Abstract
Although thousands of anti-SARS-CoV-2 monoclonal neutralizing antibodies (nAbs) have been identified and well characterized, some crucial events in the development of these nAbs during viral infection remain unclear. Using deep sequencing, we explore the dynamics of antibody repertoire in a SARS-CoV-2-infected donor, from whom the potent and broad nAb P2C-1F11 (the parent version of Brii-196) was previously isolated. Further analysis shows a rapid clonal expansion of some SARS-CoV-2-specific antibodies in early infection. Longitudinal tracing of P2C-1F11 lineage antibodies reveals that these elite nAbs were rare. Using sequence alignment, structure modeling, and bioactivity analysis based on site-mutated assay, we demonstrate that a key substitution F27I in heavy chain contributes significantly to the maturation of P2C-1F11-like antibodies. Overall, our findings elucidate the developmental process and maturation pathway of P2C-1F11, providing some important information for the design of novel immunogens to elicit more potent nAbs against SARS-CoV-2 infection.
Keywords: SARS-CoV-2, antibody repertoire, deep sequencing, P2C-1F11 lineage, neutralizing antibody
Graphical abstract
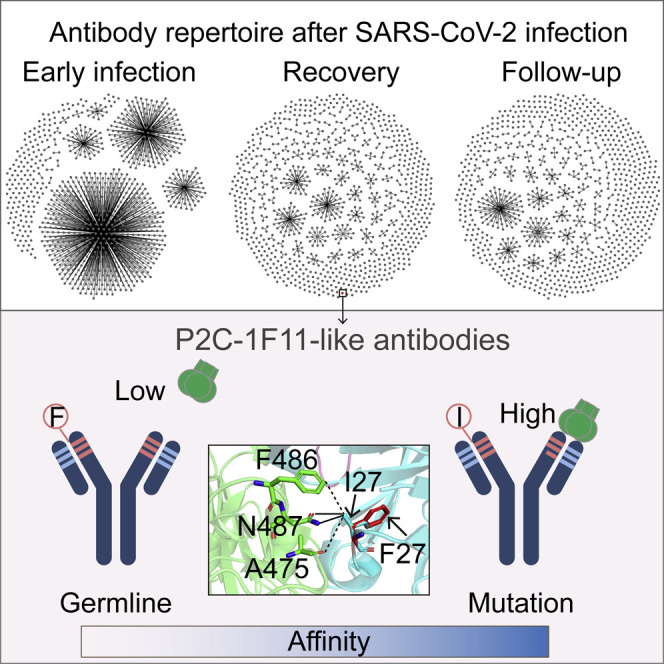
Neutralizing antibodies are essential for the prevention and treatment of SARS-CoV-2 infection. Wang et al. demonstrate dramatic changes of antibody repertoire in a COVID-19 patient throughout the whole disease process and reveal a key mutation within HCDR1 for the development of P2C-1F11-like nAbs.
Introduction
The coronavirus disease 2019 (COVID-19) caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread globally as a severe pandemic for more than 2 years (Cucinotta and Vanelli, 2020), which has already resulted in over 559 million infections and more than 6.3 million fatalities as of July 2022. To make matters worse, the adaptive evolution of SARS-CoV-2 has led to the emergence of a series of variants with higher transmissibility and/or immune evasion, such as Alpha, Beta, Gamma, Delta, and Omicron (Karim and Karim, 2021; Planas et al., 2021; Wang et al., 2021a, 2021b). The spike protein of SARS-CoV-2 is a homotrimer expressed on the surface of the virus and contains two functional subunits, named S1 and S2, which trigger the viral entry into the host cell (Lan et al., 2020). Upon the receptor-binding domain (RBD) of S1 binding to the cell receptor (angiotensin-converting enzyme 2 [ACE2]), the S1 domain is shed from the virus surface and the S2 domain is subsequently exposed to mediate the membrane fusion between the virus and host cell (Walls et al., 2019). Therefore, blocking or disturbing the binding of RBD to the ACE2 is an important intervention strategy to prevent and control the COVID-19 pandemic. Among the possible approaches, the RBD-specific monoclonal neutralizing antibodies (nAbs) are considered good candidates for the development of potential prophylactic and therapeutic agents against SARS-CoV-2.
So far, a large number of monoclonal nAbs have been identified from SARS-CoV-2-infected individuals, vaccine recipients, and immunized animals, showing the effective blockade of viral infection in vitro and in vivo (Cao et al., 2020; Ju et al., 2020; Robbiani et al., 2020; Wang et al., 2021c). Multiple neutralizing antibody drugs have also been approved for emergency use alone or in combined use, such as Regeneron (REGN10933 and REGN10987), Eli Lilly (LY-CoV555 and LY-CoV016), Vir Biotechnology (VIR-7831), AstraZeneca (AZD8895 and AZD1061), and Brii Biosciences (Brii-196 and Brii-198) (Liu et al., 2022). Current studies largely focus on the neutralizing potency and breadth of these nAbs; however, some important immunological questions remain unanswered. For example, what are the features of the monoclonal antibody (mAb) response in the early stage of SARS-CoV-2 infection? When do the broad and potent nAbs appear and what is the abundance of these nAbs? What are the key events during the development of these potent nAbs? Addressing these issues will expand our immunological insights into nAb maturation and reformation in virus acute infection such as SARS-CoV-2. Previously, we isolated a broad nAb (bnAb) with potent neutralizing activities against various SARS-CoV-2 variants, named P2C-1F11, from a COVID-19 convalescent individual (P#2), whose Fc-modified version was the aforementioned Brii-196 (Ju et al., 2020; Wang et al., 2021b). Series of blood samples of P#2 collected throughout the whole disease process give us an opportunity to study the dynamic of antibody repertoire and the development of P2C-1F11 lineage antibodies in this patient.
In this study, we performed a long-read deep sequencing and applied a repertoire analysis to study the longitudinal dynamic of B cell repertoire from the acute phase to the convalescent phase in P#2. Tens of millions of antibody sequences in total were obtained across three time points, namely day 9, day 28, and day 63 after the onset of symptoms. Antibody repertoire analysis offered a clear and detailed picture of antibody response to the SARS-CoV-2 infection. Of note, we traced the development of P2C-1F11 lineage antibodies within this patient. In summary, our repertoire analysis and function verification demonstrated the maturation pathway of P2C-1F11-like nAbs while highlighting the pivotal role of F27I mutation in heavy chain in determining the potent neutralizing activity and pointing out the direction of the novel SARS-CoV-2 vaccine design.
Results
Dynamic B cell repertoire response revealed by next-generation sequencing analysis during the SARS-CoV-2 infection
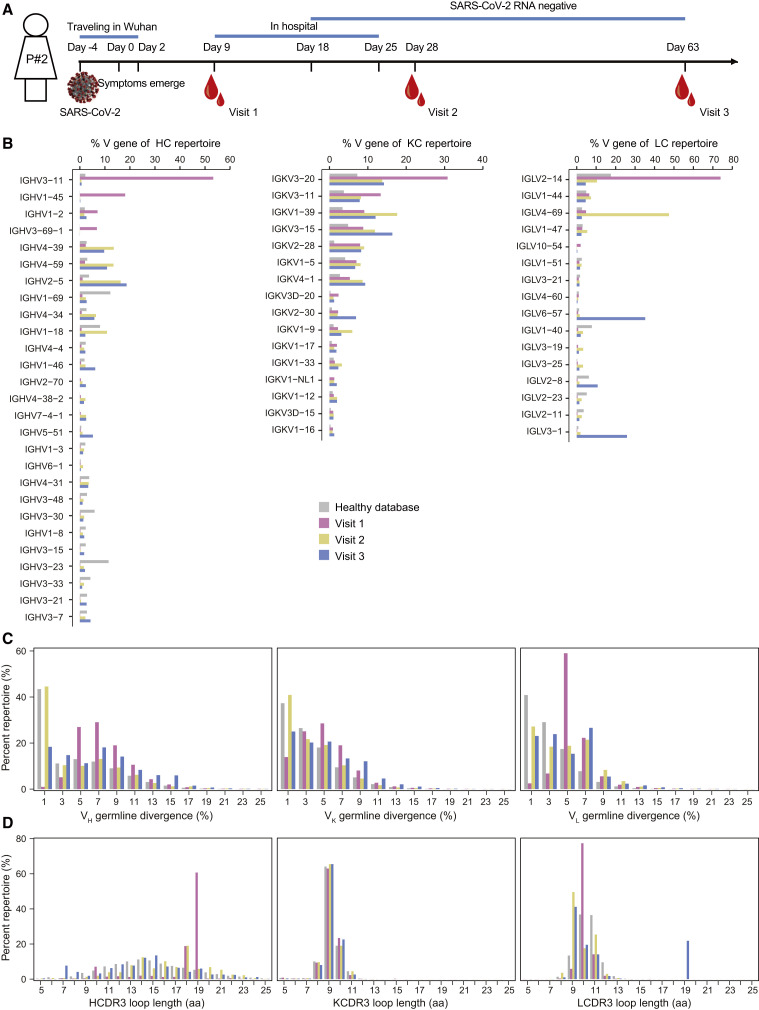
A COVID-19 patient, named P#2, was traveling in Wuhan in the first 6 days of the disease (day −4 to day 2), when symptoms of COVID-19 emerged on day 0. P#2 was admitted to hospital on day 9 and discharged on day 25. The SARS-CoV-2 RNA turned negative from day 18 after the onset of symptoms. To measure the dynamic B cell response elicited by the SARS-CoV-2 infection, peripheral blood mononuclear cells (PBMCs) were collected at three time points (visit 1: day 9, early infection; visit 2: day 28, clinical recovery; visit 3: day 63, clinical follow-up) (Figure 1A). Antibody repertoire analysis based on next-generation sequencing (NGS) has been widely used to study the complex and diverse B cell response during viral infection (Gao et al., 2019; Kong et al., 2016; Kumar et al., 2020). At each time point, total RNA was obtained from PBMCs and used to prepare the antibody libraries of heavy chain and light chains (κ and λ). We combined 5′-RACE PCR primer with 3′-specific primers targeting the constant regions of immunoglobulin G (IgG), Igκ, and Igλ to amplify the full-length variable regions of unpaired heavy and light chains for bulk sequencing. The NGS on the Illumina MiSeq (2 × 300) platform generated a total of about 13.8, 15.0, and 16.3 million raw reads for visit 1, visit 2, and visit 3, respectively (Table S1). After data processing (Figure S1), we obtained about 9.2, 10.0, and 11.6 million high-quality and full-length sequences of antibody variable regions for in-depth analysis at visit 1, visit 2, and visit 3, respectively (Table S1).
Figure 1.
Antibody repertoire profiles of donor P#2 across the SARS-CoV-2 infection
(A) Schematic representation of experimental design. Blood samples from P#2 were collected at three time points after symptom onset (visit 1: day 9, early infection; visit 2: day 28, clinical recovery; visit 3: day 63, clinical follow-up). The full-length repertoires of variable region of antibodies were obtained by targeted amplification and sequencing on the Illumina MiSeq (2 × 300 bp) platform.
(B–D) Distributions were plotted for (B) germline V gene usage (only those above 1% of the repertoire are shown), (C) degree of SHM, and (D) CDR3 loop length for heavy and light (κ/λ) chains. The antibody database of healthy individuals was downloaded and reanalyzed from the cAb-Rep database (https://cab-rep.c2b2.columbia.edu/). Color coding represents the three time points analyzed, with visit 1 shown in pink, visit 2 in yellow, and visit 3 in blue. The healthy database is shown in gray. HC, heavy chain; KC, κ chain; LC, λ chain.
See also Table S2.
In P#2, SARS-CoV-2 infection rapidly induced a strong antibody response, showing diverse and dynamic distributions in germline gene usage throughout the whole disease process (Figure 1B and Table S2). In the early stage of infection (day 9 after symptom onset), the antibody repertoires of visit 1 were dominated by a few germline genes. IGHV3-11, IGKV3-20, and IGLV2-14 occupied over 50%, 30%, and 70% of the heavy, κ, and λ chain repertoires, respectively. In the convalescent period, the heavy-chain repertoire showed a relatively steady germline distribution at visit 2 (day 28 after symptom onset) and visit 3 (day 63 after symptom onset). A similar pattern was found in the κ-chain repertoire. By contrast, IGLV4-69 and IGLV6-57 were predominantly used over 40% and 30% in the λ-chain repertoire of visit 2 and visit 3, respectively.
We next determined the degree of somatic hypermutation (SHM) of antibody repertoire at each time point. As shown in Figure 1C, antibodies at visit 1 had higher SHMs than at visit 2 and visit 3, with main distributions of around 5% for heavy-, κ-, and λ-chain repertoires. Of note, there was a significant increase in the population of germline-like antibodies at visit 2 compared with visit 1 for both heavy and light chains, whose SHMs were around or lower than 1%. This kind of limited SHM is an important characteristic of SARS-CoV-2-specific antibodies (Kreer et al., 2020; Zhang et al., 2022). In addition, we analyzed the distributions of complementary determining region 3 (CDR3) loop length in heavy- and light-chain repertoires at three time points, which is a crucial region in determining the antibody specificity (Montague et al., 2021). Due to the enormous diversity of D gene, the CDR3 of heavy chain (HCDR3) is highly variable in the loop length, yet those of κ and λ chains were relatively steady (Gao et al., 2019). As shown in Figure 1D, HCDR3 loops with 19 amino acids accounted for over 60% of the heavy-chain repertoire at visit 1. The HCDR3 lengths were evenly distributed, ranging from seven amino acids at visit 2 to 23 amino acids at visit 3. Similar to the heavy chain, there was a significantly dominant population occupying more than 70% of the λ-chain repertoire at visit 1, whose LCDR3 loop lengths were consistently 10 amino acids.
Rapid clonal expansion of SARS-CoV-2-specific antibodies in the early stage of virus infection
The above analysis of B cell repertoire revealed that the proportions of antibody sequences with the same germline gene usage and CDR3 length were rapidly increased after SARS-CoV-2 infection. We hypothesized that some B cells underwent clonal expansion in a very short time after infection. First, we summarized all antibody sequences obtained from three time points and clustered them into different clones based on the following features: identical V and J germline gene, identical CDR3 length, and the amino acid identity in the region of CDR3 ≥ 80% (Yan et al., 2021). As shown in Figure S2A, a total of 161,507, 304,798, and 68,381 clones were identified in heavy-, κ-, and λ-chain repertoires, respectively. Of note, a small number of clones were shared among the repertoires of three time points. In particular, the top 15 clones in size of heavy-chain repertoires at visit 1, visit 2, or visit 3 were scarcely detected in repertoires from the other two time points (Figure S2B).
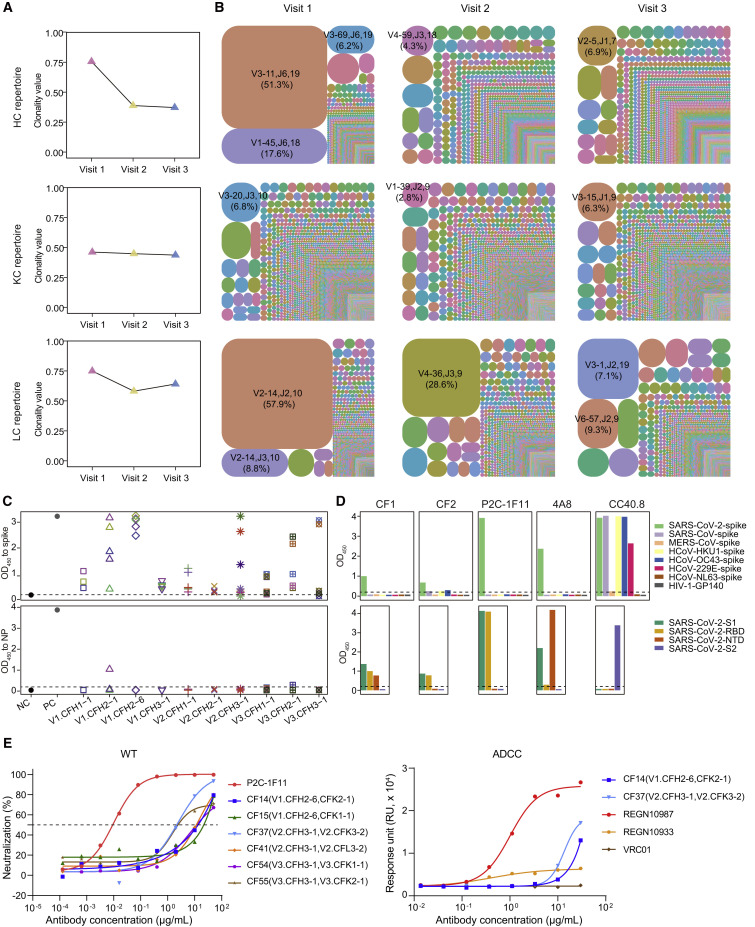
The Shannon index is often used to assess the clonality score of an immune repertoire and ranges from 0 to 1, where 0 means that each clonotype has no clonal expansion and 1 means that the group includes only one clonotype (Zhang et al., 2018). Here, we performed overall clonality analysis of the antibody repertoires at three time points. The results showed that the repertoires of heavy chain and λ chain displayed the greatest clonality at visit 1 and obviously declined over time, with Shannon indexes falling from 0.76 and 0.75 to 0.39 and 0.58 at visit 2, and 0.37 and 0.64 at visit 3, respectively (Figure 2A). To obtain a holistic view of the different antibody clonotypes at three time points, we selected the top 10,000 frequent clones of heavy, κ, and λ chains, respectively, and constructed tree-maps to represent the relative abundances at visit 1, visit 2, and visit 3 (Figure 2B). Compared with visit 2 and visit 3, the clones contained at visit 1 were obviously larger in scale. For example, clone-family 1 of heavy chain (CFH1) and clone-family 1 of λ chain (CFL1) occupied over 50% of the population of their clonotypes.
Figure 2.
Clonal expansion of antibody repertoires of donor P#2 at three time points
(A) Diversity of heavy- and light-chain repertoires in different phases. Values were calculated using the Shannon index. HC, heavy chain; KC, κ chain; LC, λ chain.
(B) V-J-CDR3 tree-map of the repertoires. Each rectangle or circle in the tree-map represents a clonotype with the same V and J gene, the same CDR3 length, and more than 80% identity between CDR3 amino acid sequences. The top 10,000 most frequent clonotypes in the antibody repertoires were used for the tree-maps. The size of each rectangle or circle denotes the relative frequency of an individual clonotype. The clonotype ID consists of V gene, J gene, and the CDR3 length, separated by commas. The top clonotypes in each repertoire are listed, and their percentages are indicated in parentheses. The colors of individual clonotypes are randomly chosen, and the colors do not match between plots.
(C) ELISA binding of representative clone-family (CF) antibodies to the SARS-CoV-2 spike (upper panel) and NP (lower panel) proteins. For each time point, selected heavy chains are represented by different shapes, and the selected light chains paired with the heavy chains are represented by different colors. An HIV-1-specific antibody, VRC01, acted as the negative control, while P2C-1F11 and rabbit anti-NP polyclonal antibody were used as positive controls for spike and NP, respectively. NC, negative control; PC, positive control. The OD450 value of ≥0.2 is considered positive.
(D) Binding specificity analysis of representative CF mAbs. CF1 and CF2 were two antibodies from the dominant clonotype in visit 1 repertoires. Three known SARS-CoV-2-spike nAbs were used as control. P2C-1F11 and 4A8, both specific for SARS-CoV-2, are anti-RBD and anti-NTD mAbs, respectively, while CC40.8 targets the S2 subunit of SARS-CoV-2 and cross-reacts with several other human coronavirus spike proteins (Chi et al., 2020; Ju et al., 2020; Zhou et al., 2022). Upper panel: ELISA binding to spike proteins of seven human coronaviruses and HIV-1-GP140 (negative control); lower panel: ELISA binding to different domains of WT SARS-CoV-2. The OD450 value of ≥0.2 is considered positive.
(E) Biological functions of representative CF mAbs. Left panel: Neutralization of six CF mAbs against WT SARS-CoV-2 pseudovirus, with P2C-1F11 as a positive control; Right panel: ADCC of two CF mAbs, with REGN10987, REGN10933, and VRC01 as strong positive control, weak positive control, and negative control, respectively (Hansen et al., 2020). ADCC, antibody-dependent cell-mediated cytotoxicity. The data presented here are means of at least two independent experiments. The curves are representatives of independent experiments with similar results. A cutoff value of 50% is indicated by a horizontal dashed line in neutralization.
See also Figures S2–S4 and Table S3.
To verify whether these expanded antibodies were SARS-CoV-2 specific and what were their biological functions, we selected and synthesized some representative sequences to evaluate their binding, neutralizing, and non-neutralizing activities against SARS-CoV-2. At each time point, we mainly verified the biological function of the top two or three expanded clones. After removing duplicate amino acid sequences, we chose ten antibody sequences with the highest abundance from each clone to calculate their phylogenetic distances. Several representative sequences of heavy and light chains were manually selected for the subsequent synthesis and antibody production (Figure S3). At visit 1, four heavy chains were paired with five light chains, respectively, and tried to produce human IgG1 mAbs for lack of the natural pairing information between heavy and light chains in bulk sequencing. Using the same strategy, a total of 21 and 18 clone-family (CF) mAbs were successfully obtained from visit 2 and visit 3, respectively (Table S3).
Firstly, we tested whether these CF mAbs could bind to the wild-type (WT) SARS-CoV-2 spike (S) protein or nucleocapsid protein (NP), which were two primary antigens examined for seroconversion (Sette and Crotty, 2021). As shown in Figure 2C, most CF mAbs showed higher affinities for binding to spike than NP, suggesting that these expanded antibodies could recognize the virus antigen to different degrees. We then selected two representative CF mAbs derived from the largest clonotypes of heavy- and light-chain repertoires to evaluate the virus specificity and determine the recognition domain of spike protein (Figure 2D). CF1 and CF2 targeting the S1 subunit were both specific for SARS-CoV-2 and did not cross-react with the spike proteins of the other six human coronaviruses (SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-229E, and HCoV-NL63), suggesting that these CF mAbs might be induced by the primary infection of SARS-CoV-2 rather than derived from the reactivation of pre-existing cross-reactive memory B cells. Finally, we evaluated the biological functions of these SARS-CoV-2 spike-specific CF mAbs including the neutralization and an important non-neutralization (antibody-dependent cell-mediated cytotoxicity [ADCC]). Compared with some approved antibody drugs (P2C-1F11 and REGN10987), these expanded mAbs displayed weaker neutralizing and ADCC activities on the whole (Figures 2E and S4; Table S3). More studies in the future are needed to determine whether these expanded mAbs identified from different stages of SARS-CoV-2 infection could play other antiviral roles or not.
Longitudinal tracing of P2C-1F11 lineage antibodies during SARS-CoV-2 infection in P#2
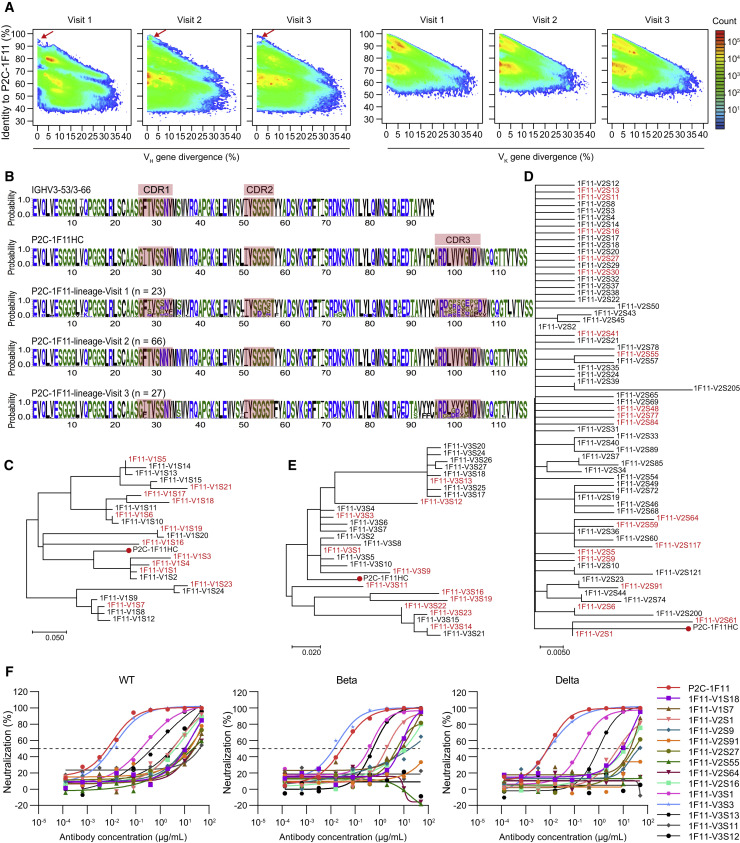
Judging from the above results, the potent neutralizing antibody, P2C-1F11, seemed not to originate from dominant germline genes or expanding clones. The heavy chain of P2C-1F11 was derived from IGHV3-66, whose abundance was lower in the antibody repertoires of P#2 at three time points (Table S2). Therefore, we used the heavy- and light-chain sequences of P2C-1F11 as bait to longitudinally trace the development of P2C-1F11 lineage antibodies in the repertoires at three time points according to our previously established methods (Kong et al., 2016; Kumar et al., 2020). Together with P2C-1F11, another six mAbs (P2B-2F6, P2C-1A3, P2C-1C10, P2B-2G4, P2A-1A8, and P2A-1A10) derived from different germline genes were also isolated from P#2 at visit 2 and effectively inhibited the infection of SARS-CoV-2 live virus (Ju et al., 2020). The heavy- and light-chain repertoires with respect to each mAb were visualized overall on identity-divergence two-dimensional (2D) plots (Figures 3A and S5). We observed some distinct or contiguous islands on these 2D plots with a high identity of over 90% and similar degrees of SHM to the reference antibody sequences, whose proportions were the largest in the repertoires of visit 2. By contrast, the dynamic lineage of antibody light chains displayed a relatively stable distribution across three time points.
Figure 3.
Lineage tracing of P2C-1F11 in the P#2 repertoires across the SARS-CoV-2 infection
(A) Identity-divergence two-dimensional (2D) plots of heavy (left panel) and light (right panel) chain repertoires of P#2 at time points visit 1, visit 2, and visit 3. The x axis indicates the sequence divergence from the putative germline gene, and the y axis indicates their identity with respect to the P2C-1F11 heavy or κ chain. Color coding denotes the density of the sequence.
(B) Sequence logo of the P2C-1F11 lineage heavy chains at visit 1 (n = 23), visit 2 (n = 66), and visit 3 (n = 27) with their CDRs highlighted.
(C–E) Phylogenetic analysis of the heavy-chain sequences of P2C-1F11 lineage antibodies at (C) visit 1, (D) visit 2, and (E) visit 3. The maximum-likelihood trees were constructed on the basis of the amino acid sequences of selected antibodies. The synthesized heavy chains paired with the P2C-1F11 light chain for being functionally tested are highlighted in red, and branches of P2C-1F11 are marked with red dots.
(F) Neutralization of representative P2C-1F11 lineage antibodies against WT SARS-CoV-2 pseudovirus and other variants (Beta and Delta). The P2C-1F11 lineage antibodies consisting of identified heavy chains and the P2C-1F11 light chain were synthesized and tested, with P2C-1F11 as the positive control. The curves are representatives of at least two independent experiments with similar results. A cutoff value of 50% is indicated by a horizontal dashed line.
See also Table S4.
Considering the high potency and wide application of P2C-1F11, we mainly investigated the dynamic of P2C-1F11 lineage antibodies. In the early stage of infection, this lineage did not appear or was not rapidly enriched. The population of these similar mAbs showed a marked expansion at visit 2, from which P2C-1F11 was isolated, and then became significantly smaller at visit 3 (Figure 3A). Furthermore, the sequence alignment among these similar antibodies revealed their great differences in detail. We selected a total of 23, 66, and 27 distinct heavy-chain sequences of P2C-1F11 lineage at visit 1, visit 2, and visit 3, respectively, according to the following process: (1) remove repetitive amino acid sequences; (2) rank by the identity to heavy chain of P2C-1F11 (P2C-1F11HC); (3) conduct phylogenetic analysis with P2C-1F11HC (Figures 3B–3E). Although the identities of these antibody sequences to P2C-1F11HC reached at least 80% or even exceeded 95%, there were obvious differences in their three CDRs. To investigate the functional variation within the P2C-1F11 lineage, we selected representative heavy-chain sequences at three time points, paired them with the P2C-1F11 light chain to produce mAbs, respectively, and then measured their neutralizing activities against WT SARS-CoV-2 and variants (Figure 3F and Table S4). Of note, most of these mAbs exhibited moderate or weak neutralizing potencies except 1F11-V3S3, which neutralized all tested pseudoviruses with potencies similar to that of P2C-1F11. These data suggested that some minor differences in sequence could lead to great divergences in the function of antibodies.
A key F27I substitution from germline contributes to the maturation of P2C-1F11-like antibodies
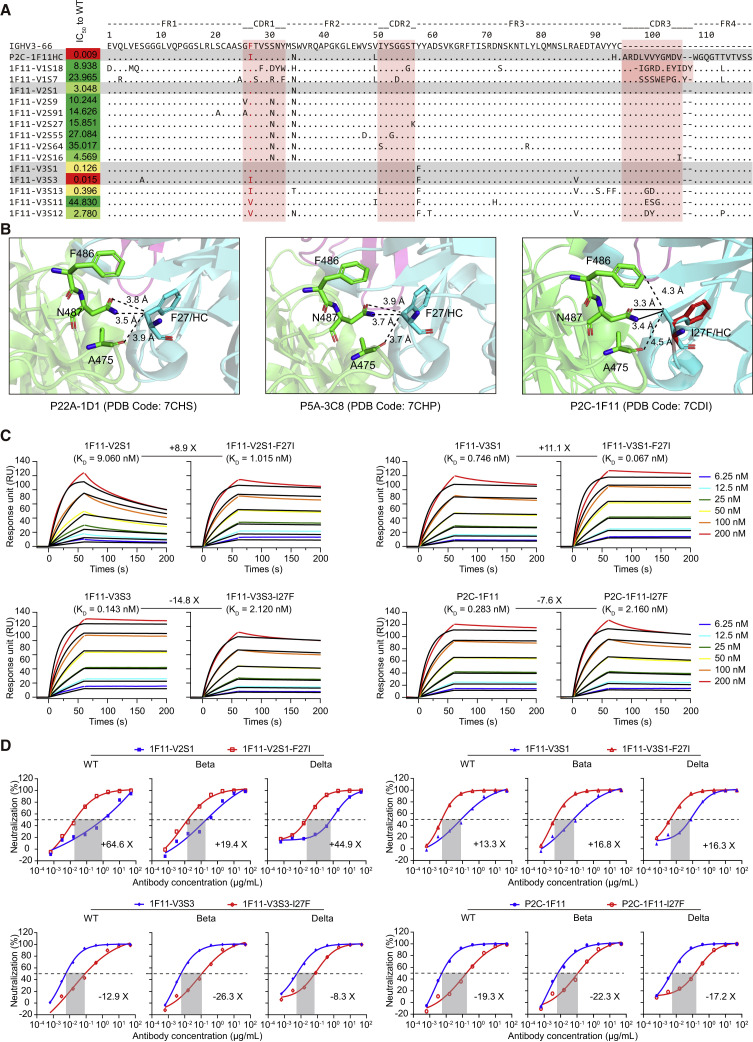
In this study, we identified a total of 14 P2C-1F11 lineage nAbs in the antibody repertoires of P#2. Sequence alignment and functional analysis showed that some key amino acid residuals might play important roles in determining the neutralization of mAbs (Figure 4A). Under the same sequences of HCDR2 and HCDR3, I27 located in HCDR1 was only shared in two potent nAbs, P2C-1F11 and 1F11-V3S3. In contrast, 1F11-V2S1 and 1F11-V3S1 still kept the F27 residue in line with the germline gene, with moderate neutralizing activities decreased by about 14-fold and 339-fold compared with P2C-1F11. Based on the crystal structure of the P2C-1F11/SARS-CoV-2 RBD complex (Ge et al., 2021; Zhang et al., 2021), the I27 residue was directly involved in the contact interface, suggesting its potential influence on recognition. In our previous study, we also identified another two IGHV3-53/3-66 public nAbs, named P22A-1D1 and P5A-3C8, and resolved their crystal structures with the RBD, whose HCDR1 sequences were identical to that of P2C-1F11HC except the F27 residue (Zhang et al., 2021). As shown in Figure 4B, F27 of the germline gene only forms three van der Waals interactions with A475, F486, and N487 residues on the SARS-CoV-2 RBD. However, the I27 builds an additional two salt bridges with N478 residue, contributing substantially to the recognition of P2C-1F11 to the RBD. This interaction in between seems to be relatively weaker when the I27 residue of P2C-1F11HC is artificially reversed to F27 of the germline gene, which (highlighted in red) is additionally displayed with reference to the structural information of P22A-1D1 and P5A-3C8.
Figure 4.
Analysis of critical residues for the functional maturation of P2C-1F11-like antibodies
(A) Sequence alignment of representative P2C-1F11 heavy-chain variants with their three CDRs highlighted in pink. The identified hotspot residue that potentially critical for the P2C-1F11 maturation is marked in red. The neutralization of each mAb against WT SARS-CoV-2 is presented as IC50 value.
(B) Structural comparison of the interactions with the WT SARS-CoV-2 RBD between P2C-1F11 and two IGHV3-53/3-66 public antibodies (P22A-1D1 and P5A-3C8) carrying germline residue F27. The germline IGHV3-53 is closely related to IGHV3-66, except for an I12V substitution within the frame region 1 (FR1), so they are often referred to collectively as IGHV3-53/3-66 antibodies (Wang et al., 2022; Yuan et al., 2020; Zhang et al., 2021). RBD is in green, while the antibody heavy chains are in cyan. The position of mutant residue I27F on HCDR1 of P2C-1F11 is inferred based on that of the two public antibodies and is highlighted in red. The distance of 4.5 Å is used as the maximal cutoff value for the intermolecular interactions. The van der Waals and salt-bridge interactions are represented by dashed and solid black lines, respectively. HC, heavy chain.
(C) Binding kinetics of mAbs to the WT SARS-CoV-2 RBD protein measured by SPR. The black lines indicate the experimentally derived curves while the colored lines represent fitted curves based on the experimental data. Binding kinetics were measured for six concentrations of the RBD at 2-fold dilution ranging from 200 to 6.25 nM. Results presented are representative of two independent experiments. The mean dissociation constant (KD) values are indicated.
(D) Neutralization of mAbs against SARS-CoV-2 pseudoviruses of WT and other variants (Beta and Delta). The prototype antibodies are marked in blue while site-mutated antibodies are in red. Results presented are representative of at least two independent experiments with similar results. A cutoff value of 50% is indicated by a horizontal dashed line.
To prove the importance of F27I mutation, the F27 residue of 1F11-V2S1 and 1F11-V3S1 was site-mutated to I27, respectively. Measured by a surface plasmon resonance (SPR) experiment, the binding affinities of 1F11-V2S1-F27I and 1F11-V3S1-F27I to the RBD were improved by 8.9-fold and 11.1-fold (Figure 4C). Meanwhile, we also mutated the I27 residue of 1F11-V3S3 and P2C-1F11 back to F27, respectively, then tested their binding activities. The results showed that the affinities of 1F11-V3S3-I27F and P2C-1F11-I27F to the RBD were largely decreased more than 7-fold compared with those of 1F11-V3S3 and P2C-1F11 (Figure 4C). Similar evidence was obtained from the neutralizing assay (Figure 4D). Whether against the WT SARS-CoV-2 or other variants, the neutralizing potencies of 1F11-V2S1-F27I and 1F11-V3S1-F27I were increased more than 13-fold. By contrast, the potencies of 1F11-V3S3-I27F and P2C-1F11-I27F were largely decreased more than 8-fold. The great differences in binding affinities and neutralizing potencies against SARS-CoV-2 between the I27 and F27 versions of these mAbs indicated that the F-to-I substitution at residue 27 of HCDR1 should be one important step in helping the development and maturation of these P2C-1F11-like mAbs.
Discussion
Since the beginning of the COVID-19 pandemic from late 2019 and early 2020, researchers immediately got to work around the clock to develop various strategies to fight against SARS-CoV-2. We and many other groups reported a number of potent nAbs (Cao et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Pinto et al., 2020; Robbiani et al., 2020; Rogers et al., 2020) that were considered as good antiviral drug candidates. Meanwhile, a large body of scientists focused on the human immune response to SARS-CoV-2 infection, especially B cell receptor repertoires (Galson et al., 2020; Montague et al., 2021; Nielsen et al., 2020; Schultheiss et al., 2020; Yan et al., 2021). Understanding this B cell response is also critical in supporting the development of COVID-19 treatments. In this study, unlike the traditional studies, we performed a rather comprehensive sequence analysis together with key function verification and drew some important conclusions.
First, we described a rapid and strong antibody response against acute SARS-CoV-2 infection in a COVID-19 patient. The germlines of IGHV3-11 and IGLV2-14 accounted for more than half of the heavy- and λ-chain repertoires, respectively, 9 days after the onset of symptoms, mainly due to the rapid clonal expansion of V1.CFH1 and V1.CFL1. We considered that this might be a SARS-CoV-2-specific antibody response, because neither CF1 nor CF2 mAbs could recognize the spike proteins of other human coronaviruses. These early antibodies did not display potent neutralizing and ADCC activities against SARS-CoV-2, so other biological functions need to be studied in the future. It should be noted that the germline gene IGHV3-66 of P2C-1F11 or its similar gene IGHV3-53 did not dominate the antibody repertoires across three time points.
Second, we traced the development of P2C-1F11 lineage antibodies in this COVID-19 patient, P#2. Unlike most of the previous studies (Galson et al., 2020; Kim et al., 2021; Montague et al., 2021; Nielsen et al., 2020; Schultheiss et al., 2020), we performed a series of strict function verification of these very similar amino acid sequences including the neutralization against WT SARS-CoV-2 and other variants. The majority of synthetic mAbs were unable or too weak to inhibit the virus infection, suggesting that this kind of potent nAbs is rare and that some key amino acid residues might play important roles in their binding neutralizing activities. Combining the potency and broad spectrum, P2C-1F11 ranked first in a total of 206 RBD-specific mAbs we isolated from eight SARS-CoV-2-infected individuals (Ge et al., 2021; Ju et al., 2020; Wang et al., 2021b; Zhang et al., 2021). This fact proved again that broadly elite nAbs originating from the P2C-1F11 lineage were uncommon, so it should be extremely important to clarify their processes of development and maturation.
Third, we demonstrated that the F27I mutation in HCDR1 was a critical factor in determining the binding affinities and neutralizing potencies of P2C-1F11-like antibodies. This mechanism was solidly supported by the sequence alignment, structure modeling, and bioactivity analysis based on a site-mutated experiment. A single substitution in the heavy chain, F27I, helped P2C-1F11-like antibodies to rapidly mature to potent nAbs from the original germline antibody. Similar to what researchers did in the study of HIV-1-specific nAbs (Huang et al., 2016; Kong et al., 2016; Umotoy et al., 2019; Wu et al., 2015), this discovery in the anti-SARS-CoV-2 antibody has at least two meanings: one is that it will largely contribute to enhancing the binding affinity and neutralization in antibody engineering, and the other is that it will also provide guidelines for the design of SARS-CoV-2 vaccine.
In conclusion, we analyzed the dynamic of antibody repertoire response to SARS-CoV-2 infection, whose pattern in the early stage was largely different from that in the convalescent stage. P2C-1F11 provided us with an opportunity to explore the early development of this lineage antibody, especially to reveal the key events in the maturation from germline. Our findings demonstrated that the human immune system successfully combated SARS-CoV-2 infection by driving the F-to-I mutation at residue 27 in HCDR1 of P2C-1F11-like nAbs, providing useful information for the rational design of next-generation nAbs and SARS-CoV-2 vaccines.
Limitations of the study
In this study, we performed 5′-RACE PCR and bulk sequencing to obtain the as many genes in the full-length antibody variable region as possible. Sufficient sequencing depth supported the analysis of antibody repertoire. However, the heavy and light chains were not naturally paired. Future study by high-throughput single B cell transcriptome sequencing with paired heavy and light chains might provide a deeper understanding of the maturation pathway of human antibodies. We demonstrated that F27I substitution was one of the key events in the development of P2C-1F11-like nAbs, but not the only one. Despite I27 also being found in the CDR1 of 1F11-V3S13, some other differences might also affect its neutralization, such as L51 in CDR2 and G100 and D101 in CDR3. More relevant mechanisms underlying the maturation of P2C-1F11-like nAbs need to be uncovered in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P2C-1F11 | Ge et al. (2021) | PDB code 7CDI |
| 4A8 | Chi et al. (2020) | PDB code 7C2L |
| CC40.8 | Zhou et al. (2022) | PDB code 7SJS |
| REGN10933 | Hansen et al. (2020) | PDB code 6XDG |
| REGN10987 | Hansen et al. (2020) | PDB code 6XDG |
| VRC01 | Zhou et al. (2013) | PDB code 4LST; RRID: AB_2491019 |
| HRP goat anti-human IgG (H + L), polyclonal | ZSGB-BIO | Cat# ZB-2304 |
| Bacterial and virus strains | ||
| E. coli DH5α | Takara | Cat# 9057 |
| SARS-CoV-2/wild-type (WT) | Ju et al. (2022) | N/A |
| SARS-CoV-2/Beta | Ju et al. (2022) | N/A |
| SARS-CoV-2/Delta | Ju et al. (2022) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| FreeStyle 293 expression medium | Gibco | Cat# 12338026 |
| SARS-CoV-2 WT S1 | Sino Biological Inc. | Cat# 40591-V08H |
| SARS-CoV-2 WT RBD | Sino Biological Inc. | Cat# 40592-V08B |
| SARS-CoV-2 WT NTD | Sino Biological Inc. | Cat# 40591-V49H |
| SARS-CoV-2 WT S2 | Sino Biological Inc. | Cat# 40590-V08B |
| SARS-CoV-2 WT spike | Sino Biological Inc. | Cat# 40589-V08B1 |
| SARS-CoV-2 WT NP | Sino Biological Inc. | Cat# 40588-V08B |
| SARS-CoV spike | Sino Biological Inc. | Cat# 40634-V08B |
| MERS-CoV spike | Sino Biological Inc. | Cat# 40069-V08B |
| HCoV-HKU1 spike | Sino Biological Inc. | Cat# 40606-V08B |
| HCoV-OC43 spike | Sino Biological Inc. | Cat# 40607-V08B |
| HCoV-229E spike | Sino Biological Inc. | Cat# 40605-V08B |
| HCoV-NL63 spike | Sino Biological Inc. | Cat# 40604-V08B |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat# 15140163 |
| Ampicillin | Amresco | Cat# 69-52-3 |
| Polyethylenimines (PEIs) 25K | PolySciences | Cat# 23966 |
| Trypsin | Gibco | Cat# 25200–072 |
| Fetal bovine serum | Gibco | Cat# 10099–141C |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965–092 |
| HEPES (1M) Buffer Solution | Gibco | Cat# 15630–080 |
| Opti-MEM Reduced Serum Medium | Gibco | Cat# 51985034 |
| Roswell Park Memorial Institute (RPMI) 1640 Medium | Gibco | Cat# 11875–101 |
| DEAE-Dextran hydrochloride | Sigma Aldrich | Cat# D9885-10G |
| Critical commercial assays | ||
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| QIAquick Gel Extraction Kit | Qiagen | Cat# 28706 |
| Bright-Lite Luciferase Assay System | Vazyme Biotech | Cat# DD1204-03 |
| Single Cell Full Length mRNA-Amplification Kit | Vazyme Biotech | Cat# N712 |
| Gold Hi EndoFree Plasmid Maxi Kit | CWBIO | Cat# CW2104M |
| TMB substrate | Sangon Biotech | Cat# E661007-0100 |
| Deposited data | ||
| Human antibody repertoire | This paper | National Genomics Data Center: HRA002366 |
| Experimental models: Cell lines | ||
| HEK293F | Gibco | Cat# R79007 |
| Human: HEK293T | ATCC | Cat# CRL-3216 |
| HEK293T expressing human ACE2 | YEASEN Biotech | Cat# 41107ES03 |
| Jurkat-FcγRIIIa-V158 Effector Cells | Vazyme Biotech | Cat# DD1301 |
| Recombinant DNA | ||
| pNL4-3.Luc.R-E− | NIH AIDS Reagent Program | Cat# 3418 |
| Software and algorithms | ||
| TrimGalore v0.6.0 | Trim Galore | https://github.com/FelixKrueger/TrimGalore/ |
| PANDAseq v2.11 | Masella et al. (2012) | https://github.com/neufeld/pandaseq |
| IgBlast v1.17.1 | Ye et al. (2013) | https://ncbi.github.io/igblast/ |
| Needleall | A program from EMBOSS package | http://emboss.sourceforge.net/apps/release/6.6/emboss/apps/needleall.html |
| MEGA v10.2.6 | Kumar et al. (2008) | https://www.megasoftware.net/ |
| PyMOL | PyMOL | http://www.pymol.org |
| Graphpad Prism 8 | GraphPad | https://www.graphpad.com/ |
| R | R | https://www.r-project.org/ |
| Other | ||
| IMGT Reference directory sets | Lefranc et al. (2005) |
http://www.imgt.org/vquest/refseqh. html#VQUEST |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zheng Zhang (zhangzheng1975@aliyun.com).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Human subjects
The blood samples were donated by a SARS-CoV-2 convalescent patient (P#2, female, 65 years old), who returned to Shenzhen from Wuhan and was hospitalized at Shenzhen Third People’s Hospital, China (Ju et al., 2020). This study was approved by the Ethics Committee of Shenzhen Third People’s Hospital (approval number: 2020–084). This participant had provided written informed consent for sample collection and subsequent analysis. Peripheral blood mononuclear cells (PBMCs) were separated from blood samples by Ficoll-Hypaque gradient (GE Healthcare) centrifugation, maintained in freezing medium, and stored in liquid nitrogen until used for antibody repertoire sequencing.
Cell lines
HEK293F (Gibco) cells were cultured in suspension using FreeStyle 293 expression medium (Gibco) at 37°C with 8% CO2 at 130 rpm. HEK293T (ATCC) and Jurkat-FcγRIIIa-V158 (Vazyme Biotech) cells were maintained in Dulbecco’s modified eagle medium (DMEM, Gibco) and Roswell Park Memorial Institute 1640 medium (RPMI-1640, Gibco), respectively, supplemented with 10% Fetal bovine serum (FBS, Gibco), 1% penicillin-streptomycin (Gibco) and 1% Hepes (1M) buffer solution (Gibco) at 37°C with 5% CO2.
Method details
Antibody libraries preparation and sequencing
The amplification of antibody variable region genes was using an improved 5′-RACE polymerase chain reaction (PCR) (Kumar et al., 2020). Total RNA was extracted from 1∼5 million PBMCs with RNeasy Plus Mini Kit (Qiagen). For antibody repertoire preparation and analysis, 5′-RACE PCR was performed with Single Cell Full Length mRNA-Amplification Kit (Vazyme) according to the manufacturer’s protocol. Three separate PCR reactions were performed for amplification of antibody VH, Vκ and Vλ (VL) regions, and the PCR products at ∼600 base pairs were gel-purified with QIAquick Gel Extraction Kit (Qiagen). The 5′-RACE primer (Universal Primer Mix, UPM) was provided in the commercial kit and the sequences of 3′-specific primers were listed here: AAGACCGATGGGCCCTTG for VH, GGGAAGATGAAGACAGATGGT for Vκ, and GGGYGGGAACAGAGTGACC for Vλ (Jiang et al., 2013; Liao et al., 2009). VH and VL libraries were prepared by BerryGenomics and further performed deep sequencing using the Illumina MiSeq platform with 2 × 300 configurations.
Bioinformatics analysis of repertoire sequencing data
TrimGalore-0.6.0 (https://github.com/FelixKrueger/TrimGalore/) was used to automate quality and adapter trimming as well as quality control on the raw FASTQ files. The 3′ ends of reads with quality scores below 20 were trimmed and reads less than 150 bp in length were discarded. The paired clean reads were assembled into full-length antibody sequences according to overlapping regions by PANDAseq v2.11 (Masella et al., 2012). Following assembling, the sequences over 300 nucleotides were annotated by IgBlast v1.17.1 (Ye et al., 2013), using reference V(D)J sequences downloaded from the IMGT database (http://www.imgt.org/) (Lefranc et al., 2005). NGS-derived sequences containing CDR3 and without stop codons or out-of-frame IGH/LJ or frameshift were retained for further analysis. As described by He et al. (He et al., 2014), the full-length read was also excluded if its V-gene alignment was less than 250 bp. After the processes above, features were carried out from antibody NGS data, such as the distributions of V(D)J germline genes, germline divergence or degree of somatic hypermutation (SHM), and CDR3 loop length.
Clonotype definition
The sequences at different time points were grouped into specific clonotypes, with thresholds of the same V and J gene, the same CDR3 length, and CDR3 amino acid sequences of 80% identity between each other (Yan et al., 2021). To quantify the overall clonality of the repertories, we used the following formula of Shannon index:
where p i is the cell frequency of clonotype i in the antibody repertoire, and S is the total number of clonotypes.
Tracing of lineage antibody
Annotated antibody sequences were subjected to compared to template antibody sequences at both the nucleotide level and the amino acid level using the needleall software with Needleman-Wunsch algorithm (Needleman and Wunsch, 1970). Here, seven reported neutralizing antibodies (P2C-1F11, P2B-2F6, P2C-1A3, P2C-1C10, P2B-2G4, P2A-1A8, and P2A-1A10) previously isolated from P#2 were used as reference (Ju et al., 2020). The sequences with amino acid identity cutoff 90% and similar CDR3 length (±1 amino acid) were clustering into the evolutionarily related to the template lineage antibodies.
Synthesis, expression and purification of monoclonal antibodies
After iterative phylogenetic analysis of the deduplicated sequences, manually selected sequences were used as group representatives for antibody synthesis and functional characterization. Gene sequences of manually selected monoclonal antibodies (mAbs) and published mAbs downloaded from the National Center of Biotechnology Information (NCBI) were synthesized and cloned into the human full-length IgG1 expression vectors (Sangon Biotech). For the published mAbs, their protein data bank (PDB) codes were P2C-1F11 (7CDI) (Ge et al., 2021), 4A8 (7C2L) (Chi et al., 2020), CC40.8 (7SJS) (Zhou et al., 2022), REGN10987 (6XDG) and REGN10933 (6XDG) (Hansen et al., 2020), and VRC01 (4LST) (Zhou et al., 2013), respectively. Paired heavy and light chains were co-transfected into 293F cells, and antibodies were purified from cell supernatants five days later using protein A columns according to the manufacturer’s instructions (Senhui Microsphere Technology). Purified mAbs were quantified using a NanoDrop spectrophotometer and stored at 4°C.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed to characterize the binding capacity of representative mAbs to WT SARS-CoV-2 spike or NP protein. Firstly, the SARS-CoV-2 protein (2 μg/mL) was coated into 96-well plates at 4°C overnight. Washed with PBST buffer, the plates were blocked with 5% skim milk and 2% bovine albumin in PBS at room temperature (RT) for 1 h. Then, the tested mAbs (10 μg/mL) were added to the plates and incubated at 37°C for 1 h. After washing again, HRP conjugated goat anti-human IgG antibodies (ZSGB-BIO) were added and incubated at 37°C for 1 h. Finally, TMB substrate (Sangon Biotech) was added and incubated at RT for 20 min and the reaction was stopped by 2M H2SO4. The readout was detected at a wavelength of 450 nm. In addition, the representative mAbs were also tested against the spike proteins of seven human coronaviruses, including SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-229E, and HCoV-NL63, as well as the different functional domains of SARS-CoV-2-spike (S1, RBD, NTD, and S2). All proteins used here were purchased from Sino Biological.
SARS-CoV-2 neutralizing assays
After HEK-293T cells inoculated in the T75 flask grew to the appropriate density, they were co-transfected with SARS-CoV-2 (WT or mutant strain) spike expression plasmid and Env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-E−) (Ju et al., 2022). After 2 days, the supernatant was collected, centrifuged, filtered to obtain pseudovirus and stored at −80°C. The continuously diluted monoclonal antibody was co-incubated with an equal-volume of pseudovirus at 37°C for 1 h and then added to the prepared 96-well plate containing HEK-293T-hACE2 cells. After 48 h post-incubation, remove the culture medium and add 100 μL bright-Lite Luciferase reagent to the medium. Leave the medium at room temperature for 2 min and then remove 90 uL into 96-well white test plate. The Luciferase activity was measured by using Varioskan™ LUX Multimode Microplate Reader. Half-maximal inhibitory concentrations (IC50) were calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model.
Antibody-dependent cellular cytotoxicity (ADCC)
ADCC activity was assessed with a Bio-Lite Luciferase assay. The antibody was evaluated by incubating the serially diluted antibodies with an equal volume of target cells (293F expressing SARS-CoV-2 spike) at 37°C for 1 h. The target cells-antibody mixture was then transferred to an all-white 96-well cell culture plate, and Jurkat-FcγRIIIa-V158 Effector Cells were subsequently added to the plates. 18 h post-incubation, 110 μL of the Bright-Lite Luciferase reagent (Vazyme Biotech) was added to the plates. After a 2-min shock incubation at RT, the cell plates were measured by luminescence using the Varioskan™LUX multimode microplate reader (Thermo Fisher Scientific).
Binding analysis by surface plasmon resonance (SPR)
The binding of mAbs to the WT SARS-CoV-2 RBD protein were evaluated using the Biacore 8K system (GE Healthcare). Briefly, RBD protein (Sino Biological) was covalently coated on the flow cell of the CM5 sensor chips with 10 mM sodium acetate buffer (pH 5.0), while the uncoated and blocked flow cell served as a control. HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween-20) was used at a flow rate of 30 μL/min. Serially diluted antibodies were injected respectively for 60 s, and the data were fitted to a 1:1 binding model using Biacore Evaluation software (GE Healthcare). Each measurement was performed in duplicate to generate the average affinity constant.
Multiple sequence alignment and structural analysis
Multiple sequence alignment (MSA) was calculated using MEGA 10.2.6 with the ClustalW algorithm (Kumar et al., 2008). To identify the “hotspot” residues critical to P2C-1F11 lineage development, the crystal structures of the SARS-CoV-2 RBD with P2C-1F11, P22A-1D1 or P5A-3C8 Fab complex were downloaded and reanalyzed from the PDB under accession number 7CDI, 7CHS, and 7CHP, respectively (Zhang et al., 2021). Here, 4.5 Å was used as the maximal cut-off value for the intermolecular interactions. Illustrations of structural models were made using PyMOL Molecular Graphics System 1.5.0.4.
Quantification and statistical analysis
The independent experiment replicates were indicated in the figure legends. Half-maximal inhibitory concentrations (IC50) of mAbs were calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model. Curves for ADCC activities were fitted after log transformation of antibody concentration using GraphPad Prism 8.0 software by non-linear regression analysis. The values of binding affinity (KD) of mAbs were calculated using Biacore Evaluation software 3.0 (GE Healthcare) by Multi-cycle kinetics/affinity model. We did not perform any statistical analysis in this study.
Acknowledgments
We thank the patient for her contribution to this study and all of the healthcare providers from Shenzhen Third People’s Hospital for the work they have done. This work was funded by the National Science Fund for Distinguished Young Scholars (82025022), the National Natural Science Foundation of China (92169204, 82002140, 82171752, 82101861), the National Key Plan for Scientific Research and Development of China (2021YFC0864500), the China Postdoctoral Science Foundation (2021M703445), the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034, 2021A1515011009, 2020A1515110656, 2019A1515011197), the Shenzhen Science and Technology Program (RCYX20200714114700046, RCBS20210706092345028), the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20220226085550001, JSGG20200207155251653, JSGG20200807171401008, JSGG20210901145200002, KQTD20200909113758004), and the Shenzhen Natural Science Foundation (JCYJ20190809115617365, JCYJ20210324115611032, JCYJ20210324131606018).
Author contributions
Z.Z. is the principal investigator of this study. Z.Z. and B.J. conceived and designed the study. M.W., Q.F., and B.Z. performed all experiments and analyzed the data together with assistance from H.Y., S.S., J.Y., L.C., and X.G. M.W., B.J., and Z.Z. wrote the manuscript, and all authors read and approved this version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 23, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111335.
Supplemental information
Data and code availability
The sequencing data generated in this study have been deposited in the National Genomics Data Center (https://bigd.big.ac.cn/) under accession number: HRA002366, and the data will be publicly available as of the date of publication. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson J.D., Schaetzle S., Bashford-Rogers R.J.M., Raybould M.I.J., Kovaltsuk A., Kilpatrick G.J., Minter R., Finch D.K., Dias J., James L.K., et al. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. Front. Immunol. 2020;11:605170. doi: 10.3389/fimmu.2020.605170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Lin X., He L., Wang R., Wang H., Shi X., Zhang F., Yin C., Zhang L., Zhu J., Yu L. Development of a potent and protective germline-like antibody lineage against zika virus in a convalescent human. Front. Immunol. 2019;10:2424. doi: 10.3389/fimmu.2019.02424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Sok D., Azadnia P., Hsueh J., Landais E., Simek M., Koff W.C., Poignard P., Burton D.R., Zhu J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci. Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Kang B.H., Ishida E., Zhou T., Griesman T., Sheng Z., Wu F., Doria-Rose N.A., Zhang B., McKee K., et al. Identification of a CD4-binding-site antibody to HIV that evolved near-Pan neutralization breadth. Immunity. 2016;45:1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., He J., Weinstein J.A., Penland L., Sasaki S., He X.S., Dekker C.L., Zheng N.Y., Huang M., Sullivan M., et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 2013;5:171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhou B., Song S., Fan Q., Ge X., Wang H., Cheng L., Guo H., Shu D., Liu L., Zhang Z. Potent antibody immunity to SARS-CoV-2 variants elicited by a third dose of inactivated vaccine. Clin. Transl. Med. 2022;12:e732. doi: 10.1002/ctm2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.H., et al. Stereotypic neutralizing VH antibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci. Transl. Med. 2021;13:eabd6990. doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., He L., de Val N., Vora N., Morris C.D., Azadnia P., Sok D., Zhou B., Burton D.R., Ward A.B., et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:1663–1673. doi: 10.1016/j.cell.2020.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Ju B., Shapero B., Lin X., Ren L., Zhang L., Li D., Zhou Z., Feng Y., Sou C., et al. A VH1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite. Sci. Adv. 2020;6:eabb1328. doi: 10.1126/sciadv.abb1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lefranc M.P., Pommié C., Kaas Q., Duprat E., Bosc N., Guiraudou D., Jean C., Ruiz M., Da Piédade I., Rouard M., et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K., et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., Neufeld J.D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague Z., Lv H., Otwinowski J., DeWitt W.S., Isacchini G., Yip G.K., Ng W.W., Tsang O.T.Y., Yuan M., Liu H., et al. Dynamics of B cell repertoires and emergence of cross-reactive responses in patients with different severities of COVID-19. Cell Rep. 2021;35:109173. doi: 10.1016/j.celrep.2021.109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.Y., Rustagi A., Rogers A.J., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., et al. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53:442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umotoy J., Bagaya B.S., Joyce C., Schiffner T., Menis S., Saye-Francisco K.L., Biddle T., Mohan S., Vollbrecht T., Kalyuzhniy O., et al. Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity. 2019;51:141–154.e6. doi: 10.1016/j.immuni.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yuan M., Lv H., Peng J., Wilson I.A., Wu N.C. A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2. Immunity. 2022;55:1105–1117.e4. doi: 10.1016/j.immuni.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Z., Schramm C.A., Joyce M.G., Kwon Y.D., Zhou T., Sheng Z., Zhang B., O'Dell S., McKee K., et al. Maturation and diversity of the VRC01-antibody lineage over 15 Years of chronic HIV-1 infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., He P., Huang X., Luo K., Zhang Y., Yi H., Wang Q., Li F., Hou R., Fan X., et al. Germline IGHV3-53-encoded RBD-targeting neutralizing antibodies are commonly present in the antibody repertoires of COVID-19 patients. Emerg. Microbes Infect. 2021;10:1097–1111. doi: 10.1080/22221751.2021.1925594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Ju B., Ge J., Chan J.F.W., Cheng L., Wang R., Huang W., Fang M., Chen P., Zhou B., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat. Commun. 2021;12:4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan Q., Luo K., He P., Hou R., Zhao X., Wang Q., Yi H., Liang H., Deng Y., et al. Analysis of B Cell receptor repertoires reveals key signatures of the systemic B cell response after SARS-CoV-2 infection. J. Virol. 2022;96:e0160021. doi: 10.1128/jvi.01600-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yuan M., Song G., Beutler N., Shaabani N., Huang D., He W.T., Zhu X., Callaghan S., Yong P., et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl. Med. 2022;14:eabi9215. doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.Y., Joyce M.G., et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study have been deposited in the National Genomics Data Center (https://bigd.big.ac.cn/) under accession number: HRA002366, and the data will be publicly available as of the date of publication. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.