In both healthy and cancer-bearing individuals, Th1/Tc1 response of spike protein–specific memory T cells is associated with susceptibility to SARS-CoV-2 infection.
Abstract
Vaccination against coronavirus disease 2019 (COVID-19) relies on the in-depth understanding of protective immune responses to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). We characterized the polarity and specificity of memory T cells directed against SARS-CoV-2 viral lysates and peptides to determine correlates with spontaneous, virus-elicited, or vaccine-induced protection against COVID-19 in disease-free and cancer-bearing individuals. A disbalance between type 1 and 2 cytokine release was associated with high susceptibility to COVID-19. Individuals susceptible to infection exhibited a specific deficit in the T helper 1/T cytotoxic 1 (Th1/Tc1) peptide repertoire affecting the receptor binding domain of the spike protein (S1-RBD), a hotspot of viral mutations. Current vaccines triggered Th1/Tc1 responses in only a fraction of all subject categories, more effectively against the original sequence of S1-RBD than that from viral variants. We speculate that the next generation of vaccines should elicit Th1/Tc1 T-cell responses against the S1-RBD domain of emerging viral variants.
Significance:
This study prospectively analyzed virus-specific T-cell correlates of protection against COVID-19 in healthy and cancer-bearing individuals. A disbalance between Th1/Th2 recall responses conferred susceptibility to COVID-19 in both populations, coinciding with selective defects in Th1 recognition of the receptor binding domain of spike.
See related commentary by McGary and Vardhana, p. 892.
This article is highlighted in the In This Issue feature, p. 873
Introduction
The emergence and spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), have resulted in devastating morbidities and socioeconomic disruption. The development of community protective immunity relies on long-term B- and T-cell memory responses to SARS-CoV-2. This can be achieved through viral infection (1) or by vaccination (2–4). Reports on rapidly decreasing spike- and nucleocapsid (NC)-specific antibody titers post–SARS-CoV-2 infection (5) or reduced neutralizing capacity of vaccine-induced antibodies against viral escape variants compared with the ancestral SARS-CoV-2 strain (6, 7) have shed doubts on the importance of humoral immunity as a standalone response. In contrast, T-cell immunity was identified as an important determinant of recovery and long-term protection against SARS-CoV-1, even 17 years after infection (8–11).
The Th1 versus Th2 concept suggests that modulation of the relative contribution of Th1 or Th2 cytokines regulates the balance between immune protection against microbes and immunopathology (12–14). Th1 cells (as well as cytotoxic T cells with a similar cytokine pattern, referred to as Tc1 cells) produce IFNγ, IL2, and TNFα as well as promote macrophage activation, antibody-dependent cell cytotoxicity, delayed type hypersensitivity, and opsonizing and complement-fixing IgG2a antibody production (12). Therefore, Th1/Tc1 cells drive the phagocyte-dependent host response and are pivotal for antiviral responses (13, 14). In contrast, Th2 (and Tc2) cells produce IL4, IL5, IL10, and IL13, providing optimal help for both humoral responses and mucosal immunity, through the production of mast cell and eosinophil growth and differentiation factors, thus contributing to antiparasitic and allergic reactions. Naïve T-cell differentiation to distinct Th fates is guided by inputs integrated from TCR affinity, CD25 expression, costimulatory molecules, and cytokines (15).
SARS-CoV-2–specific T-cell immunity plays a key role during acute COVID-19 and up to eight months after convalescence (16–20). Indeed, functional T-cell responses remain increased in both frequency and intensity up to six months postinfection (5). They are mainly directed against spike, membrane, and NC proteins and have been studied in greater detail by single-cell sequencing in a limited number of patients (21). Memory Th1/Tc1 T cells specific for SARS-CoV-2 and follicular T helper (Tfh) cells have been detected in mild cases (21). However, cases of reinfection have been reported (22), raising questions on the clinical significance of T-cell polarization and peptide repertoire specificities against current viral variants. Moreover, pioneering reports suggest that, before SARS-CoV-2 became prevalent (i.e., before 2020), some individuals exhibited immune responses, mainly among CD4+ T cells, against SARS-CoV-1 NC and ORF1a/b, or common cold coronaviruses (CCC) spike and NC proteins that are cross-reactive with SARS-CoV-2 (9, 23–25). However, the relevance of CCC or SARS-CoV-1–specific memory T cells for effective protection against the current pandemic remains questionable (21, 26). The current study was designed to correlate preexisting T-cell responses to clinical protection against COVID-19, in healthy individuals and patients with cancer, who are more susceptible to severe infections, and by extension to reinfection and breakthrough infection. Moreover, COVID-19 lethality was not predicted by oncologic features in patients with cancer (27), but was associated with virus-induced lymphopenia (28).
In this report, we studied SARS-CoV-2– and CCC-specific T-cell responses in 383 subjects with and without cancer, and prospectively followed up 203 COVID-19–free individuals to understand which T-cell polarity and peptide repertoire may convey resistance to COVID-19. We found that a SARS-CoV-2–specific IL2/IL5 lymphokine ratio <1 conferred susceptibility to SARS-CoV-2 infection in both health care workers (HCW) and patients with cancer, coinciding with defective Th1/Tc1 recognition of the RBD of the spike protein, likely affecting viral evolution by selecting for new antigenic variants. Moreover, vaccine-induced T-cell immunity against the S1-RBD reference strain significantly decreased against the RBD sequences of viral variants of concern in healthy subjects and patients with cancer.
Results
Effector and Memory T-cell Responses against Coronaviruses during SARS-CoV-2 Infection
We conducted a cross-sectional analysis of the functional T-cell responses across several cohorts of healthy individuals and patients with cancer enrolled during the first surge of the pandemic with the final aim of determining T-cell correlates with clinical protection against COVID-19 diagnosed until March 2021 (Fig. 1A; Supplementary Tables S1–S3; ref. 28). First, we focused on the quality of SARS-CoV-2–specific T-cell responses detected in 191 patients with cancer who stayed COVID-19–free between mid-April and September 2020, which we then compared with 19 and 28 patients with cancer in the acute and convalescence phases of SARS-CoV-2 infection, respectively (Supplementary Table S1A and S1B; Fig. 1A). In parallel, we analyzed 22 controls (COVID-19–free and cancer-free) from 15 distinct families at the same time as their 28 family members who were in the convalescent phase for COVID-19 (Supplementary Table S3; Fig. 1A). Moreover, leukocytes frozen between 1999 and 2018 in the pre–COVID-19 era belonging to either cancer-free donors from the blood bank (n = 37) or patients with cancer (n = 29) recruited in clinical trials (29–32) were used as controls of the contemporary period (Fig. 1A).
Figure 1.
SARS-CoV-2 T-cell responses in COVID-19 and unexposed individuals. A, Graphical representation of the prospective patient and healthy cohorts used for the study (refer to Supplementary Table S1A and S1B). B, First experimental in vitro stimulation assay of PBLs using cross-presentation of viral lysates by autologous DCs. Twelve plex flow-cytometric assay to monitor cytokine release in replicates. Mo-DC, monocyte-derived dendrtic cell; PBMC, peripheral blood mononuclear cell. C and D, Mean fold changes (log2 FC) between SARS-CoV-2–specific cytokine secretions of acute COVID-19 patients and convalescent COVID-19 individuals and controls (C). The columns represent the mean fold change and the adjusted P value for each cytokine between COVID-19–positive, sex- and age-matched contemporary COVID-19–negative controls (C; also refer to Supplementary Fig. S1C). Ratios of cytokine secretion between PBLs stimulated with DCs pulsed with SARS-CoV-2 (or the other CCC lysates) versus VeroE6 (or versus CCC respective control cell lines), at the acute or convalescent phase of COVID-19 (D). One typical example is outlined in Supplementary Fig. S1A. Each dot represents the mean of replicate wells for one patient (controls, n = 279, in blue; convalescent COVID-19, n = 56, in green; acute COVID-19, n = 19, in red). Statistics used the two-sided Wilcoxon–Mann–Whitney test. E, Idem as in D comparing CoV-2/VeroE6 ratios of the most relevant cytokines in cancer (gold) versus cancer-free (dark blue) convalescent individuals. Statistics used the two-sided Wilcoxon–Mann–Whitney test.
T-cell responses directed against viral lysates from the reference SARS-CoV-2 strain IHUMI846 (CoV-2) isolated in early 2020 or two endemic CCC, OC43 and 229E, were evaluated by an in vitro stimulation assay (IVS) depicted in Fig. 1B. This first 48-hour IVS assay was aimed at monitoring T-cell recall responses to viral antigens pulsed onto autologous dendritic cells (DC). Cytokine secretion was analyzed by a 12-plex flow cytometry–based bead assay (Supplementary Fig. S1A). In this cross-presentation assay, SARS-CoV-2–related cytokine release from peripheral blood lymphocytes (PBL) depended on MHC class I and II molecules, as shown using specific neutralizing antibodies (Supplementary Fig. S1B). We calculated the ratio of cytokine release by dividing interleukin concentrations following exposure to viral lysates by those obtained with the respective control supernatants, to ascribe the specificity of the reactivity to SARS-CoV-2 or to CCC antigens for each subject. First, we characterized the intensity and the quality of PBL responses elicited at the acute phase of SARS-CoV-2 infection (day of symptom onset and/or first positive qPCR of the oropharyngeal swab and/or serology), between mid-April and mid-May 2020 in 19 interpretable tests performed on COVID-19–positive subjects compared with a cohort of 279 controls (Supplementary Table S1A and S1B). Fifty percent, 36%, and 14% manifested mild, moderate, and severe disease, respectively (Supplementary Table S1A). Robust SARS-CoV-2–specific IL2 and IFNγ release, most likely caused by Th1/Tc1 cells, and the secretion of IL4 and IL5, most likely mediated by Th2/Tc2 effector T cells, were detectable (Fig. 1C and D). Of note, SARS-CoV-2 infection did not reactivate CCC-specific T-cell responses (Fig. 1D). We next examined the polarization of SARS-CoV-2–specific memory T-cell responses between mid-April and September 2020 in 56 convalescent COVID-19 individuals (median time lapse between PCR-negative and T-cell assay: 85 days, range, 13–106 days) compared with contemporary controls (Fig. 1A; Supplementary Table S1B). A mixed SARS-CoV-2–specific memory Th1/Th2 response leading to IL2, IFNγ, and IL5 was observed in most convalescent subjects within the next two to three months after acute infection (Fig. 1C and D). Differences in memory T-cell responses between COVID-19–positive individuals and unexposed controls could not be attributed to age, gender, or cancer status, as they were still statistically significant for IL2 and IL5 (as well as TNFα) in a separate analysis matching 56 convalescent patients to 56 control patients using a propensity score adjusting for age, gender, and cancer status (Supplementary Fig. S1C). More specifically, SARS-CoV-2–specific IL2 and IL5 secretion levels were comparable in cancer and cancer-free COVID-19 patients during the recovery phase independently of their comorbidities (Fig. 1E; Supplementary Table S1C–S1F). Flow-cytometric analyses of SARS-CoV-2–reactive T cells revealed central memory (TCM) Th1 (CD3+CD4+CD45RA−CCR7+T-bet+GATA3−CD69+Ki-67−) and effector memory (TEM) Tc1 (CD3+CD8+CD45RA−CCR7−T-bet+CD25+Ki-67+) phenotypes (Supplementary Fig. S1D).
Of note, SARS-CoV-2–specific IL2 release at recovery correlated with proxies of humoral immunity. Indeed, IL2+ recall responses coincided with higher frequencies of circulating nonactivated Tfh cells (Supplementary Fig. S1E; ref. 28), as well as SARS-CoV-2 NC IgG antibody titers (reported to be stable for 8 months; ref. 5), but not IgG and IgA antibodies targeting the SARS-CoV-2 S1-RBD domain (Supplementary Fig. S1F and S1G). SARS-CoV-2–specific IL5 release in COVID-19 patients correlated with calprotectin, a serum hallmark of severity (Supplementary Fig. S1H; ref. 33).
Hence, SARS-CoV-2 infection elicited memory responses leading to virus-specific release of Th1 cytokines (in 53% cases for IL2 and 26% cases for IFNγ; Fig. 2A) and of the prototypic Th2 cytokine IL5 (in 14% cases; Fig. 2A) that were detectable in both healthy subjects and patients with cancer to a comparable extent and stably over time (Supplementary Fig. S1I).
Figure 2.
Unexposed individuals susceptible to COVID-19 exhibited a SARS-CoV-2–specific Th2 profile during the first surge of the pandemic. A, Percentage and number of patients in each cohort—pre–COVID-19 era [yes (+)/no(−)], cancer [yes (+)/no(−)], and COVID-19 [yes (+)/no(−)]—who had a SARS-CoV-2–specific cytokine release (for the prototypic cytokines) compared with VeroE6 (control, n = 279; convalescent, n = 56; Supplementary Table S1B). Fisher exact test to compare the number of cytokine-positive patients across groups. B, Outline of the prospective collection of blood samples used to identify COVID-19–resistant (yellow) versus susceptible (red) patients with cancer (B, top; Supplementary Table S2A and S2B). Bottom, outline of the prospective collection of blood samples used for the comparison of T-cell responses in the cohort of cancer-free individuals who lived in the same household with family members who tested positive for COVID-19 during the 2020 lockdown (G and I). Pie chart (C) indicating the absolute numbers (and percentage) of patients reported as contact (resistant) or infected (susceptible) or unexposed (green) during 1-year follow-up (D). Number of positive cytokines released by SARS-CoV-2–specific PBLs during the cross-presentation assay (Fig. 1B and C) in each group (unexposed, n = 153; resistant, n = 42; susceptible, n = 19). E and F, SARS-CoV-2–specific IL2 (left) and IL5 (right) secretion contrasting resistant (yellow) versus infected (red) cancer cases. E, Each dot represents the ratio of the replicate wells in one individual, and the box plots indicate medians as well as 25th and 75th percentiles for each cancer patient subset. F, The bar plots represent the percentage of positive patients (resistant, n = 42; susceptible, n = 19). Fisher exact test to compare the number of cytokine-positive patients across groups. G and H, SARS-CoV-2–specific IL2/IL5 ratios (means ± SEM) in the different subsets of healthy individuals and patients with cancer presented in B. Refer to Supplementary Fig. S3A for the waterfall plots to visualize variations in the percentages of individuals with IL2/IL5 ratios > or < 1 according to subject category. All group comparisons were performed using the two-sided Wilcoxon–Mann–Whitney test, and P < 0.05 indicates statistically significant differences. I and J, Validation cohort investigating eight additional HCW from Hospices Civils de Lyon and 10 patients with cancer from Gustave Roussy investigated in cross-presentation assays with the dual-color IFNγ/IL5 ELISpot. I, Prototypic photograph of IFNγ and IL5 dual-color ImmunoSpot of a DC/SARS-CoV-2 or VeroE6 PBL coculture (or OKT3 as positive control) for one representative resistant (left) and susceptible (right) HCW. SFC, spot-forming colony counted per 105 PBLs. J, Percentages of SARS-CoV-2–specific Th1 or Th2 cell responses determined by dual ELISPOT assay (CoV-2/VeroE6 >1.5 increase in IFNγ+ (left) or IL5+ (middle) SFC, respectively. Calculation of the IFNγ+/IL5+ SFC ratio per individual in VeroE6 or SARS-CoV-2 condition, and percentages of patients with an increased (>2×) ratio in the SARS-CoV-2 condition, in both resistant versus susceptible groups (right). Fisher exact test to compare the number of positive patients between both groups.
Clinical Relevance of Preexisting Th1/Th2 Immunity to Predict SARS-CoV-2 Infection
Unsupervised hierarchical clustering considering 12 cytokines monitored in 355 subjects did not segregate contemporary unexposed individuals from convalescent patients (Supplementary Fig. S2A). As previously described (21, 23, 24, 34, 35), contemporary COVID-19–negative subjects also harbored spontaneous (cross-reactive) SARS-CoV-2–specific IL2, IFNγ, and IL5 release in 17.8%, 12.6%, and 13.1% cases, respectively (Fig. 2A), as well as polyfunctional memory responses that appear to preexist in patients with cancer and healthy individuals in the pre–COVID-19 era, even prior to outbreaks of SARS-CoV-1 and Middle East respiratory syndrome (MERS; Fig. 1A; Supplementary Fig. S2A and S2B). Preexisting frequencies of SARS-CoV-2–specific IL2+ and IL5+ T-cell responses were comparable in individuals with or without cancer, with no impact of cancer staging, hematologic versus solid malignancy, therapy, or comorbidities (Supplementary Fig. S2C and S2D; Supplementary Table S1C–S1F).
To determine the clinical significance of these memory T-cell responses monitored in unexposed patients with cancer from mid-April to mid-May 2020 to predict susceptibility or resistance to SARS-CoV-2 infection, we called 214 patients with cancer (Fig. 2B) to discover contact cases (n = 61) and infections (n = 19) diagnosed by qPCR or serology during the successive surges of this viral pandemic in fall 2020 and winter 2021. Hence, about 28.5% of the initially COVID-19–free individuals became contact cases, and 31.1% among these contact cases were diagnosed with SARS-CoV-2 infection by specific qRT-PCR or serology (Fig. 2C; Supplementary Table S2A). Five patients developed moderate or severe COVID-19 according to the World Health Organization (WHO) criteria (Supplementary Table S2A). The polyfunctionality of T-cell responses failed to segregate the two categories of patients with cancer (Fig. 2D; Supplementary Fig. S2B). However, distinct SARS-CoV-2–specific cytokines appeared relevant to predict resistance or susceptibility to SARS-CoV-2 (Fig. 2E and F). Indeed, both the levels of IL2 in the recall response and the proportions of individuals exhibiting IL2-polarized T-cell memory responses were associated with resistance to SARS-CoV-2 infection (Fig. 2E and F; P = 0.017, two-sided Wilcoxon–Mann–Whitney test, and P = 0.048, Fisher exact test). In contrast, IL5 levels in recall responses tended to be associated with increased susceptibility to SARS-CoV-2 infection (Fig. 2E; P = 0.057, two-sided Wilcoxon–Mann–Whitney test).
Consequently, we analyzed the clinical significance of the ratio between SARS-CoV-2–specific IL2 and IL5 release. The IL2/IL5 recall response ratio was significantly higher in patients with cancer who were SARS-CoV-2 resistant (Fig. 2G and H) and in convalescent patients (Fig. 2G; Supplementary Fig. S3A). The vast majority of patients with cancer deemed to be infected with SARS-CoV-2 exhibited an IL2/IL5 ratio ≤1, with the two severe COVID-19 cases displaying an IL2/IL5 ratio <0.1 (Fig. 2H).
The SARS-CoV-2–specific IL2/IL5 recall response ratio was also clinically significant in a cohort of cancer-free individuals who were locked down with their COVID-19–positive family members (Fig. 2B; Supplementary Table S3). Individuals who did not get infected harbored IL2/IL5 ratios >1 reaching mean values comparable with those achieved in convalescent individuals (Fig. 2G; Supplementary Fig. S3A). We next utilized the double-color IFNγ/IL5 ELISpot assay to enumerate cytokine-producing T cells in blood from cancer (n = 8) and cancer-free (n = 10) individuals drawn in March 2020 and followed up for 12 months for the COVID-19 diagnosis (Fig. 2B; Supplementary Table S2B). Although six of nine resistant subjects (who did not develop COVID-19) exhibited a SARS-CoV-2–specific 2-fold increase in IFNγ+/IL5+ spot ratios, none of the nine susceptible subjects (who developed asymptomatic or mild CeOVID-19) did so (Fig. 2I and J). Moreover, the frequency of IL5-secreting cells detected in ELISpot assays correlated with the IL5 levels monitored in the first IVS ELISA assay and with the proliferation of CD8+CCR4+T-bet− during the cross-presentation assay (Supplementary Fig. S3B and S3C). Finally, the transcription profile of PBLs in the cross-presentation assays leading to IL2/IL5 ratios > versus <1 performed in 18 patients (8 with an IL2/IL5 ratio >1 and 10 with an IL2/IL5 ratio <1) was enriched in genes expressed in Th1/Tc1 (e.g., IFNG and GZMB) versus Th2/Tc2 (e.g., CXCR5 and CD79A), respectively (Supplementary Fig. S3D; Supplementary Table S4).
In contrast to preexisting SARS-CoV-2–specific memory T cells, CCC-specific cross-reactive T cells did not allow us to differentiate susceptible from resistant individuals (Supplementary Fig. S3E), although IL5 (not IL2) stood out as the strongest correlate between SARS-CoV-2– and OC43-specific T-cell responses among 156 individuals (Supplementary Fig. S3F). Of note, titers of IgG antibodies directed against the spike of the seasonal beta coronaviruses OC43 and HKU1 (but not the alphacoronavirus 229E and NL63) were higher in individuals susceptible to SARS-CoV-2 compared with resistant individuals (Supplementary Fig. S3G).
We next compared the T-cell polarization of healthy multicontact COVID-19–free individuals (Supplementary Table S5), resistant patients with cancer (Supplementary Table S2A) or SARS-CoV-2–reinfected (Supplementary Table S5) patients toward the original SARS-CoV-2 strain (IHUMI846) with that directed toward the United Kingdom (IHUMI3076, B.1.1.7), South Africa (IHUMI3147, B.1.351), and Brazil (IHUMI3191, P.1; ref. 25) viral variants of concern (VOC) in the cross-presentation assay. Some individuals lost the Th1/Tc1 profile and acquired a Th2/Tc2 profile (IL4, IL5, IL10), depending on the strain (Supplementary Fig. S4A). We also compared the immunogenicity of the original IHUMI846 strain with that of the Danish (IHUMI2096, 20A.EU2, B.1.367, GH) and North African (IHUMI2514, 20C, B.1.160, GH) strains isolated at the end of 2020 (25). T cells lost their capacity to produce IL2 in response to the IHUMI2096 and IHUMI2514 viral variants (Supplementary Fig. S4B).
We conclude that an imbalanced Th1/Tc1 versus Th2/Tc2 polarity of SARS-CoV-2–specific memory T-cell responses determines susceptibility to infection, with an IL2/IL5 ratio >1 indicating resistance to SARS-CoV-2 infection.
Defects in the Th1/Tc1 Response against the SARS-CoV-2 RBD of Spike Glycoprotein in Susceptible Individuals
In hosts affected by viral infections or cancer, the breadth of T-cell epitope recognition is a prerequisite for protective immunity (36–38). We analyzed the diversity of SARS-CoV-2 T-cell responses by single peptide mapping using 187 peptides with 9 to 51 amino acids corresponding to 146 nonoverlapping or poorly overlapping epitopes of the SARS-CoV-2 Open Reading Frame peptidome (ORFeome; among which 25 epitopes were shared with SARS-CoV-1), encompassing in the 5′UTR to the 3′UTR sequence order, spike, ORF3a, membrane, ORF8, NC, and ORF10 structural proteins, plus 41 epitopes covering the SARS-CoV-1 ORFeome of immunologic relevance (among which eight epitopes were shared with SARS-CoV-2), as well as a series of positive controls, namely, epitopes from influenza virus, Epstein–Barr virus (EBV), and cytomegalovirus (CMV), phytohemagglutinin (PHA), and anti-CD3ε (OKT3) antibody (Supplementary Table S6). IFNγ responses against the 187 peptides were evaluated in 211 individuals (124 patients with cancer, 63 cancer-free individuals, 24 pre–COVID-19 era, 27 convalescent patients; Supplementary Table S7). To enable the detection of low-frequency SARS-CoV-2 peptide–specific T cells, we used an in vitro 7-day-long, IL2 + IL15–enriched IVS assay in the presence of each individual peptide (Fig. 3A). We chose to monitor IFNγ, a proxy for Th1/Tc1 responses, as opposed to IL2, in the 7-day coculture supernatants by ELISA because recombinant human IL2 was already added to the IVS assay to maintain T-cell viability. The overall recognition patterns of these peptides across various patient populations, and their individual frequencies are detailed in Fig. 3B and C and Supplementary Fig. S5. About 10% of convalescent individuals recognized more than 15% of our peptide selection within the SARS-CoV-2 ORFeome (Fig. 3B). T-cell responses in unexposed patients, in particular in the pre–COVID-19 era, covered large specificities, as suggested by previous reports (refs. 9, 21, 24; Fig. 3C and D, right; Supplementary Fig. S5). In accordance with the literature (9, 24), the T-cell repertoire of convalescent COVID-19 patients was larger than that of unexposed individuals, mainly directed against spike, membrane, and NC and to a lesser extent against ORF3a, ORF8, and ORF10 (Fig. 3E, left). The breadth of the peptide recognition coverage was not significantly reduced in patients with cancer compared with others (Fig. 3D, left; Fig. 4A). In a limited number of individuals, we measured not only IFNγ but also IL5, IL9, and IL17 by ELISA. The recognition profile specific to the spike (and more specifically the RBD) as well as ORF8 was more geared toward Th1/Tc1 (IFNγ) than Th2 (IL5), Th9 (IL9), or Th17 (IL17) production (Supplementary Fig. S6A–S6C). The membrane- and NC-specific repertoire was strongly Th17-oriented (Supplementary Fig. S6B).
Figure 3.
Peptide repertoire breadth does not predict resistance to COVID-19. A, Experimental setting for the 187 peptide-based in vitro stimulation assay. B, Bicolor map of peptide recognition (positive in salmon, negative in purple, not determined in gray). Patients (n = 148) were ordered in columns by unsupervised hierarchical clustering, and peptides were ordered in rows according to the 5′ to 3′ sequence location in the ORFeome with a distinct color code for each protein. SARS-CoV-1 peptides are aligned at the end in gray. The upper line indicates the frequency of positive individuals for each peptide in the 187 peptide list. C, Peptide frequencies within unexposed and convalescent (with history of COVID-19) patients with cancer compared with unexposed cancer-free subjects. Also refer to Fig. 4A. D and E, Percentages of positive peptides in individuals from the pre–COVID-19 era (n = 24) versus contemporary controls (n = 97; D, right) and in cancer (n = 111) versus cancer-free contemporary individuals (n = 10; D, left) and in uninfected [control (contemporary), n = 97] versus convalescent (n = 27; E, left) and resistant individuals (noninfected contact cases, n = 44) versus susceptible (infected, n = 18) individuals (E, right). Group comparisons within D and E were performed using the two-sided Wilcoxon–Mann–Whitney test.
Figure 4.
Spike receptor binding domain (S1-RBD)–directed Th1/Tc1 recall responses predict resistance to COVID-19. A, Statistically significant peptide signatures in the peptide-based IVS assay (Fig. 3B) using a multivariable logistic regression analysis adjusted for period (pre–COVID-19 era or contemporary patients), COVID-19 history, and cancer (refer to Supplementary Table S7). The left column shows variables, and the x-axis indicates the significant peptides (P < 0.05). The magnitude of the log (odds ratio) is indicated in the red/blue color code, whereas that of the P value is represented by the circle size. B, Linear regression analysis of the relative contribution (t-value corresponding to the regression coefficient) of each peptide to SARS-CoV-2–specific Th1/Tc1 responses (measured as IL2 secretion in response to whole virus lysate in Fig. 1D), as determined in the peptide-specific IFNγ secretion assay in 123 COVID-19–negative individuals. Statistically significant peptides (P < 0.05) are annotated with asterisks (left). Peptides colored in blue reportedly harbor at least one mutation within SARS-CoV-2 variants (Supplementary Table S12). Peptide set enrichment analysis plot (right). The contribution of each peptide to the SARS-CoV-2–specific IL2 secretion was used to rank 164 peptides. The enrichment score of S1-RBD peptides suggested that this peptide set presented lower t-values than randomly expected (P = 0.048; right). C, Volcano plot showing statistical significance (P values) and magnitude of change in odd ratios of IFNγ secretion in response to SARS-CoV-1 (sarbecovirus) and SARS-CoV-2 peptides belonging to distinct viral proteins (each scatter plot) between susceptible versus resistant individuals. D–H, Percentages of patients recognizing at least one of the 11 S1-RBD peptides in the IFNγ ELISA of the peptide IVS assay across patients’ groups (D) or convalescent versus reinfected patients (G) or vaccinees experiencing breakthrough infection (H; Supplementary Table S8), or recognizing at least one peptide from the pre–COVID-19 (E) or convalescent (F) signature identified in the logistic regression analyses of A in the IFNγ ELISA in the peptide IVS assay. Fisher exact test to compare the number of positive patients for each signature between groups.
Using logistic regression analyses, we determined the Th1/Tc1 peptide recognition fingerprint significantly associated with each patient category (Fig. 4A). The hallmark repertoire of the pre–COVID-19 era consisted of a stretch of peptides covering part of the SARS-CoV-1 genome (spike, membrane, ORF3a, NC), some peptide residues sharing high or complete homology with SARS-CoV-2, as well as numerous ORF8 sequences (Supplementary Table S6). Of note, the recognition pattern of these SARS-CoV-1 epitopes highly correlated with responses directed against ORF8 peptides. In contrast, the COVID-19–associated blueprint encompassed many NC peptides (NC_1, residues 1–15), NC_6–7, (residues 76–105), the HLA-A2–restricted nonamer (RLNQLESKV) NC_226–234 from SARS-CoV-1 (sharing high structural homology with the SARS-CoV-2 epitope RLNQLESKM) and another SARS-CoV-1 NC nonamer peptide (NC_345–361), three peptides residing in ORF8, and two epitopes belonging to the spike region [“SPIKE29” found in the S1-RBD region at high frequency across subjects (17.8%), as well as “SPIKE84” (residues 1246–1260) from the C-terminal portion; Fig. 4A].
Next, we investigated the ORFeome peptide repertoire associated with SARS-CoV-2–specific IL2 (supposedly protective) memory responses in 148 unexposed and convalescent individuals by means of linear regression analysis (Fig. 4B, left). Among the nine peptides associated with a positive contribution to IL2 secretion, one nonamer (KLPDDFMGCV in the SARS-CoV-1 genome and KLPDDFTGCV in the SARS-CoV-2 genome) resided in the RBD region that constitutes the binding site for its cellular receptor angiotensin-converting enzyme 2 (ACE2; ref. 39), whereas, among the 13 peptides associated with a hole in the Th1 response, five resided within the RBD of the spike glycoprotein. More specifically, there was a statistically significant enrichment of RBD-related peptides within this Th1/Tc1 hole (Fig. 4B, right).
In order to validate the clinical significance of the Th1/Tc1 repertoire hole and the assumption that a defect in the Th1/Tc1 recognition pattern of the RBD sequence could be a risk factor for COVID-19, we annotated the presence of at least one positive peptide selected from the RBD region spanning amino acid 331–525 residues (called “SPIKE23” to “SPIKE35” in Supplementary Table S6), versus other regions of the ORFeome in each of the 98 individuals who were comprehensively explored in the peptide-based IVS assay, 45 resistant (contact) individuals, 18 infected persons (susceptible), as well as 35 controls (unexposed lockdown and/or unknown) in addition to 24 individuals from the pre–COVID-19 era (Supplementary Table S7) using the IFNγ ELISA. The volcano plot assigning significant odd ratios of Th1/Tc1 reactivities to different SARS-CoV1/CoV2 amino acid sequences between susceptible versus resistant individuals highlighted that anti–S1-RBD Th1/Tc1 reactivity selectively correlated with resistance to infection (Fig. 4C and D). In accordance with the immunodominance of S1-RBD, the other signatures indicated by our logistic regression analysis (Fig. 4A), namely, the convalescent or the pre–COVID-19 era–related blueprints were not significantly associated with resistance to SARS-CoV-2 infection (Fig. 4E and F). Although susceptible patients with cancer exhibited a significant defect in the RBD-related Th1/Tc1 repertoire (Fig. 4D), up to 25% of the patients with resistant cancer harbored robust Th1/Tc1 responses to the 331–525 amino acid residues of RBD (Fig. 4D; P = 0.049, Fisher exact test).
Next, we analyzed peripheral blood mononuclear cells (PBMC) in a series of cancer-free individuals (n = 17) who were diagnosed with COVID-19 during the first surge of the SARS-CoV-2 pandemic and then were reinfected with viral variants prevailing during the later outbreak occurring in fall 2020 or winter 2021 (Supplementary Table S5). The RBD-specific Th1/Tc1 responses were almost undetectable in patients who got infected twice with SARS-CoV-2, whereas they could be measured in 50% of convalescent COVID-19 patients (Fig. 4G; P = 0.011, Fisher exact test), which is in accordance with a recent report highlighting the immunodominance of the S346–365 region (corresponding to our “SPIKE24” epitope) in convalescent individuals (40). Third, patients diagnosed with COVID-19 breakthrough infections more than 1 month after complete vaccination (Supplementary Table S8) harbored a major defect in S1-RBD–specific Th1/Tc1 cell responses (Fig. 4H). Of note, neutralizing antibody titers were above the detection limit in 66% of COVID-19 patients infected once versus 40% of reinfected patients. In contrast, in vaccinees experiencing breakthrough infection, IgG antibody titers against trimeric spike assessed within 2 months after second vaccine were comparable to levels measured in unaffected vaccinees (Supplementary Fig. S6D). Thus, the cellular anti–S1-RBD Th1/Tc1 response might be a better predictor of protection against SARS-CoV-2 infection than the humoral response against trimeric spike.
IFNγ and IL5 T-cell responses to S1-RBD peptides were evaluated in 67 patients. About 10% of individuals harbored S1-RBD–specific Th2/Tc2 responses (Supplementary Table S9). Long-lived Th2 clones could be derived from two patients exhibiting robust spontaneous or breakthrough SARS-CoV-2 infection or SPIKE25-specific IL5 release (Supplementary Fig. S7A–S7F). Of note, there was a robust concordance of the polarization status of patients between the two (cross-priming and peptide-based) IVS assays (P = 2.2e−16 for the Th1/Tc1 cytokines IL2 and IFNγ release; p < 1e−16 for the Th2/Tc2 factor IL5, McNemar test).
Vaccine-Induced S1-RBD Th1 Immunity Observed in Patients with Solid Cancer Is Reduced in Hematologic Malignancies
During the course of this study, SARS-CoV-2 mRNA and DNA vaccines were approved by the FDA and the European Medicines Agency (EMA) based on reports that they prevent SARS-CoV-2 infection with an efficacy of >90% (3, 41). Using a simple 22-hour whole-blood stimulation assay allowing the quantitative measurement of IFNγ using the enzyme-linked fluorescent assay technique in an automated platform (VIDAS IFNγ RUO; ref. 42), we analyzed RBD-specific T-cell reactivities before and/or after one or two shots of vaccination with BNT162b2 mRNA (BioNTech/Pfizer) and/or AZD1222 adenovirus (AstraZeneca) in 368 patients (Supplementary Table S10)—259 cancer-free and 109 patients with cancer—including >50 convalescent individuals before and/or after one vaccine (Fig. 5A and B; Table 1). First, we used our 11 S1-RBD nonoverlapping peptide pool (“PEPwtRBD”; Fig. 5C; Supplementary Table S11). PBL reactivities to these peptide pools were MHC class I and II–dependent (Supplementary Fig. S8A). As a positive control of memory responses against SARS-CoV-2 (43), we used a pool of 18 15-mer epitopes, “PEPOrf,” comprising not only different stretches of overlapping S1-RBD peptides but also peptides spanning Spike S1 and S2 as well as membrane and NC sequences (Fig. 5C; Supplementary Table S11). At day 180 after vaccine initiation, about 40% of HCW (with no history of COVID-19 or cancer) mounted PEPwtRBD-specific Th1/Tc1 responses, whereas >80% responded to PEPOrf, reaching similar levels as individuals with a history of COVID-19 and one course of vaccination (Fig. 5D, top; Table 1). The magnitude of PEPwtRBD-specific IFNγ release after vaccination (day 90) was maintained up to day 180 in both patient subsets (Fig. 5D, bottom). Although vaccination could elicit Th1/Tc1 immune responses against S1-RBD in patients with solid cancer independently of concomitant chemotherapy, immunotherapy, Eastern Cooperative Oncology Group (ECOG) performance status, and staging (Supplementary Fig. S8B–S8F), the percentages and magnitude of responses against PEPOrf and PEPwtRBD were significantly reduced only in hematologic malignancies (Table 1) as compared with cancer-free individuals and patients bearing solid cancers in univariate analysis (Fig. 5E). Multivariate analyses concluded that administration of two vaccines or SARS-CoV-2 infection followed by one vaccine elicited significant Th1/Tc1 immune responses against S1-RBD independently of age, gender, and time of sampling but was reduced in patients with hematologic cancer compared with healthy subjects (Fig. 5F; P = 0.025). We acknowledge that the current study enrolled too few patients diagnosed with hematologic malignancies to allow fair comparisons in the magnitude of vaccine-induced S1-RBD–specific IFNγ release between solid and hematologic malignancies. Of note, the titers of S1-RBD IgG antibodies poorly correlated with PEPwtRBD-specific T-cell IFNγ secretions in 232 cancer-free vaccinees without a history of COVID-19 (Fig. 5G, left; R = 0.2, P = 0.019) as well as in patients with cancer (Fig. 5G, right; R = 0.13, P = 0.32).
Figure 5.
Patients with cancer (except hematologic malignancies) could mount S1-RBD–specific Th1/Tc1 immune responses during the prime–boost vaccination rollout. A, Description of cohorts of vaccinees in cancer-free individuals and patients with cancer (refer to Supplementary Table S10; Table 1). B, Experimental setting for the peptide pool–based ex vivo stimulation assays. C, Amino acid sequence coverage of the three peptide pools utilized in the high-throughput T-cell screening assay (refer to Supplementary Table S11). D and E, High-throughput screening T-cell assay using the ELISA technique in an automatic platform monitoring IFNγ levels in whole-blood samples from several independent cohorts of HCW (D) or patients with cancer [E, solid or hematologic malignancies (hemato cancer)] with (D) or without (D and E) COVID-19 history, pre- and/or per (after 1 immunization, day 21) and/or post-vaccination (day 90, day 180 for D; only after two shots of vaccines for E) using different peptide pools (C). Monitoring of IFNγ release (bottom) and percentages of individuals with IFNγ levels greater than the threshold of detection (top). The standard errors have been computed with their confidence intervals for these estimates, with each interval most probably containing the genuine percentage. F, Forest plot depicting the impact of the each covariate on the PEPwtRBD IFNγ secretion levels (refer to Table 1 for statistics). Specimens were not systematically paired in the kinetic study. The log10-normalized IFNγ secretions for all peptide stimulation were pooled to model simultaneously their dynamics from the first vaccine to day 180 using linear mixed-effect regression adjusted for patient age, sex, cancer status, type of cancer, COVID history, and vaccine schedule. G, Spearman correlation between serum S1-RBD–specific IgG titers (expressed in arbitrary units) and IFNγ release in the VIDAS IFNγ RUO platform in all cancer-free (left) and cancer vaccinees (right) monitored in Fig. 5D. Each dot represents one sample at one time point. Most individuals have been drawn only once at any time point. H, Percentages and absolute numbers of mutations contained in our S1-RBD peptide list reported in the current SARS-CoV-2 variants (refer to Supplementary Table S12). The difference of the probability of mutation in the S1-RBD region and in other regions was evaluated using logistic regression (odds ratio = 0.21; 95% confidence interval, 0.06–0.68; P = 0.01). I, Paired analysis of the differential magnitude of Th1/Tc1 reactivity against PEPwtRBD versus PEPmutRBD in 343 cancer-free vaccinees with no history of COVID-19. Each line represents one patient sample. Group comparisons were performed using the two-sided paired Wilcoxon–Mann–Whitney test.
Table 1.
Efficacy of FDA/EMA-approved vaccines to elicit anti-WT– versus mutRBD–specific Th1/Tc1 responses: multivariate analyses
| PEPorf | PEPwtRBD | PEPmutRBD | ||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | n | % | Multivariable estimate (95% CI) | P | Multivariable estimate (95% CI) | P | Multivariable estimate (95% CI) | P |
| Gender | ||||||||
| Female | 259 | 70 | Reference | Reference | Reference | |||
| Male | 109 | 30 | −0.072 (−0.163–0.026) | 0.159 | −0.041 (−0.104–0.033) | 0.264 | −0.073 (−0.162–0.012) | 0.123 |
| Age (years) mean ± SEM | 43 ± 14 | 0.003 (−0.001–0.007) | 0.086 | 0 (−0.003–−0.002) | 0.878 | 0 (−0.003–0.004) | 0.831 | |
| Range (18–86) | ||||||||
| Sampling time (May 10, 2021–September 8, 2021) | 0.002 (0–0.005) | 0.049 | 0.001 (−0.001–0.003) | 0.551 | −0.001 (−0.003–0.001) | 0.433 | ||
| Range (days): (0–135) | ||||||||
| COVID history and vaccine rounds | ||||||||
| Pre-vacc | 103 | 24 | Reference | Reference | Reference | |||
| No COVID-19 history; 1 round | 60 | 14 | 0.282 (0.173–0.395) | <0.001 | 0.118 (−0.006–0.247) | 0.068 | 0.044 (−0.034–0.121) | 0.291 |
| No COVID-19 history; 2 rounds | 195 | 45 | 0.442 (0.332–0.560) | <0.001 | 0.197 (0.098–0.292) | <0.001 | −0.016 (−0.116–0.08) | 0.751 |
| Convalescent; no round | 45 | 11 | 0.347 (0.207–0.502) | <0.001 | 0.087 (−0.039–0.234) | 0.185 | −0.064 (−0.183–0.044) | 0.257 |
| Convalescent; 1 round | 27 | 6 | 0.603 (0.422–0.766) | <0.001 | 0.175 (0.028–0.322) | 0.014 | 0.011 (−0.171–0.166) | 0.902 |
| Cancer-free | 259 | 70 | Reference | Reference | Reference | |||
| Cancer | 109 | 30 | — | — | — | — | — | — |
| Solid malignancies | 88 | 81a | 0.004 (−0.154–0.176) | 0.963 | −0.047 (−0.167–0.076) | 0.453 | 0.045 (−0.1–0.175) | 0.532 |
| Breast | 30 | 28a | — | — | — | — | — | — |
| Gastrointestinal | 19 | 17a | — | — | — | — | — | — |
| Lung | 7 | 6a | — | — | — | — | — | — |
| Head and neck | 9 | 8a | — | — | — | — | — | — |
| neurologic tumor | 1 | 1a | — | — | — | — | — | — |
| Melanoma | 7 | 6a | — | — | — | — | — | — |
| Gynecologic | 7 | 6a | — | — | — | — | — | — |
| Genitourinary | 5 | 5a | — | — | — | — | — | — |
| Sarcoma | 1 | 1a | — | — | — | — | — | — |
| Thymus | 1 | 1a | — | — | — | — | — | — |
| Unknownb | 1 | 1a | — | — | — | — | — | — |
| Hematologic malignancies | 21 | 19a | −0.361 (−0.594–−0.124) | 0.002 | −0.172 (−0.312–−0.024) | 0.025 | −0.038 (−0.212–0.138) | 0.681 |
| B-cell malignancies | 12 | 11a | — | — | — | — | — | — |
| Myeloid malignancies | 4 | 4a | — | — | — | — | — | — |
| Multiple myeloma | 4 | 4a | — | — | — | — | — | — |
| Others | 1 | 1a | — | — | — | — | — | — |
| Tumor stage | ||||||||
| Localized | 13 | 12 | — | — | — | — | — | — |
| Locally advanced | 21 | 19 | — | — | — | — | — | — |
| Metastatic | 70 | 64 | — | — | — | — | — | — |
| Unknown | 5 | 5 | — | — | — | — | — | — |
| Therapies | ||||||||
| Not treated | 22 | 20 | — | — | — | — | — | — |
| Chemotherapy | 42 | 39 | — | — | — | — | — | — |
| Hormonotherapy | 4 | 4 | — | — | — | — | — | — |
| Immunotherapy | 21 | 19 | — | — | — | — | — | — |
| Radiotherapy/radiofrequency | 2 | 2 | — | — | — | — | — | — |
| Targeted therapy | 33 | 30 | — | — | — | — | — | — |
| ECOG performance status | ||||||||
| 0–1 | 102 | 94 | — | — | — | — | — | — |
| ≥2 | 7 | 6 | — | — | — | — | — | — |
Abbreviations: CI, confidence interval; Pre-vacc, prevaccination.
aPercentages in cancer group (n = 109).
bCancer diagnostic phase.
Given that immunoselection may drive antigenic drift of viruses as well as the evolution of viral phylogeny, we analyzed the coincidence of mutations (mutations occurring in at least 75% of emergent variants or predicted to decrease antibody neutralizing activity) in the SARS-CoV-2 ORFeome (44) with T-cell memory patterns of clinical significance (Supplementary Table S12). Significantly higher mutation frequencies were detected within the S1-RBD–specific Th1 response (62%) compared with other regions of the SARS-CoV-2 ORFeome (25.5%; odds ratio = 0.21; 95% confidence interval, 0.06–0.68; P = 0.01; Fig. 5H).
Finally, we analyzed T-cell responses directed against S1-RBD sequences of the viral VOCs that were recently renamed by WHO as alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2). Indeed, these strains predominantly mutate in the S gene compared with the reference (Wuhan-Hu-1) strain and more precisely within the S1-RBD peptide residues of the “PEPwtRBD” pool. Therefore, we generated a fourth peptide pool, “PEPmutRBD,” encompassing the 14 mutations described within the S1-RBD sequences of VOC (Supplementary Table S11) that we tested in 343 individuals. Th1/Tc1 cell reactivity tended to be higher against PEPwtRBD than PEPmutRBD in univariate analyses [n = 33 positive/83 (39.7%) vs. 20/71 (28%) at day 180 in HCW, P = 0.337; n = 7 positive/11 (63%) vs. 3/8 (37%) at day 180, P = 0.69 in COVID-19 convalescent patients], coinciding with a significant drop in the magnitude of IFNγ secretion levels in cancer-free individuals (Fig. 5I; P < 0.001). In multivariate analyses, vaccines failed to elicit significant Th1/Tc1 immune responses cross-reactive against VOC (Table 1). The difference in T-cell reactivity between PEPwtRBD and PEPmutRBD could not be ascribed to nonmutated peptide residues missing in the PEPmutRBD pool (such as the immunodominant spike 29, which was recognized in <3% of vaccinees when tested separately in this high-throughput screening T-cell assay (P = 0.4; Supplementary Fig. S8G).
Of note, the binding affinity of S1-RBD peptides to MHC class I and II proteins could be calculated using the NetMHCpan algorithm. This approach predicted strong binding to MHC class I HLA-A, -B, and -C alleles for the RBD epitopes “SPIKE25” (residues 361_375), “SPIKE27” (residues 391–405), and “SPIKE31” (residues 451–465). In contrast, “SPIKE33” (residues 481_495) was estimated to have a low affinity for HLA-B and no affinity for HLA-C alleles (Supplementary Table S13A). Only “SPIKE24,” “SPIKE25,” and “SPIKE31” were predicted to bind with a high affinity to MHC class II HLA-DR alleles (Supplementary Table S13B), as already reported for the immunodominant S346–365 region (40).
Altogether, these results suggest that defects in the Th1/Tc1 repertoire affecting the recognition of the SARS-CoV-2 S1-RBD, mostly observed in patients with hematologic malignancies rather than solid cancer or cancer-free individuals, are associated with susceptibility to infection or reinfection by SARS-CoV-2. T-cell responses against S1-RBD from VOCs appear to be reduced in vaccinees as of August 2021, commensurate with the fact that this antigenic region mutates more than other regions of the SARS-CoV-2 ORFeome.
Discussion
Identifying immune correlates of protection from SARS-CoV-2 is critical to predict the efficacy of existing and future vaccines and to follow a potential decay in immune protection imposing repeated immunizations. Thus, the titers of neutralizing antibodies that correlate with IgG antibodies against trimeric S or RBD represent a good proxy of protection against breakthrough infections (45, 46). The landscape of prevalence and immunodominance of SARS-CoV-2 T-cell epitopes—supposedly associated with protection during the acute phase—has been thoroughly investigated (43). Using 40-mer peptide pools covering regions of membrane, NC, ORF3a, ORF7/8, and spike proteins, Tan and colleagues observed a statistically significant correlation between the early appearance of SARS-CoV-2 peptide–reactive cells and shorter duration of infection (47). Here, we unravel the first “prospective” correlation between preexisting (before the first surge) SARS-CoV-2–specific Th2/Tc2 immune responses and susceptibility to infection with SARS-CoV-2 or reinfection with viral variants, based on three independent cohorts and two different methods to monitor Th1/Tc1 and Th2/Tc2 cytokines (ELISA and ELISpot). In both healthy individuals and cancer subjects, the best immunologic correlate for the susceptibility to infection with SARS-CoV-2 was undistinguishably a recall response characterized by a low ratio of Th1/Th2 lymphokines (and more precisely an IL2/IL5 ratio <1) secreted upon exposure to the reference SARS-CoV-2 viral strain. The IL5 memory response coincided with a hole within the Th1/Tc1 cell repertoire affecting the RBD of the spike protein. Five lines of evidence argue in favor of the clinical significance and protective effect against the infection of Th1/Tc1 immune responses directed against anti–S1-RBD for the current pandemic. Th1/Tc1 responses were undetectable in individuals from the prevaccine era who were susceptible to infection by SARS-CoV-2, in reinfected persons, and in subjects manifesting breakthrough infections after vaccination and were reduced against the S1-RBD–mutated sequences from VOC in vaccinated HCW. Finally, given the high rate of mutations residing in the immunologically and clinically relevant sequence of interest (331–525 amino acid residues of the spike protein), we are tempted to conclude that an immune-driven selection process of viral phylogeny is currently occurring, as already discussed (48, 49).
Reportedly, CD4+ Th1 and Th2 responses are induced during the primary phase of viral infection, and both Th1 and Th2 can generate an anamnestic response upon rechallenge with the same virus (50). Survivors from SARS-CoV-1 infection developed polyfunctional T cells producing Th1 cytokines and long-term CD8+ T-cell responses as late as 11 years after infection (9). The Th1 cytokine IL2 (which correlated with circulating nonactivated Tfh cells in convalescent patients in our study) was the pivotal factor distinguishing resistant from susceptible individuals. Signaling via the high-affinity IL2 receptor (which requires CD25/IL2Rα expression) favors the generation of CXCR5− T effector cells, and this is associated with Th1 responses sustained by the transcription factor TBX21. Moreover, the development of IFNγ-producing effector memory T cells depends upon CD25 (15). Accordingly, upon infection with lymphocytic choriomeningitis (LCMV), CD25-deficient CD4+ T cells largely fail to form IFNγ-producing T effector cells in secondary lymphoid organs and to generate lung tissue resident memory T cells (51). In contrast, increased Th2 cytokine release correlated with poor outcome in patients, a finding corroborated in mouse studies of SARS-CoV-1 (52, 53) and SARS-CoV-2 (54). During SARS-CoV-2 infection, Th2-associated blood markers, such as eosinophilia and circulating IL5, IL33, eotaxin-2, and eotaxin-3, are correlated with COVID-19 severity (55). Even though cancer-specific Th2 responses have been described (56, 57), SARS-CoV-2– or S1-RBD–specific Th2/Tc2 recall responses were not more frequent in patients with cancer versus cancer-free subjects, regardless of their staging, therapies, or comorbidities that influenced COVID-19 severity and the systemic inflammatory tonus (27, 58).
TCR signaling plays a major role in CD4+ polarization and can vary according to the TCR affinity, the amount of peptide/MHC II complexes perceived by a TCR, or the length of time a T cell spends proofreading peptide/MHC II complexes (15). Several authors reported cross-reactivities between CCC and SARS-CoV-2 (9, 20, 23, 24, 34, 35, 59, 60). However, such cross-reactive T cells may correlate with poor clinical outcome (61–66). Indeed, according to one report (21), preexisting CCC-specific memory CD4+ T cells exhibit low TCR avidity in almost all unexposed individuals and are strongly expanded in severe but not mild COVID-19. Moreover, CCC/SARS-CoV-2–cross-reactive T-cell clones shared among convalescent and infected individuals harbored lower functional avidity than non–cross-reactive clones, suggesting antigenic imprinting of the TCR repertoire by previous exposure to CCC (26, 67). Of note, these spike-specific cross-reactive CD4+ T cells might reexpand not only during infection but also following vaccination. In line with this possibility, we detected a strong positive correlation between CCC and SARS-CoV-2–specific IL5 release by memory T cells in unexposed individuals. Moreover, CCC-specific IgG titers were higher in susceptible compared with resistant individuals. Finally, the SARS-CoV-1 and ORF8-specific T-cell repertoire prevailing in the pre–COVID-19 era failed to be clinically relevant for the avoidance of COVID-19, and such a repertoire was frequently detected in reinfected individuals during their convalescence phase. Of note, we generated S1-RBD–specific IL4- or IL5-producing T-cell lines and CD4+CD8+ T-cell clones from one HCW presenting a breakthrough infection after vaccination. Hence, we cannot rule out the possibility that a preexisting Th2 immunity (that we monitored in about 10% individuals), for instance, directed against S1-RBD sequences shared by sarbecoviruses (9) could increase the susceptibility to, and severity of, SARS-CoV-2 infection (52, 53, 55, 68).
Our data fuel the theory that (i) robust Th1 memory immune responses against RBD might restrain viral infection, thus exerting a selective pressure on the virus, obliging it to generate escape variants by mutation of RBD and (ii) preexisting Th2 antiviral responses might not only be incapable of eliminating SARS-CoV-2–infected cells but actually favor (re)infection with SARS-CoV-2, ultimately increasing the viral reservoir, thus favoring the emergence of viral variants. Hence, immunization strategies should aim to trigger Th1/Tc1 (rather than Th2/Tc2) responses against S1-RBD. The efficacy of cellular immune response relies on three components: (i) the antigen, (ii) the adjuvant, and (iii) the dynamics of viral evolution (69). Immunization with inactivated SARS-CoV-1 or with the whole spike (S) protein caused eosinophilic infiltration following viral reexposure in mice (70, 71). Unfortunately, the efficacy of the vaccines composed of inactivated virus produced by Sinovac Biotech (CoronaVac) and Sinopharm (BBIBP-CorV) against VOCs has not yet been reported. In contrast, at least in the case of SARS-CoV-1, immunization with RBD induced neutralizing antibodies in the absence of Th2/Tc2 responses (72). Vaccine adjuvants can stimulate Th1/Tc1-favorable innate immunity, as this is the case for multiple viral vectors, virus-like particles, and mRNA-containing nanoparticles (67, 73). Finally, virus adaptation to the host has to be outcompeted. One might infer from our data that the currently protective immunodominant regions generating a Th1/Tc1 profile may be the focus of the future antigenic drift of SARS-CoV-2, in which case, vaccines would have to be updated regularly (74). In countries with a broad vaccine coverage, it may be advantageous to screen the population for IFNγ responses against S1-RBD to determine the need of each individual for booster vaccination. In particular, although solid cancer–bearing patients could mount Th1 immune responses against PEPOrf that may be able to protect them against COVID-19 severity (as previously discussed; refs. 75, 76), patients with hematologic malignancies were less capable of doing so. Indeed, patients with solid cancer could get efficiently immunized against the S1-RBD region, regardless of staging and types of therapies. There are some limitations to our study due to its nature (cross-sectional rather than longitudinal), enrolling solid more than hematologic malignancies, mostly during their therapies rather than at the remission status. Despite these limitations, our data are in line with previous reports showing that vaccinees bearing hematologic neoplasms had lower rates of seroconversion and an increased risk of breakthrough infections compared with vaccinated matched controls (77–79). In fact, a recent phase I trial administering a third boost of the BNT162b2 (NCT04936997) vaccine in patients with cancer undergoing therapy could not increase their specific Th1 immune responses while augmenting neutralizing antibody titers (80). Given the fifth wave of this pandemic, to win the race against emerging variants, we might consider an expedited worldwide vaccination rollout ensuring an immunization en masse against more relevant epitopes, in particular the entire RBD region of the current omicron VOCs and sarbecovirus (68) or the virus polymerase (81) with vaccine formulations ensuring Th1/Tc1 responses (and not Th2/Tc2 responses). Finally, current efforts to decipher HLA haplotypes associated with maladaptive S1-RBD Th1 responses may open an avenue for more personalized vaccine design (82–84).
Methods
Patient and Cohort Characteristics
All clinical studies were conducted after written informed consent in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki. Cohort and subset characteristics are detailed in Supplementary Tables S1–S3, S5, S7, S8, and S10 and Figs. 1A, 2B, and 5A. Two cohorts of patients with cancer (from the pre–COVID-19 era and from the COVID-19 era) and three cohorts of healthy volunteers (from the pre–COVID-19 era and from the COVID-19 period), including two cohorts of cancer or cancer-free vaccinees (ONCOVID, CoV3-APHP, respectively), were analyzed for the translational research program. PBMCs were provided by the Gustave Roussy Cancer Campus (Villejuif, France) and IHU Méditerranée Infection (Marseille, France; see “Blood Analyses” section). Three tables present a detailed enumeration of subject samples utilized for each immunologic assay (Supplementary Tables S1B, S7, and S10).
Contemporary Clinical Studies (COVID-19 Era)
ONCOVID clinical trial and regulatory approvals.
The protocol is available at https://clinicaltrials.gov/ct2/show/NCT04341207. The Gustave Roussy Cancer Center sponsored the trial named “ONCOVID” and collaborated with the academic authors on the trial design and on the collection, analysis, and interpretation of the data. Sanofi provided trial drugs. Protocol approval was obtained from an independent ethics committee (ethics protocol number EudraCT: 2020-001250-21). For details, refer to a previous report (28).
Samples for Translational Research.
PBMCs were isolated less than 8 hours after the blood collection (at patient inclusion and at every hospital visit) and kept frozen at −80°C.
PROTECT-Cov Clinical Trial and Regulatory Approvals.
Principles.
IHU Méditerranée Infection sponsored the PROTECT-Cov trial and collaborated with the academic authors on the trial design and on the collection, analysis, and interpretation of the data. Protocol approval was obtained from an independent ethics committee (ethics protocol number ANSM: 2020-A01546-33). The trial was conducted in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki. All patients provided written informed consent.
Subjects.
PROTECT-Cov eligible subjects were members of the same family/home comprised of two or more people and selected from the microbiology laboratory register on SARS-Cov-2 tests performed between March 23 and April 10, 2020.
Trial design.
Members of the same family/home who had at least one (a)symptomatic COVID-19–positive member (qRT-PCR <35 Ct values for SARS-CoV-2 on nasopharyngeal swabs) and at least one member with negative RT-qPCR for SARS-CoV-2 (≥35 Ct) were screened. A telephone interview was conducted in order to confirm and complete the list of family circles in connection with the positive case. The compliant subjects that were finally selected were invited to come back to the IHU Méditerranée Infection hospital, where they were included in the trial and had a blood test.
COVID-SER Clinical Trial and Regulatory Approvals.
Principles.
The COVID-SER trial was conducted at the Hospices Civils de Lyon, France. Protocol approval was obtained from an independent ethics committee (the national review board for biomedical research, Comité de Protection des Personnes Sud Méditerranée, ID-RCB-2020-A00932-37). The clinical study was registered on ClinicalTrials.gov (NCT04341142). For details, refer to Mouton and colleagues (42). Written informed consent was obtained from all participants and for the study. Blood sampling was performed before vaccination and 4 weeks after receiving one or two doses of vaccine for naïve and convalescent HCWs, respectively. According to French procedures, a written nonopposition to the use of donated blood for research purposes was obtained from healthy volunteers. The donors’ personal data were anonymized before transfer to our research laboratory. We obtained approval from the local ethical committee and the French ministry of research (DC-2008-64) for handling and conservation of these samples. Human biological samples and associated data were obtained from NeuroBioTec (CRB Hospices Civils de Lyon; Biobank BB-0033-00046) and Virginie Pitiot.
COV3AP-HP Clinical Trial and Regulatory Approvals.
BioMérieux S.A. is the promoter of the COV3AP-HP trial, which was approved by the local ethical committee (number ID-RCB: 2021-A00304-37). The trial was conducted in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki at Gustave Roussy and Cochin Institute, France. All subjects provided written informed consent.
Principles of Follow-up in COVID-19–Negative Patients
Two main prospective studies were conducted—one in Villejuif-Grand Paris at Gustave Roussy in patients with cancer from April 15, 2019, to January 2020 (ONCOVID trial; ref. 28), and one in HCW at Hospices Civils de Lyon (parallel study COVID-Ser reported by Pozzetto and colleagues; ref. 85) from March 2019 to February 2020—to address the clinical relevance of spontaneous T-cell responses directed against SARS-CoV-2 or CCC viral lysates or RBD peptides for the susceptibility or resistance to COVID-19 in the subsequent waves of the pandemic. In these two prospective studies, SARS-CoV-2–specific PCR on nasopharyngeal swabs and/or SARS-CoV-2–specific serologies were performed in all patients with cancer entering the ancillary study at each medical visit (every 3 weeks or so) at Gustave Roussy and in all HCWs in contact with COVID-19–positive patients at Lyon, respectively. Phone call inquiries were performed to follow these individuals longitudinally up to manuscript finalization. A third study called PROTECT-Cov was a retrospective study performed in the same household composed of two or more people selected on the basis of SARS-CoV-2 tests performed between March 23 and April 10, 2020. Members of the same family who had at least one (a)symptomatic COVID-19–positive (RT-qPCR <35 Ct values for SARS-CoV-2 on nasopharyngeal swabs) and at the same time, one or several family members who remained negative by RT-qPCR for SARS-CoV-2 (≥35 Ct) were retained in the study. A telephone interview was conducted in order to confirm the person's health status up to the manuscript preparation.
Clinical Studies from the Pre–COVID-19 Era
Series of patients with cancer.
This cohort is composed of different Gustave Roussy Cancer Campus (GRCC) clinical trials. Patients were included, and blood was collected and banked between 1999 and 2018 (pre–COVID-19 era). Clinical studies have been described in previous reports (refs. 30, 32, 86; CALEX protocol, no. 1 ID RCB 2007-A01074-49, date 29 February 2008). (Study code « Dex2 »: NCT01159288, date 19 December 2005.) (Study code « LUD 99 003 »: N-CSET: 99/090/752, date 1 December 1999.) [Phase I IMAIL-2 trial approved by the Kremlin Bicêtre Hospital Ethics Committee (no. 07–019) and the Agence Française de Sécurité Sanitaire des Produits de Santé (no. A70385–27); EudraCT No.:2007–001699–35 in 2007.]
Series of patients without cancer.
Peripheral blood was obtained from healthy volunteers at the Etablissement Français du Sang (Paris France, no. 18EFS031, date September 24, 2018).
Blood Analyses
Blood samples (for serum and PBL) were drawn from patients enrolled in the different cohorts presented in the cohort description section above. Whole human peripheral blood was collected into sterile vacutainer tubes.
Anti–SARS-CoV-2 Immunoglobulin Measurements.
Serum was collected from whole blood after centrifugation at 600 × g for 10 minutes at room temperature and transferred to a −80°C freezer to await analysis. Serologic analysis of SARS-CoV-2–specific IgA, IgM, and IgG antibodies was measured in 119 serum samples from 87 patients with The Maverick SARS-CoV-2 Multi-Antigen Serology Panel (Genalyte) according to the manufacturer's instructions. The Maverick SARS-CoV-2 Multi-Antigen Serology Panel (Genalyte) is designed to detect antibodies to five SARS-CoV-2 antigens: NC, Spike S1-RBD, Spike S1S2, Spike S2, and Spike S1 or seasonal HCoV-NL-63 NC, -OC-43, -229E, and -HK-U1 Spike in a multiplex format based on photonic ring resonance technology. This system detects and measures with good reproducibility changes in resonance when antibodies bind to their respective antigens in the chip. The instrument automates the assay. Briefly, 10 μL of each serum sample was added in a sample well plate array containing required diluents and buffers. The plate and chip were loaded in the instrument. First, the chip was equilibrated with the diluent buffer to get baseline resonance. A serum sample was then charged over the chip to bind specific antibodies to antigens present on the chip. Next, the chip was washed to remove low-affinity binders. Finally, specific antibodies of patients were detected with anti-IgG, anti-IgA, or anti-IgM secondary antibodies.
Isolation of PBMCs from Fresh Blood Sampling.
Venous blood samples (10–30 mL) were collected in heparinized tubes (BD Vacutainer LH 170 U.I.). On the same day, blood was processed in a biosafety level 2 laboratory at Gustave Roussy Institute, Villejuif, France, or in IHU Méditerranée Infection, Marseille, France. PBMCs were freshly isolated by the lymphocyte separation medium (Eurobio Scientific) density gradient centrifugation according to the manufacturer's instructions (Leucosep tubes, Greiner; Biocoll, Bio&SELL). PBMCs were then collected, washed once with phosphate-buffered saline solution (PBS), and aliquoted in 1 mL of cryopreservation medium (CryoStor, STEMCELLS Technologies) in cryovials (two cryovials per patient). Cryovials (Cryotube vials, Thermo Fisher Scientific) were conserved for 24 hours at −80°C in a cryo-freezing container (Mr. Frosty, Thermo Fisher Scientific) before storage in liquid nitrogen.
Serum and serologies.
Specific anti–SARS-CoV-2 IgG antibodies were detected by the Liaison XL automated chemiluminescent immunoassay (CLIA; Diasorin Inc.) according to the manufacturer's recommendations. Seroneutralization was performed as already described (87). For Supplementary Fig. S6, we used the bioMérieux VIDAS SARS-COV-2 IgG II (9COG) kit measuring IgG directed against S1-RBD (reference 424114).
Reagents: Culture Media, Cytokines, ELISA, and Multiplex Assays
PBMC isolation.
Blood samples were collected in heparinized tubes, BD Vacutainer LH 170 U.I., from Dutscher (cat. #367526), diluted in PBS 1× purchased from Eurobio Scientific (cat. #CS3PBS01-01) and transferred in Leucosep–50 mL purchased from Greiner Bio-One (cat. #227290). Blood was centrifuged using MF48-R centrifuge from AWEL Industries (cat. #20023001). PBMCs were collected in a centrifuge tube, 50 mL, TPP from Dutscher (cat. #91050), washed with PBS 1×, resuspended in CryoStor CS10 purchased from STEMCELL Technologies (cat. #5100-0001), and transferred in CryoTube vials from Thermo Fisher Scientific (cat. #377267). Samples were finally conserved for 24 hours at −80°C in a cryo-freezing container (Mr. Frosty, Thermo Fisher Scientific) before storage in liquid nitrogen.
Cross-presentation Assay or PBL Stimulation with Autologous mo-DCs.
Frozen PBMCs were thawed, washed, and resuspended in RPMI 1640 (1×) purchased from GIBCO (cat. #31870-025). Counting and viability were evaluated using Vi-CELL XR Cell Viability Analyzer from Beckman Coulter (cat. #AV13289). To separate adherent and nonadherent cell populations, PBMCs were transferred to a 6- or 24-well flat-bottom sterile tissue culture testplate, TPP purchased from Dutscher (cat. #92006/92024), and cultured in complex medium (Complex Medium 1) containing human AB serum (cat. #201021334), purchased from Institut de Biotechnologies), RPMI 1640 (1×; cat. #31870-025), sodium pyruvate (cat. #11360–039), penicillin/streptomycin (cat. #15140–122), L-glutamine (200 mmol/L; cat. #25030-024), HEPES buffer solution (cat. #15630-056), and MEM NEAA (cat. #1140-035) purchased from GIBCO/Thermo Fisher Scientific. The nonadherent fraction was cultured in another complex medium (Complex Medium 2) containing human AB serum, Iscove's modified Dulbecco's medium (IMDM; cat. #I3390), from Sigma-Aldrich, sodium pyruvate (cat. #11360-039), penicillin/streptomycin (cat. #15140-122), L-glutamine (200 mmol/L; cat. #25030-024), HEPES buffer solution (cat. #15630-056), and MEM NEAA (cat. #1140-035) from GIBCO/Thermo Fisher Scientific and recombinant human IL2 (PHAR000306) from Gustave Roussy Institute Pharmacy. The adherent fraction was differentiated into monocyte-derived dendritic cells (mo-DC) in mo-DC differentiating media constituted with Complex Medium 1 supplemented with Recombinant Human GM-CSF Premium purchased from Miltenyi (cat. #130-093-867) and human IFNα-2b (Introna) purchased from MSD (France; cat. #PHAR008943). For activation and maturation, DCs were stimulated with LPS purchased from Invivogen (cat. #tlrl-3pelps) and GM-CSF purchased from Miltenyi Biotec (cat. #130-093–867). PBLs and mo-DCs were finally cocultured in a 96-well V bottom Sterile Nunc plate, VWR purchased from Dutscher (cat. #92097). For positive control, PBLs were stimulated with Dynabeads Human T-Activator CD3/CD28 purchased from GIBCO/Thermo Fisher Scientific (cat. #11131D). All cell cultures were performed at 37°C in 5% CO2 in a Heraus incubator purchased from Kendro Laboratory Products, Thermo Fisher Scientific (cat. #BB 6220), and supernatants were transferred to a 96-well V bottom sterile Nunc plate, VWR purchased from Dutscher (cat. #734-0491) and frozen.
Peptide-Based assay.
The 96-well V bottom sterile Nunc plates were coated with peptides at 2 μg/mL in RPMI 1640 (1×; cat. #31870-025) supplemented with 1% penicillin/streptomycin (cat. #15140-122), and conserved at −80°C. PBMCs were then thawed and plated in a plate containing peptides in RPMI 1640 (1×; cat. #31870-025) supplemented with 1% penicillin/streptomycin (cat. #15140-122) supplemented with recombinant human IL15 premium grade from Miltenyi Biotec (cat. #130-095-765) and recombinant human IL2 (PHAR000306) from Gustave Roussy Hospital. For positive control, PBMCs were stimulated with functional-grade CD3 (OKT3) purchased from Thermo Fisher Scientific (cat. #16-0037-85). Cell cultures were then supplemented with human AB serum (cat. #201021334) purchased from Institut de Biotechnologies Jacques Boy (France) and cultured at 37°C in 5% CO2.
Cytokine monitoring.
Supernatants from cultured cells from the cross-presentation assay were monitored using the human MACS-Plex Cytokine 12 Kit purchased from Miltenyi Biotec (cat. #130-099-169). Acquisitions and analyses were performed on CytoFLEX S purchased from Beckman Coulter (cat. #B75442)/FACSAria Fusion purchased from BD Biosciences and FlowJo Software from Treestar, respectively. Supernatants from cultured cells from the peptide-based assay were monitored using ELISA tests purchased from BioLegend: ELISA MAX Deluxe Set Human IFNγ (cat. #430104), ELISA MAX Deluxe Set Human IL17 (cat. #433914), and ELISA MAX Deluxe Set Human IL9 (cat. #434705).
Viral Studies
Biosafety levels for in vitro experiments.
Frozen PBMCs from patients with a confirmed negative RT-qPCR for SARS-CoV-2 genome at the time of blood drawing were processed in a biosafety level 2 laboratory at Gustave Roussy Institute, Villejuif, France. All samples from patients with positive RT-qPCR were processed in a biosafety level 3 laboratory at Henri Mondor Hospital, Créteil, France. When a patient was sampled at different time points, samples were processed together in the same laboratory.
RT-qPCR analysis.
SARS-CoV-2 diagnostic testing of clinical nasopharyngeal swabs or other samples by RT-qPCR was conducted from March 14 to 23, 2020, at an outside facility using the Charité protocol. From March 23, 2020, testing was performed internally at Gustave Roussy. The cycle thresholds were collected only for assays performed at Gustave Roussy. Nasopharyngeal swab samples were collected using flocked swabs (Sigma Virocult) and placed in viral transport media. SARS-CoV-2 RNA was detected using one of two available techniques at Gustave Roussy: the GeneFinder COVID-19 Plus RealAmp kit (ELITech Group) targeting three regions (RdRp gene, NC, and envelope genes) on the ELITe InGenius (ELITech Group) or the multiplex real-time RT-PCR diagnostic kit (the Applied Biosystems TaqPath COVID-19 CE-IVD RT-PCR Kit) targeting three regions (ORF1ab, NC, and spike genes) with the following modifications. Nucleic acids were extracted from specimens using automated Maxwell instruments following the manufacturer's instructions (Maxwell RSC simplyRNA Blood Kit; AS1380; Promega). Real-time RT-PCR was performed on the QuantiStudio 5 Dx Real-Time PCR System (Thermo Fisher Scientific) in a final reaction volume of 20 μL, including 5 μL of extracted nucleic acids, according to the manufacturer's instruction.
Viral Lysates and Their Production.
SARS-CoV-2 IHUMI2, IHUMI845, IHUMI846, IHUMI847 (early 2020 episode), IHUMI2096 (20A.EU2, B.1.160), IHUMI2514 (20C, B.1.367; ref. 25), IHUMI3076 (20I/501Y.V1, B.1.1.7), IHUMI3147 (20H/501Y.V2, B.1.351), and IHUMI3191 (20J/501Y.V3, P.1) strains were isolated from human nasopharyngeal swabs as previously described (25) and grown in VeroE6 cells (ATCC CRL-1586) in Minimum Essential Medium (MEM) with 4% fetal calf serum (FCS) and 1% L-glutamine. Influenza strains H1N1 (0022641132) and H3N2 (8091056304) were isolated and then produced from human nasopharyngeal swabs in MDCK cells (ATCC CCL-34) in MEM with 10% FCS and 1% L-glutamine. All these clinical isolates were characterized by whole viral genome sequencing from culture supernatants. Coronavirus OC43 (ATCC vr-1558) was grown in HCT8 cells (ATCC CCL-244) in RPMI with 10% FCS. Coronavirus 229E (ATCC vr-740) was grown in MRC5 cells (ATCC CCL-171) in MEM with 10% FCS. All reagents for culture were from Thermo Fisher Scientific, and all cultures were incubated at 37°C under 5% CO2 without antibiotics. All viral strains were produced in 125-cm2 cell culture flasks. When destruction of cell monolayer reached approximately 80%, between 2 and 7 days according to cell line and viral strain, culture supernatant was harvested. After low-speed centrifugation to remove cells and debris (700 × g for 10 minutes), supernatants were filtered through 0.45- and then 0.22-μm pore-sized filters. These viral suspensions were then inactivated for 1 hour at 65°C before use. Batches of scrapped control uninfected cells were rinsed twice in PBS and then finally resuspended in 5 mL of PBS at 5 × 105 cells/mL. All cells and antigens were tested negative for Mycoplasma before use.
In Vitro Stimulation Assays
Cross-presentation Assay or PBL Stimulation with Autologous mo-DCs.
Frozen PBMCs were thawed, washed, and resuspended in RPMI 1640 media (GIBCO). Viability and count were evaluated using a Vi-Cell XR Cell Counter (Beckman Coulter). PBMCs were then cultured in RPMI 1640 supplemented with 10% human AB serum, 1 mmol/L glutamine, 1% sodium pyruvate, 1% HEPES, and 1% penicillin/streptomycin at a cell density of 0.5M cells/cm2 for 2 hours at 37°C in 5% CO2 and separated into adherent and nonadherent cell populations. Nonadherent cells, containing PBL, were collected and cultured 4 days at 37°C in 5% CO2 in IMDM (Sigma-Aldrich) supplemented with 10% human AB serum (Institut de Biotechnologies Jacques Boy, France), 1 mmol/L glutamine (GIBCO/Thermo Fisher Scientific) 1% sodium pyruvate (GIBCO/Thermo Fisher Scientific), 1% HEPES (GIBCO/Thermo Fisher Scientific), 1% penicillin/streptomycin (GIBCO/Thermo Fisher Scientific), and 200 UI/mL rhIL-2 (Miltenyi). The adherent cell population was cultured for 3 days at 37°C in 5% CO2 in mo-DC differentiating media containing RPMI 1640 supplemented with 10% human AB serum, 1 mmol/L glutamine, 1% sodium pyruvate, 1% HEPES, 1% penicillin/streptomycin, 1,000 UI/mL rhGM-CSF (Miltenyi), and 250 UI/mL human IFNα-2b (Introna, MSD France). At day 3, adherent cells were slowly detached by pipetting after 20 minutes of incubation at 4°C, and 20,000 cells were seeded in a 96-well round-bottom plate and were pulsed, or not (control condition), overnight at 37°C in 5% CO2 with 1/10 heat-inactivated viral lysates or their respective control (see “Viral lysates and their production” section). Spinoculation (800 g for 2 hours, Centrifuge 5810R, Eppendorf) was next performed to ensure synchronized capture of the viral particles by mo-DCs. For activation and maturation, adherent cells were stimulated with LPS (10 ng/mL, Thermo Fisher) and GM-CSF (1,000 UI/mL). After 6 hours, mo-DCs were washed twice to remove LPS from the media and 100,000 PBL/well were seeded onto mature mo-DCs. PBL alone served as negative control, and PBL stimulated with anti-CD3 and anti-CD28 microbeads (1 μL/mL, Dynabeads T-Activator, Invitrogen) served as a positive control. mo-DC–PBL coculture was incubated at 37°C in 5% CO2 for 48 hours, and supernatants were harvested and stored at −20°C.
Multiplex Cytokine Analysis or Bead-Based Multiplex Assays.
mo-DC–PBL coculture supernatants were analyzed using bead-based multiplex kit assays (MACSplex cytokine 12 human, Miltenyi) according to the manufacturer's protocol. Briefly, 50 μL of supernatants were used with a MACSPlex cytokine 12 capture beads (Miltenyi) to measure the concentration of 12 cytokines (GM-CSF, IFNα, IFNγ, IL10, IL12, IL17A, IL2, IL4, IL5, IL6, IL9, and TNFa). Bead fluorescence was acquired on a CytoFLEX flow cytometer (Beckman Coulter) for samples processed at the Gustave Roussy Institute and on a FACSAria Fusion (Becton Dickinson) for samples processed in the biosafety level 3 laboratory at Henri Mondor Hospital. FlowJo (Treestar) software was used for analysis.
Positivity Threshold Determination for Cytokine Concentration Using Multiplex Assays and Commercial ELISA.
For multiplex assays (or ELISA), a four-parameter logistic regression was fitted for each cytokine based on the APC mean fluorescent intensity (or optical density) of standard dilution samples using nlpr (v0.1-7). This model was then used to calculate the concentration of each sample of unknown concentration. For multiplex assays, a ratio was computed for each cytokine using the cytokine concentration measured in response to each virus (SARS-CoV-2, HCoV-229E, and HCoV-OC43) divided by the median concentration of their respective biological controls (Vero 81, MRC5, and HCT8). A positivity threshold was set up based on the ratio for each cytokine. A ratio of above 1.5 minimum was requested to consider the supernatant “positive” for a cytokine. When necessary, a higher threshold was set up as such, median cytokine concentration of the biological controls + 2 times the standard deviation of the biological control concentrations divided by the median concentration. For ELISA, a ratio was computed as the concentration of the sample divided by the mean concentration of the negative controls.
ELISpot assay.
The enumeration of antigen-specific IFNγ- and IL5-producing T cells was performed using the ImmunoSpot human IFNγ/IL-5 double-color enzymatic ELISPOT kit (Cellular Technology Limited, CTL). PBLs were stimulated with autologous mo-DCs loaded with SARS-CoV-2 lysates or their respective controls (see “In Vitro Stimulation Assays,” “Cross-presentation assay” section). After 48 hours, cells were resuspended in serum-free testing medium (CTL) containing 1 mmol/L GlutaMAX (Gibco) and 1% penicillin/streptomycin (GIBCO) at a final volume of 200 μL/well and seeded in a 96-well nitrocellulose plate coated with human IFNγ and IL5 capture antibody. Plates were incubated for 18 hours at 37°C in 5% CO2. ELISPOT assays were then performed according to the manufacturer's instructions. Spots were counted by CTL ImmunoSpot Analyzer using ImmunoSpot software.
Flow-Cytometric analyses.
Sample preparation.
Cells from the cross-presentation assays (PBL + DC loaded with viral lysates or VeroE6 supernatants) were stained for viability with Zombie Aqua (BioLegend; cat. #423102) for 20 minutes at +4°C and then washed in staining buffer (PBS 1×, BSA 2%, 2 mmol/L EDTA). Then cells were stained with a panel of antibodies (as indicated in the table for Supplementary Methods) for 20 minutes at room temperature in staining buffer with Brillant Strain Buffer (BD; cat. #563794). Cells were then washed, fixed, and permeabilized (Foxp3/Transcription Factor Staining Buffer Set; eBiosciences; cat. #00-55-23-00) for 40 minutes at +4°C before being stained with intracellular antibodies for 30 minutes at +4°C.
Data acquisition.
Samples were acquired on a BD LSRFortessa X-20 flow cytometer.
Data analysis.
Analysis was performed with FlowJo software (Tree Star).
Whole-transcriptome RNA sequencing.
PBLs from 11 resistant and seven susceptible patients as well as eight and 10 patients for whom cross-presentation assays revealed an IL2/IL5 ratio > and <1, respectively, were used for the RNA sequencing (RNA-seq) of PBLs at 48 hours after incubation with DCs loaded with viral lysates. Cells from 18 wells after stimulation with SARS-CoV-2 or VeroE6 were analyzed. The RNA integrity (RNA integrity score ≥7.0) was checked on the Agilent 2100 Bioanalyzer (Agilent), and the quantity was determined using Qubit (Invitrogen). The SureSelect Automated Strand Specific RNA Library Preparation Kit was used according to the manufacturer's instructions with the Bravo Platform. Briefly, 30 to 100 ng of total RNA sample was used for poly-A mRNA selection using oligo(dT) beads and subjected to thermal mRNA fragmentation. The fragmented mRNA samples were subjected to cDNA synthesis and were further converted into double-stranded DNA using the reagents supplied in the kit, and the resulting dsDNA was used for library preparation. The final libraries were bar-coded, purified, pooled together in equal concentrations, and subjected to paired-end sequencing (2 × 100 bp) on a Novaseq-6000 sequencer (Illumina) at Gustave Roussy.
Peptide-Based assays.
Rationale of peptide selection and peptide synthesis (Supplementary Table S11).
The peptides from the spike and NC proteins were selected by dividing the sequences of the SARS-CoV-2 spike protein (RefSeq ID QHD43416.1) and of the NC protein (RefSeq ID QHD43423.2) in nonoverlapping 15 amino acid segments. The peptides from the membrane protein were selected by dividing the sequence of two potential immunogenic regions of the SARS-CoV-2 (RefSeq ID QHD43422.1) membrane protein in overlapping 15 amino acid segments. The peptides from the ORF8 and ORF10 proteins were selected by dividing the sequences of the SARS-CoV-2 ORF8 protein (RefSeq ID QHD43422.1) and of the ORF10 protein (RefSeq ID QHI42199.1) in overlapping 15 amino acid segments. The peptides from ORF3 and some for ORF8 were selected based on a previous study (88). The SARS-CoV-1 peptides were peptides found to be immunogenic in previously reported studies (11, 52, 89–93). The peptides were synthesized by peptides & elephants GmbH. The peptide pools for the controls for influenza, EBV, and CMV were acquired from peptides & elephants GmbH (Berlin, Germany), with the order numbers LB01774, LB01361, and LB01232, respectively.
185 single peptides in 96-well plates.
Lyophilized peptides were dissolved in sterile water and used at 2 μg/mL in RPMI 1640 glutamax media (GIBCO) supplemented with 1% penicillin/streptomycin (GIBCO). Single peptides (185) were plated in duplicates in 96-well round-bottom TPP-treated culture plates. Peptide plates were then stored at −80°C until use. The day of the experiment, peptide plates were thawed at room temperature. Frozen PBMCs were thawed, washed, and resuspended in RPMI 1640 media (GIBCO). Viability and count were evaluated using a Vi-Cell XR Cell Counter (Beckman Coulter). PBMCs were then plated in RPMI 1640 glutamax media (GIBCO) supplemented with 1% penicillin/streptomycin (GIBCO), with 200 UI/mL rhIL2 (Miltenyi) and 200 UI/mL rhIL15 (Miltenyi) at a cell density of 10 × 103 cells and incubated with each peptide at 37°C in 5% CO2. PBMCs were stimulated with 60 ng/mL OKT-3 antibody (Thermo Fisher Scientific, clone OKT3) or with 10 μg/mL phytohemagglutinin as positive controls, and PBMCs alone served as negative controls. After 6 hours, 20 μL of human AB serum was added to each well and plates were incubated at 37°C in 5% CO2 for 6 additional days. On day 7, supernatants were harvested and frozen at −80°C. The concentration of IFNγ, IL9, IL5, and IL17A in the culture supernatant was determined using a commercial ELISA kit (ELISA Max Deluxe set human IFNγ, BioLegend).
Peptide pools and COVID IGRA bioMérieux assay utilized for the COV3AP-HP clinical trial vaccinees (42).
Fresh blood collected in heparinized tubes was stimulated for 22 hours at 37°C under 5% CO2 with peptide pools spanning distinctive genomic sequences of the SARS-CoV-2 ORFeome (PEPOrf) or the WT or mutated 331–525 amino acid RBD sequence (Fig. 5C; Supplementary Table S11; bioMérieux) diluted in IFA solution (bioMérieux). The IFA solution was used as a negative control, and a mitogen (MIT) was used as a positive control. The peptides PEPOrf and PEPovRBD (15-mer) encompassed distinct genomic sequences from the SARS-CoV2 ORFeome and the whole RBD protein sequence and overlapped by five residues (Supplementary Table S11). In the second phase of the study, we used nonoverlapping 15-mer peptides covering the WT or mutated 331–535 RBD region (Supplementary Table S11; Fig. 5D–F). The concentration of IFNγ in the supernatant was measured using the VIDAS automated platform (VIDAS IFNγ RUO, bioMérieux). The positivity range was 0.08 to 8 IU/mL, and IFA positivity thresholds were defined at 0.08 IU/mL. The IFNγ response was defined as positive when the IFNγ concentration of the test was above threshold and the negative control was below threshold or when the IFNγ concentration of the test minus the IFNγ concentration of the negative control was above threshold. All positive controls were ≥8 IU/mL.
Generating Th2 Cell Lines
Generating SARS-CoV-2 Lysate–Specific Clones.
Ten million PBLs from a healthy donor with a history of SARS-CoV-2–specific IL5 release (refer to Fig. 1D) were stimulated with autologous mo-DCs loaded with SARS-CoV-2 lysates (see Cross-presentation assay section). After 18 hours, cells were harvested, and CD137+ cells were isolated using CD137 MicroBead Kit, human (Miltenyi) according to the manufacturer's instructions. Limiting dilution of CD137+ cells was performed by seeding 100 μL of CD137-positive cellular suspension at a 10 cells/mL concentration in 96-well round-bottom plates in sterile conditions. Feeder cells were generated by isolating CD14-positive cells using CD14 MicroBead Kit, human (Miltenyi). Isolated feeder cells were cocultured with CD137-positive cells at a 1,000:1 ratio and cultivated in IMDM (Sigma-Aldrich) supplemented with 10% human AB serum (Institut de Biotechnologies Jacques Boy, France), 1 mmol/L glutamine (GIBCO/Thermo Fisher Scientific), 1% sodium pyruvate (GIBCO/Thermo Fisher Scientific), 1% HEPES (GIBCO/Thermo Fisher Scientific), and 1% penicillin/streptomycin (GIBCO/Thermo Fisher Scientific), supplemented with 100 UI/mL IL7 (Miltenyi) and 100 UI/mL IL15 (Miltenyi). The medium was changed every 2 to 3 days. Clones were screened for IFNγ and IL5 secretion by quantification of the accumulation of these cytokines in supernatants between days 7 and 13 using commercial ELISA kits. Ninety-three clones of interest were identified and screened for specificity against SARS-CoV-2 lysates by quantifying IFNγ and IL5 secretion after restimulation with autologous mo-DCs loaded with SARS-CoV-2 lysate or its respective control at day 21. Three rounds of IVS were performed over 3 weeks. Clones were starved in cytokine-free media 2 days before restimulation. Six SARS-CoV-2–specific cell lines could be identified, and their MHC I/II recognition dependency was assessed by monitoring IFNγ and IL5 production after stimulation with autologous mo-DCs loaded with SARS-CoV-2 lysate or its respective control in the presence or absence of neutralizing anti–HLA-ABC and HLA-DR, DP, and DQ antibodies (W6/32 and Tü39) at day 28. Flow-cytometric determination of CD4, CD8, T-bet, and GATA3 was performed on the IL5-producing SARS-CoV-2–specific cell lines according to methods already reported (28).
Generating spike 25–specific cell lines.
PBMCs from a healthy donor with a history of breakthrough COVID-19 infection after complete vaccination were stimulated using 186 peptides spanning the ORFeome of SARS-CoV-2 (Fig. 3A). IFNγ and IL5 were monitored in supernatants after 7 days of culture using commercial ELISA kits to identify IL5-restricted reactivity. One Spike 25–specific IL5-producing (but IFNγ negative) T-cell line was identified and further expanded using mo-DCs pulsed with Spike 25 for 1 week at a concentration of 1 μg/mL in RPMI supplemented with 10% human AB serum (Institut de Biotechnologies Jacques Boy, France), 1 mmol/L glutamine (GIBCO/Thermo Fisher Scientific), 1% sodium pyruvate (GIBCO/Thermo Fisher Scientific), 1% penicillin/streptomycin (GIBCO/Thermo Fisher Scientific), IL2 200 UI/mL (Miltenyi), and IL15 (Miltenyi). After the third week, the T-cell line was restimulated with Spike 25–loaded DCs in the presence or absence of neutralizing anti–HLA-ABC and HLA-DR, DP, and DQ antibodies (W6/32; Tü39) in duplicate wells to monitor cytokine release using the 12 plex assay and stained with CD3-, CD4-, CD8-, GATA3-, T-bet–specific antibodies to assess phenotypical characteristic by flow cytometry (refer to “Flow-Cytometric Analyses”).
Statistical Analyses
All calculations, statistical tests, and data visualization were performed using R v4.0.3. All analyses were performed on independent samples, except when the presence of replicates is mentioned. The associations between continuous variables were evaluated using Spearman correlation. Group comparisons were performed using nonparametric test with the wilcox.test R function: the Wilcoxon–Mann–Whitney test for independent samples and the Wilcoxon signed rank test for paired samples. When the number of replicates was unbalanced between the individuals, the Wilcoxon signed rank test for paired comparisons of clustered data was performed with the clusWilcox.test function of the R package clusrank. The comparison of categorical data was performed using the Fisher exact test with the fisher.test R function. Hierarchical clustering was performed with the package hclust, using the Euclidean distance. Linear and logistic regressions were performed with the lm and the glm R base functions, respectively. A peptide set enrichment analysis was performed with the R package fgsea (version 1.14.0), using as statistic the t-value of the coefficient of univariable linear regressions of the logarithm-normalized IL2 secretion on the different peptides. All hypothesis tests (including those of regression coefficients) were two-sided and considered statistically significant when P < 0.05. Graphical illustrations were drawn using the standard R packages dedicated to the data visualization (ggplot2, ggpubr, corrplot, complexheatmap, circlize, and Hmisc).
RNA-seq data analysis.
Quality control was done on raw FastQ files with FastQC (v0.11.9; ref. 94). Quality reports were gathered with MultiQC (v1.9; ref. 95). Abundance estimation was performed with Salmon (v0.9.0; ref. 96) using GENCODE (GRCh38, v34) annotation (97). Quantification results were aggregated with tximport (v1.14.0), and differential gene analysis was performed with DESeq2 (v1.30.0), according to the procedure by Soneson and colleagues (98). The whole pipeline was powered by both Snakemake (99) and SnakemakeWrappers. Gene set enrichment analysis on DESeq2 results was performed with GSEA software (v4.1.0, preranked based on Wald test statistic, 1,000 permutations, weighted enrichment statistic) and immunologic signature gene sets coming from MSigDB (C7, v7.4; ref. 100).
Multivariate Analyses of Peptide Pool–Specific T-cell Responses According to Covariates.
We pooled the log10-normalized IFNγ secretion measurements obtained with the three peptide pools to model simultaneously their dynamics from the first shot of vaccine using linear mixed-effect regression adjusted for the patient age, sex, cancer status (yes/no), COVID history, and vaccine schedule. To identify the differences between the dynamics of each panel, we adjusted the model for the peptide pool (representing baseline differences) and added interaction terms between the peptide pool and each covariable (including the time since the first vaccine). Intrapatient and intrapanel correlations were considered by adding patient–peptide random effect for the intercept. A statistically significant interaction indicates that the covariable has an impact on the peptide-specific IFNγ measurement that is statistically different from its impact on the reference peptide pool (Fig. 5 and Table 1).
Data Availability
Expression profile data used in this article are publicly available at EGA under accession number EGAD00001008538. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Prof. Antoine TESNIERE (MESRI) for his support in launching Cov3-APHP as well as the ET-EXTRA team (Biological Resource Center (NF 96-600) and the microbiology team for technical help. We thank the staff from health and safety of the GRCC for helping to set up the translational research studies. We are thankful to Genalyte for their supportive help. We are thankful to Jeanne Magnan, Marie Malige, and Roxanne Birebent for their technical help. The Lyon COVID Study Group and authors thank the Hospices Civils de Lyon and Fondation des Hospices Civils de Lyon and all the personnel of the occupational health and medicine department of Hospices Civils de Lyon who contributed to the sample collection. We thank Anne Florin for her help in including HCW from Gustave Roussy.
The Lyon COVID study group members are as follows: Sophie Assant, William Mouton, Christelle Compagnon, Kahina Saker, Soizic Daniel, Xavier Lacoux, Guy Oriol, Sophia Djebali, Franck Berthier, Jacqueline Marvel, Thierry Walzer, Karen Brengel-Pesce, Jean-Baptiste Fassier, and Amelie Massardier-Pilonchery.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
J.-E. Fahrner reports other support from Transgene outside the submitted work. I. Lahmar and A-G. Goubet were supported by Fondation pour la Recherche Médicale (FRM). D. Drubay reports consulting fees from Chugai and Roche. The Lyon COVID Study Group reports personal fees and nonfinancial support from bioMérieux during the conduct of the study; personal fees and nonfinancial support from bioMérieux outside the submitted work; and some members of the group are bioMérieux employees or received grants from bioMérieux to perform experiments. E. de Sousa reports grants from Fundação para a Ciência e Tecnologia (FCT) in the context of the project UIDB/04443/2020 and grants from the European Regional Development Fund (ERDF) in the context of projects LISBOA-01-0145-FEDER-022231 and LISBOA-01-0246-FEDER-000007 during the conduct of the study, as well as a patent for EP21171378.9 pending and a patent for EP21171380.5 pending. A. Geraud reports other support from Novartis outside the submitted work. A. Geraud, A. Bernard-Tessier, and A. Marabelle, as part of the Drug Development Department (DITEP), are Principal/sub-Investigators of Clinical Trials for AbbVie, Adaptimmune, Aduro Biotech, Agios Pharmaceuticals, Amgen, Argen-X Bvba, Arno Therapeutics, Astex Pharmaceuticals, AstraZeneca, AstraZeneca Ab, Aveo, Bayer Healthcare Ag, Bbb Technologies Bv, Beigene, Bioalliance Pharma, BioNTech Ag, Blueprint Medicines, Boehringer Ingelheim, Boston Pharmaceuticals, Bristol Myers Squibb, Bristol Myers Squibb International Corporation, Ca, Celgene Corporation, Cephalon, Chugai Pharmaceutical Co., Clovis Oncology, Cullinan-Apollo, Daiichi Sankyo, Debiopharm S.A., Eisai Limited, Eli Lilly, Exelixis, Forma Therapeutics, Gamamabs, Genentech, Gilead Sciences, GlaxoSmithKline, Glenmark Pharmaceuticals, H3 Biomedicine, Hoffmann-La Roche Ag, Incyte Corporation, Innate Pharma, Institut De Recherche Pierre Fabre, Iris Servier, Janssen Cilag, Janssen Research Foundation, Kura Oncology, Kyowa Kirin Pharm. Dev., Lilly France, Loxo Oncology, Lytix Biopharma As, MedImmune, Menarini Ricerche, Merck Kgaa, Merck Sharp & Dohme Chibret, Merrimack Pharmaceuticals, Merus, Millennium Pharmaceuticals, Molecular Partners Ag, Nanobiotix, Nektar Therapeutics, Nerviano Medical Sciences, Novartis Pharma, Octimet Oncology Nv, Oncoethix, Oncomed, Oncopeptides, Onyx Therapeutics, Orion Pharma, Oryzon Genomics, Ose Pharma, Pfizer, Pharma Mar, Philogen S.P.A., Pierre Fabre Medicament, Plexxikon, Rigontec GmbH, Roche, Sanofi Aventis, Sierra Oncology, Sotio A.S, Syros Pharmaceuticals, Taiho Pharma, Tesaro, Tioma Therapeutics, Wyeth Pharmaceuticals France, Xencor, and Y's Therapeutics; reports research grants from AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Janssen Cilag, Merck, Novartis, Pfizer, Roche, and Sanofi; and reports nonfinancial support (drug supplied) from AstraZeneca, Bayer, Bristol Myers Squibb, Boehringer Ingelheim, Johnson & Johnson, Lilly, MedImmune, Merck, NH TherAGuiX, Pfizer, and Roche. M. Picard was supported by the Italian Ministry of Health (grants Ricerca CorrenteLinea 1, 1 “Infezioni Emergenti e Riemergenti,” projects COVID-2020-12371675 and COVID-2020-12371817). J.R. Lerias reports grants from FCT in the context of the project UIDB/04443/2020 and grants from ERDF in the context of projects LISBOA-01-0145-FEDER-022231 and LISBOA-01-0246-FEDER-000007 during the conduct of the study, as well as a patent for EP21171378.9 pending and a patent for EP21171380.5 pending. M. Miyara and G. Gorochov were supported by the ANR Flash COVID-19 program and the SARS-CoV-2 Program of the Faculty of Medicine from Sorbonne University ICOViD programs (principal investigator: G. Gorochov). G. Gorochov is a member of the scientific board for Luxia Scientific and reports consultancy for Pileje and Luxia Scientific outside the submitted work. F. Barlesi reports personal fees from AstraZeneca, Bayer, Bristol Myers Squibb, Boehringer Ingelheim, Eli Lilly Oncology, Hoffmann-La Roche Ltd., Novartis, Merck, Mirati, MSD, Pierre Fabre, Pfizer, Seattle Genetics, and Takeda outside the submitted work. P. Lavaud reports grants from ESMO and Servier, and personal fees from AstraZeneca, Pfizer, Astellas, and Sanofi outside the submitted work. E. Deutsch reports grants and personal fees from Roche/Genentech, Servier, AstraZeneca, Bristol Myers Squibb, MSD, Merck Serono, and Boehringer, personal fees from Amgen, and grants from AWS, Servier, Bristol Myers Squibb, and MSD and was supported by INCa 2018-1-PL BIO-06-1 outside the submitted work. J.-P. Spano reports nonfinancial support from the CARE committee during the conduct of the study, as well as personal fees from MSD, Gilead, Pfizer, Novartis, AstraZeneca, Daiichi Sankyo, GlaxoSmithKline, LeoPharma, Biogaran, Mylan, Lilly, ViiV, Roche, Bristol Myers Squibb, and PFO, grants from Bristol Myers Squibb and MSD Avenir, and other support from Pierre Fabre Oncology outside the submitted work. M. Merad is a member of the Gates Foundation's innate immunity advisory group and is supported by the Champalimaud Foundation through funds from grants UIDB/04443/2020, LISBOA-01-0145-FEDER-022231 and LISBOA-01-0246-FEDER-000007. F. Scotté reports personal/consulting fees from Pfizer, Bristol Myers Squibb, MSD, Roche, Pierre Fabre Oncology, Leo Pharma, Bayer, Mylan/Viatris, Mundi Pharma, Astellas, Vifor Pharma, Amgen, Arrow, Biogaran, and Helsinn and nonfinancial support from Pierre Fabre Oncology outside the submitted work. A. Marabelle reports grants from Fondation Gustave Roussy during the conduct of the study, as well as personal fees from Clover Biopharmaceuticals outside the submitted work. Over the last 5 years: A. Marabelle has been a Principal Investigator of Clinical Trials from the following companies: Roche/Genentech, Bristol Myers Squibb, Merck (MSD), Pfizer, Lytix Pharma, Eisai, AstraZeneca/MedImmune, Tesaro, Chugai, OSE Immunotherapeutics, SOTIO, Molecular Partners, IMCheck, Pierre Fabre, Adlai Nortye. A. Marabelle has been a member of clinical trial steering committees for NCT02528357 (GlaxoSmithKline) and NCT03334617 (AstraZeneca), and a member of the Data Safety and Monitoring Board for NCT02423863 (sponsor: Oncovir) and NCT03818685 (sponsor: Centre Léon Bérard). A. Marabelle has been a compensated member of the following scientific advisory boards: Merck Serono, eTheRNA, Lytix Pharma, Kyowa Kirin Pharma, Novartis, Bristol Myers Squibb, Symphogen, Genmab, Amgen, Biothera, Nektar, Tesaro/GlaxoSmithKline, Oncosec, Pfizer, Seattle Genetics, AstraZeneca/MedImmune, Servier, Gritstone, Molecular Partners, Bayer, Partner Therapeutics, Sanofi, Pierre Fabre, RedX pharma, OSE Immunotherapeutics, Medicxi, HiFiBio, IMCheck, MSD, iTeos, Innate Pharma, Shattuck Labs, Medincell, Tessa Therapeutics, and Deka Biosciences. A. Marabelle has provided compensated teaching/speaker activities for Roche/Genentech, Bristol Myers Squibb, Merck (MSD), Merck Serono, AstraZeneca/MedImmune, Amgen, Sanofi, and Servier. A. Marabelle has been compensated for scientific and medical consulting for Roche, Pierre Fabre, Onxeo, EISAI, Bayer, Genticel, Rigontec, Daichii Sankyo, Imaxio, Sanofi/BioNTech, Molecular Partners, Pillar Partners, BPI, Faron, and Applied Materials. A. Marabelle has benefited from nonfinancial support (travel expenses) from AstraZeneca, Bristol Myers Squibb, Merck (MSD), and Roche. A. Marabelle is a shareholder in Pegascy SAS, Centessa Pharmaceuticals, HiFiBio, and Shattuck Labs. A. Marabelle has received preclinical and clinical research grants (institutional funding) from Merus, Bristol Myers Squibb, Boehringer Ingelheim, Transgene, Fondation MSD Avenir, Sanofi, and AstraZeneca. B. La Scola has received founding from the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU. Y. Blay reports grants from Lyrican during the conduct of the study, as well as grants from AstraZeneca outside the submitted work. J.-C. Soria reports other support from Amgen, AstraZeneca, Gritstone bio, and Relay Therapeutics during the conduct of the study; other support from Amgen, AstraZeneca, Gritstone bio, and Relay Therapeutics outside the submitted work; was a full-time employee of AstraZeneca between September 2017 and December 2019; and reports consultancy for Relay Therapeutics and Gritstone Oncology and shares in Gritstone, AstraZeneca, and Daiichi Sankyo outside the submitted work. M. Merad reports grants from Regeneron, Genentech, Takeda, and Boehringer and personal fees from Compugen, Genenta, Asher Bio, DrenBio, Morphic Therapeutic Inc., Myeloid Therapeutics Inc., and Innate Pharma outside the submitted work. F. André reports grants from AstraZeneca, Daiichi, Roche, Eli Lilly, Pfizer, and Novartis outside the submitted work. M.F. Chevalier reports personal fees from MSD outside the submitted work. E. Guttman-Yassky is a consultant for AbbVie, Almirall, Amgen, Arena Pharmaceuticals, Asana Biosciences, AstraZeneca, Boehringer Ingelheim, Bristol Meyers Squibb, Cara Therapeutics, Celgene, DBV, DS Biopharma, Eli Lilly, EMD Serono, Galderma, Ichnos Sciences (Glenmark), Incyte, Janssen Biotech, Kyowa, Kirin, Leo Pharmaceuticals, Pandion Therapeutics, Pfizer, RAPT Therapeutics, Regeneron, Sanofi, UCB, Union Therapeutics, Connect, Biopharm, SATO, Siolta Therapeutics, Target Pharma Solutions, Ventyx, Novartis, Boston Pharmaceuticals, Evidera, Principia, and Bluefin Biomedicine, and reports grants from Almirall, Amgen, AnaptysBio, Asana Bioscience, AstraZeneca, Bristol Meyers Squibb, Cara Therapeutics, DS Biopharma, Galderma, Innovaderm, Janssen Biotech, Kiniska, Kyowa Kirin, Leo Pharmaceuticals, Pfizer, Regeneron, UCB, and KAO. G. Kroemer reports grants from Daiichi Sankyo, Eleor, Kaleido, Lytix Pharma, PharmaMar, Samsara, Sanofi, Sotio, Vascage, and Vasculox/Tioma outside the submitted work; is on the Board of Directors for the Bristol Myers Squibb Foundation France; and is a scientific cofounder of EverImmune, Samsara Therapeutics, and Therafast Bio. B. La Scola reports grants from Agence nationale de la recherche (ANR) during the conduct of the study. M. Maeurer reports patents for EP21171378.9 and EP 21171380-5 pending. L. Derosa has received support from the Philanthropia Fondation Gustave Roussy; reports grants from Malakoff Humanis, Izipizi, Ralph Lauren, Agnès b, and the Dassault family during the conduct of the study; grants from Philantropia Fondation, nonfinancial support from AstraZeneca, and personal fees from Bristol Myers Squibb outside the submitted work; and a patent for EP 21171378.9 pending. L. Zitvogel reports nonfinancial support and other support from bioMérieux, grants from the French Government and Malakoff Humanis, and other support from Transgene and Champalimaud Foundation during the conduct of the study; nonfinancial support from EverImmune, grants from Kaleido and Daiichi Sankyo, and personal fees from Transgene outside the submitted work; and a patent for EP 21171378.9 pending. L. Zitvogel and G. Kroemer are cofounders of EverImmune, a biotech company devoted to the use of commensal microbes for the treatment of cancers. L. Zitvogel and G. Kroemer were supported by RHU Torino Lumiére (ANR-16-RHUS-0008), the ONCOBIOME H2020 network, the SEERAVE Foundation, the Ligue contre le Cancer (équipe labelisée), Agence Nationale de la Recherche—Projets blancs, ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases, Association pour la recherche sur le cancer (ARC), Cancéropôle Ile-de-France, FRM, a donation by Elior, the European Research Council (ERC), Fondation Carrefour, the High-end Foreign Expert Program in China (GDW20171100085 and GDW20181100051), Institut National du Cancer (INCa), Inserm (HTE), Institut Universitaire de France, LeDucq Foundation, the LabEx Immuno-Oncology, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE), the CARE network (directed by Prof. Mariette, Kremlin Bicêtre AP-HP), and SIRIC Cancer Research and Personalized Medicine (CARPEM). The clinical study on COVID-19 (ONCOVID; NCT04341207) has been supported by the Fondation Gustave Roussy, the Dassault family, Malakoff Humanis, Agnès b., Izipizi, and Ralph Lauren. No disclosures were reported by the other authors.
Authors’ Contributions
J.-E. Fahrner: Conceptualization, data curation, software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. I. Lahmar: Conceptualization, data curation, software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. A.-G. Goubet: Formal analysis, investigation, visualization, writing–original draft, writing–review and editing. Y. Haddad: Data curation, investigation, visualization, writing–original draft, writing–review and editing. A. Carrier: Data curation, investigation, visualization, writing–original draft, writing–review and editing. M. Mazzenga: Data curation, investigation, writing–original draft, writing–review and editing. D. Drubay: Formal analysis, writing–original draft, writing–review and editing. C. Alves Costa Silva: Data curation, investigation, visualization, writing–original draft, writing–review and editing. Lyon COVID Study Group: Data curation, investigation, writing–original draft, writing–review and editing. E. de Sousa: Resources, methodology, writing–original draft, writing–review and editing. C. Thelemaque: Data curation, investigation, visualization, writing–original draft, writing–review and editing. C. Melenotte: Resources, writing–original draft, writing–review and editing. A. Dubuisson: Investigation, visualization, writing–original draft, writing–review and editing. A. Geraud: Resources, data curation, writing–original draft, writing–review and editing. G. Ferrere: Investigation, writing–original draft, writing–review and editing. R. Birebent: Data curation, investigation, writing–review and editing. C. Bigenwald: Resources, investigation, writing–review and editing. M. Picard: Investigation, visualization, writing–original draft, writing–review and editing. L. Cerbone: Resources, writing–original draft, writing–review and editing. J.R. Lerias: Resources, writing–original draft, writing–review and editing. A. Laparra: Resources, writing–original draft, writing–review and editing. A. Bernard-Tessier: Resources, writing–original draft, writing–review and editing. B. Kloeckner: Data curation, investigation, writing–original draft, writing–review and editing. M. Gazzano: Investigation, writing–original draft, writing–review and editing. F.-X. Danlos: Resources, writing–original draft, writing–review and editing. S. Terrisse: Resources, writing–original draft, writing–review and editing. E. Pizzato: Data curation, investigation, writing–original draft, writing–review and editing. C. Flament: Investigation, writing–original draft, writing–review and editing. P. Ly: Investigation, writing–original draft, writing–review and editing. E. Tartour: Resources, writing–original draft, writing–review and editing. N. Benhamouda: Investigation, writing–original draft, writing–review and editing. L. Meziani: Writing–original draft, writing–review and editing. A. Ahmed-Belkacem: Resources, writing–original draft, writing–review and editing. M. Miyara: Investigation, writing–original draft, writing–review and editing. G. Gorochov: Investigation, writing–original draft, writing–review and editing. F. Barlesi: Methodology, writing–original draft, writing–review and editing. A. Trubert: Data curation, investigation, writing–original draft, writing–review and editing. B. Ungar: Resources, writing–original draft, writing–review and editing. Y. Estrada: Resources, writing–original draft, writing–review and editing. C. Pradon: Resources, writing–original draft, writing–review and editing. E. Gallois: Resources, writing–original draft, writing–review and editing. F. Pommeret: Resources, writing–original draft, writing–review and editing. E. Colomba: Resources, writing–original draft, writing–review and editing. P. Lavaud: Resources, writing–original draft, writing–review and editing. M. Deloger: Formal analysis, writing–original draft, writing–review and editing. N. Droin: Investigation, writing–original draft, writing–review and editing. E. Deutsch: Writing–original draft, writing–review and editing. B. Gachot: Resources, writing–original draft, writing–review and editing. J.-P. Spano: Writing–original draft, writing–review and editing. M. Merad: Resources, writing–original draft, writing–review and editing. F. Scotté: Resources, writing–original draft, writing–review and editing. A. Marabelle: Resources, methodology, writing–original draft, writing–review and editing. F. Griscelli: Investigation, writing–original draft, writing–review and editing. J.-Y. Blay: Writing–original draft, writing–review and editing. J.-C. Soria: Writing–original draft, writing–review and editing. M. Merad: Resources, writing–original draft, writing–review and editing. F. André: Writing–original draft, writing–review and editing. J. Villemonteix: Investigation, writing–original draft, writing–review and editing. M.F. Chevalier: Investigation, writing–original draft, writing–review and editing. S. Caillat-Zucman: Investigation, writing–original draft, writing–review and editing. F. Fenollar: Resources, writing–original draft, writing–review and editing. E. Guttman-Yassky: Resources, writing–original draft, writing–review and editing. O. Launay: Resources, methodology, writing–original draft, writing–review and editing. G. Kroemer: Writing–original draft, writing–review and editing. B. La Scola: Resources, writing–original draft, writing–review and editing. M. Maeurer: Conceptualization, resources, supervision, validation, methodology, writing–original draft, project administration, writing–review and editing. L. Derosa: Conceptualization, data curation, supervision, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. L. Zitvogel: Conceptualization, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol 2020;20:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak Ret al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021;384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart Set al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020;383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman Aet al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med 2020;383:2439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bilich T, Nelde A, Heitmann JS, Maringer Y, Roerden M, Bauer Jet al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci Transl Med 2021;13:eabf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BMet al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021;184:2372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moore JP. Approaches for optimal use of different COVID-19 vaccines: issues of viral variants and vaccine efficacy. JAMA 2021;325:1251. [DOI] [PubMed] [Google Scholar]
- 8. Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol 2014;88:11034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia Aet al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020;584:457–62. [DOI] [PubMed] [Google Scholar]
- 10. Ng O-W, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti Aet al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 2016;34:2008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DKet al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 2016;44:1379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol 1989;7:145–73. [DOI] [PubMed] [Google Scholar]
- 13. Romagnani S. Biology of human TH1 and TH2 cells. J Clin Immunol 1995;15:121–9. [DOI] [PubMed] [Google Scholar]
- 14. Romagnani S, Maggi E. Th1 versus Th2 responses in AIDS. Curr Opin Immunol 1994;6:616–22. [DOI] [PubMed] [Google Scholar]
- 15. Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4+ T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu Rev Immunol 2020;38:705–25. [DOI] [PubMed] [Google Scholar]
- 16. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CEet al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021;371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Habel JR, Nguyen THO, van de Sandt CE, Juno JA, Chaurasia P, Wragg Ket al. Suboptimal SARS-CoV-2−specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype. Proc Natl Acad Sci U S A 2020;117:24384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oxford Immunology Network Covid-19 Response T cell Consortium, ISARIC4C Investigators, Peng Y, Mentzer AJ, Liu G, Yao Xet al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol 2020;21:1336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CDet al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 2021;184:169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson Aet al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020;183:158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky Eet al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity 2020;53:1258–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JS, Kim SY, Kim TS, Hong KH, Ryoo NH, Lee Jet al. Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clin Infect Dis 2021;73:e3002–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth Fet al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020;587:270–4. [DOI] [PubMed] [Google Scholar]
- 24. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CRet al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181:1489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fournier PE, Colson P, Levasseur A, Devaux CA, Gautret P, Bedotto Met al. Emergence and outcomes of the SARS-CoV-2 ‘Marseille-4’ variant. Int J Infect Dis 2021;106:228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dykema AG, Zhang B, Woldemeskel BA, Garliss CC, Cheung LS, Choudhury Det al. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J Clin Invest 2021;131:e146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinato DJ, Zambelli A, Aguilar-Company J, Bower M, Sng CCT, Salazar Ret al. Clinical portrait of the SARS-CoV-2 epidemic in European patients with cancer. Cancer Discov 2020;10:1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goubet AG, Dubuisson A, Geraud A, Danlos FX, Terrisse S, Silva CACet al. Prolonged SARS-CoV-2 RNA virus shedding and lymphopenia are hallmarks of COVID-19 in cancer patients with poor prognosis. Cell Death Differ 2021;28:3297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice Pet al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet North Am Ed 2002;360:295–305. [DOI] [PubMed] [Google Scholar]
- 30. Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard Det al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. OncoImmunology 2016;5:e1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borg C, Terme M, Taïeb J, Ménard C, Flament C, Robert Cet al. Novel mode of action of c-kit tyrosine kinase inhibitors leading to NK cell–dependent antitumor effects. J Clin Invest 2004;114:379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wemeau M, Kepp O, Tesnière A, Panaretakis T, Flament C, De Botton Set al. Calreticulin exposure on malignant blasts predicts a cellular anticancer immune response in patients with acute myeloid leukemia. Cell Death Dis 2010;1:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silvin A, Chapuis N, Dunsmore G, Goubet A-G, Dubuisson A, Derosa Let al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 2020;182:1401–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JMet al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020;370:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden Met al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol 2021;22:74–85. [DOI] [PubMed] [Google Scholar]
- 36. Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham Met al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med 2014;6:238ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij MAJet al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 2019;25:89–94. [DOI] [PubMed] [Google Scholar]
- 38. Yager EJ, Ahmed M, Lanzer K, Randall TD, Woodland DL, Blackman MA. Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J Exp Med 2008;205:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara Het al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020;581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Low JS, Vaqueirinho D, Mele F, Foglierini M, Jerak J, Perotti Met al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science 2021;372:1336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Voysey M, Costa Clemens SA, Madhi SA, Weckx LY, Folegatti PM, Aley PKet al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet 2021;397:881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mouton W, Compagnon C, Saker K, Daniel S, Lacoux X, Pozzetto Bet al. A novel whole-blood stimulation assay to detect and quantify memory T-cells in COVID-19 patients. medRxiv 2021.03.11.21253202 [Preprint]. 2021. Available from: https://doi.org/10.1101/2021.03.11.21253202. [Google Scholar]
- 43. Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu EDet al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med 2021;2:100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama Met al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021;591:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bergwerk M, Gonen T, Lustig Y, Amit S, Lipsitch M, Cohen Cet al. Covid-19 breakthrough infections in vaccinated health care workers. N Engl J Med 2021;385:1474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hall VJ, Foulkes S, Charlett A, Atti A, Monk EJM, Simmons Ret al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet 2021;397:1459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran Ket al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 2021;34:108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol 2009;9:503–13. [DOI] [PubMed] [Google Scholar]
- 49. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKPet al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol 2020;21:1293–301. [DOI] [PubMed] [Google Scholar]
- 50. Whitmire JK, Asano MS, Murali-Krishna K, Suresh M, Ahmed R. Long-term CD4 Th1 and Th2 memory following acute lymphocytic choriomeningitis virus infection. J Virol 1998;72:8281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hondowicz BD, Kim KS, Ruterbusch MJ, Keitany GJ, Pepper M. IL-2 is required for the generation of viral-specific CD4+ Th1 tissue-resident memory cells and B cells are essential for maintenance in the lung. Eur J Immunol 2018;48:80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li CK, Wu H, Yan H, Ma S, Wang L, Zhang Met al. T cell responses to whole SARS coronavirus in humans. J Immunol 2008;181:5490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Page C, Goicochea L, Matthews K, Zhang Y, Klover P, Holtzman MJet al. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J Virol 2012;86:13334–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Donlan AN, Sutherland TE, Marie C, Preissner S, Bradley BT, Carpenter RMet al. IL-13 is a driver of COVID-19 severity. JCI Insight 2021;6:150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yale IMPACT Team, Lucas C, Wong P, Klein J, Castro TBR, Silva Jet al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020;584:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SFet al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov 2011;1:54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shiao SL, Ruffell B, DeNardo DG, Faddegon BA, Park CC, Coussens LM. TH2-polarized CD4(+) T cells and macrophages limit efficacy of radiotherapy. Cancer Immunol Res 2015;3:518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dettorre GM, Dolly S, Loizidou A, Chester J, Jackson A, Mukherjee Uet al. Systemic pro-inflammatory response identifies patients with cancer with adverse outcomes from SARS-CoV-2 infection: the OnCovid inflammatory score. J Immunother Cancer 2021;9:e002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meckiff BJ, Ramírez-Suástegui C, Fajardo V, Chee SJ, Kusnadi A, Simon Het al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 2020;183:1340–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman Het al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020;5:eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann Set al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 2019;176:1340–55. [DOI] [PubMed] [Google Scholar]
- 62. Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto Met al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med 2018;10:eaan2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto Set al. Human CD8+ T cell cross-reactivity across influenza A, B and C viruses. Nat Immunol 2019;20:613–25. [DOI] [PubMed] [Google Scholar]
- 64. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman Wet al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 2013;19:1305–12. [DOI] [PubMed] [Google Scholar]
- 65. Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses: heterologous immunity between viruses. Immunol Rev 2010;235:244–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Woodland DL, Blackman MA. Immunity and age: living in the past? Trends Immunol 2006;27:303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Saini SK, Hersby DS, Tamhane T, Povlsen HR, Amaya Hernandez SP, Nielsen Met al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci Immunol 2021;6:eabf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan CW, Chia WN, Young BE, Zhu F, Lim BL, Sia WRet al. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N Engl J Med 2021;385:1401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Iwasaki A, Omer SB. Why and how vaccines work. Cell 2020;183:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris Met al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 2011;85:12201–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RLet al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 2012;7:e35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jiang S, Bottazzi ME, Du L, Lustigman S, Tseng CTK, Curti Eet al. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines 2012;11:1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Martins KA, Bavari S, Salazar AM. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev Vaccines 2015;14:447–59. [DOI] [PubMed] [Google Scholar]
- 74. Jo WK, Drosten C, Drexler JF. The evolutionary dynamics of endemic human coronaviruses. Virus Evol 2021;7:veab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fendler A, Shepherd STC, Au L, Wilkinson KA, Wu M, Byrne Fet al. Adaptive immunity and neutralizing antibodies against SARS-CoV-2 variants of concern following vaccination in patients with cancer: the CAPTURE study. Nat Cancer 2021;2:1321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson Jet al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med 2021;27:1280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mittelman M, Magen O, Barda N, Dagan N, Oster HS, Leader Aet al. Effectiveness of the BNT162b2mRNA covid-19 vaccine in patients with hematological neoplasms. Blood 2021;blood.2021013768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thakkar A, Pradhan K, Jindal S, Cui Z, Rockwell B, Shah APet al. Patterns of seroconversion for SARS-CoV2-IgG in patients with malignant disease and association with anticancer therapy. Nat Cancer 2021;2:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cavanna L, Citterio C, Toscani I. COVID-19 vaccines in cancer patients. seropositivity and safety: systematic review and meta-analysis. Vaccines 2021;9:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shroff RT, Chalasani P, Wei R, Pennington D, Quirk G, Schoenle MVet al. Immune responses to two and three doses of the BNT162b2 mRNA vaccine in adults with solid tumors. Nat Med 2021;27:2002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Swadling L, Diniz MO, Schmidt NM, Amin OE, Chandran A, Shaw Eet al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022;601:110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Routhu NK, Cheedarla N, Bollimpelli VS, Gangadhara S, Edara VV, Lai Let al. SARS-CoV-2 RBD trimer protein adjuvanted with Alum-3M-052 protects from SARS-CoV-2 infection and immune pathology in the lung. Nat Commun 2021;12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sun S, Cai Y, Song TZ, Pu Y, Cheng L, Xu Het al. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res 2021;31:1011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parker R, Partridge T, Wormald C, Kawahara R, Stalls V, Aggelakopoulou Met al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep 2021;35:109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pozzetto B, Legros V, Djebali S, Barateau V, Guibert N, Villard Met al. Immunogenicity and efficacy of heterologous ChAdOx1-BNT162b2 vaccination. Nature 2021;600:701–6. [DOI] [PubMed] [Google Scholar]
- 86. Chaput N, Flament C, Locher C, Desbois M, Rey A, Rusakiewicz Set al. Phase I clinical trial combining imatinib mesylate and IL-2: HLA-DR+ NK cell levels correlate with disease outcome. OncoImmunology 2013;2:e23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jaafar R, Boschi C, Aherfi S, Bancod A, Le Bideau M, Edouard Set al. High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS-CoV-2. Viruses 2021;13:2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. de Sousa E, Ligeiro D, Lérias JR, Zhang C, Agrati C, Osman Met al. Mortality in COVID-19 disease patients: correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS-CoV-2) variants. Int J Infect Dis 2020;98:454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Janice OHL, Ken-En Gan S, Bertoletti A, Tan YJ. Understanding the T cell immune response in SARS coronavirus infection. Emerg Microbes Infect 2012;1:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ren Y, Zhou Z, Liu J, Lin L, Li S, Wang Het al. A strategy for searching antigenic regions in the SARS-CoV spike protein. Genomics Proteomics Bioinformatics 2003;1:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang J, James E, Roti M, Huston L, Gebe JA, Kwok WW. Searching immunodominant epitopes prior to epidemic: HLA class II-restricted SARS-CoV spike protein epitopes in unexposed individuals. Int Immunol 2009;21:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. He Y, Zhou Y, Siddiqui P, Niu J, Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol 2005;43:3718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. He Y, Zhou Y, Wu H, Kou Z, Liu S, Jiang S. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol 2004;42:5309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. de Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 2019;8:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland Jet al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 2018;34:3600. [DOI] [PubMed] [Google Scholar]
- 100. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MAet al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression profile data used in this article are publicly available at EGA under accession number EGAD00001008538. Other data that support the findings of this study are available from the corresponding author upon reasonable request.



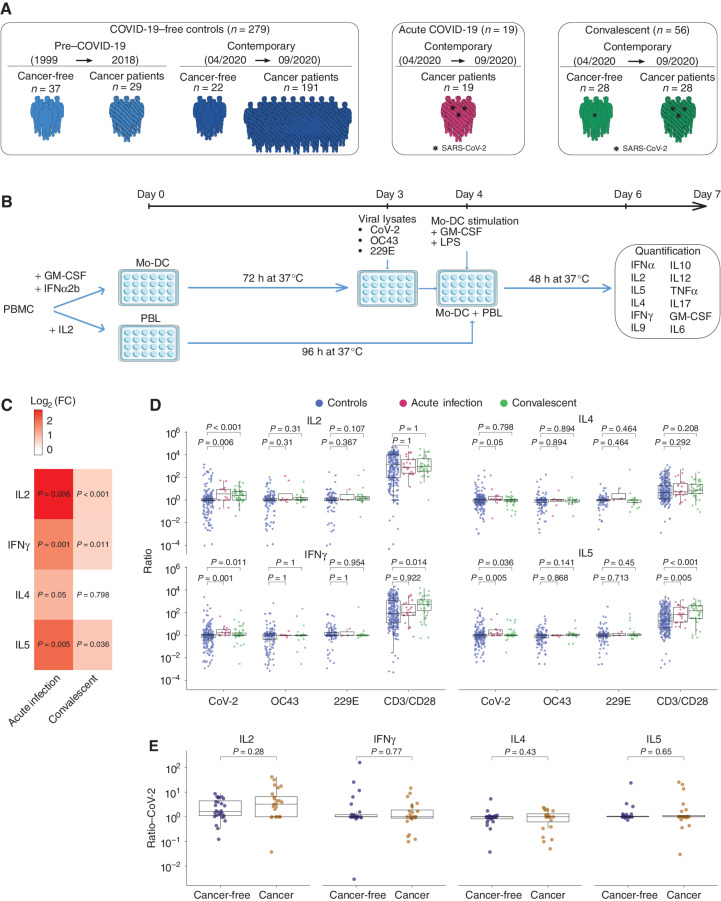
![Figure 2. Unexposed individuals susceptible to COVID-19 exhibited a SARS-CoV-2–specific Th2 profile during the first surge of the pandemic. A, Percentage and number of patients in each cohort—pre–COVID-19 era [yes (+)/no(−)], cancer [yes (+)/no(−)], and COVID-19 [yes (+)/no(−)]—who had a SARS-CoV-2–specific cytokine release (for the prototypic cytokines) compared with VeroE6 (control, n = 279; convalescent, n = 56; Supplementary Table S1B). Fisher exact test to compare the number of cytokine-positive patients across groups. B, Outline of the prospective collection of blood samples used to identify COVID-19–resistant (yellow) versus susceptible (red) patients with cancer (B, top; Supplementary Table S2A and S2B). Bottom, outline of the prospective collection of blood samples used for the comparison of T-cell responses in the cohort of cancer-free individuals who lived in the same household with family members who tested positive for COVID-19 during the 2020 lockdown (G and I). Pie chart (C) indicating the absolute numbers (and percentage) of patients reported as contact (resistant) or infected (susceptible) or unexposed (green) during 1-year follow-up (D). Number of positive cytokines released by SARS-CoV-2–specific PBLs during the cross-presentation assay (Fig. 1B and C) in each group (unexposed, n = 153; resistant, n = 42; susceptible, n = 19). E and F, SARS-CoV-2–specific IL2 (left) and IL5 (right) secretion contrasting resistant (yellow) versus infected (red) cancer cases. E, Each dot represents the ratio of the replicate wells in one individual, and the box plots indicate medians as well as 25th and 75th percentiles for each cancer patient subset. F, The bar plots represent the percentage of positive patients (resistant, n = 42; susceptible, n = 19). Fisher exact test to compare the number of cytokine-positive patients across groups. G and H, SARS-CoV-2–specific IL2/IL5 ratios (means ± SEM) in the different subsets of healthy individuals and patients with cancer presented in B. Refer to Supplementary Fig. S3A for the waterfall plots to visualize variations in the percentages of individuals with IL2/IL5 ratios > or < 1 according to subject category. All group comparisons were performed using the two-sided Wilcoxon–Mann–Whitney test, and P < 0.05 indicates statistically significant differences. I and J, Validation cohort investigating eight additional HCW from Hospices Civils de Lyon and 10 patients with cancer from Gustave Roussy investigated in cross-presentation assays with the dual-color IFNγ/IL5 ELISpot. I, Prototypic photograph of IFNγ and IL5 dual-color ImmunoSpot of a DC/SARS-CoV-2 or VeroE6 PBL coculture (or OKT3 as positive control) for one representative resistant (left) and susceptible (right) HCW. SFC, spot-forming colony counted per 105 PBLs. J, Percentages of SARS-CoV-2–specific Th1 or Th2 cell responses determined by dual ELISPOT assay (CoV-2/VeroE6 >1.5 increase in IFNγ+ (left) or IL5+ (middle) SFC, respectively. Calculation of the IFNγ+/IL5+ SFC ratio per individual in VeroE6 or SARS-CoV-2 condition, and percentages of patients with an increased (>2×) ratio in the SARS-CoV-2 condition, in both resistant versus susceptible groups (right). Fisher exact test to compare the number of positive patients between both groups.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bb56/9394394/2af58aa6432e/958fig2.jpg)
![Figure 3. Peptide repertoire breadth does not predict resistance to COVID-19. A, Experimental setting for the 187 peptide-based in vitro stimulation assay. B, Bicolor map of peptide recognition (positive in salmon, negative in purple, not determined in gray). Patients (n = 148) were ordered in columns by unsupervised hierarchical clustering, and peptides were ordered in rows according to the 5′ to 3′ sequence location in the ORFeome with a distinct color code for each protein. SARS-CoV-1 peptides are aligned at the end in gray. The upper line indicates the frequency of positive individuals for each peptide in the 187 peptide list. C, Peptide frequencies within unexposed and convalescent (with history of COVID-19) patients with cancer compared with unexposed cancer-free subjects. Also refer to Fig. 4A. D and E, Percentages of positive peptides in individuals from the pre–COVID-19 era (n = 24) versus contemporary controls (n = 97; D, right) and in cancer (n = 111) versus cancer-free contemporary individuals (n = 10; D, left) and in uninfected [control (contemporary), n = 97] versus convalescent (n = 27; E, left) and resistant individuals (noninfected contact cases, n = 44) versus susceptible (infected, n = 18) individuals (E, right). Group comparisons within D and E were performed using the two-sided Wilcoxon–Mann–Whitney test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bb56/9394394/b6e32e2836d9/958fig3.jpg)
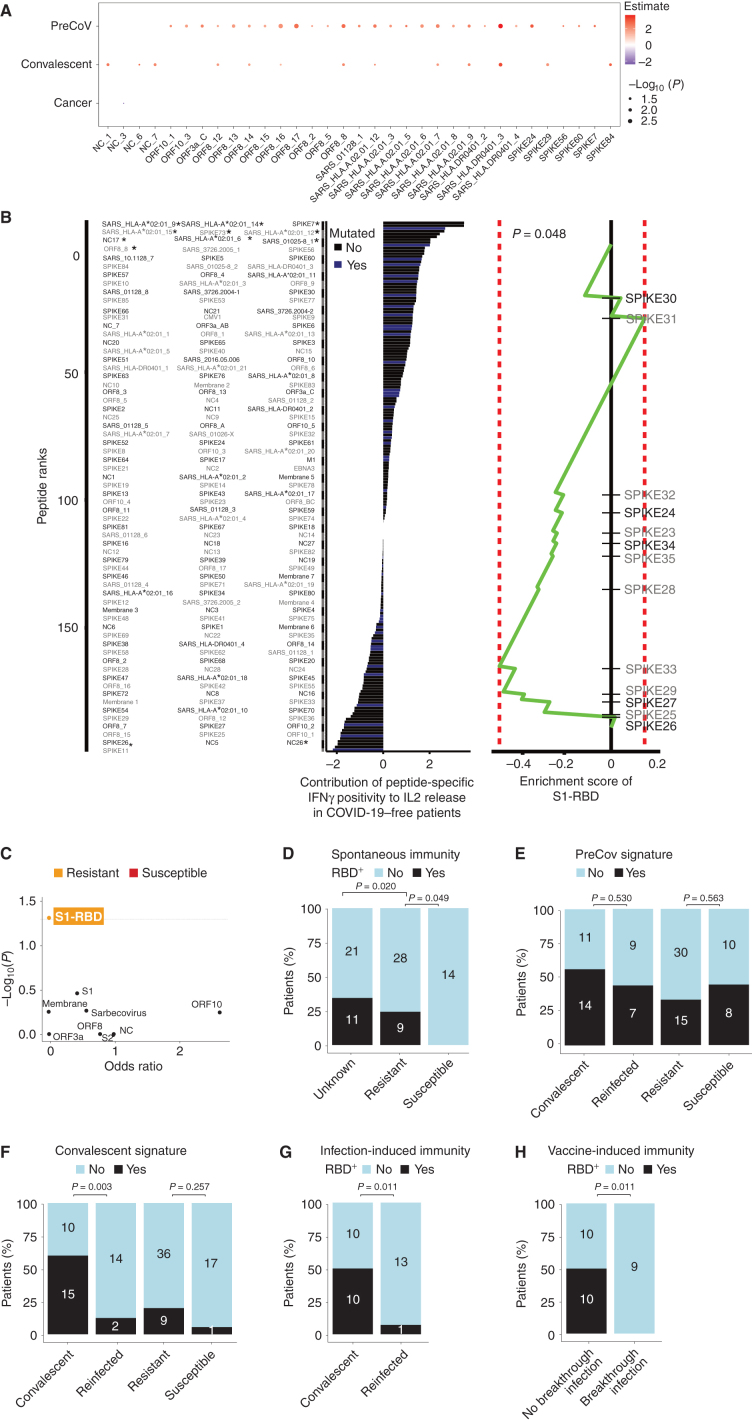
![Figure 5. Patients with cancer (except hematologic malignancies) could mount S1-RBD–specific Th1/Tc1 immune responses during the prime–boost vaccination rollout. A, Description of cohorts of vaccinees in cancer-free individuals and patients with cancer (refer to Supplementary Table S10; Table 1). B, Experimental setting for the peptide pool–based ex vivo stimulation assays. C, Amino acid sequence coverage of the three peptide pools utilized in the high-throughput T-cell screening assay (refer to Supplementary Table S11). D and E, High-throughput screening T-cell assay using the ELISA technique in an automatic platform monitoring IFNγ levels in whole-blood samples from several independent cohorts of HCW (D) or patients with cancer [E, solid or hematologic malignancies (hemato cancer)] with (D) or without (D and E) COVID-19 history, pre- and/or per (after 1 immunization, day 21) and/or post-vaccination (day 90, day 180 for D; only after two shots of vaccines for E) using different peptide pools (C). Monitoring of IFNγ release (bottom) and percentages of individuals with IFNγ levels greater than the threshold of detection (top). The standard errors have been computed with their confidence intervals for these estimates, with each interval most probably containing the genuine percentage. F, Forest plot depicting the impact of the each covariate on the PEPwtRBD IFNγ secretion levels (refer to Table 1 for statistics). Specimens were not systematically paired in the kinetic study. The log10-normalized IFNγ secretions for all peptide stimulation were pooled to model simultaneously their dynamics from the first vaccine to day 180 using linear mixed-effect regression adjusted for patient age, sex, cancer status, type of cancer, COVID history, and vaccine schedule. G, Spearman correlation between serum S1-RBD–specific IgG titers (expressed in arbitrary units) and IFNγ release in the VIDAS IFNγ RUO platform in all cancer-free (left) and cancer vaccinees (right) monitored in Fig. 5D. Each dot represents one sample at one time point. Most individuals have been drawn only once at any time point. H, Percentages and absolute numbers of mutations contained in our S1-RBD peptide list reported in the current SARS-CoV-2 variants (refer to Supplementary Table S12). The difference of the probability of mutation in the S1-RBD region and in other regions was evaluated using logistic regression (odds ratio = 0.21; 95% confidence interval, 0.06–0.68; P = 0.01). I, Paired analysis of the differential magnitude of Th1/Tc1 reactivity against PEPwtRBD versus PEPmutRBD in 343 cancer-free vaccinees with no history of COVID-19. Each line represents one patient sample. Group comparisons were performed using the two-sided paired Wilcoxon–Mann–Whitney test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bb56/9394394/c39b7c788736/958fig5.jpg)