Abstract
Objective
This study aimed to model the precision of SARS-CoV-2 seroprevalence estimates.
Methods
Sample size and precision estimates were calculated using the normal approximation to the binomial distribution. The relationship between sample size and precision was visualized across a range of assumed SARS-CoV-2 seroprevalence from 2% to 75%.
Results
The calculation found that 2% precision was attainable by taking moderately sized sample sets when the expected seroprevalence of SARS-CoV-2 infection exceeds 2%. In populations with a low incidence of SARS-CoV-2 infection and an expected seroprevalence of less than 2%, larger samples are required for precise estimates.
Conclusions
Taking a sample of 177–1000 participants can provide precise prevalence estimates of SARS-CoV-2 infection in vaccinated and unvaccinated populations. Larger sample sizes are only necessary in low prevalence settings.
Keywords: Prevalence, SARS-CoV-2, Precision, Study design, Serological survey, Epidemiology
Surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections has evolved throughout the COVID-19 pandemic but remains a critical public health tool to estimate risk and inform containment, mitigation, and treatment strategies.1 In the first year of the pandemic, many countries used nucleic acid amplification tests for routine surveillance of SARS-CoV-2 infection counts in their populations.2 Overtime, the use of molecular tests was scaled down in many jurisdictions due to a combination of factors, including cost, patient hesitancy, and availability of take-home rapid antigen tests. In the second year of the pandemic, surveillance programs transitioned to using serological testing and novel methods such as environmental sampling of wastewater to estimate SARS-CoV-2 infection rates.3 Going forward, the use of serological testing for surveillance is likely to increase, as it can be used to estimate SARS-CoV-2 point-prevalence and measure antibody responses post COVID-19 vaccination and fluctuations in anti-SARS-CoV-2 antibody levels over time.4
In response to the increased need to design and implement SARS-CoV-2 serosurveillance studies, we provide a simple approximation of the sample size required for precise estimates.
Accurately estimating the point prevalence of a disease in a population requires a precision-based sample size calculation. The precision of an estimate refers to half the width of the desired confidence interval (CI).5 For example, if the CI equals five units, then the precision would equal two and a half units. Calculating the desired precision in the design stage of the study has several benefits, as it requires that the investigators think about the width of the CI and whether it exceeds the expected prevalence value. For example, if a disease has an expected prevalence of 3%, then a study with a CI of 10% lacks sufficient precision to observe the true prevalence in the population of interest: Prevalence = 3%, 95% CI: –2% to 8%, the CI includes zero. Therefore, precision should be derived from the expected prevalence estimate, and studies should not be conducted before making a precision-based sample size calculation.
The simplest (frequentist) methods to perform a precision-based sample size calculation assume that the test being used to classify disease or evidence of infection possesses perfect sensitivity and specificity. This assumption is not impractical, as many studies use a test with high diagnostic accuracy, reporting 95% CIs accommodates for error and corrections for instrument bias can be made postestimation by the Rogan-Gladen method.6 Biological confounders of test results should also be taken into consideration, for example, interpretation of serological test results should always be done in reference to the time of infection and disease severity, as antibodies wane overtime and those with mild symptoms may have a less robust serological response. We used a sample size formula derived from the normal approximation of the binomial distribution to estimate the required sample size for precise SARS-CoV-2 serological surveys.7 Notation of formula to estimate precision is as follows:
where n is the sample size, N represents population size∗, p represents prevalence, h represents precision, and z is Z-score†. A population∗ of one million and Z-score† of 1.96 was specified.
We used the above formula to calculate the necessary sample size to achieve precise estimates across a range of expected SARS-CoV-2 prevalence from 2% to 75% (Fig. 1 ; Table S1).
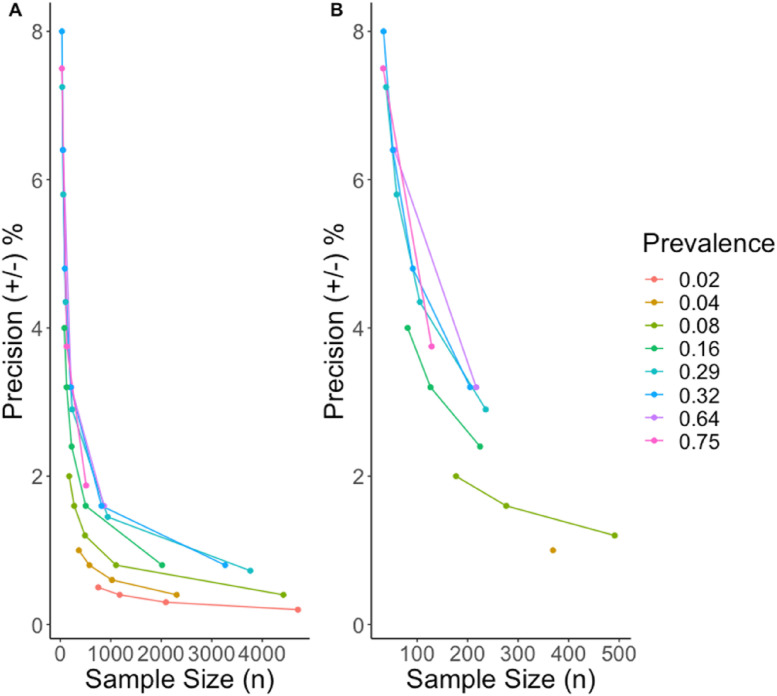
Fig. 1.
Calculated estimates of precision by sample size assuming varying prevalence. (A) X-axis is scaled from 0 to 5000; (B) X-axis is scaled from 0 to 500.
The precision curve (Fig. 1) shows that sample sizes ranging from 177 to 1000 possess adequate precision even when the expected prevalence equals 2% (Fig. 1, Table S1). In the fall of 2021, during the fourth epidemiological phase of the COVID-19 pandemic, high seroprevalence estimates were observed in unvaccinated persons living in South Africa, anti-spike 68.4% (95% CI, 67.2–69.6%), and antinucleocapsid 39.7% (95% CI, 38.4–41.0%).8 In the United States, similar trends were reported with antinucleocapsid seroprevalence increasing from 2021 to 2022 from 33.5% (95% CI, 33.1–34.0%) to 57.7% (95% CI, 57.1–58.3%).9 Assuming that, early 2022 seroprevalence rates exceed approximately 30%, a sample of 817 provides an estimate with <2% precision (Table S1). In health-based research, a precision of 2–5% is recommended for most applications, we recommend that the investigator select the appropriate precision on a case-to-case basis.10 Of note, if a serosurvey is planning to use multiple antigenic targets in its study design, the estimated sample size for high precision results should be calculated using the assay target that is expected to yield lower seroprevalence estimates (e.g. such as the antinucleocapsid antibodies in the case of SARS-CoV-2, that are known to wane over time).11
Investigators should consider the precision afforded by their sample size in the design phase of a SARS-CoV-2 seroprevalence study (or any other pathogen of interest), samples size greater than 1000 are only necessary in low prevalence settings. In a high prevalence but low incidence environment, a smaller sample size can be used to estimate the number of people infected but may not reliably measure changes overtime. We recommend taking several randomly selected stratified samples overtime to reduce bias and observe longitudinal trends.
Author statements
Ethical approval
No ethical approval was sought for the study, as it does not involve the recruitment of participants, data collection, or measurement of an intervention. The analysis describes calculations, which does not rely on observational or experimental data of any kind.
Funding
A.M.N. (under the supervision of A.N.J.) was awarded a Frederick Banting and Charles Best Canada Graduate Scholarship from the Canadian Institutes of Health Research to pursue doctoral studies (#434951). I.S. and A.N.J. obtained funding from Public Health Agency of Canada through the COVID-19 Immunity Taskforce (2021-HQ-000141).
Competing interests
The authors have no conflicts of interest to declare.
Author contributions
A.M.N., I.S., and A.N.J. conceived and designed the study. Data analysis was performed by A.M.N. A.M.N., I.S., and A.N.J. responded to the peer review. All authors interpreted the data, contributed to writing and editing the manuscript, and provided their approval for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.puhe.2022.08.008.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ibrahim N.K. Epidemiologic surveillance for controlling Covid-19 pandemic: types, challenges and implications. J Infect Public Health [Internet] 2020 Nov;13(11):1630–1638. doi: 10.1016/j.jiph.2020.07.019. https://linkinghub.elsevier.com/retrieve/pii/S1876034120306031 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z., Azman A.S., Chen X., Zou J., Tian Y., Sun R., et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet [Internet] 2022 Apr 28;54(4):499–507. doi: 10.1038/s41588-022-01033-y. https://www.nature.com/articles/s41588-022-01033-y Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farkas K., Hillary L.S., Malham S.K., McDonald J.E., Jones D.L. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr Opin Environ Sci Heal [Internet] 2020 Oct;17:14–20. doi: 10.1016/j.coesh.2020.06.001. https://linkinghub.elsevier.com/retrieve/pii/S2468584420300404 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz N.A., Corman V.M., Echterhoff A.K.C., Müller M.A., Richter A., Schmandke A., et al. Seroprevalence and correlates of SARS-CoV-2 neutralizing antibodies from a population-based study in Bonn, Germany. Nat Commun [Internet] 2021 Dec 9;12(1):2117. doi: 10.1038/s41467-021-22351-5. http://www.nature.com/articles/s41467-021-22351-5 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pourhoseingholi M.A., Vahedi M., Rahimzadeh M. Sample size calculation in medical studies. Gastroenterol Hepatol from bed to bench [Internet] 2013;6(1):14–17. http://www.ncbi.nlm.nih.gov/pubmed/24834239 Available from: [PMC free article] [PubMed] [Google Scholar]
- 6.Kritsotakis E.I. On the importance of population-based serological surveys of SARS-CoV-2 without overlooking their inherent uncertainties. Public Heal Pract [Internet] 2020 Nov;1 doi: 10.1016/j.puhip.2020.100013. https://linkinghub.elsevier.com/retrieve/pii/S2666535220300124 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson M.A. Sample size estimation in Veterinary epidemiologic research. Front Vet Sci [Internet] 2021 Feb 17;7:539573. doi: 10.3389/fvets.2020.539573. https://www.frontiersin.org/articles/10.3389/fvets.2020.539573/full Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madhi S.A., Kwatra G., Myers J.E., Jassat W., Dhar N., Mukendi C.K., et al. Population immunity and Covid-19 severity with Omicron variant in South Africa. N Engl J Med [Internet] 2022 Apr 7;386(14):1314. doi: 10.1056/NEJMoa2119658. http://www.nejm.org/doi/10.1056/NEJMoa2119658 26. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke K.E.N., Jones J.M., Deng Y., Nycz E., Lee A., Iachan R., et al. MMWR Morb Mortal Wkly Rep [Internet]; 2022 Apr 29. Seroprevalence of infection-induced SARS-CoV-2 antibodies — United States, September 2021–February 2022.http://www.cdc.gov/mmwr/volumes/71/wr/mm7117e3.htm?s_cid=mm7117e3_w 71(17):606–8. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Mesa J., González-Chica D.A., Bastos J.L., Bonamigo R.R., Duquia R.P. Sample size: how many participants do I need in my research? An Bras Dermatol [Internet] 2014 Jul;89(4):609–615. doi: 10.1590/abd1806-4841.20143705. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0365-05962014000400609&lng=en&tlng=en Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosado J., Pelleau S., Cockram C., Merkling S.H., Nekkab N., Demeret C., et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study. Lancet Microbe [Internet] 2021 Feb;2(2):e60–e69. doi: 10.1016/S2666-5247(20)30197-X. https://linkinghub.elsevier.com/retrieve/pii/S266652472030197X Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.