Abstract
Background
Omicron was detected in South Africa for the first time at the month of November 2021, from then it expanded swiftly over the world, outcompeting other SARS-CoV-2 variants such as Delta. The toxicity, resistance to antiviral medicines, transmissibility, and vaccine-induced immunity of newly developed SARS-CoV-2 variants are major worldwide health concerns.
Aim of study
This study investigates the comprehensive explanation of all mutations and their evolutionary linkages between the Omicron variant and recently discovered SARS‐CoV‐2 variants.
Method
On Illumina MiniSeq Machine, 31 RNA isolates from clinical specimens were sequenced utilizing next-generation sequencing technique. Different bioinformatics tools have been used to analyze the mutations in omicron variant. A phylogenetic tree was constructed to determine Omicron's evolutionary relationships with other variants.
Results
In our investigation, we discovered 79 distinct types of mutations in 31 fully vaccinated COVID-19 positive samples. Mostly mutations were found in non-spike region. According to the NJ approach of phylogenetic tree revels, the nearest variants were in the order listed, based on sequence identity: Omicron, Gamma, Alpha, Delta, Mu and Beta. On the other hand as per UPGMA approach, the Omicron variation creates a novel monophyletic clade that is distinct from previous SARS-CoV-2 variants.
Conclusion
Despite the fact that some of the mutations are prevalent in Omicron and other VOCs, there are several unique mutations that have been connected to the virus's transmissibility and immune evasion, indicating a substantial shift in SARS-CoV-2 evolution.
Key Words: Spike, Phylogenetic, Lineage, Non-structural Protein
Introduction
The Coronavirus infectious disease 2019 (COVID-19) outbreak presented a severe threat to international public health, with over 572 million confirmed cases and more than 6 million deaths worldwide. In India alone, there were more than 43 million confirmed cases and 526,258 deaths reported by August 1, 2022 (1). The original wild-type strain of SARS-CoV-2 was identified in Wuhan at the end of 2019, and since then, it has undergone constant mutations, leading to the emergence of multiple variants. According to the recent World Health Organization (WHO) 2022 update report, there are now two Variants of Concern (VOC): (B.1.617.2) Delta and (B.1.1.529) Omicron. On November 26th, 2021, the WHO designated Omicron as a VOC (2).
A novel SARS-CoV-2 strain B.1.1.529/BA.1 was identified on November 24, 2021, raising concern throughout the world (3). On November 26, the (WHO) declared B.1.1.529 Omicron as the fifth VOC (4). B.1.1.529 was first detected in Botswana on November 9, 2021 and the number of cases quickly increased dramatically in Gauteng and South Africa (3).
Since the November 30, 2021, Omicron has been detected in 17 countries, including South Africa, Australia, several countries in Asian, the United Kingdom, Canada, the United States and Hong Kong, and the list continues to increase (5). In India, the first case of Omicron was detected on December 2, 2021, in Karnataka. The confirmation was made by genome sequencing at an Indian SARS-CoV-2 Genomics Consortium (INSACOG) laboratory (6). As the viral transmission reached its peak in the middle of December 2021, there was an increasing need of using sequencing to determine the specific variant of SARS-CoV-2 responsible for the steep rise in the number of cases.
Initially, the Omicron variant of SARS-CoV-2 consisted of three sub-lineages, namely BA.1, BA.2, and BA.3. BA.1 (EPI ISL 6640916) was detected in Botswana on November 11, 2021, BA.2 (EPI ISL 6795834) in Gauteng on November 17, 2021 and BA.3 (EPI ISL 7605713) in northwestern South Africa on November 18, 2021. The spike protein in BA.1 has 37 mutations, in BA.2 it has 31 mutations and there are 33 mutations in the spike protein of BA.3 during the fourth wave of the pandemic in South Africa. At the beginning of 2022, two more sub-lineages of Omicron were detected, namely BA.4 and B.5. BA.4 (EPI ISL 10860989) was reported in Gauteng on January 26, 2022 with 33 spike protein mutations, while BA.5 (EPI ISL 11017528) was reported in KwaZulu-Natal, in South Africa, on February 25, 2022, with 34 spike protein mutations (7).
In a comparison of the spike protein mutations, 16 specific mutations (A67V, H69del, V70del, T95I, V143del, Y144del, Y145del, N211I, L212del, ins214EPE, S371L, G446S, G496S, T547K, N856K and L981F) in BA.1 and 10 specific mutations in the BA.2 lineage (T19I, L24S, P25del, P26del, A27S, V213G, T376A and R408S) were reported. However, the BA.1 lineage requires mutations at the S371, T547K, N856K, and L981F sites for viral oligomerization interface, as an essential element for viral fusion to the host cell and formation of the fission pore. Whereas the BA.3 lineage shares 21 common mutations with both the BA.1 and BA.2 lineages, with two specific mutations in BA.2: D405F and S371F and ten specific mutations in BA.1: A67V, H69del, V70del, T95I, V143del, Y144del, Y145del, N211I, L212del, and G446S (8). The BA.4 and BA.5 subvariants spiked worldwide and spread faster than any other previous subvariant of Omicron, causing an increase in COVID-19 cases in the first half of 2022. These two Omicron subvariants are more closely related to the BA.2 lineage than to the BA.1 lineage, which was discovered in late 2021. The BA.4 and BA.5 variants have two distinct mutations in the spike protein, L452R and F486V, which alter its capacity to attach to the host cells and avoid certain immune responses (9). Through the recombination of the existing variants, three new co-variants of Omicron evolved, commonly known as XD, XE, and XF. XD and XF are recombinants of the Delta variant and BA.1 lineage of Omicron. On the other hand, XE is the recombination of Omicron BA.1 and BA.2 variants that was initially reported in the United Kingdom. The recombinant location of mutations in XE from BA.1 is NSP1-6, while the remainder mutations involved the NSP6 gene from the BA.2. lineage. The XE variant has three peculiar mutations that are not identical to any of the recombinant variants (BA.1 or BA.2), namely C3241T, V1069I at NSP3, and C14599T at NSP12. These mutations are responsible for the cleavage of viral polyproteins during replication. Their influence on viral transmission and immune evasion is still under investigation (10).
The Spike protein is the key component of the virus that determines its infection rate and antigenicity. In general, it contains 30–35 mutations, 15 of which were discovered in the Receptor Binding Domain (RBD) of the spike protein. These alterations include practically all of the previous VOCs’ important mutations like K417N, E484A, and N501Y and other known changes that alter the virus’ sensitivity to the action of protective antibodies (11). It is thought that the spike protein's complicated alterations allow it to overcome the immunity generated by prior infection or vaccination, resulting in a high number of breakthrough infections or re-infections by mutant virus strains.
Several issues accompany the emergent nature of the Omicron variant, including the effect of Omicron mutations on vaccine response, the role of mutations on host immune regulation, the source of emergence, Omicron spreading potency, clinical implications and mortality. This study attempted to establish the evolutionary connections of the Omicron genome. Various approaches were used to attain the best fit of alignment of complete genomes.
Materials and Method
Clinical Specimens
Viral transport medium containing nasopharyngeal and oropharyngeal swabs from SARS-CoV-2-positive samples with E-gene: Ct value ranging from 17–25 and RdRp gene: Ct value ranging from 17–25, were obtained from the Virology Laboratory of All India Institute of Medical Science, Raipur, Chhattisgarh in January of 2022. Fresh RNA was extracted from the samples to be used for next-generation sequencing.
Next-Generation Sequencing of RT-PCR-Positive Samples
RNA isolates from positive clinical specimens were subjected to whole genome sequencing of SARS-CoV-2, using a Next Generation sequencer (Illumina MiniSeq). The RNA libraries were constructed with the Enhanced QIAseq Direct SARS-CoV-2 library preparation kit. The method begins with random-primed cDNA synthesis, followed by the use of multiplexed primer pools to prepare two pools of 225–275 bp SARS-CoV-2 overlapping amplicons. The two enriched pools from each sample were then combined into a single tube and cleaned with beads. All enriched samples were amplified and sample-indexed with unique dual indices (UDIs), and subsequently cleaned with beads. The RNA libraries that had been amplified were quantified in a Qubit 4 Fluorometer and thereby normalized before being uploaded into the Next generation sequencing machine.
Genome Data Analysis
The CLC genome workbench V12 software was used for genome analysis, using reference-based mapping to acquire the entire genome sequence of SARS-CoV-2. The nucleotide variation of the sequences studied was obtained using variant detection tools in CLC software. All sequenced samples were uploaded to the Global Initiative on Sharing All Influenza Data (GISAID), a public genomic repository.
Phylogenetic Tree Construction. To determine Omicron's evolutionary relationships with other variants, 10 higher-quality sequences having good 399X coverage were selected from among the dataset of samples used in the study (n = 31). The lineages obtained from these samples were also compared with recent, higher quality and good coverage sequences of the SARS-CoV-2 variants Alpha, Beta, Gamma, Delta, and Mu (n = 5) sequences each (5 × 5 = 25). GISAID was used to acquire the whole genome sequence of SARS-CoV-2 variants other than the Omicron variant and aligned via Multiple Alignment using the Fast Fourier Transform (MAFFT) program. A pairwise comparison was performed with gap analysis, differences in sequence and mutations were recorded to create an identity matrix, which was further used to construct a phylogenetic tree using Molecular Evolutionary Genetics Analysis X (MEGAX) software. The Phylogenetic tree was created using ultrametric and metric clustering methods via unweighted pair group method with arithmetic mean (UPGMA), using the kimura-2 parameter model where-in nucleotide substitution was permitted via nucleotide transition and transversion, with 1000 bootstrap. The Neighbor-joining method, with the evolutionary distance calculated by Jukes-Cantor models was also used to build the phylogenetic tree.
Results
Brief Description of the Circulating Omicron Variants
The whole genome sequence of >95% of the extracted RNA was obtained from 31 SARS-CoV-2 positive clinical specimens collected randomly in January 2022.
Clinically, all these patients had moderate COVID symptoms. There were 19 males and 12 females with a median age of 37 years. All patients were residents of Raipur city and had received two doses of the COVID vaccines provided in India. In this study, 18 patients had received Covaxin (BBV152; India's first indigenous COVID-19 vaccine; a killed, whole-virus vaccine) and 13 patients received Covishield (ChAdOx1-nCOV or AZD1222; a recombinant, replication-deficient chimp adenovirus vector containing the SARS-CoV-2 Spike glycoprotein vaccine manufactured at the Serum Institute of India).
All samples were aligned using MAFFT version 7. A multiple sequence alignment application operates on a Unix-like operating system and provides a variety of alignment methods. With the exception of a few samples from Delta variants from the year 2021, all sequences were submitted to GISAID with submission IDs ranging from EPI ISL 10716772–EPI ISL 10716815. The sequences were also submitted to Nextstrain: real-time tracking of pathogen evolution, to check the name of the clade and lineage. This revealed that 29 samples belonged to the clade 21L Omicron group and the same samples belonged to the pangolin BA.2 lineage, while the remaining two samples belonged to clade 21K in Nextstrain and the pangolin BA.1 lineage. Finally, all of the sequences were examined in GISAID, including one sample from the G clade, two from the GR, and 28 from the GRA. On February 25th, 2022, all clade and lineage nomenclature was finalized. However, this may be subject to change as the sites are constantly and dynamically updated.
Characterization of Mutations found in Omicron Variants
A broad range of mutations have accumulated in the BA.1 and BA.2 lineages of the Omicron variant. Both spike and non-spike regions were the target of mutations in these lineages. There are some mutations which are common in Omicron and other VOCs or VOIs (Variant of Interest). A total of 25 out of 98 reported mutations were determined to be present in at least one other variant of SARS-CoV-2. As a result, there are 75 mutations that were not discovered in any VOCs or VOIs and were identified and labelled as “Omicron Specialized Mutations”. The majority of the non-spike protein mutations found in Omicron were not detected in any of the previous VOCs or VOIs.
In the study, 79 distinct types of mutations in thirty-one COVID-19 positive samples were found. The samples were collected from the clinical care centre at All India Institute of Medical Science, in Raipur, Chhattisgarh, in January of 2022, with a genome coverage of >95%. 28 of these mutations were shared by the Omicron lineages BA.1 and BA.2 lineages. In contrast, 30 mutations were unique to the BA.2 lineage and 21 were unique to the BA.1 lineage (Supplementary Table 1).
Mutations in S-Glycoprotein
Thirty-four different mutations were detected in the S-glycoprotein, with the signature mutations of the BA.1 and BA.2 lineages being found in the majority of the samples.
There were 13 distinct mutations (T19I, L24S, P25del, P26del, A27del, G142D, V143del, Y144del, Y145del, N211I, L212del, V213G, and ins214EPE), of which six (T19I, L24S, P25del, P26del, A27del and V213G) were unique to the BA.2 lineage. Five mutations (V143del, Y144del, Y145del, N211I and L212del) were common to the BA.1, BA.1.1, and BA.3 lineages and one unique mutation (214EPE) was detected in both the BA.1 and BA.1.1 lineages. G142D mutations were found universally across all lineages.
The Receptor Binding Domain (RBD) of the spike protein includes sequences extending from 319–541. Eight mutations were found in this region (S371F, S371L, S373P, S375F, T376A, D405N, R408S, and K417N), three of which (S373P, S375F and K417N) were shared by all the lineages of the Omicron variant. A single S371L mutation was shared by the BA.1 and BA.1.1 lineages. S371F and D405N mutations were found in both the BA.2 and BA.3 lineages, while two mutations (T376A and R408S) were unique to the BA.2 lineage.
The Receptor Binding Motif (RBM) of the spike protein contains the region extending from 437–508, which is necessary for binding of the virus to the human ACE2 receptor. However, only two mutations (N440K and G446S) were detected in this region because of the low coverage of this region, even though ten different types of mutations occurred in the entire lineage of the Omicron variant. The N440K mutation was common throughout all lineages, whereas the G446S mutation was specific to the BA.2 lineage.
The spike SD1/SD2 cleavage junction comprises three kinds of mutations (T547K, D614G and H655Y), with the T547K mutation being unique to the BA.1 lineage and the other two mutations being shared by all lineages of the Omicron variant. Three mutations (N679K, P681H and N764K) were shared by the BA.1 and BA.2 lineages at the spike polybasic cleavage site. The Spike C-terminal Domain (CTD) had five substitutions (D796Y, N856K, Q954H, N969K and L981F); two mutations N856K and L981F were specific to the BA.1 and BA.1.1 lineages and the remaining three were shared by all lineages of the Omicron variant.
Mutations in ORF
In the Omicron variant, there are 32 distinct types of mutations detected in the ORF 1ab region, which is a Non-Structural Protein (NSP), 29 of which were detected in the present study. The NSP1 region had two mutations (P62T and S135R), with P62T being shared by both the BA.2 and BA.3 lineages and S135R being unique to the BA.2 lineage in this study. There were two mutations (V364G and S378F) in the NSP2 region, both of which are rare and exclusive to the BA.2 lineage.
NSP3 contained 10 types of mutations (T24I, K38R, G489S, S1265del, L1266I, A1892T, E642K, C1392F, L689F and A150V), 5 of which (E642K, L689F, A150V, G489S and T24I) were specific to the BA.2 lineage and rest were shared by both the BA.1 and BA.1.1 lineages. The NSP4 region contained 5 mutations (L264F, T327I, L438F, L447F and T492I), with one (T492I) being shared by all the Omicron lineages and the other four being unique to the BA.2 lineage.
There was a single mutation found in the NSP5 region at position P132H that is shared by all the Omicron lineages. A total of 5 mutations were discovered in the NSP6 region: three of which (S106del, G107del and F108del) were shared by all lineages, and the remaining two (I189V and L106del) being specific to the BA.1 and BA.1.1 lineages. The NSP12 region had two mutations (P323L and Q822E), with P323L being the most common and Q822E being a new mutation specific to the BA.2 lineage. There were two mutations in the NSP14 region (I42V and D301E), with the I42V mutation being common to all lineages and the D301E mutation being unique to the BA.1 lineage. There was a single mutation in the NSP15 region (T112I), and it was unique to the BA.2 lineage.
Two mutations (T223I and L65F) were found in the ORF3a region, which is common to the BA.2 and BA.3 lineages of Omicron. In the BA.2 lineage there was one rare mutation at position L65F. We observed a single rare mutation (L17F) in the ORF7a gene, which is found in the BA.2 and BA.1 lineages.
Structural Protein Mutations
A total of 13 types of mutations were found in structural proteins. Two mutations (ET9I and L18I) in the envelope protein were common to both the BA.1 and BA.2 lineages, as well as one rare mutation (L18I) found in the Gamma variant. Nine mutations were found in the nucleocapsid protein (S413R, S33del, E31del, P13L, R32del, R203K, R203M, N G215C, and 204R), six of which were shared by all Omicron lineages and two which were unique to the BA.2 lineage. Two membrane protein mutations (Q19E, A63T) were found to be shared by both lineages.
Phylogenetic Analysis
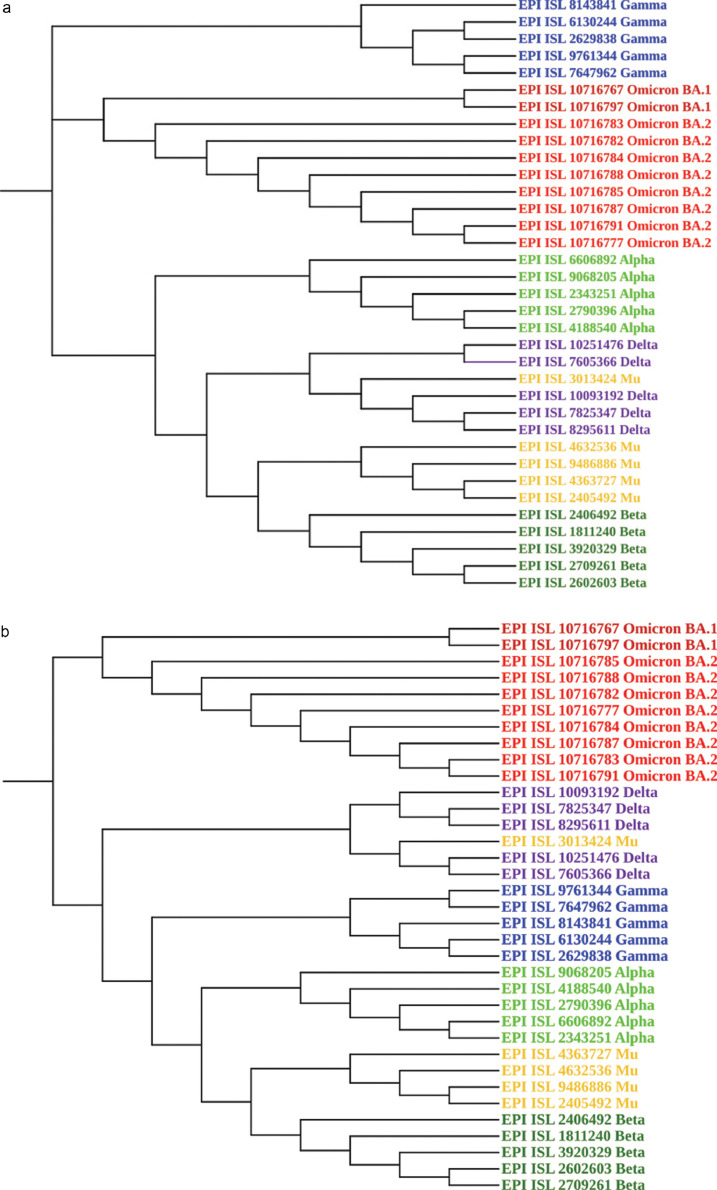
In a neighbor-joining phylogenetic tree, constructed usingt the Jukes-cantor model with 1000 bootstrap, the Omicron variants found in our study showed close relationship with the recently-discovered Gamma and a variants. The evolutionary tree based on sequence alignment, alteration and gaps, demonstrated there are few common nucleotide alterations amongst the Omicron BA.1 lineage and the Gamma variant. On the other hand, the BA.2 lineage shared common nucleotide variations with the Alpha variant. Because of the striking similarities that exist between the BA.1 and BA.2 Omicron lineages and the Gamma and Alpha variants, respectively, it may Beta concluded that the Omicron variant was already in circulation before it was identified in the laboratory. Based on the sequence similarities between Omicron and other variants, the most closely related variants were found to be Gamma, Alpha, Delta, Mu, and Beta, as shown in Figure 1 A.
Figure 1.
A. Phylogenetic tree utilizing Neighbor‐Joining technique and Jukes-cantor substitution model. The figure was created with MegaX & iTOL software. B. Phylogenetic tree utilizing UPGMA algorithm and kimura-2 parameter substitution model. The figure was created with MegaX & iTOL software.
In a UPGMA tree, where ultrametric distances between variants and equal rates of evolutionary change across branches were used to calculate the relationship between other variants and Omicron, the Omicron variant was phylogenetically distinct from other variants, but the results showed a different picture where we can see Omicron in a monophyletic clade. The evolutionary distance was calculated using the kimura-2 parameter model and was measured in base substitutions per site. The Omicron variant stood out from the others in Figure 1B because the UPGMA algorithm combines sequence pairings with a relatively minimal gap.
Discussion
Detailed Description of Each Mutation Present in the Omicron Variant
In comparison to all other SARS-CoV-2 variants, the number of mutation in the spike glycoprotein is substantially greater in the Omicron variant. All Omicron lineages shared 21 common mutations in the spike region. BA.1.1 includes a unique (R346K) mutation in the spike region that distinguishes it from its sub-lineage BA.1. When comparing spike glycoprotein mutations in both of the Omicron lineages, 16 unique mutations were found in the BA.1 lineage and 10 distinct mutations in the BA.2 lineage. A total of 34 mutations were discovered in the spike protein genome in this study, 13 of which were shared by both the BA.1 and BA.2 lineages, 12 which were unique to the BA.1 lineage and 9 which were specific to the BA.2 lineage.
Mutations in the Spike RBD
Because the RBD binds with hACE2 receptors on human cells, mutations in this region may have a significant influence on coronavirus fitness. A combination of three mutations; K417N, E484K and N501Y in a receptor binding domain showed the enhanced affinity for the ACE2 protein, as these mutations dramatically change RBD structure (12). However, Omicron favours the cathepsin-dependent pathway for entering the host cell, in contrast to Delta and other variants. There are 12 mutations in the spike protein of Omicron which involve basic amino-acid substitutions, which generate more amino groups, thereby enhancing protonation and increasing susceptibility to endosomal hydrolytic enzymes like cathepsin. As a result, the Omicron spike protein prefers the endocytic route for entering the host cell, while Delta and other variants use membrane-bound serine proteinases like TMPRSS2 via cell surface fusion (13). According to Peacock TP, et al. Omicron can use either of these means of entry to infect the cell, but favours endosomal fusion over cell surface fusion. Omicron's capacity to infect cells through either route may significantly increase the number of infected cells, thereby enhancing it's transmissibility (14).
The study by Alouane, T et al. found that the G339D mutation may reduce ACE2 binding affinity (15). Fratev F, et al. showed that the R346K mutation has little effect on antibody binding and affects only class 2 monoclonal antibodies (mAbs) (16). Mutations at the S371 position include S371P, S371T, S371A and S371F reported in previous variants of SARS-CoV-2. The S371 residue can be found in a cryptic binding epitope matched with a conserved hinge location. Furthermore, there were no glycosylated sites in the S371-F541 region. Mutations in the S371 amino acid residue, on either side, lead to a reduced RBD detection by antibodies (17). RBD stability is also reduced by the S371L mutation reported in the BA.1 lineage, which may compromise antibody identification of the spike protein epitope (18).The mutation at S371F may have the same impact. The S373P mutation may aid in evading neutralizing antibodies. The S375F mutation was most likely first seen in Omicron. It may modestly affect RBD conformation through the substitution of serine for phenylalanine (19). The D504N/Y mutation enables the RBD to evade monoclonal antibody binding. In the RBD, both R408 and D405 play a significant role in the RBD-opening transition (20). Consequently, it is important to examine these residues in order to better understand their involvement in viral cell entry and how mutations may alter this capacity.
Ten mutations have been reported in the RBM, but only 2 were found in this study: N440K and G446S. The N440K mutation changes the binding free energy, and can thus potentially increase binding affinity to the ACE2 receptors. The substitution of glycine for serine in the G446S mutation reduces the stability of the RBD, whereas ACE2 binding affinity increases (21). It may help to avoid neutralising antibodies (22). The S477N mutation confers resistance to monoclonal antibody neutralisation, as well as increased ACE2 receptor binding. As a result, this mutation makes the virus more transmissible. At the same time, this mutation was capable of decreasing binding affinity to the neutralizing antibodies. The T478K mutation reappeared multiple times on its own. It may enhance ACE2 receptor binding affinity because it is commonly found (23). Some neutralizing antibodies were unable to bind because of the E484A mutation. The Q493R mutation decreased protein sensitivity to several monoclonal antibodies in a neutralisation experiment (24). G496 is an essential amino acid that interacts with the ACE2 receptor and, therefore, mutations at this position G496W/Y result in one of the most significant binding energy changes among all RBD mutations. The combination of Q498R and N501Y mutations improves ACE2 receptor affinity. The Y505 residue is a significant region, with Y505H being the most likely substitution (22). However, this mutation has the potential to lower ACE2 receptor binding affinity.
Mutations in other Spike Protein Domains
T19 has a high mutation rate and is located in the N-terminal domain of the spike protein gene. NTD may be used as an antibody epitope since it is so close to the RBD and, as a result, mutations at this location can aid in the avoidance of neutralising antibodies (25). An LPPA24S mutation causes a nucleotide loss at locations 21633–21641 (TACCCCCTG), resulting in a substitution of LPPA24S. This mutation helps the virus to avoid identification by neutralizing antibodies. With the A67V mutation, where alanine is substituted for valine, new hydrophobic bonds occur with the I100, F79 and A263 residues. As a result, it significantly changes the structure of 3–4 loops (25). The deletion of 69–70 may affect S1 conformation allosterically and thus be favourable to RBD mutations. Such deletions have been associated with enhanced viral proliferation as well as antibody resistance. Some neutralizing antibodies have been shown to be resistant to the spike protein with the Y145D mutation. The “ins214EPE” insertion (nucleotides GAGCCAGAA) was detected in 86.3% of BA.1 and BA.1.1 samples of the Omicron variant. Given its proximity to RDR3, just like other insertions throughout this position, it may have an effect on SARS-CoV-2 infectivity (26,27).
T547K mutation cases have already been recruited, however they were initially resolved via Omicron BA.1 and BA.1.1. The D614G mutation initially appeared in March 2020 and it may boost transmissibility by promoting proteolysis at the furin cleavage site. The P681H mutation was common to the Alpha, Mu and Omicron variants, but Omicron has weak cleavage band so a mutation near the furin cut site (H655Y, N679K) may increase transmission. In the Delta variant, the P681R mutation was observed to result in higher fusogenicity and pathogenicity, while the Mu and Omicron variants were weaker than wild type SARS-CoV-2 strains despite the P681H mutation (13). The N679K and P681H mutations in Omicron are located at the S1/S2 junction and may boost the virus's capacity to spread (28,29). The transition from a neutrally charged to a positively charged amino acid improves coupling between the furin cleavage site and the proteolytic enzyme, resulting in enhanced cleavage at the spike S1/S2 site. A single N679K substitution is insufficient to improve transmission (30). Omicron was the first to have the N764K mutation repaired. It decreases the protein's stability and may impair its function (18). D796Y happened independently multiple times, including examples of intra-host evolution in immunocompromised (HIV-positive) individuals (31). The positively charged amino acid changes caused by N856K mutations may change the structure of this area and boost its affinity for certain proteins. The alterations caused by the two mutations N969K and Q954H in the Heptad Repeat 1 (HR1) region, which is involved in host cell fusion, might have an impact on infectivity (32). The Omicron and Mu variants have a major spike gene mutation that allows them to immunologically evade the effects of existing vaccinations (13).
Non-Spike Mutations
In comparison to other VOCs and VOIs, the Omicron variant has a significantly larger number of mutations in the non-spike region. Noncoding sequence, non-structural protein, and other structural protein mutations have received less attention. The retention of NSPs mutations is more difficult, and verified mutations or deletions in polyprotein1a/1ab are infrequent, therefore it is critical to investigate mutations in these areas because it plays a significant role in SARS-CoV-2 replication and regulation.
Mutation in ORF1ab and its Significance
Impact of Genetic Changes in NSP1
Non-structural Protein1 interacts with the ribosomal 40S subunit, restricting the expression of the host gene and circumventing detection by the immune system. NSP1 also increases viral gene expression by destroying host mRNA (33,34). NSP1 also helps in viral gene translation by recognizing the RNA 5’-UTR region of SARS-CoV-2. Most of the mutations in NSP1 weakened the protein and enhanced its flexibility. Because of its functionality, it can be used as a vaccine target and a drug design target (35).
Impact of Genetic Changes in NSP2
NSP2 is a replicase product that is required for viral replication proofreading. NSP2 interacts with the host's prohibitin (PHB and PHB2) and modifies the host cell's survival signalling system (36).
Impact of Genetic Changes in NSP3
NSP3 was speculated to collaborate with NSP4 and NSP6 and create Double-Membrane Vesicles (DMVs), which act as a key element of Replication-Transcription complexes (RTCs) (37). It also helps in the binding of the nucleocapsid by interacting with the C-Terminal Domain of the nucleocapsid and RTC (38). NSP3 suppresses NF-Kappa-signalling and inhibits the immunological response to type1 interferon (39). Mutations in NSP3 played a crucial role in the selection of Beta-Coronavirus (40).
Impact of Genetic Changes in NSP4
NSP4 is associated with the formation of cytoplasmic DMVs as well as with viral replication (40,41).
Impact of Genetic Changes in NSP5
NSP5 is a cysteine protease also known as 3C-like protease (3CLpro), and is essential for the cleavage of viral polyproteins into pp1a and pp1ab, which results in the production of 12 distinct functional proteins (42). As a result, such a protein might be a target for anti-coronavirus treatment. By suppressing the Mitochondrial Antiviral Signalling (MAVS) protein and IFN production, NSP5 may help in preventing the innate immune response (43). Furthermore, mutations in the NSP5 protein can impair its catalytic activity, thus allowing the virus to succumb to the host's immune response.
Impact of Genetic Changes in NSP6
NSP6 collaborates with NSP3 and NSP4 to build DMVs containing replication-transcription complexes. The presence of phenylalanine residues in the NSP6 protein demonstrated its affinity for the endoplasmic reticulum. The NSP6 deletion mutation from 105–107 may help in escaping from the innate immune response by lowering the host cells’ capacity to eliminate viral particles (44).
Impact of Genetic Changes in NSP12 or RNA-dependent RNA Polymerase
From the very beginning of the COVID-19 pandemic in China and throughout the globe, RdRp was thought to be a mutation hotspot. (45,46). The P323L mutation was associated with the rise in genetic variations and influenced RdRp's interaction with cofactors, resulting in less effective proofreading activity and giving rise to multiple SARS-CoV-2 variations (47). Antiviral drugs were designed to dock onto a hydrophobic cleft around the P323L mutation region in an in-silico analysis. This mutation served to decrease RdRp's affinity for currently available antiviral medicines (45).
Impact of Genetic Changes in NSP14
In RNA replication, proofreading by 3’–5’ exoribonuclease is accomplished by NSP14. This protein also has a secondary role as a N7-methyltransferase (C-terminal domain). As a result, any mutation in the NSP14 protein will impact the proofreading or stability of the newly generated viral mRNAs (48). Interferon signalling can be suppressed (49).
Impact of Genetic Changes in NSP15
The viral RNA's 5’-polyuridines are chelated by NSP15, an endoribonuclease, which enables the virus to elude the innate immune response by decreasing dsRNA sensor activation.(42). The viral life cycle depends on the NSP15 protein. Mutations in this protein gene resulted in a fast antiviral response within macrophages, allowing the infection to be controlled within a short period of time (50).
Impact of Genetic Changes in ORF3a
An ion channel is created by ORF3a which induces host cell lysis, enabling new viral particles to be shed. Autophagy inhibition and lysosome disruption are also implicated. As a result, it is a protein that is essential for the viral cycle (51). Because of its position on the membrane's surface, ORF3a can elicit both humoral and cellular immune responses from affected patients (52).
Impact of Genetic Changes in ORF6
ORF6 has been shown to reduce interferon signalling by reducing the creation of primary interferons. ORF6 can nonetheless disrupt the activation of the innate immune system. According to a molecular docking and dynamics simulation study, the C-terminus of the ORF6 protein can establish hydrophobic interactions with the transcription factor IRF3. As a result, ORF6 acts as an antagonist to IFNs (53).
Impact of Genetic Changes in ORF7a
ORF7a has been polyubiquitinated at Lys 119 (K119), which may decrease the IFN-I (type-I interferon) response through STAT2 (Signal Transducer and Activator of Transcription 2). ORF7a also participates in protein transport within the endoplasmic reticulum and Golgi complex. Any changes in this region can alter the virus-host relationship (42,54).
Mutations in Structural Proteins
Impact of Genetic Changes in E-Protein
T9I and L18I mutations in the transmembrane domain impact upon the envelope protein configuration and enhance viral membrane anchoring. Any mutation in the E-protein affects its structural configuration and hence the activities associated with it, such as viral assembly and transmission, and its virulence (55,56).
Impact of Genetic Changes in M-Protein
The membrane protein is the most prevalent conserved structural protein. It possesses the ability to interact with nucleocapsid phosphoprotein, spike-protein and viral RNA (57). M-protein mutations may have an impact upon the relationship with the host cell (58).
Impact of Genetic Changes in N-Protein
The nucleocapsid phosphoprotein interacts with viral RNA through the N-terminal and C-terminal domains. It also attaches to the M-protein during viral assembly. According to the WHO categorization of VOCs and VOIs of SARS-CoV-2 variants, the viral genome should have at least one mutation in the 199–205 region, with >50% prevalence rate, and this highlighted the importance of mutations in this N-protein region (59).
Phylogenetic Relation of Omicron
Bioinformatics and phylogenetic tools have become the gold standards for microbial evolutionary information and drug development against specific molecular targets (60). The genetic content of SARS-CoV-2 showed the dynamics influencing viral evolution (61). A variety of approaches were used in this study to obtain insight into the evolutionary history of the Omicron variants. According to the evolutionary analysis of Omicron variants, using UPGMA with a Kimura 2-parameter model, the result was a separate emerging group which does not originate from other variants, whereas, the neighbor-joining model indicated a strong link between the Omicron BA.1 lineage and the Gamma variant and between the Omicron BA.2 lineage and the Alpha variants. Due to the intricacy of viral development and differences, even within single gene, a single evolutionary model cannot be proposed to account for viral evolution.
Conclusion
The emergent nature of the Omicron variant raises a number of questions, such as how Omicron mutations affect vaccination response, how the hosts’ immunity is affected by the mutations and the infectivity and transmissibility of the variant. In the Omicron BA.1 and BA.2 lineages, a very large number of mutations have accumulated. These mutations have spread across the whole genome, affecting both spike and non-spike regions. More than 30 changes were discovered in the spike protein, 13 of which result from basic amino-acid substitutions which create more amino groups, improving protonation and thus making the protein more susceptible to endosomal hydrolytic enzymes like cathepsin, while also increasing the virus’ transmissibility. The non-spike region has more than 40 mutations or deletions, some of these are uncommon and may boost infectivity cumulatively when compared to other SARS-CoV-2 variants. Since vaccines induce immunity, and the humoral immunity elicited by earlier SARS-CoV-2 variants does not provide any protection against the current Omicron variant, it has been shown that all fully-vaccinated individuals contracted the infection relatively quickly. However, due to the rapid evolution of SARS-CoV-2 variants and the threat they pose to public health, very fit variants are expected to evolve in the future. Consequently, it is necessary to constantly monitor the emergence of new variants.
Funding
We did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Editor: Ana Carolina Sepúlveda Vildósola
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.arcmed.2022.08.006.
Appendix. Supplementary materials
References
- 1.WHO Coronavirus (COVID-19) Dashboard. Published 2022. https://covid19.who.int/table. (Accessed August 1, 2022).
- 2.WHO, Tracking SARS-CoV-2 variants. Published 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants. (Accessed July 31, 2022).
- 3.Callaway E. Heavily mutated coronavirus variant puts scientists on alert. Nature. 2021:1–5. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 4.WHO . WHO; 2021. Classification of Omicron (B.1.1.529): SARS- CoV-2 Variant of Concern.https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (Accessed July 31, 2022) [Google Scholar]
- 5.Wang Y, Zhang L, Li Q, et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.India's first Omicron cases detected in Karnataka. Hindustantimes. https://www.hindustantimes.com/india-news/indias-first-omicron-cases-detected-in-karnataka-101638445884205.html. (Accessed December 2, 2021).
- 7.GISAID. GISAID. Published 2021. https://www.gisaid.org/hcov19-variants. (Accessed July 15, 2022).
- 8.Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022;94:1808–1810. doi: 10.1002/jmv.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaway Ewen. What the Latest Omicron Subvariants Mean for the Pandemic. Nature. 2022;606:848–849. doi: 10.1038/d41586-022-01730-y. [DOI] [PubMed] [Google Scholar]
- 10.Ma K, Chen J. Omicron XE emerges as SARS-CoV-2 keeps evolving. Innov. 2022;3 doi: 10.1016/j.xinn.2022.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-beltran WF, Lam EC, Denis KS, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity Graphical. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.013. 2372–2383.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson G, Buzko O, Spilman P, et al. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escap. bioRxiv. 2021 https://www.biorxiv.org/content/10.1101/2021.01.13.426558v1%0Ahttps://www.biorxiv.org/content/10.1101/2021.01.13.426558v1.abstract Published online2021.01.13.426558. (Accessed July 15, 2022) [Google Scholar]
- 13.Du X, Tang H, Gao L, et al. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther. 2022;7:5–7. doi: 10.1038/s41392-022-00903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peacock TP, Brown JC, Zhou J, et al. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 2022 https://www.biorxiv.org/content/10.1101/2021.12.31.474653v1 Published online. (Accessed July 15, 2022) [Google Scholar]
- 15.Alouane T, Laamarti M, Essabbar A, et al. Genomic diversity and hotspot mutations in 30,983 SARS-CoV-2 genomes: Moving toward a universal vaccine for the “confined virus”? Pathogens. 2020;9:1–19. doi: 10.3390/pathogens9100829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratev F. R346K Mutation in the Mu Variant of SARS-CoV-2 Alters the Interactions with Monoclonal Antibodies from Class 2: A Free Energy Perturbation Study. J Chem Inf Model. 2022;62:627–631. doi: 10.1021/acs.jcim.1c01243. [DOI] [PubMed] [Google Scholar]
- 17.Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS–CoV-2 neutralizing antibodies 2. bioRxiv. 2021 doi: 10.1038/s41586-021-04385-3. https://www.biorxiv.org/content/10.1101/2021.12.07.470392v2 Published online. (Accessed July 15, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Thambiraja TS, Karuppanan KSG. Omicron and Delta variant of SARS-CoV-2 A comparative computational study of spike protein. J Med Virol. 2022;94:1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Wang Y, Zhu Y, et al. Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections. Nat Commun. 2021;12:264. doi: 10.1038/s41467-020-20465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sztain T, Ahn S, Bogetti AT, et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma J, Subbarao N. Insilico study on the effect of SARS-CoV-2 RBD hotspot mutants’ interaction with ACE2 to understand the binding affinity and stability. Virology. 2021;561:107–116. doi: 10.1016/j.virol.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Wang R, Wang M, et al. Mutations Strengthened SARS- CoV-2 Infectivity Jiahui. J Mol Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giorgi FM, Mercatelli D, Rakhimov A. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS - CoV - 2) Spike mutation T478K. J Med Virol. 2021:5638–5643. doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Focosi D, Novazzi F, Genoni A, et al. Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19. Emerg Infect Dis. 2021;27:17–20. doi: 10.3201/eid2710.211538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinakis A, Cournia Z, Rampias T. N-terminal domain mutations of the spike protein are structurally implicated in epitope recognition in emerging SARS-CoV-2 strains. Comput Struct Biotechnol J. 2021;19:5556–5567. doi: 10.1016/j.csbj.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resende PC, Naveca FG, Lins RD, et al. The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N) -terminal domain of the spike. Virus Evol. 2021;7:1–11. doi: 10.1093/ve/veab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey WT, Carabelli AM, Jackson B, et al. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin. J Virus Erad. 2021;12:517–522. doi: 10.1101/2021.12.10.472102. [DOI] [Google Scholar]
- 28.Tiecco G, Storti S, Antoni MD, et al. Omicron Genetic and Clinical Peculiarities That May Overturn SARS-CoV-2 Pandemic: A Literature Review. Int J Mol Sci. 2022;23:1–15. doi: 10.3390/ijms23041987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan J, He X, Ren Y, et al. Structural and computational insights into the SARS-CoV-2 Omicron RBD-ACE2 interaction. bioRxiv. 2022 doi: 10.1038/s41422-022-00644-8. https://www.biorxiv.org/content/10.1101/2022.01.03.474855v1 Published online. (Accessed July 15, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alpha B, Lubinski B, Maureen HV, et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant. ISCIENCE. 2022;25:1–16. doi: 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallm J, Bundschuh C, Kim H, et al. Local emergence and decline of a SARS-CoV-2 variant with mutations L452R and N501Y in the spike protein. Published online 2021. https://www.medrxiv.org/content/10.1101/2021.04.27.21254849v1. (Accessed July 15, 2022).
- 32.Sarkar R, Lo M, Saha R, et al. S glycoprotein diversity of the Omicron variant. medRxiv. 2021 https://www.medrxiv.org/content/10.1101/2021.12.04.21267284v1 Published online. (Accessed july 15, 2022) [Google Scholar]
- 33.Jauregui AR, Savalia D, Lowry VK, et al. Identification of Residues of SARS-CoV nsp1 That Differentially Affect Inhibition of Gene Expression and Antiviral Signaling. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert K, Karousis ED, Jomaa A, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 35.Mou K, Mukhtar F, Khan MT, et al. Emerging Mutations in Nsp1 of SARS-CoV-2 and Their Effect on the Structural Stability. Pathogens. 2021;2:1–11. doi: 10.3390/pathogens10101285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornillez-ty CT, Liao L, Iii JRY, et al. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 2 Interacts with a Host Protein Complex Involved in Mitochondrial Biogenesis and Intracellular Signaling. 2009;83:10314–10318. doi:10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed]
- 37.Sakai Y, Kawachi K, Terada Y, et al. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan MT, Zeb MT, Ahsan H, et al. SARS-CoV-2 nucleocapsid and Nsp3 binding: an in silico study. Arch Microbiol. 2021;203:59–66. doi: 10.1007/s00203-020-01998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angeletti S, Benvenuto D, Bianchi M, et al. COVID-2019:The role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020;92:584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forni D, Cagliani R, Mozzi A, et al. Extensive Positive Selection Drives the Evolution of Nonstructural Proteins in Lineage C Betacoronaviruses. J Virol. 2016;90:3627–3639. doi: 10.1128/jvi.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.V'kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arya R, Kumari S, Pandey B, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:1–25. doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Qin C, Rao Y, et al. SARS-CoV-2 Nsp5 Demonstrates Two Distinct Mechanisms Targeting RIG-I and MAVS to Evade the Innate Immune Response. MBio. 2021;12:1–21. doi: 10.1128/mBio.02335-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benvenuto D, Angeletti S, Giovanetti M, et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pachetti M, Marini B, Benedetti F, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R, Hozumi Y, Yin C WG. Decoding SARS-CoV‑2 Transmission and Evolution.pdf. J Chem Inf Model. 2020;60:5853–5865. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majumdar P, Niyogi S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol Infect. 2021;149:1–7. doi: 10.1017/S0950268821001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuen CK, Lam JY, Wong WM, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng X, Hackbart M, Mettelman RC, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci USA. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kern DM, Sorum B, Mali SS, et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol. 2021;28:573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu B, Tao L, Wang T, et al. Humoral and cellular immune responses induced by 3a DNA vaccines against severe acute respiratory syndrome (SARS) or SARS-like coronavirus in mice. Clin Vaccine Immunol. 2009;16:73–77. doi: 10.1128/CVI.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timmers LFSM, Peixoto JV, Ducati RG, et al. SARS‑CoV‑2 mutations in Brazil: from genomics to putative clinical conditions. Sci Rep. 2021;11:1–14. doi: 10.1038/s41598-021-91585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z, Xiao F, Chen J. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14+ monocytes. iScience. 2021;24:1–19. doi: 10.1016/j.isci.2021.102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Q, Zhang Y, Lü H, et al. The E protein is a multifunctional membrane protein of SARS-CoV. Genomics. proteomics Bioinforma /Beijing Genomics Inst. 2003;1:131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regla-Nava JA, Casta C, Fernandez-delgado R, et al. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Troyano-Hernáez P, Reinosa R, Holguín Á. Evolution of sars-cov-2 envelope, membrane, nucleocapsid, and spike structural proteins from the beginning of the pandemic to september 2020: A global and regional approach by epidemiological week. Viruses. 2021;13:1–16. doi: 10.3390/v13020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jakhmola S, Indari O, Kashyap D, et al. Mutational analysis of structural proteins of SARS-CoV-2. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06572. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Syed AM, Taha TY, Tabata T, et al. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science. 2021;374:1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kandeel M, Mohamed MEM, Abd El-Lateef HM, et al. Omicron variant genome evolution and phylogenetics. J Med Virol. 2022;94:1627–1632. doi: 10.1002/jmv.27515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:1–28. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.