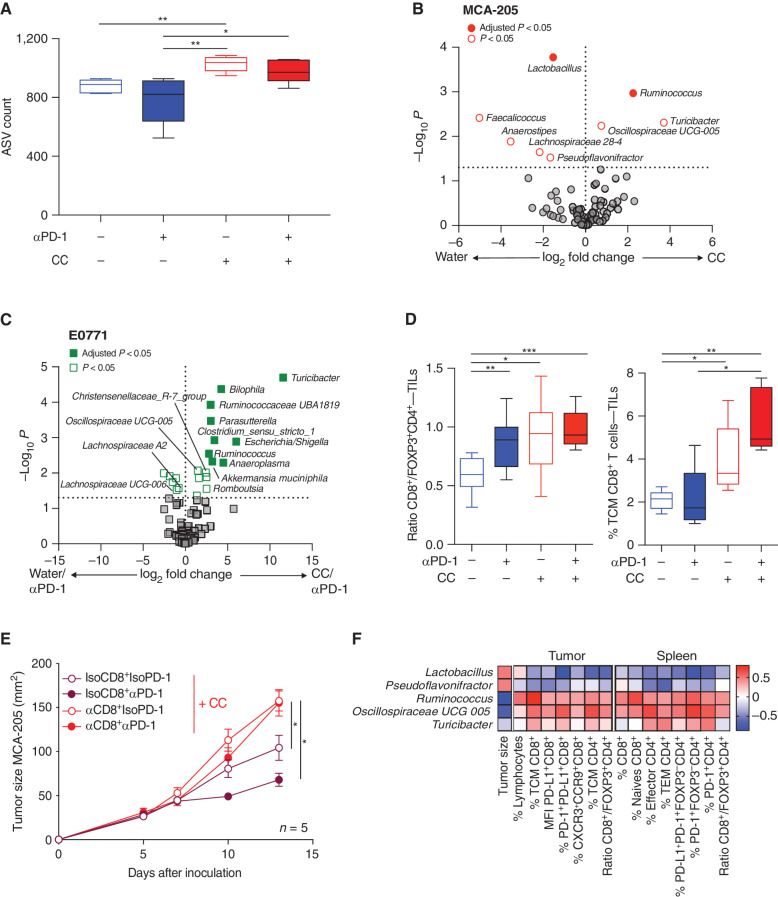
To elucidate the impact of CC on the gut microbiome composition, we characterized the microbiota composition in the MCA-205 murine model treated with CC without patient FMT (Fig. 1A). CC treatment was associated with increased alpha diversity compared with water/IsoPD-1, independent of αPD-1 therapy (Fig. 2A; Supplementary Fig. S3A). qRT-PCR with universal 16S rRNA gene primers confirmed an increased bacterial abundance in the CC group compared with water (Supplementary Fig. S3B). The Bray–Curtis index revealed that oral gavage with CC led to distinct bacterial clusters compared with pre-CC treatment (P = 0.006; Supplementary Fig. S3C), whereas oral gavage with water/αPD-1 showed no separation of the microbiome into distinct clusters (P = 0.16; Supplementary Fig. S3D). Differential abundance analysis showed that Ruminococcus bacteria at the genus level were overrepresented in the CC groups compared with the water groups (adjusted P < 0.05; Fig. 2B). Moreover, Ruminococcus and Turicibacter were the only genera that were consistently more abundant in both CC/IsoPD-1 and CC/αPD-1 groups compared with the water/IsoPD-1 group (Supplementary Fig. S3E and S3F). In parallel, profiling of the gut microbiome using 16S rRNA sequencing in the E0771 tumor model showed that Ruminococcaceae UBA1819, Turicibacter, Parasutterella, Ruminococcus, A. muciniphila (adjusted P < 0.05) as well as Christensenellaceae R-7 group and Oscillospiraceae UCG-005 (P < 0.05) were increased in mice treated with CC/αPD-1 compared with water/αPD-1 (Fig. 2C).
Figure 2.
Impact of CC on microbiome composition and association with CD8+ T-cell antitumor activity. A, 16S rRNA analysis of the fecal samples from mice from the four groups in the MCA-205 experiments, and representation of the alpha diversity measured by the amplicon sequence variant (ASV) count in each group. B and C, Volcano plot representation of differential abundance analysis comparing (B) pooled CC vs. water (αPD-1 and IsoPD-1) groups in the MCA-205 model and (C) the water/αPD-1 vs. CC/αPD-1 groups in the E0771 model (n = 10 and 5 mice/group, respectively). Bacteria enriched in each group are represented using adjusted P value (fill shape symbol) and P value (no fill shape symbol; false discovery rate: 0.05). D, Flow cytometry analysis of the ratio of CD8+/FOXP3+CD4+ T cells and CD8+ T central memory (TCM) cell (CD45RB−CD62L+) subpopulations in MCA-205 TILs in the four experimental groups (n = 10 mice/group). E, Effects of anti-CD8 (αCD8) depletion or its isotype control (IsoCD8) on MCA-205 tumor growth kinetics with daily oral gavage of CC (n = 5 mice/group) treated or not with αPD-1 or its isotype control. F, Pairwise Spearman rank correlation heat map between significantly different bacteria enriched in CC/IsoPD-1 vs. water/IsoPD-1 groups with positively correlated TILs or splenocyte cytometry and matching tumor size in the MCA-205 experiment. MFI, mean fluorescence intensity; TEM, T effector memory. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further define how CC modifies the microbiome, we analyzed the fecal samples of “avatar” mice that received FMT followed by treatment with CC ± IsoPD-1. At the genus level, addition of CC/IsoPD-1 to FMT NR avatar mice was associated with an increase in the relative abundance of Bifidobacterium (P < 0.001) as well as Ruminococcaceae and Ruminococcaceae UCG-009 (P = 0.054 and P = 0.056, respectively; Supplementary Table S2).
Altogether, these results demonstrate that CC induced a shift in the gut microbiota composition, irrespective of tumor model and αPD-1 administration. These results highlighted commonalities between the bacteria enriched in FMT from R patients with those of mice who received CC.
Antitumor Activity of CC Is CD8+ T-cell Dependent and Correlates with Immunogenic Bacterial Species
We next characterized the immune-potentiating effect of CC by assessing flow cytometry surrogate markers within tumor-infiltrating lymphocytes (TIL). In the MCA-205 model without FMT (Fig. 1A), analyses revealed that the ratio of CD8+/FOXP3+CD4+ regulatory T cells (Treg) in the three groups that showed antitumor activity (CC/IsoPD-1, CC/αPD-1, or water/αPD-1) was increased compared with water/IsoPD-1 (Fig. 2D). We further examined the subpopulations of TILs and found that CC/IsoPD-1 increased central memory (TCM) CD8+ T cells (CD45RB−CD62L+) compared with water/IsoPD-1. To verify that the antitumor activity associated with CC was mediated by CD8+ T cells, we administered an anti-CD8+ monoclonal antibody in combination with CC in MCA-205–bearing mice to deplete the CD8+ T-cell subpopulation. Results showed increased tumor growth compared with CC/IsoCD8+ (control), indicating that the microbiome-mediated effect of CC was CD8+ T-cell dependent (Fig. 2E).
We further explored the interactions between the microbiome and the systemic immune response on tumor size related to CC. In the MCA-205 murine model, pairwise comparisons using nonparametric Spearman correlations between bacteria enriched in CC/IsoPD-1 versus water/IsoPD-1 groups were performed and characterized by intratumoral cytometry immune markers and tumor size. Increase in the proportion of PD-L1+CD8+ T cells, TCM CD8+ cells, and ratio of CD8+/FOXP3+CD4+ T cells in MCA-205 TILs were associated with bacteria enriched by CC, such as Ruminococcus and Oscillospiraceae, and downregulation of Lactobacillus and Pseudoflavonifractor (Fig. 2F). Similarly, in splenocytes of the CC/IsoPD-1 group, CD8+ T cells and CD8+/FOXP3+CD4+ T-cell ratio correlated with Ruminoccocus and Oscillospiraceae UCG-005 compared with water/IsoPD-1.
In parallel, we analyzed the TILs in the E0771 tumor model posttreatment. CC combined with αPD-1 induced activation of intratumoral CD8+ T cells, which was represented by an increase in the mean fluorescent intensity of ICOS+CD8+ T cells when compared with water/αPD-1 (Supplementary Fig. S4A). Spearman rank correlations were performed between bacteria upregulated in water/αPD-1 and CC/αPD-1 groups with regard to immune markers and tumor size. The results further demonstrated a positive correlation between the CD8+/FOXP3+CD4+ T-cell ratio and upregulation of ICOS+FOXP3−CD4+ T-cell infiltration. This was associated with the abundance of Ruminococcus, Christensenellaceae R-7 group, Bilophila and A. muciniphila, which correlated with a reduced tumor size (Supplementary Fig. S4B).
Castalagin Is the Bioactive Polyphenol Molecule from CC That Is Responsible for Its Antitumor Effect
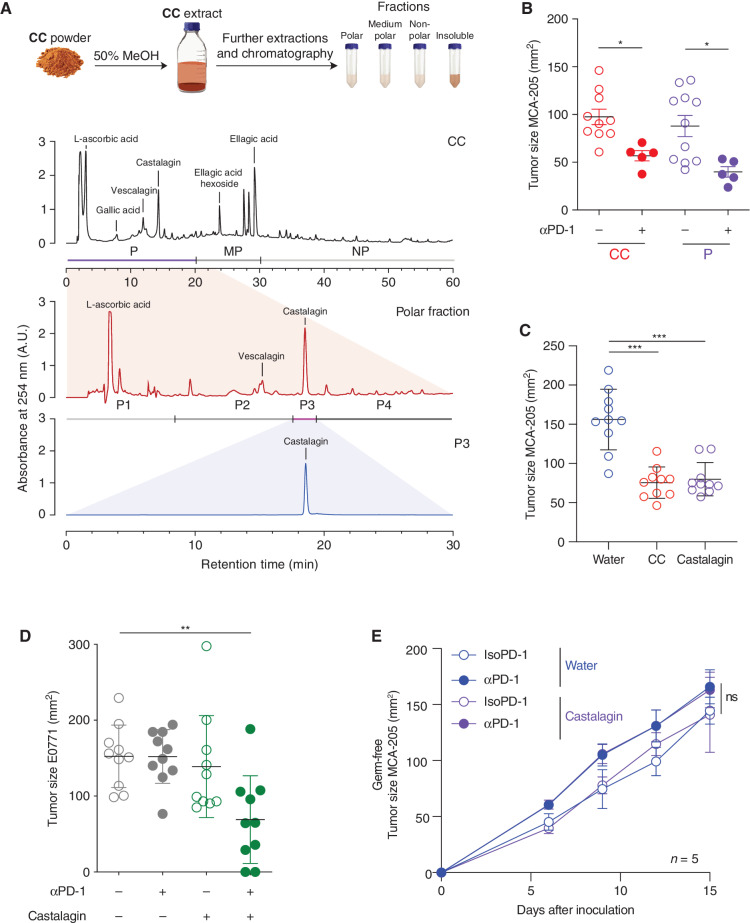
CC is composed of more than 50 compounds, including multiple polyphenols that can all contribute to its prebiotic properties (22). We aimed to identify the bioactive molecule responsible for the observed microbiome-dependent antitumor activity of CC. Using high-performance liquid chromatography (HPLC), we separated the raw extract from CC according to its molecular polarity: polar (P), medium polarity (MP), and nonpolar (NP) as well as an insoluble fraction (INS; Fig. 3A; Supplementary Fig. S5A and S5B). These four fractions were independently delivered by oral supplementation in the MCA-205 tumor model as previously described and compared with CC and water. Among the four different fractions, only the P fraction was associated with an antitumor effect similar to CC (Fig. 3B; Supplementary Fig. S6). We further separated the P fraction into four subfractions according to retention times (P1–P4). The P1 fraction was mostly composed of ascorbic acid; P2 was composed of the polyphenol vescalagin and gallic acid; P3 was composed of the polyphenol castalagin; and P4 was composed of different complex polyphenols (Supplementary Fig. S7A and S7B). We tested these four subfractions in the MCA-205 model and found that P3, which is 95% castalagin, was the only fraction that was associated with an antitumor effect similar to CC (Supplementary Fig. S7C).
Figure 3.
Identification of castalagin as the bioactive compound of CC with similar antitumor activity. A, Schematic of the CC fractionation procedure and summary of chromatograms (λ = 254 nm) showing the isolation of bioactive fractions from CC: CC extract (top chromatogram) containing fractions polar (P), medium polar (MP), and nonpolar (NP). Representation of the polar fraction (middle chromatogram) containing subfractions P1–4 and polar subfraction 3 (P3; bottom chromatogram). B, Tumor size at sacrifice of mice bearing MCA-205 treated with daily oral gavage with CC or P fraction (n = 10 mice/group). C, Tumor size at sacrifice in the MCA-205 model after daily oral supplementation with CC, castalagin at the standard concentration (0.85 mg/kg per mouse), or water (n = 10 mice/group). D, Tumor size at sacrifice of E0771-bearing SPF mice after sequential injections of αPD-1 or IsoPD-1 and a daily oral gavage with water or castalagin at the standard concentration (0.85 mg/kg per mouse; n = 10 mice/group). E, MCA-205 tumor kinetics in mice reared under GF conditions treated with sequential injections of αPD-1 or IsoPD-1 and a daily oral gavage with water or castalagin (n = 5 mice/group). Means ± SEM are represented in B–E. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To validate that castalagin was responsible for the activity of subfraction P3, we obtained pure castalagin from different sources: (i) an analytical standard manufacturer and (ii) oak wood extracts. Analysis of the various sources of castalagin by liquid chromatography–mass spectrometry (LC-MS) and 2-D nuclear magnetic resonance spectroscopy confirmed that subfraction P3 and castalagin isolated from oak were identical to the analytical standard (Supplementary Fig. S8A–S8F). Using the analytical standard or castalagin from oak at similar concentrations present in fraction P (0.85 mg/kg per mouse), we obtained similar tumor inhibition in the MCA-205 and E0711 models when used alone or in combination with αPD-1, respectively (Fig. 3C and D). Next, we performed a proof-of-principle experiment to confirm the microbiome-dependent effect of castalagin whereby oral gavage of castalagin in GF mice resulted in no antitumor effect even in combination with αPD-1 (Fig. 3E).
Castalagin Demonstrates Antitumor Activity through Shift in Microbiome Composition and Metabolite Production
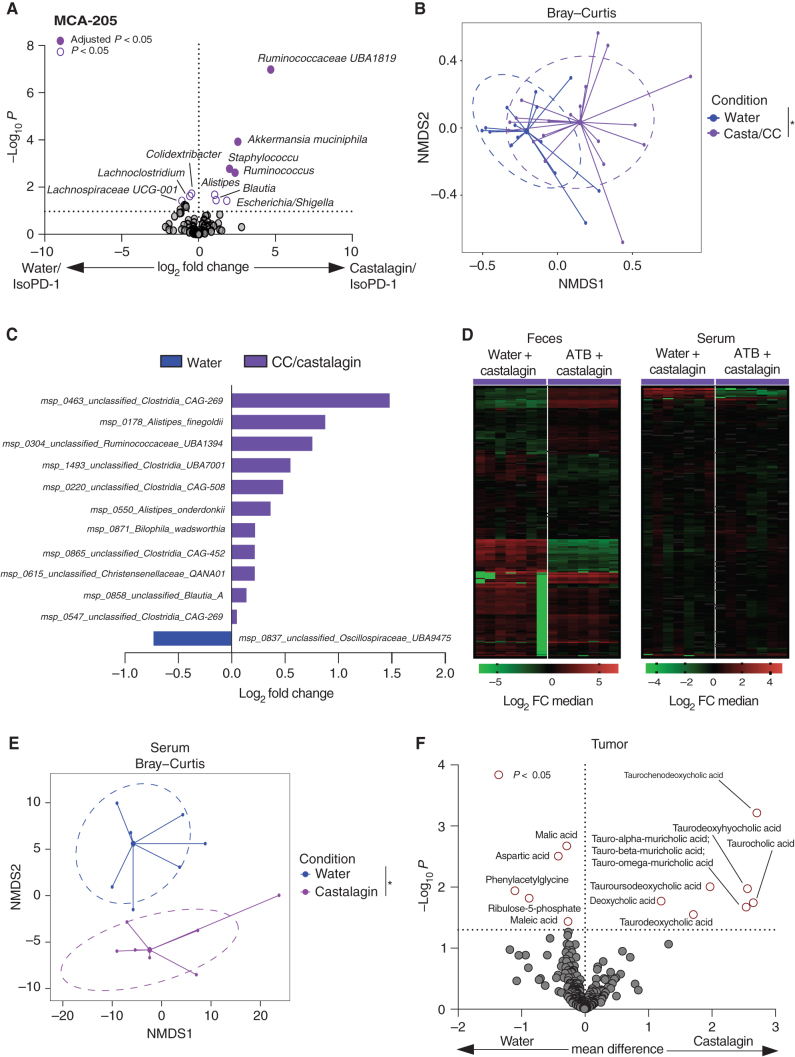
We next addressed the impact of castalagin on the microbiome in SPF mice by 16S rRNA microbiome profiling in the MCA-205 model. Castalagin increased alpha diversity after oral supplementation (Supplementary Fig. S9A). At the taxa level, castalagin led to the enrichment of Ruminococcaceae UBA1819, A. muciniphila, Ruminococcus, Blautia, and Alistipes, whereas feces from the water group were enriched with Lachnospiraceae UCG-001 and Lachnoclostridium (Fig. 4A). Combining 16S rRNA results from all SPF experiments regardless of the tumor model, we showed Ruminoccocus, Alistipes, Parasutterella, and Oscillospiraceae UCG 005 post–CC and castalagin supplementation (Supplementary Fig. S9B).
Figure 4.
Castalagin influences the microbiome composition and metabolite production. A, Volcano plot representation of differential abundance analysis results after 16S rRNA sequencing analysis of water/IsoPD-1 vs. castalagin/IsoPD-1 groups of the MCA-205 model (n = 10 mice/group). Bacteria enriched in each group are represented using adjusted P value (fill shape symbol) and P value (no fill shape symbol; false discovery rate: 0.05). B, Metagenomics sequencing with representation of species richness in fecal samples from MCA-205 experiment in water (IsoPD-1 and αPD-1) vs. castalagin/CC (IsoPD-1 and αPD-1) groups. Bray–Curtis representation for beta diversity. C, Bar plot representation of differential abundance analysis results after metagenomics sequencing analysis between water (IsoPD-1 and αPD-1) and castalagin/CC (IsoPD-1 and αPD-1) groups. D, Heat map of change in metabolite relative abundance in the feces and serum. Hierarchical clustering (Euclidean distance, Ward linkage method) of the metabolite abundance is shown (n = 7 mice/group). E, Metabolites’ principal coordinate analysis between water and castalagin in serum. F, Volcano plot representation for the differential metabolite difference from the tumor of mice receiving water or castalagin (n = 7 mice/group). The horizontal dotted black line shows where P = 0.05 with points above indicating metabolites with significantly different abundance (P < 0.05). *, P < 0.05.
To confirm these changes in microbiome composition, we performed metagenomics shotgun sequencing of murine feces. Shannon and Simpson indexes were significantly more elevated posttreatment with CC and castalagin, whereas observed species were numerically increased but not significant (Supplementary Fig. S9C). Supplementation with CC/castalagin demonstrated unique bacterial cluster compared with water control and was associated with enrichment of Ruminococcaceae UBA1394, Alistipes finegoldii, Alistipes onderdonki, Christensenellaceae QANA01, and Bilophila wadsworthia post–CC and castalagin supplementation (Fig. 4B and C). In addition to an enrichment of Ruminococcaceae UBA 1394, the castalagin/IsoPD-1 group showed that Ruminococcaceae UBA 3818, Ruthenibacterium lactatiformans, and A. muciniphila were also increased post-castalagin (Supplementary Fig. S9D). Of note, Ruthenibacterium lactatiformans would correspond to the Ruminococcaceae UBA1819 signal identified by 16S rRNA according to the SILVA rRNA database.
Based on our results showing a shift in microbiome composition post-castalagin, we investigated the impact of castalagin on metabolite production. Mice were treated with either 14 days of broad-spectrum ATB conditioning or water. MCA-205 tumor cells were then inoculated, and each group was subdivided and treated with castalagin or PBS. Metabolite extraction and measurement by a combination of chromatographic and mass spectrometric methods was performed on the serum, feces, tumor, liver, ileum, and colonic contents. Metabolite production post–CC supplementation was divergent in water compared with ATB groups in the feces and serum, supporting the microbiome-dependent mechanism of castalagin (Fig. 4D). Next, we evaluated the mice that did not receive ATB and found a significant metabolomic shift in the serum of mice treated with castalagin compared with water control (Fig. 4E). More specifically, castalagin was associated with an upregulation of taurine-conjugated bile acids in the tumor and serum (Fig. 4F; Supplementary Fig. S10A). Conversely, secondary taurinated bile acids were decreased in feces post-castalagin (Supplementary Fig. S10B and S10C). These results revealed that castalagin alone was able to shift the gut microbiome composition, translating into a distinct metabolic phenotype and providing further evidence that it represents the active microbiome-dependent component of CC.
Similar to CC, Systemic Immune Effects of Castalagin Are CD8+ T-cell Dependent
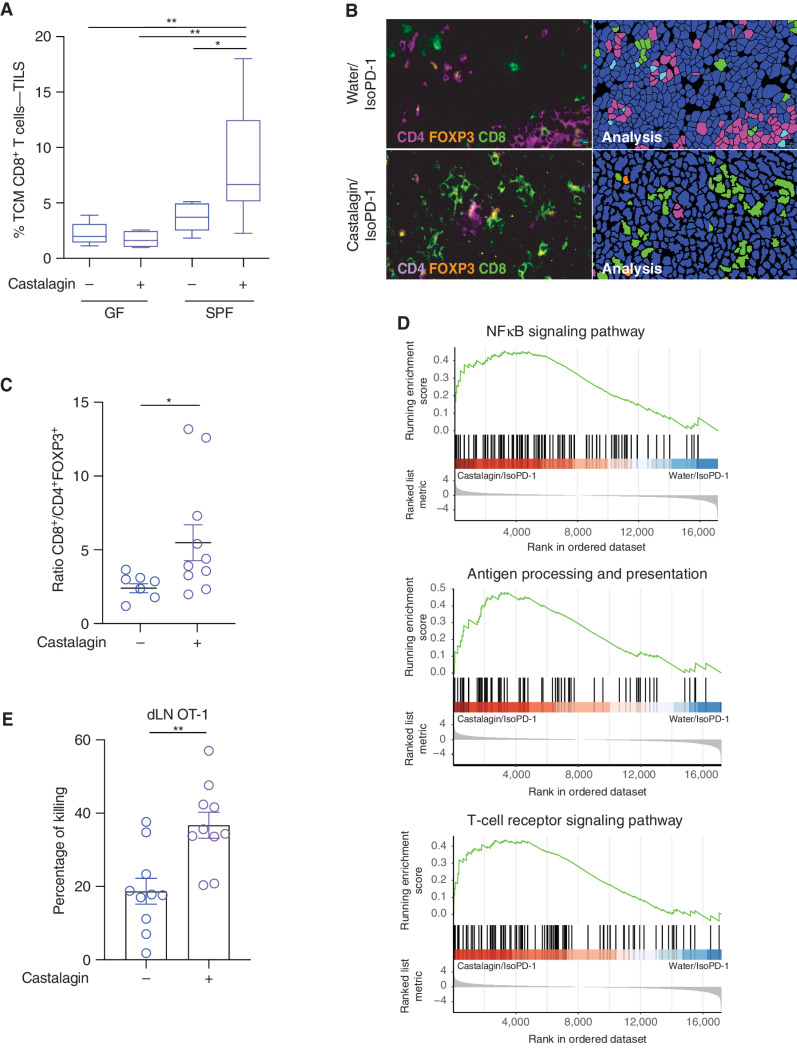
We next examined whether castalagin could also reproduce the systemic immune response of CC, using five approaches. First, flow cytometry analysis in MCA-205 experiments confirmed that the polyphenol upregulated TCM CD8+ T cells in TILs from SPF mice, whereas no difference in frequency was observed in GF mice (Fig. 5A). In the E0771 model, castalagin was also associated with an increase in the frequency of memory CD8+ T cells (CD44hiCD62L−CD8+ T cells) in combination with αPD-1 in both TILs and splenocytes when compared with water/αPD-1 (Supplementary Fig. S11A). Second, immunofluorescence staining of tumors further demonstrated an increase of the CD8+/CD4+FOXP3+ T-cell ratio in the castalagin/IsoPD-1 group compared with water/IsoPD-1 (Fig. 5B and C). Third, to support our demonstration that castalagin modulates TILs, tumor transcriptome profiling was performed through RNA sequencing (RNA-seq) to define differentially expressed pathways in the MCA-205 models. We observed an upregulation of gene pathways including antigen processing and presentation, T-cell receptor signaling pathway, as well as NF-κB signaling associated with a more inflamed tumor microenvironment in the castalagin/IsoPD-1 group compared with water/IsoPD-1 control (Fig. 5D). Within these pathways, transcriptome analysis showed an overexpression of CD3ε, CD8α, IFNγ, and MAPK11 in the castalagin group (Supplementary Fig. S11B). Fourth, using the CIBERSORT program to estimate immune cells from RNA-seq, castalagin was associated with a numerically higher frequency of CD8+ T cells compared with water groups (Supplementary Fig. S11C). Lastly, an in vivo CD8+ T-cell killing assay was performed after mice received an intravenous injection of carboxyfluorescein succinimidyl ester (CSFE)–labeled target cells as previously described (23). After injection of these OT-1 cells, mice were immunized with CpG/OVA. CSFElo pulsed OVA OT-I splenocytes and CFSEhi no-pulsed OVA OT-I splenocytes were then transferred to the mice. We observed a lower proportion of CFSElo CD8+ T cells in the draining lymph nodes in mice in the castalagin group versus the water group corresponding to higher percentage of killing, pointing to the CD8+ T cell–dependent mechanism of castalagin (Fig. 5E).
Figure 5.
Immune-potentiating effect of castalagin. A, Flow cytometry analysis of MCA-205 TIL and CD8+ TCM cell (CD45RB− CD62L+) subpopulation in the GF and SPF experiments comparing castalagin vs. water (n = 10 mice/group). B, Representative immunofluorescence images of MCA-205 tumors for CD8+, CD4+, and FOXP3+ cells in castalagin/IsoPD-1 and water/IsoPD-1 groups. C, Tumor immunofluorescence for CD8+/FOXP3+CD4+ cell ratio results for both groups shown in B. Each circle represents one tumor. D, Pathway classification from RNA-seq results using gene set enrichment analysis based on the KEGG database comparing castalagin/IsoPD-1 vs. water/IsoPD-1 groups of MCA-205 tumors. E, Percentage of killing of CD8+ T OT-1 cells from the draining lymph node (dLN) in mice treated with castalagin or water and immunized with CpG/OVA (n = 10 mice/group). *, P < 0.05; **, P < 0.01.
Given that CC and castalagin induced similar shifts in the gut microbiome composition, translating into a modification in the TILs, and that castalagin is the only polyphenol enriched in fraction P3 that induced an antitumor response, we deduced that castalagin is the compound responsible for the antitumor activity of CC.
Castalagin Can Circumvent αPD-1 Resistance and Has a Dose-Dependent Antitumor Effect That Correlates with Ruminococcaceae Abundance
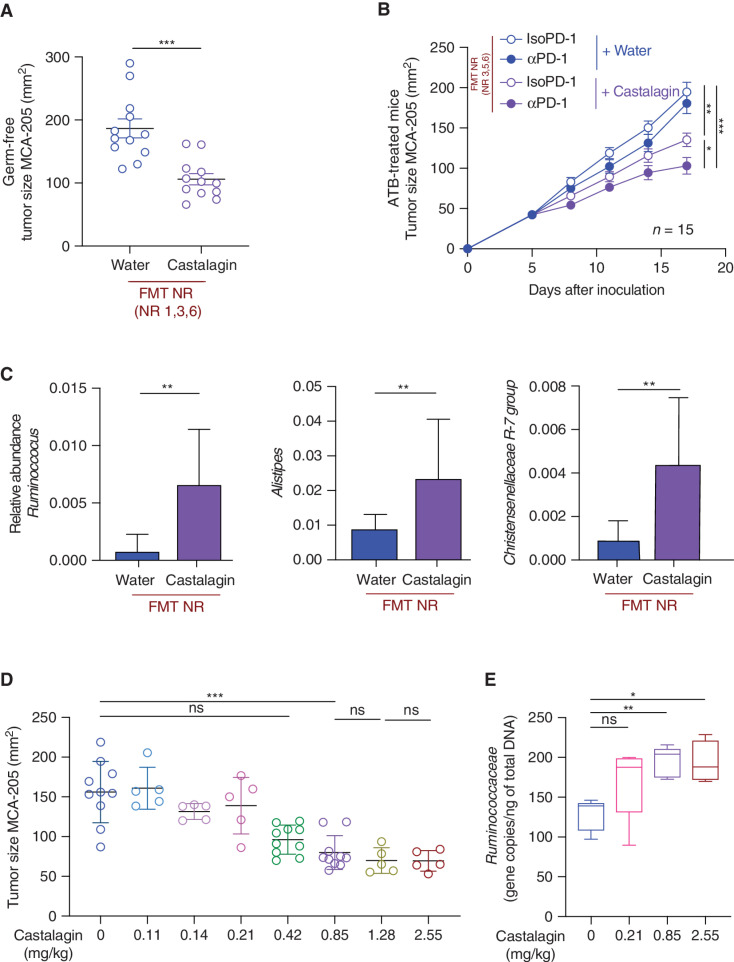
As previously performed with our avatar mice, we tested the therapeutic effects of castalagin following FMT using feces from four NR patients with NSCLC in ATB-treated and GF mice. The addition of castalagin alone was able to exert antitumor activity but also increased αPD-1 activity (Fig. 6A and B). Profiling of fecal samples from these FMT experiments using 16S rRNA further confirmed that castalagin treatment increased the relative abundance of Ruminococcus, Alistipes, Christensenellaceae R-7 group, and Paraprevotella compared with the water control group (Fig. 6C; Supplementary Fig. S12A). Conversely, Lachnosclostridium was underrepresented after castalagin treatment.
Figure 6.
Castalagin circumvents αPD-1 resistance in mice conferred by FMT from NR patients with NSCLC and influences the microbiome composition in a dose-dependent manner. A, FMT of stool sample from three NR patients with NSCLC (NR 1, 3, and 6) was performed in GF C57BL/6 mice (n = 12/group). Two weeks later, MCA-205 sarcoma cells were inoculated, and mice received a daily oral gavage with castalagin or water. B, MCA-205 tumor growth kinetics in the ATB-avatar model after FMT from three NR patients (NR 3, 5, and 6) with daily oral supplementation with castalagin or water in combination with αPD-1 or IsoPD-1. C, Relative abundance analysis results after 16S rRNA sequencing analysis of Ruminococcus, Alistipes, and Christensenellaceae R-7 group between castalagin and water groups in the NR FMT experiments. D, MCA-205 tumor size at sacrifice of mice that received daily oral gavage of castalagin with increasing concentrations from 0 mg/kg to 2.55 mg/kg. E, Real-time PCR of DNA extracted from mouse feces after 6 days of oral gavage with water or castalagin at 0.21, 0.85, and 2.55 mg/kg, using specific primers for Ruminococcaceae detection in the MCA-205 experiment (n = 5/group). Means ± SEM are represented. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine if castalagin activity was dose dependent, we performed oral gavage in mice using different concentrations of castalagin from 0 to 2.55 mg/kg. The antitumor activity became significant only at 0.85 mg/kg, plateauing at higher concentrations (Fig. 6D; Supplementary Fig. S12B). More-over, to determine if the abundance of Ruminococcaceae correlated with castalagin activity, we performed qRT-PCR with Ruminococcaceae-specific primers on the feces from the dose-dependent experiments. Ruminococcaceae was not increased in the 0.21 mg/kg concentration dose of castalagin (with no antitumor effect) compared with the control (0 mg/kg; Fig. 6E). However, Ruminococcaceae abundance was significantly increased following supplementation with castalagin at 0.85 mg/kg (with antitumor effect), and no further incrementation of Ruminococcaceae abundance was observed beyond this concentration.
Castalagin Binds Preferentially to the Cellular Envelope of Ruminococcus
In the gut, castalagin is hydrolyzed to castalin and ellagic acid, which is further converted into urolithins by intestinal bacterial species (ref. 24; Supplementary Fig. S13). We next sought to elucidate if the metabolites of castalagin or its diastereomer, vescalagin (Supplementary Fig. S14A–S14G), were involved in its antitumor activity and detectable in fecal samples. In contrast to castalagin, no antitumor effects were observed in our MCA-205 model with vescalagin or with the downstream metabolites castalin, ellagic acid, and urolithin A (Fig. 7A). This result suggests that bypassing the metabolism by gut bacteria does not lead to an antitumor effect and that the metabolism of castalagin by gut bacteria might be required for the microbiota composition shift.
Figure 7.
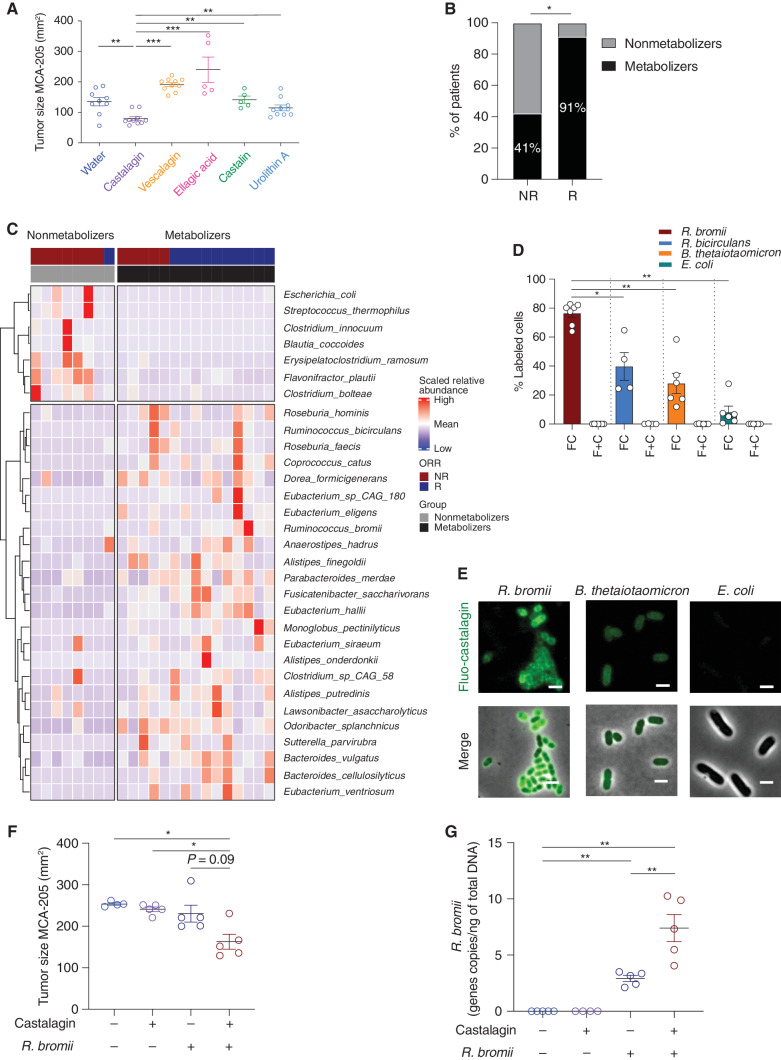
Castalagin and not its metabolites provides antitumor activity and directly interacts with the cellular envelope of Ruminococcus. A, Tumor size at sacrifice of mice that received castalagin or its downstream metabolites or its diastereomer vescalagin. Structures of these compounds are shown in Supplementary Fig. S13. B, Relative proportion of castalagin metabolizers and nonmetabolizers based on ex vivo castalagin metabolism assays using fecal samples from NSCLC ICI-responder (R; N = 11) and ICI-nonresponder (NR; N = 12) patients. Associations between ex vivo castalagin metabolism and patient response status were determined using the Fisher exact test on the number of patients in each category (P = 0.0272). Detailed assignments of metabolic phenotypes (metabotypes) for patients are reported in Supplementary Fig. S15B. C, Heat map representation of metagenomic microbiome sequencing from 23 patients with NSCLC amenable to ICI and segregated between castalagin nonmetabolizer and metabolizers. ORR, objective response rate. D, Labeling experiments on R. bromii, R. bicirculans, E. coli, and B. thetaiotaomicron. Cells were incubated with either 2 μmol/L fluo-castalagin (FC) or 2 μmol/L free fluorescein and free castalagin (F + C) for 1 hour at 37°C. Each circle represents one independent experiment. E, Representative epifluorescence microscopy images of fluo-castalagin–labeled R. bromii, E. coli, and B. thetaiotaomicron. Merge represents superimposed images of fluorescence and phase contrast. Scale bar, 2 μm. F, Tumor size at sacrifice of GF mice bearing MCA-205 treated with castalagin or water in combination with oral gavage with R. bromii or PBS. Each circle represents one animal. G, Real-time qPCR of DNA extracted from mouse feces using specific primers for R. bromii detection in GF mice bearing MCA-205 tumors. Mean ± SEM represented. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We were not able to detect castalagin using HPLC analysis on feces and serum of mice treated with up to 20× = 17 mg/kg of daily castalagin. Therefore, to assess if castalagin was metabolized differently by individual patients with cancer, we collected fecal samples from 23 patients with NSCLC treated with ICI (12 NR and 11 R; Supplementary Table S3) and performed ex vivo castalagin metabolism assays. Bacteria present in the feces from 91% of R patients compared with 41% in NR were capable of metabolizing castalagin to end-product metabolites such as urolithin A, isourolithin A, and urolithin B (P < 0.05; Fig. 7B; Supplementary Fig. S15A and S15B). We then analyzed metagenomic microbiome profiling between metabolizers and nonmetabolizers and found that the metabolizers showed a higher alpha diversity and distinct clusters (Supplementary Fig. S15C and S15D) as well increased abundance of Ruminococcus bicirculans, Ruminococcus bromii, Alistipes finegoldii, Alistipes onderdonki, and Eubacterium (Fig. 7C).
In both murine and human experiments, Ruminococcus was one of the bacteria consistently enriched post–castalagin supplementation. Therefore, we sought to demonstrate an interaction between castalagin and Ruminococcus, which is known to be beneficial for patients with cancer amenable to ICI (25). Although Ruminococcus are fastidious bacteria, R. bromii and R. bicirculans have been successfully cultivated and were enriched in metabolizer patients. We compared the growth curves of R. bromii and R. bicirculans against different control bacteria with castalagin and did not observe any beneficial effect on growth (Supplementary Fig. S16).
To test for a potential interaction between castalagin and Ruminococcus, we labeled castalagin with fluorescein (fluo-castalagin; Supplementary Fig. S17A), which was incubated in the presence of R. bromii and R. bicirculans as well as control bacteria Bacteroides thetaiotaomicron and Escherichia coli in anaerobic conditions. Flow cytometry analysis revealed no cell labeling with free fluorescein, whereas 79%, 47%, and 32% fluo-castalagin labeled with R. bromii, R. bicirculans, and B. thetaiotaomicron cells, respectively (Fig. 7D and E; Supplementary Fig. S17B). In contrast, only 7% of E. coli cells were labeled. Moreover, the addition of an excess (100×) of unlabeled castalagin significantly decreased R. bromii labeling efficiency, providing evidence of a competitive effect of free castalagin with the fluo-castalagin probe (Supplementary Fig. S17C). We used fluorescence microscopy to examine labeled bacteria and observed that fluo-castalagin was not internalized by R. bromii cells but remained associated with the cell envelope (Fig. 7E). Importantly, the fluorescence was weaker for B. thetaiotaomicron and barely detectable for E. coli cells compared with R. bromii, similar to our flow cytometry results. To further demonstrate an interaction between R. bromii and castalagin, we inoculated MCA-205 in GF mice. Oral supplementation of neither R. bromii nor castalagin had an effect, whereas the combination of both demonstrated antitumor activity (Fig. 7F). This additive antitumor effect of castalagin and R. bromii was then validated in SPF mice (Supplementary Fig. S18A). Importantly, qRT-PCR showed that the combination translated to a significant increase in Ruminococcaceae and R. bromii relative abundance in both GF and SPF mice feces (Fig. 7G; Supplementary Fig. S18B–S18D).
Finally, in a proof-of-principle experiment, we analyzed the feces of two non–cancer patients enrolled in a clinical trial (NCT04058392) addressing the safety of oral administration of CC (26). Preliminary fecal metagenomics revealed a positive trend in diversity and toward enrichment of R. bromii, consistent with the results obtained in CC-treated mice (Supplementary Fig. S18E).
Discussion
The gut microbiome has emerged as a key player in shaping tumor immunosurveillance and potentiating ICI efficacy. Harnessing the gut microbiome is thus considered a novel and promising strategy for adjuvant therapy in cancer. CC was previously shown to exert strong microbiome-dependent activity against obesity-linked metabolic syndrome, mainly through the enrichment of A. muciniphila (19).
In this study, we discovered that the polyphenol castalagin, an abundant ellagitannin in CC, is the principal bioactive molecule that carries the antitumor activity of this prebiotic extract. In two tumor models, we showed that, similar to CC, the antitumor effect of castalagin induced a beneficial shift in microbiome composition, independent of αPD-1 administration. Metagenomics sequencing corroborated that castalagin treatment reproduced the effect that CC had on the abundance of Ruminoccocacceae, Alistipes, and Ruthenibacterium lactatiformans (corresponds to the 16S rRNA signal of Ruminococcaceae UBA1819), which correlated with the recruitment of CD8+, PD-L1+CD8+, and CD8+/FOXP3+CD4+ T cells within the tumor microenvironment. In addition, we showed that these gut microbial/immune changes induced by castalagin were accompanied by an increase in taurine-conjugated bile acids. Furthermore, we have developed an efficient extraction and purification protocol using oak wood, ensuring a readily available and accessible source of castalagin for future therapeutic trials.
We also show that the action of castalagin is specific because other closely related metabolites and its diastereomer vescalagin did not exhibit antitumor activity in vivo. Castalagin metabolism likely gives consuming bacteria a competitive advantage that leads to their expansion, causing a global shift in composition, which in turn shifts the production of metabolites and exerts an antitumor effect, increasing αPD-1 activity and circumventing resistance. Moreover, secondary taurinated bile acids are known to inhibit colon adenoma (27) and are decreased after ATB treatment, which has also been associated with ICI resistance (28). Furthermore, using fluorescent castalagin, we demonstrated a non–energy-dependent but specific interaction between castalagin and the cell wall of certain bacteria, preferentially with R. bromii compared with R. bicirculans. This suggests that, even in the absence of direct metabolism, castalagin itself may promote the growth of R. bromii and possibly other beneficial bacteria in the gut. Although the castalagin–bacterial interaction did not provide a competitive advantage in nutrient-rich media in vitro, it may provide some advantage within a complex and nutrient-restricted environment such as the gut. Ellagitannins are known for their ability to scavenge reactive radicals (29), and the administration of grape polyphenols in a mouse model of metabolic disease was shown to decrease gut reactive oxygen species (ROS; ref. 30). Thus, bacteria that associate with castalagin may be protected from ROS present at the oxic–anoxic interface of the intestinal epithelium (31, 32). Polyphenols can also chelate ferric iron via their catechol moieties, which may provide an advantage in an iron-deprived environment (33). Importantly, our finding that NSCLC responders enriched with Ruminococcus were able to metabolize castalagin and our observation of an increase in Ruminococcus post-CC treatment further suggest that castalagin has a propensity to interact with specific and beneficial bacteria.
The present study validates the importance of specific immunogenic gut bacteria in modulating response to ICI. In patients with advanced melanoma, higher baseline diversity and overrepresentation for Ruminococcaceae including R. bromii were associated with an increased proportion of CD8+ T cells in the tumor and peripheral blood and correlated with a beneficial response (12). In addition, Ruminococcus bacterium cv2 is one of 11 beneficial bacteria present in the VE800 probiotic capsule that was linked to increased IFNγ+CD8+ T cells in the colon and tumors and is currently under investigation in a clinical trial (NCT04208958; ref. 14). Specific bacteria modulating ICI outcomes are further demonstrated with A. muciniphila, which predicted the response in 153 patients with advanced NSCLC and renal cell cancer amenable to ICI (10). This observation was recently validated in a prospective trial of 338 patients with NSCLC (25). In this study, R. bromii was the most abundant bacteria in patients with detectable A. muciniphila in their feces. Moreover, in the first clinical trials combining FMT with ICI, Ruminococcaceae spp. and A. muciniphila were both enriched post-FMT in R patients with melanoma refractory to ICI (18, 34).
Altogether, this is the first study demonstrating the microbiome-dependent antitumor effect of a single polyphenol as a prebiotic, paving the way for future clinical trials using strategies with castalagin as an ICI adjuvant in patients with cancer.
Methods
Murine Studies
All animal studies were approved by the Institutional Animal Care Committee (CIPA) and carried out in compliance with the Canadian Council on Animal Care guidelines (Ethics numbers: C18029BRs and C18025BRs). Murine experiments were conducted using seven-week-old female C57BL/6 mice, obtained from Charles River. GF female C57BL/6 mice were purchased from the International Microbiome Centre Germ-Free Facility (University of Calgary, Canada) and maintained at the Centre de recherche du Centre hospitalier de l'Université de Montréal (CRCHUM) GF Facility.
Cell Culture, Reagents, and Tumor Cell Lines
MCA-205 fibrosarcoma cells and E0771 mammary adenocarcinoma, class I MHC H-2b syngeneic cell lines for C57BL/6 mice were used for this study. MCA-205 and E0771 cells were cultured as previously described (10, 20). All cultures were checked for Mycoplasma using PlasmoTest Mycoplasma Detection Kit (InvivoGen).
Subcutaneous Model of MCA-205 Sarcoma and E0771 Breast Cancer
Syngeneic C57BL/6 mice were implanted subcutaneously with 0.8 × 106 MCA-205 cells or 0.5 × 106 E0771 cells. When tumors reached 20 to 35 mm2 in size, mice were treated four times (or two times for flow cytometry analysis; see section below) intraperitoneally every three days with αPD-1 mAb (250 μg/mouse; clone RMP1-14, Bio X Cell) or isotype control (clone 2A3, Bio X Cell). Upon treatment initiation, mice received a daily oral gavage with the following products: Myrciaria dubia, CC raw extract (Sunfood; 200 mg/kg per mouse); fractions from extraction round 1 (P, MP, NP, and INS; 40.18 mg/kg per mouse); fractions from extraction round 2 (P1, P2, P3, and P4; equivalent to fraction P dose of 40.18 mg/kg per mouse); vescalagin (extracted from CC; see isolation process below; 0.85 mg/kg per mouse); ellagic acid (0.85 mg/kg per mouse; Sigma-Aldrich); urolithin A (0.85 mg/kg per mouse; Sigma-Aldrich); castalin (0.5 mg/kg per mouse; PhytoProof, Sigma-Aldrich); and castalagin (PhytoProof, Sigma-Aldrich, or isolated from food grade oak; see isolation process below). In the control group, mice received daily oral gavage with water (100 μL). Tumor area was routinely monitored every three days by means of a caliper. In the CD8+ depletion experiments, we used the anti-CD8 mAb (150 μg/mouse; clone 53-6.7, Bio X Cell) or isotype control (clone 2A3, Bio X Cell).
Antibiotic Treatments
For experiments with ATB, mice were treated with an ATB solution containing ampicillin (1 mg/mL), streptomycin (5 mg/mL), and colistin (1 mg/mL; Sigma-Aldrich), which was added to the sterile drinking water of mice (10). ATB activity was confirmed as previously described (10). For the FMT experiments in SPF-reared mice, mice received three days of the same combination of ATB prior to FMT. For the metabolomic experiments, mice received one week of the same combination of ATB for the ATB group, then tumor cells were injected subcutaneously. For the ATB groups, the mice continued to receive ATB after tumor cell implantation, throughout the experiment until sacrifice. When tumors reached 20 to 35 mm2 in size, mice were treated with a daily oral gavage of castalagin or water for six days as described above. The stools and blood (for serum) were collected three days after the start of oral gavage, and the liver, tumor, ileum, and colon contents were collected at sacrifice and snap-frozen for metabolomic analysis described in Supplementary Materials and Methods.
FMT Experiments
Appropriate ethics approval was obtained at the CRCHUM in Montreal (Ethics 17.035). As previously published (10), FMT was performed by thawing fecal material from ten different patients with NSCLC amenable to ICI after patient records were retrospectively analyzed to identify their response status (Supplementary Table S1). Two weeks after FMT, tumor cells were injected subcutaneously, and mice were treated with αPD-1 or isotype control with or without CC, castalagin, or water as described above.
Flow Cytometry Analysis of Immune Cells
Tumors and spleens were harvested at 9 days after the first injection of αPD-1 in mice bearing MCA-205 tumors and at 11 days after the first injection of αPD-1 in mice bearing E0771 tumors. TILs and splenocyte suspensions were prepared as previously published (10). Two million tumor cells or splenocytes were preincubated with purified anti-mouse CD16/CD32 (clone 93; eBioscience) for 30 minutes at 4°C before membrane staining with anti-mouse antibodies listed in Supplementary Table S4. For intracellular staining, the FOXP3 staining kit (eBioscience) was used. Dead cells were excluded using the Live/Dead Fixable Aqua Dead Cell Stain Kit (Life Technologies). Samples were acquired on a BD Fortessa 16-color cytometer (BD), and analyses were performed with FlowJo software (BD).
Immunofluorescence Staining
Murine tumors preserved in optimal cutting temperature compound were cut (5-μm-thick sections) and adhered to microscope slides and stored at −80°C. At the start of the experiment, slides were air-dried and washed with cold acetone. To prevent nonspecific binding to biotin, the Endogenous Avidin Biotin Blocking Kit (Thermo Fisher) was used. Additionally, tissues were incubated with 10% donkey serum in order to reduce background staining. Primary antibodies used were anti-CD4, anti-CD8, and anti-FOXP3. Donkey anti-goat and donkey anti-rat conjugated to AF-488 were used as secondary antibodies, and slides were incubated with Cy3-streptavidin to detect biotinylated antibodies (Supplementary Table S5). Nuclei were visualized by counterstaining with DAPI (Thermo Fisher). Images were generated using the whole slide scanner Olympus BX61VS (20 × 0.75NA objective with a resolution of 0.3225 mm).
VIS software from Visiopharm was used for whole tissue immunofluorescence analysis. The protocol for the analysis is detailed in Supplementary Materials and Methods.
CC Extraction and Analysis
Polyphenols in CC were extracted in 50% and 90% MeOH and analyzed by LC-MS according to the procedure of Fracassetti and colleagues (22). The supernatants from both extracts were combined and filtered prior to analysis. We compared the relative retention times at 254 nm and negative ion mass spectra from our LC-MS analysis to those from a published characterization of CC polyphenols (22).
Identification of Castalagin as the Active Compound in CC
To discover which compounds in CC were responsible for its antitumor activity, CC was fractionated by extraction (0, 50, and then 90% MeOH) or by reverse-phase chromatography (see Supplementary Materials and Methods for detailed descriptions). Fractions were iteratively screened in the MCA-205 sarcoma mouse model to determine the active fractions.
Fluo-Castalagin Synthesis and Purification
Castalagin was monofunctionalized with fluorescein via a transesterification reaction with 5/6-carboxyfluorescein succinimidyl ester (fluorescein-NHS). Briefly, castalagin (3.2 μmol/L) was dissolved in 200 μL dimethylformamide and then reacted with fluorescein-NHS (2 eq.) in the presence of triethylamine (2 eq.) and 4-dimethyaminopyridine (Sigma). Following the workup with DOWEX 50WX8 resin, the crude mixture was analyzed by LC-MS. Peaks corresponding to monofunctionalized fluo-castalagin were isolated by HPLC.
Bacterial Strains and Culture Conditions
All bacteria were grown under anaerobic conditions at 37°C in an anaerobic chamber (Coy Laboratory Products, 5% H2, 20% CO2, 75% N2). Ruminococcus bromii (strain ATCC 27255) and Ruminococcus bicirculans (strain CRCHUM12T) were grown on anaerobe basal broth (ABB; Oxoid) agar plates. Escherichia coli (strain CRCHUM10) and Bacteroides thetaiotaomicron (strain CRCHUM9) were grown on tryptone soya agar with 5% sheep blood (Oxoid). R. bicirculans, B. thetaiotaomicron, and E. coli were isolated in our laboratory from the feces of patients with melanoma. A colony of each bacterium was inoculated into fresh ABB media and grown for 48 hours prior to the start of growth experiments.
Fluo-Castalagin Labeling Experiments on Bacterial Isolates
Stationary-phase cultures were diluted into fresh ABB media (4 mL/condition) and grown to early–mid exponential phase (OD600 ≥ 0.2). Bacteria were washed in a defined minimum medium (MM; see recipe in Supplementary Materials and Methods) and then resuspended in 176 μL of MM.
For labeling experiments at 0°C, bacteria were incubated for one hour in an ice bath prior to the addition of fluo-castalagin. For competition experiments, castalagin was added to a final concentration of 200 μmol/L prior to the addition of fluo-castalagin. The fluo-castalagin probe was added to a final concentration of 2 μmol/L and incubated with bacteria for one hour at 37°C (or 0°C for the ice condition). After incubation, bacteria were washed twice in PBS and then analyzed by flow cytometry and epifluorescence microscopy (detailed below).
Flow Cytometry Acquisition for Fluo-Castalagin Labeling Experiments
Flow cytometry analysis of fluo-castalagin–labeled bacteria was performed on an LSRFortessa cytometer (BD Biosciences) equipped with a blue (488 nm) laser. The blue 695/40-area channel was used as a background channel. A gate was established above the unstained population in the blue 530/30-area channel, and the signal for 100,000 events was measured. Bacteria within the gate were considered to be labeled by fluo-castalagin.
Epifluorescence Microscopy Acquisition for Fluo-Castalagin Labeling Experiments
Fluo-castalagin–labeled bacteria were loaded onto a slide with 0.2% Gelzan (Sigma) pads. Epifluorescence microscopy imaging of fluo-castalagin–labeled bacteria was performed using a Nikon Ti2 microscope equipped with a 60× CFI Plan Apochromat phase contrast objective with a numerical aperture of 1.40. Fluorescein imaging used a SpectraX illuminator (Lumencor) with an FITC filter set (470/24 nm excitation, 525/50 nm emission). Images were captured with a Hamamatsu Orca Fusion CMOS camera. Images were processed in FIJI as follows: images were cropped, the background from the fluorescence channel was subtracted, and an identical lookup table was applied to the fluorescence channels.
16S rRNA Analysis
Isolated DNA was analyzed using 16S rRNA gene sequences to investigate the microbial composition in fecal samples. The detailed procedure is described in Supplementary Materials and Methods.
Shotgun Sequencing Analysis
Isolated DNA was analyzed using shotgun sequencing to investigate the microbial composition in fecal samples. The detailed procedure is described in Supplementary Materials and Methods.
CC Trial in Non–Cancer Patients
As part of a CC clinical trial study (NCT04058392), two participants living with HIV with undetectable viremia were invited to take CC capsules (total 1 g, Natural Traditions) per day for four weeks in addition to their antiretroviral therapy. Feces were collected by each participant before and after four weeks of CC intake, and were kept at 4°C for a maximum of 16 hours before freezing at −80°C.
The CC study was approved by Health Canada, the Natural and Non-prescription Health Products Directorate, and the research ethics board of the McGill University Health Centre (study number 2020-5903). Each participant provided written informed consent before entering the study. The study is conducted in accordance with the Declaration of Helsinki.
Statistical Analysis
The Mann–Whitney U test was used to determine significant differences among the different groups using alpha diversity, which shows the diversity in each individual sample. DESeq2 (35) was used to perform differential abundance analysis at the genus level. All P values of differential abundance analyses have been adjusted for multiple comparisons with the Benjamini–Hochberg method. The Spearman rank correlation test was obtained using GraphPad Prism 8 and was used to compare continuous variables between the flow cytometry analysis parameters and the significant bacteria identified with differential abundance analysis in the water versus CC, water/αPD-1 versus CC/αPD-1, water/αPD-1 versus castalagin/αPD-1, and water/IsoPD-1 versus castalagin/IsoPD-1 groups for the MCA-205 and E0771 tumor models. P values were two-sided with 95% confidence intervals: *, P < 0.05; **, P < 0.01; ***, P < 0.001. The castalagin antitumor dose–response curve was constructed by fitting data to a baseline-adjusted variable slope dose–response equation in GraphPad Prism 8. Fisher exact test was used for the analysis of the frequency of metabolizers and nonmetabolizers of castalagin in the NR and R cohort patients in GraphPad Prism 8.
Data Availability Statement
The raw read data generated from the RNA-seq, metagenomic sequencing, and 16S rRNA sequencing performed in this study are deposited under BioProject ID's PRJNA783624 (RNA-seq and metagenomic sequencing) and PRJNA706824 (16S rRNA sequencing). The newly constructed 5.0M nonredundant mouse gut microbial gene catalog and associated Metagenomic Species Pan-genomes reconstructed thereof are available through dataverse INRAE-MGP (https://doi.org/10.15454/L11MXM).
Supplementary Material
Acknowledgments
We thank Liliane Meunier and Véronique Barrès of the molecular pathology core facility of the CRCHUM for performing the microscopy slide scanning. We thank the CRCHUM animal facility, Montreal Clinical Research Institute (IRCM) animal facility, and Aurélie Cleret-Buhot of the experimental imaging core facility of the CRCHUM for performing the image analysis and immunofluorescence images for figures. In addition, we thank Thibeault Varin of IBIS (Institut de Biologie Intégrative et des Systèmes, Université Laval) for his help with 16S rRNA sequencing. This study was financially supported by Institut du Cancer de Montréal (to M. Messaoudene and B. Routy); Canadian Institute for Health Research (to B. Routy and B. Castagner); Prefontaine Family and Fondation du CHUM (to B. Routy); Canada Research Chair in Therapeutic Chemistry (to B. Castagner); Réseau de Thérapie cellulaire, Tissulaire et Génique du Québec (ThéCell; to B. Routy); Canada 150 Research Chair in Bacterial Cell Biology (to Y.V. Brun); Fonds de la Recherche Québec-Santé (FRQ-S): Réseau SIDA/Maladies infectieuses, Thérapie cellulaire, and the Vaccines and Immunotherapies Core of the CIHR Canadian HIV Trials Network (CTN; grant CTN 247; to S. Isnard); Foundation scheme grant from CIHR and by the Fondation J.A. De Sève (to A. Marette); Pfizer research fund to study the pathogenesis of diabetes and cardiovascular diseases (to A. Marette); Fonds de recherche du Québec–Santé (FRQS), FRQS junior I career awards (to B. Routy); FRQS postdoctoral research scholarship (303637; to C. Richard); European Union Oncobiome (to G. Kroemer); and Seerave Foundation (to G. Kroemer).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Messaoudene reports a patent for G17004-00006-AD (use of castalagin or analogs thereof for anticancer efficacy and to increase the response to immune-checkpoint inhibitors) pending. R. Pidgeon reports grants and personal fees from Canadian Institutes of Health Research, grants from Weston Family Foundation and Canadian Glycomics Network (GlycoNet), and personal fees from Fonds de recherche du Quebec (Santé; FRQS) during the conduct of the study; other support from Micropredictome (cofounder from 2018–2019) outside the submitted work; and a patent for G17004-00006-AD (use of castalagin or analogs thereof for anticancer efficacy and to increase the response to immune-checkpoint inhibitors) pending. S. Isnard reports grants from Canadian HIV Trials Network during the conduct of the study. D. Raoult reports other support from Eurofins and Culturetop outside the submitted work. G. Kroemer reports grants from Agence Nationale de Recherche, Institut Nationale du Cancer, Ligue National contre le Cancer, and Cancéropôle Ile de France during the conduct of the study; grants from Daiichi Sankyo, Eleor, Kaleido, Lytix Pharma, PharmaMar, Samsara, Sanofi, Sotio, Vascage, and Vasculox/Tioma outside the submitted work; and is on the Board of Directors for the Bristol Myers Squibb Foundation France and is a scientific cofounder of Everimmune, Samsara Therapeutics, and Therafast Bio. A. Elkrief reports other support from Canadian Institutes of Health Research, Royal College of Physicians and Surgeons of Canada, and the Henry R. Shibata Award outside the submitted work. A. Marette reports grants from Weston Foundation during the conduct of the study. B. Castagner reports a patent for G17004-00006-AD (use of castalagin or analogs thereof for anticancer efficacy and to increase the response to immune-checkpoint inhibitors) pending. B. Routy reports grants from Davoltera, Kaleido, and Vedanta outside the submitted work, as well as a patent for G17004-00006-AD (use of castalagin or analogs thereof for anticancer efficacy and to increase the response to immune-checkpoint inhibitors) pending. No disclosures were reported by the other authors.
Authors’ Contributions
M. Messaoudene: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. R. Pidgeon: Resources, formal analysis, and methodology. C. Richard: Formal analysis, validation, and methodology. M. Ponce: Investigation. K. Diop: Investigation. M. Benlaifaoui: Investigation. A. Nolin-Lapalme: Formal analysis. F. Cauchois: Investigation. J. Malo: Investigation. W. Belkaid: Investigation. S. Isnard: Resources. Y. Fradet: Writing–review and editing. L. Dridi: Investigation. D. Velin: Formal analysis, investigation, writing–review and editing. P. Oster: Formal analysis and investigation. D. Raoult: Resources. F. Ghiringhelli: Resources, writing–review and editing. R. Boidot: Investigation and methodology. S. Chevrier: Investigation. D.T. Kysela: Investigation. Y.V. Brun: Investigation, writing–review and editing. E. Falcone: Resources. G. Pilon: Investigation. F. Plaza Onate: Software, investigation, methodology. O. Gitton-Quent: Software, investigation, methodology. E. Le Chatelier: Software, investigation, and methodology. S. Durand: Resources, formal analysis, investigation, and methodology. G. Kroemer: Resources, methodology, writing–review and editing. A. Elkrief: Writing–review and editing. A. Marette: Resources, investigation, methodology, writing–review and editing. B. Castagner: Resources, formal analysis, investigation, methodology, writing–review and editing. B. Routy: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Elkrief A, Joubert P, Florescu M, Tehfe M, Blais N, Routy B. Therapeutic landscape of metastatic non–small-cell lung cancer in Canada in 2020. Curr Oncol Tor Ont 2020;27:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis Fet al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 3. Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TKet al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 2018;378:1277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy Bet al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity 2016;44:1255–69. [DOI] [PubMed] [Google Scholar]
- 5. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. [DOI] [PubMed] [Google Scholar]
- 6. Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 2018;15:382–96. [DOI] [PubMed] [Google Scholar]
- 7. Elkrief A, El Raichani L, Richard C, Messaoudene M, Belkaid W, Malo Jet al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology 2019;8:e1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilson BE, Routy B, Nagrial A, Chin VT. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: a systematic review and meta-analysis of observational studies. Cancer Immunol Immunother CII 2020;69:343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi Het al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol 2018;29:1437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère Ret al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 11. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-Let al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TVet al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli Eet al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol 2020;78:195–206. [DOI] [PubMed] [Google Scholar]
- 14. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Yet al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019;565:600–5. [DOI] [PubMed] [Google Scholar]
- 15. Hakozaki T, Richard C, Elkrief A, Hosomi Y, Benlaïfaoui M, Mimpen Iet al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res 2020;8:1243–50. [DOI] [PubMed] [Google Scholar]
- 16. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med 2019;25:377–88. [DOI] [PubMed] [Google Scholar]
- 17. Daillère R, Derosa L, Bonvalet M, Segata N, Routy B, Gariboldi Met al. Trial watch: the gut microbiota as a tool to boost the clinical efficacy of anticancer immunotherapy. Oncoimmunology 2020;9:1774298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz Let al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371:602–9. [DOI] [PubMed] [Google Scholar]
- 19. Anhê FF, Nachbar RT, Varin TV, Trottier J, Dudonné S, Le Barz Met al. Treatment with camu camu (Myrciaria dubia) prevents obesity by altering the gut microbiota and increasing energy expenditure in diet-induced obese mice. Gut 2019;68:453–64. [DOI] [PubMed] [Google Scholar]
- 20. Bourgeois-Daigneault M-C, St-Germain LE, Roy DG, Pelin A, Aitken AS, Arulanandam Ret al. Combination of paclitaxel and MG1 oncolytic virus as a successful strategy for breast cancer treatment. Breast Cancer Res 2016;18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament Cet al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fracassetti D, Costa C, Moulay L, Tomás-Barberán FA. Ellagic acid derivatives, ellagitannins, proanthocyanidins and other phenolics, vitamin C and antioxidant capacity of two powder products from camu-camu fruit (Myrciaria dubia). Food Chem 2013;139:578–88. [DOI] [PubMed] [Google Scholar]
- 23. Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer Cet al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut 2021Jul 21 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selma MV, Beltrán D, Luna MC, Romo-Vaquero M, García-Villalba R, Mira Aet al. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid. Front Microbiol 2017;8:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derosa L, Routy R, Maltez Thomas A, Iebba V, Zalcman G, Friard Set al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small cell lung cancer. Nat Med 2022Feb 3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Isnard S, Fombuena B, Ouyang J, Royston L, Lin J, Bu Set al. Camu Camu Study Group. Camu Camu effects on microbial translocation and systemic immune activation in ART-treated people living with HIV: protocol of the single-arm non-randomised Camu Camu prebiotic pilot study (CIHR/CTN PT032). BMJ Open 2022;12:e053081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Heumen BWH, Roelofs HMJ, Te Morsche RHM, Marian B, Nagengast FM, Peters WHM. Celecoxib and tauro-ursodeoxycholic acid co-treatment inhibits cell growth in familial adenomatous polyposis derived LT97 colon adenoma cells. Exp Cell Res 2012;318:819–27. [DOI] [PubMed] [Google Scholar]
- 28. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu Met al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018;360:eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haslam E. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J Nat Prod 1996;59:205–15. [DOI] [PubMed] [Google Scholar]
- 30. Kuhn P, Kalariya HM, Poulev A, Ribnicky DM, Jaja-Chimedza A, Roopchand DEet al. Grape polyphenols reduce gut-localized reactive oxygen species associated with the development of metabolic syndrome in mice. PLoS One 2018;13:e0198716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imlay JA. Where in the world do bacteria experience oxidative stress? Environ Microbiol 2019;21:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khademian M, Imlay JA. Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci U S A 2017;114:E6922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perron NR, Brumaghim JL. A review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell Biochem Biophys 2009;53:75–100. [DOI] [PubMed] [Google Scholar]
- 34. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RMet al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science 2021;371:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw read data generated from the RNA-seq, metagenomic sequencing, and 16S rRNA sequencing performed in this study are deposited under BioProject ID's PRJNA783624 (RNA-seq and metagenomic sequencing) and PRJNA706824 (16S rRNA sequencing). The newly constructed 5.0M nonredundant mouse gut microbial gene catalog and associated Metagenomic Species Pan-genomes reconstructed thereof are available through dataverse INRAE-MGP (https://doi.org/10.15454/L11MXM).