Abstract
The evidence of rising numbers of multidrug-resistant organisms requires the implementation of effective stewardship programs. However, this should be informed by evidence-based knowledge of local antimicrobial resistance patterns. The current study aims to establish the prevalence of common pathogenic microbes including their antimicrobial susceptibility patterns and distribution in the Cape Coast Metropolis. This was a retrospective study where microbial culture and antimicrobial susceptibility records for 331 patients were reviewed from January to December 2019, at a private health centre. All data were analysed using Excel (Microsoft Office, USA), SPSS and GraphPad Prism 8 software programs. Among the samples tested, 125 (37.76%) were positive for microbes with high vaginal swab (HVS) samples recording the highest number of pathogens (44%), followed by urine (40%) and both pleural and semen samples having the least (0.3% each). Again, gram-negative isolates were more prevalent than the gram-positive isolates. The prevalence of antimicrobial resistance was very significant with isolates resistant to more than one antibiotic (P < 0.05). Escherichia coli showed the highest level of resistance, followed by Citrobacter spp. These were followed by Klebsiella spp., Staphylococcus spp., Coliforms, Pseudomonas spp., Commensals and Candida spp. The high resistance pattern suggests an inevitable catastrophe requiring continuous monitoring and implementation of effective antibiotic stewardship.
Subject terms: Biochemistry, Genetics, Microbiology
Introduction
Antimicrobial resistance (AMR) arises when microbes advance mechanisms that guard them from the effects of antimicrobials1. However, antibiotic resistance (AR) is when bacteria develop the ability to survive exposure to antibiotics1. Resistant microbes are difficult to treat, requiring higher doses, or alternative medications that may prove more toxic and expensive. Whereas microbes that are resistant to multiple antimicrobials are called multidrug-resistant (MDR), those that are known as extensively drug-resistant (XDR) or totally drug-resistant (TDR) are also called “superbugs”2. Resistance can occur naturally due to chance mutations. However, protracted use of antimicrobials encourages selection for mutations which can make antimicrobials ineffective. Furthermore, the lack of swift and proper identification of pathogens especially in patients with critical infection leads to broad-spectrum antibiotic overuse. Therefore, the prevention of antibiotic misuse can lead to a significant reduction in antibiotic resistance3. Narrow-spectrum antibiotics are favored over broad-spectrum antibiotics due to their effectiveness and accuracy in targeting specific organisms with less side effects4. For those who engage in self-medication, education about the detrimental effects of their actions is required. Health care providers can engage in antimicrobial stewardship to decrease the heavy load of antibiotic resistance5. Rising drug resistance is caused mainly by the use of antimicrobials in humans and other animals, and the spread of resistant strains between the two. Increasing resistance has also been associated with the discarding of inadequately treated wastes from the pharmaceutical industry, particularly in countries where majority of drugs are manufactured6. Antimicrobial resistance is increasing globally because of greater access to antibiotic drugs in developing countries7. A recent study estimates that 700,000 to several million deaths result per year and continues to pose a major public health threat worldwide due to bacterial resistance8. According to the world health organization (WHO) estimates, 350 million deaths could be caused by AMR by 20501, thereby calling on the public for global collective action to address the threat that includes proposals for international treaties on antimicrobial resistance, as poorer countries with weaker healthcare systems are often more affected3.
Accurate information on the use of antibiotics is crucial to address the problem of antibiotic overuse and resistance9. Constant assessment of antibiotic use is necessary to preserve the efficacy of antibiotics and reduce harm to patients. The WHO recommends the surveillance of antibiotic use as a strategy for improving antibiotic use among patients and also for controlling antibiotic resistance1. Since health facilities are prone to nosocomial infections caused by hostile pathogens, the current study aimed to establish the prevalence of common pathogenic bacteria including their antimicrobial susceptibility patterns and distribution in the Cape Coast Metropolis through a 1-year retrospective study.
Results
Study population and demographics
In this study, patients were characterized into their sex and age groups respectively. Out of 331 patients recruited in the study, there were 105 males (31.7%) and 226 females (68.3%) represented in Table 1. On the tests conducted at the facility, urine analysis was observed to be the highest (51.1%), followed by high vaginal swab (HVS), (26.9) with pleural and semen analyses being the lowest (0.3% each).
Table 1.
Number of tests conducted.
| Test | Female | Male | Total number | Percentage (%) |
|---|---|---|---|---|
| Blood | 8 | 4 | 12 | 3.6 |
| Cervical | 6 | – | 6 | 1.8 |
| HVS | 89 | – | 89 | 26.9 |
| Pleural | 1 | – | 1 | 0.3 |
| Semen | – | 1 | 1 | 0.3 |
| Stool | 9 | 8 | 17 | 5.1 |
| Throat | – | 1 | 1 | 0.3 |
| Urethral | 1 | 13 | 14 | 4.2 |
| Urine | 100 | 69 | 169 | 51.1 |
| Wound | 12 | 9 | 21 | 6.3 |
| Total | 226 (68.3%) | 105 (31.7%) | 331 | 100 |
Prevalence of microbial isolates
The overall number of individual isolates and the prevalence of pathogens during the study period of 1 year (P < 0.05) were established and compared using one-way ANOVA analysis. Candida spp. were the most abundant, followed by E. coli, with the least being both Enterobacter and Micrococcus (Fig. 1 and Table 2). In terms of samples, HVS recorded the highest prevalence of bacteria followed by urine and wound. However, no bacteria were detected in the urethra, throat, semen and pleural (Fig. 2, One-way ANOVA, P < 0.05).
Figure 1.
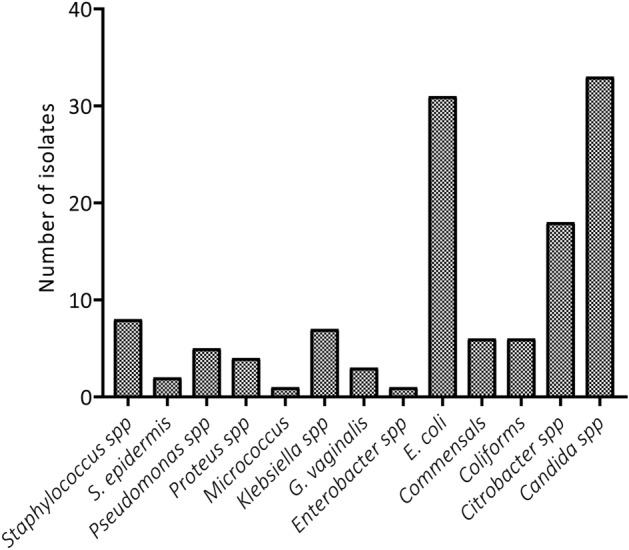
Type and frequency of microorganisms isolated from the patient visiting the health facility.
Table 2.
Prevalence of Microorganisms in samples.
| Microbe | Number of samples of tests within which microbes were identified | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Blood | Cervical | HVS | Pleural | Semen | Stool | Throat | Urethra | Urine | Wound | |
| Candida spp. | – | 1 | 24 | – | – | – | – | – | 8 | – |
| Citrobacter spp. | – | – | 7 | – | – | – | – | – | 10 | 1 |
| Coliforms | – | – | – | – | – | – | – | – | – | 6 |
| Commensals | – | – | 6 | – | – | – | – | – | – | – |
| E. coli | – | 1 | 7 | – | – | – | – | – | 23 | – |
| Enterobacter spp. | – | – | – | – | – | – | – | – | 1 | – |
| G. vaginalis | – | – | 3 | – | – | – | – | – | – | – |
| Klebsiella spp. | – | – | – | – | – | – | – | – | 6 | 1 |
| Micrococcus | – | – | – | – | – | – | – | – | 1 | – |
| Proteus spp. | – | – | 1 | – | – | 1 | – | – | 1 | 1 |
| Pseudomonas spp. | – | – | – | – | – | – | – | – | – | 5 |
| S. epidermis | 1 | – | 1 | – | – | – | – | – | – | – |
| Staphylococcus spp. | – | – | 6 | – | – | – | – | – | – | 2 |
Figure 2.
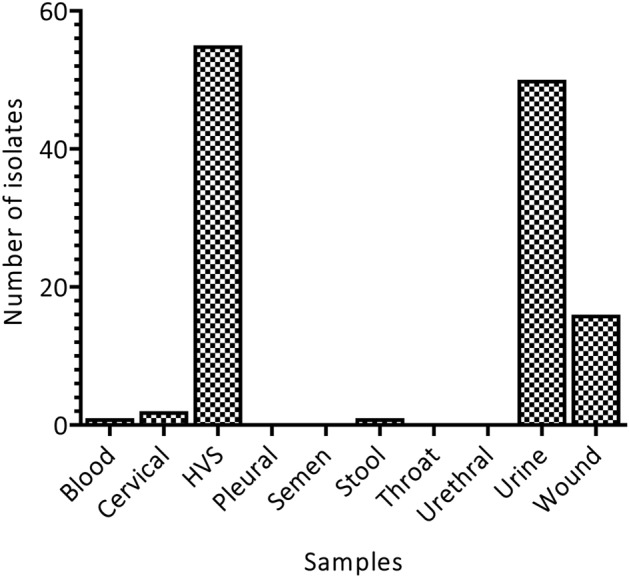
Distribution of different clinical isolates among the various samples.
Distribution of microbes according to demographics
To identify the group that was more susceptible to bacterial infections, isolates were distributed according to age and sex of which Citrobacter spp., E. coli, Klebsiella spp., Proteus spp., among others were identified in both sexes. On the other hand, however, Enterobacter spp. was seen only in males whereas Candida spp., G. vaginalis, and Micococcus were identified in females only. Candida spp. and E. coli were the most common isolates (Fig. 3, Two-way ANOVA, P < 0.05). Other Coliform and Commensal bacteria were present but few.
Figure 3.
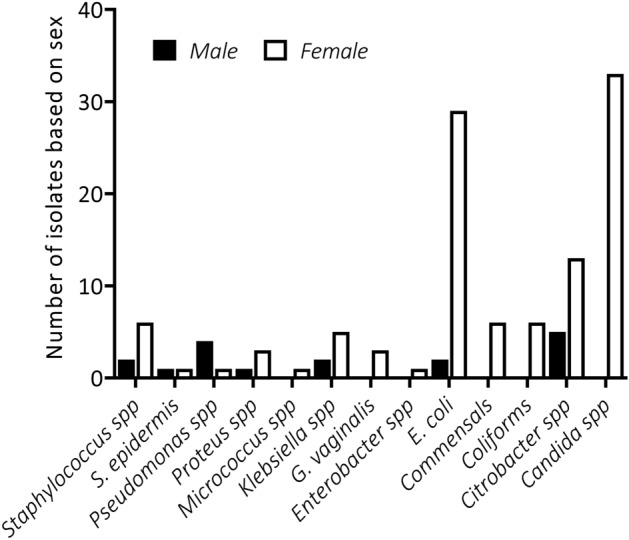
Gender-specific distribution of microbes.
Participants aged 26–40 years recorded the highest number of microbes followed by age groups 16 to 25 years and 60 years and above (Table 3). In addition, the distribution of the microbes was not in an age-specific manner. As indicated earlier, Candida spp. and E. coli were observed to be the bacteria with the highest prevalence. Candida spp. was more in the 16–25 and 26–40 age groups, whereas E. coli was prominent in the 26–40 and ≥ 60 years age groups (Table 4). Again, high prevalence of Candida spp. and E. coli infections in the age group 26–40 years were observed.
Table 3.
Prevalence of microorganisms among age groups.
| Age | Number of samples | ||
|---|---|---|---|
| Total | Microbe absent | Microbe present | |
| 1–5 | 8 | 7 | 1 |
| 6–15 | 9 | 6 | 3 |
| 16–25 | 51 | 27 | 34 |
| 26–40 | 145 | 96 | 49 |
| 41–60 | 41 | 31 | 10 |
| Above 60 | 67 | 35 | 32 |
Table 4.
Individual bacteria identified in samples per age groups.
| Microbe | Age groups | |||||
|---|---|---|---|---|---|---|
| 1–5 | 6–15 | 16–25 | 26–40 | 41–60 | > 60 | |
| Candida spp. | 0 | 0 | 12 | 19 | 1 | 1 |
| Citrobacter spp. | 0 | 2 | 4 | 3 | 2 | 7 |
| Coliforms | 0 | 0 | 0 | 2 | 3 | 1 |
| Commensals | 0 | 0 | 1 | 3 | 1 | 1 |
| E. coli | 0 | 2 | 3 | 14 | 2 | 10 |
| Enterobacter spp. | 0 | 0 | 0 | 0 | 1 | 0 |
| G. vaginalis | 0 | 0 | 1 | 1 | 0 | 1 |
| Klebsiella spp. | 0 | 0 | 1 | 3 | 0 | 3 |
| Micrococcus | 0 | 0 | 1 | 0 | 0 | 0 |
| Proteus spp. | 0 | 0 | 0 | 1 | 0 | 3 |
| Pseudomonas spp. | 0 | 0 | 0 | 2 | 1 | 2 |
| S. epidermis | 1 | 0 | 0 | 1 | 0 | 0 |
| Staphylococcus | 0 | 0 | 2 | 3 | 0 | 3 |
Antimicrobial susceptibility profiles of microbes
All isolates were tested for their susceptibility or resistance to the most commonly used antimicrobials at the diagnostic centre during the one-year period. All bacteria isolates showed resistance to at least two antibiotics (Fig. 4, Supplementary Table S1a,b, P < 0.05, Two-way ANOVA), hence, a disturbing level of antimicrobial resistance was registered in the study. E. coli showed the highest resistance level among all the pathogens, followed by Citrobacter spp., Klebsiella spp., Staphylococcus spp., Coliforms, Pseudomonas spp., Commensals and then lastly Candida spp. Interestingly, Enterobacter spp. was not susceptible to any of the antibiotics. The microbes were remarkably resistant to Cloxacillin with significantly low susceptibility. Interestingly, microbes showed significantly high susceptibility and relatively low resistance to Amikacin. It is worthy of note that only one Enterobacter was resistant to 12 out of the 14 antibiotics tested (Supplementary Table S1b). The prevalence of antibiotic resistance was very significant among both gram-negative and gram-positive organisms. This high resistance pattern foreshadows an inevitable catastrophe that requires continuous monitoring and implementation of effective antibiotic policies.
Figure 4.
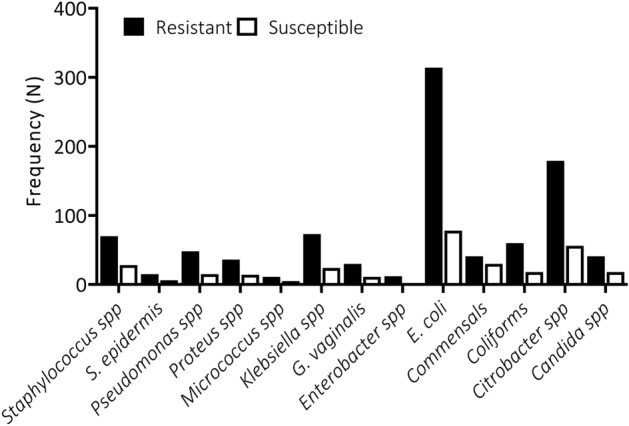
Prevalence of antimicrobial resistance among the specific microbe isolates.
Discussion
Understanding the distribution of microbial pathogens and their associated infections is required to control infectious diseases and monitor antimicrobial resistance. The current study aimed at establishing the prevalence of common pathogenic microorganisms including their antimicrobial susceptibility patterns and distribution according to specimens, age groups and sex at a private diagnostic Centre in the Cape Coast Metropolis.
The excessive use of antibiotics among other factors has led to extensive antimicrobial resistance. If this trend continues unabated, then all other antibiotic options will be exhausted making the treatment of associated infections extremely difficult. Hence, the WHO identified it as an international health problem of prime concern10–12. To control this rising predicament, all-inclusive antibiotic and other relevant stewardship especially in poor countries are essential. However, enough data concerning antimicrobial resistance are inaccessible to exactly measure the degree of the problem. The few available studies regarding results on microbiological samples suggest that there are hotbeds of emerging high-level resistance10.
In this study, gram-negative bacteria were more prevalent than gram-positive isolates, similar to reports by Newman and colleagues, and Fahim10,13. Most of the isolates were recovered from HVS samples representing 44%, followed by urine samples which recorded 40% of the total samples that contained pathogens unlike the results of Fahim who reported higher recovery from blood, followed by urine specimens10. Gram-negative bacteria cause various infections including pneumonia, bloodstream infections, wound or surgical site infections, and meningitis among others. Gram-negative bacteria are resistant to multiple drugs with suggested development of resistance to most of the available antibiotics. This observation can be attributed to their in-built abilities to find alternative ways to develop resistant and thus, causing significant morbidity and mortality worldwide14.
The high prevalence of microbial isolates reported in this study highlights the need for effective monitoring and surveillance of microbial infections in resource-limited health care facilities15.
Among all microorganisms isolated, Candida spp. was seen to be most abundant followed by E. coli, with the least being both Enterobacter and Micrococcus. Among the gram-negatives, E. coli represented the most isolated pathogen while Enterobacter spp. was the least whereas in the gram-positive isolates, Candida spp. (fungus) represented the most isolated pathogen whiles Micrococcus spp. was the least. In Nigeria, Osifo and Aghahowa reported that E. coli and Klebsiella pneumoniae were the most frequently isolated pathogens16. Invasive Candida infections are often associated with high rates of morbidity and mortality17, therefore, the high levels observed in the current study is a cause for concern. While E. coli is a normal resident of the healthy gut, it is also an important and widespread pathogen which has been associated with human infections including diarrhoea, urinary tract infections and meningitis.
To identify the most vulnerable group from bacterial infections, isolates were distributed according to sex and age groups of the patients. There were more pathogens in females (226, representing 68.3%) than in males (105 representing 31.7%) and in some cases found in only females (Candida spp. and G. vaginalis spp.) with the exception of Pseudomonas spp., which was higher in males (4 representing 80%) than females (1 representing 20%). This observation deviates from what has been reported elsewhere, where the distribution in males and females were virtually the same, thus, 51% and 49% respectively15 but similar to that of Mapanguy and colleagues who reported a significantly higher prevalence among females (61%) than males (39%)18. Also, a significant number of bacteria were isolated from the age group 26–40 years, followed by 60 and above with age group 1–5 years recording the least. The study recruited more adults than children hence the observation that more isolates were obtained from adults corresponds with the high number of adult clients recorded.
We report a high prevalence of microorganisms with variable susceptibility patterns to key antimicrobials. All microorganisms isolated showed resistance to more than one antimicrobial agent. Cotrimoxazole, Erythromycin, Vancomycin, Chloramphenicol and Cefuroxime were among the top five antimicrobials with a high prevalence of resistance. However, Amikacin, Gentamicin and Nitrofurantoin were the three most effective antibiotics. This is similar to an earlier report where amikacin was among the group with lowest resistance13. Furthermore, Fahim also reported in Egypt that gram-negative isolates exhibited high resistance to almost all the classes of antibiotic in use with the least frequency recorded against nitrofurantoin, amikacin, followed by imipenem and meropenem10.
Escherichia coli was the pathogen with the highest resistance and the highest resistance was toward cefuroxime, chloramphenicol, meropenem, vancomycin and erythromycin. The next resistant microbe was Citrobacter spp., which was highly resistant only to chloramphenicol. Conversely, E. coli and Citrobacter spp. were highly susceptible to amikacin. This is similar to a study in Congo, where E. coli was the highly resistant ceftazidime, followed by amoxicillin, piperacillin-tazobactam, ofloxacin, and azithromycin18. Also, a previous report among healthy individuals in an Indian population showed similar patterns of resistance19. Interestingly, a high prevalence of resistance to ceftazidime was reported in a study in Uganda20 and amoxicillin in Nigeria21.
Apart from E. coli and Citrobacter spp., Klebsiella spp., Staphylococcus spp., other coliforms, Pseudomonas spp., Candida spp. and other commensals were among the most resistant microbes isolated. Other studies have reported similar results where the most prevalent organisms in the collection included E. coli, S. aureus, Klebsiella spp., Pseudomonas aeruginosa, Citrobacter spp. and Enterobacter spp.13,22.
Factors that may have contributed to the emergence and prevalence of resistance, includes uncontrolled use of these drugs, non-compliance with treatment and geographical location/unsanitary environment. Another significant factor for increased resistance to antibiotics is the use of substandard and counterfeit drugs, and the unauthorized sale of antibiotics without prescription18,23,24. Interestingly, Enterobacter spp. was not susceptible to any of the antibiotics whereas the majority of the microbes were remarkably resistant to Cloxacillin with lower susceptibility levels. In contrast, Amikacin showed high activity towards these microorganisms. This means that amikacin is the antibiotic effective against the greatest number of microorganisms characterized in this study. Correspondingly in another study, a lower percentage of resistance was observed for ceftriaxone, ciprofloxacin, and amikacin13.
It is significant to note that this study provides a general overview of the current shocking situation in the area under study. This implies that urgent action needs to be taken to halt this catastrophic menace by starting an effective action plan for its containment.
Conclusion
Gram-negative isolates were the most common bacteria isolated from patients attending this referral laboratory service compared to the gram-positive isolates. Of these, E. coli represented the most isolated pathogen while Enterobacter spp. However, in the gram-positive isolates, Candida spp. represented the most isolated pathogen whiles Micrococcus spp. was the least. The prevalence of antimicrobial resistance was very significant among the isolated pathogens. The highest resistance was found in Escherichia coli and the highest resistance was toward cefuroxime, chloramphenicol, meropenem, vancomycin and erythromycin. The increased antimicrobial resistance reported in the study could be due to the unreasonable use of antibiotics by the populace. Nonetheless, to fight against antimicrobial resistance, a localized epidemiological surveillance program is required to help establish evidence-based guidelines for the treatment and management of microbial infections. The observed high resistance pattern also requires continuous monitoring and implementation of effective antibiotic stewardship.
Method
Study area and design
The study was conducted at a Private Diagnostic Centre in the Cape Coast Metropolis, Central Region, Ghana. The facility serves as a referral diagnostic centre in the Central Region of Ghana. This study is a retrospective analysis of routine recovered bacterial isolates subjected against a panel of antibiotics for susceptibility testing25. The study spun from January to December 2019.
Data extraction
A retrospective audit of records for three hundred and thirty-one (331) participants’ bacterial culture and susceptibility testing results from the month of January 2019 to December 2019 from a private diagnostic centre was conducted. This Private diagnostic centre is a referral unit where laboratory tests from various hospitals within the Cape Coast Metropolis are sent. By using a Convenient sampling technique, records of specimens such as urine, stool, blood, and various body sites (cervical, wound, etc) were all included.
Processing and identification of isolates
Sample processing and identification of the isolates were performed per the standard operating procedures (SOPs) of the laboratory. The samples were cultured on the routinely used microbiological media and incubated for 24 h at 37 °C10. If no growth, the plates were incubated for a total of 48 h. The identification of the isolates was done according to colony morphology, gram stain, and standard confirmatory biochemical tests. Gram-positive bacteria were identified by testing the hemolytic activity on blood agar and further identification using different biochemical tests such as catalase reaction, slide and tube coagulase tests, culture on DNase agar, bile esculin, in addition to different differentiating antibiotic discs such as optochin and bacitracin. For gram-negative bacteria, identification was conducted by biochemical tests such as oxidase, triple sugar iron, motility indole ornithine, citrate, lysine iron arginine, and urease tests10.
Antimicrobial susceptibility
Antimicrobial susceptibility tests of the isolates were performed using the Kirby–Bauer disk diffusion method and interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines26. Briefly, a standardized is swabbed onto the surface of MH agar. Since reproducibility depends on the log growth phase of organisms, fresh subcultures are used. Filter paper disks impregnated with a standardized concentration of an antimicrobial agent were placed on the surface, and the size of the zone of inhibition around the disk is measured after overnight incubation. Specific incubation time ranges were outlined in the Clinical and Laboratory Standards Institute [CLSI] documents27.
Data analysis
All data were analysed using statistical package for social sciences (SPSS) computer software (Version 25) and GraphPad Prism 8 software (San Diego, CA, USA). Graphs were used to show the prevalence and distribution of the isolated bacteria. In addition, a frequency table expressed in percentages and absolute numbers were used to display the susceptibility patterns of the commonly isolated bacteria against the commonly used antibiotics. A statistically significant difference was considered at a P-value of ≤ 0.05.
Ethical approval
Ethical approval was obtained from the Cape Coast Teaching Hospital Ethical Review Committee (CCTHERC) with reference number CCTHERC/EC/2021/013 for data acquisition. All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from all subjects and/or their legal guardian(s).
Supplementary Information
Acknowledgements
We are grateful to the Laboratory staff at the Private Diagnostic Centre for their support.
Abbreviations
- BSI
Bloodstream infection
- FBC
Full blood count
- HIV
Human immunodeficiency virus
- HVS
High vaginal swab
- IBI
Invasive bacterial infection
- NTS
Non-typhoidal salmonella
- UTI
Urinary tract infection
- VDRL
Venereal Disease Research Laboratory
- WBC
White blood count
- WHO
World Health Organization
Author contributions
G.G.K. and A.S.A. developed the idea. G.G.K., E.B., B.G.B., B.A. and L.L.A. wrote the manuscript. E.B., S.D.B. and B.G.B. analysed the data. A.S.A., S.D.B., Y.K.O., L.L.A., B.A. and G.G.K. edited the manuscript. All authors read and approved the final version of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article [and its Supplementary Information files].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-18595-w.
References
- 1.Organization, W. H. WHO|Antimicrobial resistance: Global report on surveillance 2014. In Antimicrobial Resistance: Global Report on Surveillance 2014 (2016).
- 2.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 3.Zilahi G, McMahon MA, Povoa P, Martin-Loeches I. Duration of antibiotic therapy in the intensive care unit. J. Thorac. Dis. 2016;8:3774–3780. doi: 10.21037/jtd.2016.12.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber JS, et al. Association of broad- vs narrow-spectrum antibiotics with treatment failure, adverse events, and quality of life in children with acute respiratory tract infections. JAMA J. Am. Med. Assoc. 2017;318:2325–2336. doi: 10.1001/jama.2017.18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Changing Markets, E. Impacts of Pharmaceutical Pollution on Communities and Environment in India. (Nordea Asset Management, 2016).
- 6.Gullberg E, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dramé O, et al. Antimicrobial resistance of campylobacter in broiler chicken along the food chain in Canada. Foodborne Pathog. Dis. 2020;17:512–520. doi: 10.1089/fpd.2019.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’neill, J. Tackling drug-resistant infections globally: Final report and recomendations. Straits Times (2018).
- 9.Ceyhan M, Yildirim I, Ecevit C, Aydogan A, Ornek A, Salman N, Somer A, Hatipoǧlu N, Camcioglu Y, Alhan E, Celik U, Hacimustafaoglu M, Celebi S, Inan D, Kurt N, Oner AF, Gulumser O, Gunes A, Coskun Y. Inappropriate antimicrobial use in Turkish pediatric hospitals: A multicenter point prevalence survey. Int. J. Infect. Dis. 2010;14(1):e55–e61. doi: 10.1016/j.ijid.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Fahim NAE. Prevalence and antimicrobial susceptibility profile of multidrug-resistant bacteria among intensive care units patients at Ain Shams University Hospitals in Egypt—A retrospective study. J. Egypt. Public Health Assoc. 2021;96:7. doi: 10.1186/s42506-020-00065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(WHO). W. H. O. Antimicrobial resistance: global report on surveillance. (2014).
- 12.Levy SB. The challenge of antibiotic resistance. Sci. Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 13.Newman MJ, Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infect. Drug Resist. 2011;4:215–220. doi: 10.2147/IDR.S21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breijyeh Z, Jubeh B, Karaman R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25:1340. doi: 10.3390/molecules25061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumwenda P, et al. Prevalence, distribution and antimicrobial susceptibility pattern of bacterial isolates from a tertiary Hospital in Malawi. BMC Infect. Dis. 2021;21:34. doi: 10.1186/s12879-020-05725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osifo OD, Aghahowa SE. Audit of antibiotic therapy in surgical neonates in a tertiary hospital in Benin City. Niger. Afr. J. Paediatr. Surg. 2011;8:23–28. doi: 10.4103/0189-6725.78664. [DOI] [PubMed] [Google Scholar]
- 17.Fridkin SK. Candidemia is costly—Plain and simple. Clin. Infect. Dis. 2005;41:1240–1241. doi: 10.1086/496935. [DOI] [PubMed] [Google Scholar]
- 18.Mapanguy CCM, et al. High prevalence of antibiotic-resistant Escherichia coli in Congolese students. Int. J. Infect. Dis. 2021;103:119–123. doi: 10.1016/j.ijid.2020.09.1441. [DOI] [PubMed] [Google Scholar]
- 19.Purohit MR, Lindahl LF, Diwan V, Marrone G, Lundborg CS. High levels of drug resistance in commensal E. coli in a cohort of children from rural central India. Sci. Rep. 2019;9:6682. doi: 10.1038/s41598-019-43227-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odongo I, Ssemambo R, Kungu JM. Prevalence of Escherichia coli and its antimicrobial susceptibility profiles among patients with UTI at Mulago Hospital, Kampala, Uganda. Interdiscip. Perspect. Infect. Dis. 2020;2020:8042540. doi: 10.1155/2020/8042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuben RC, Owuna G. Antimicrobial resistance patterns of Escherichia coli O 157: H 7 from Nigerian fermented milk samples in Nasarawa State, Nigeria. Int. J. Pharm. Sci. Invent. 2013;2:38–44. [Google Scholar]
- 22.Saba CKS, Gonzalez-Zorn B. Microbial food safety in Ghana: A meta-analysis. J. Infect. Dev. Ctries. 2012;6:828–835. doi: 10.3855/jidc.1886. [DOI] [PubMed] [Google Scholar]
- 23.Elton L, et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob. Resist. Infect. Control. 2020;9:145. doi: 10.1186/s13756-020-00800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues CF. Self-medication with antibiotics in Maputo, Mozambique: Practices, rationales and relationships. Palgrave Commun. 2020;6:6. doi: 10.1057/s41599-019-0385-8. [DOI] [Google Scholar]
- 25.Inusah A, Quansah E, Fosu K, Dadzie I. Resistance status of bacteria from a health facility in Ghana: A retrospective study. J. Pathog. 2021;2021:6648247. doi: 10.1155/2021/6648247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Institute, C. A. L. S. Performance standards for antimicrobial susceptibility testing. CLSI (2018).
- 27.Christenson, J. C., Korgenski, E. K. & & Relich, R. F. In Principles and Practice of Pediatric Infectious Diseases (Fifth Edition) (eds Long, S. S., Prober, C. G., & Fischer, M.) 1422–1434.e1423 (Elsevier Inc, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its Supplementary Information files].


