Abstract
Key message
We have established a DNA-free genome editing method via ribonucleoprotein-based CRISPR/Cas9 in cultivated tomato and obtained mutant plants regenerated from transfected protoplasts with a high mutation rate.
Abstract
The application of genome editing as a research and breeding method has provided many possibilities to improve traits in many crops in recent years. In cultivated tomato (Solanum lycopersicum), so far only stable Agrobacterium-mediated transformation carrying CRISPR/Cas9 reagents has been established. Shoot regeneration from transfected protoplasts is the major bottleneck in the application of DNA-free genome editing via ribonucleoprotein-based CRISPR/Cas9 method in cultivated tomato. In this study, we report the implementation of a transgene-free breeding method for cultivated tomato by CRISPR/Cas9 technology, including the optimization of protoplast isolation and overcoming the obstacle in shoot regeneration from transfected protoplasts. We have identified that the shoot regeneration medium containing 0.1 mg/L IAA and 0.75 mg/L zeatin was the best hormone combination with a regeneration rate of up to 21.3%. We have successfully obtained regenerated plants with a high mutation rate four months after protoplast isolation and transfection. Out of 110 regenerated M0 plants obtained, 35 (31.8%) were mutated targeting both SP and SP5G genes simultaneously and the editing efficiency was up to 60% in at least one allele in either SP or SP5G genes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00299-022-02893-8.
Keywords: Solanum lycopersicum, Mesophyll protoplast regeneration, CRISPR/Cas9, Ribonucleoprotein, SP and SP5G genes
Introduction
Variations of CRISPR/Cas9 technology have been applied for genome editing in recent years (Gao 2021). This technology has surpassed the other genome editing tools, such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), as it is more easily and cheaply customized and yields high mutation efficiency in some species (Lowder et al. 2015). So far, successful applications using CRISPR/Cas9 have been reported in model plants, such as Arabidopsis thaliana (Li et al. 2013; Yan et al. 2015) and Nicotiana benthamiana (Li et al. 2013; Nekrasov et al. 2013), and many commercial crops, such as potato (Wang et al. 2015; Andersson et al. 2017), wheat (Upadhyay et al. 2013; Zhang et al. 2016), rice (Jiang et al. 2013; Zhou et al. 2014), maize (Liang et al. 2014; Char et al. 2016), tomato (Brooks et al. 2014; Ito et al. 2015) and many others (Gao 2021).
In general, CRISPR/Cas9 reagents, usually as DNA plasmids, can be delivered to cell-wall-free protoplasts by polyethylene glycol (PEG) transformation, or to plant tissues by stable Agrobacterium-mediated transformation, or by other means, such as particle bombardment and electroporation (Chen et al. 2019). To avoid foreign DNA integrated into plant cells, Woo et al. (2015) were the first to develop the delivery of Ribonucleoprotein (RNP) complexes into plant protoplasts using in vitro preassembled complexes of purified Cas9 protein and guide RNA (gRNA) in Arabidopsis, tobacco, lettuce and rice. Subsequently, studies on the application of RNPs for genome editing in plant species have been reported in crop plants, such as maize (Svitashev et al. 2016), wheat (Liang et al. 2017), potato (Andersson et al. 2018) and canola (Sidorov et al. 2021).
Tomato (Solanum lycopersicum) is an important commercial agricultural crop which is extensively cultivated all over the world as well as being a model plant used in scientific research due to its simple diploid genetics (2n = 2x = 24) and short life cycle (Ito et al. 2015). It has been demonstrated that the CRISPR/Cas9 technology can be used to generate mutated tomato plants for crop improvement such as improved disease resistance (Ito et al. 2015; Pan et al. 2016). Hitherto, most reports about CRISPR/Cas9 applied to tomato were based on stable Agrobacterium tumefaciens-mediated transformation and usually the homozygous mutation rate was low and segregation in the next generation needed to eliminate foreign DNA integrated into the plant genome. An attractive alternative technology would be the use of RNPs to deliver the CRISPR/Cas9 reagents into protoplasts, resulting in transgene-free plants. Although a high editing efficiency has been reported on tomato calli from transfected protoplasts of cultivated tomato, the shoot regeneration from RNP-transfected protoplasts is a bottleneck (Nicolia et al. 2021a). Very recently, (Lin et al. 2022) reported the successful protoplast regeneration of wild tomato (Solanum peruvianum) harboring CRISRP/Cas9 mutations, with a mutation rate varying from 8.3% to 63.6%. However, for cultivated tomato, there are only a few old reports published on plant regeneration from unedited protoplasts (Morgan and Cocking 1982; Sakata et al. 1987; Tan et al. 1987). Thus, establishing a protocol with high editing efficiency and regeneration rate in cultivated tomato would be beneficial regarding genetic studies as well as for breeding purposes. From this aspect, such a protocol could also be further adapted to wild relatives of the tomato, that represent a precious source of variability (e.g., Solanum pennellii, Solanum pimpinellifolium), to speed up programs of “de novo” domestication and/or introgression (Li et al. 2018).
The vegetative-to-reproductive phases in tomato are altered in the sympodial shoots and the switch between those two phases is controlled by the flowering repressor gene SELF PRUNING (SP). Genetic variation and mutations in this gene yields tomato genotypes that are classified into two categories: 'determinate' and 'indeterminate' varieties due to different growth habits (Pnueli et al. 1998; Carmel-Goren et al. 2003). Another flowering repressor SELF PRUNING 5G (SP5G), which is a paralog of the SP gene, is mainly responsible for flower repression in primary and canonical axillary shoots (Soyk et al. 2017). Tomato plants with mutation in either the SP gene or both the SP and SP5G genes showed the determinate phenotype, which resulted in acceleration of flowering, short internodes, bushy appearance and rapid life cycling (Soyk et al. 2017; Kwon et al. 2020). Those mutated plants would be suitable for urban vertical farming and greenhouse cultivation since the agricultural productivity can be increased due to their fast growth habit and compact size, especially in a confined environment. It is also beneficial for open field cultivation in that they grow as small bushes that need less attention compared to indeterminate varieties needing support. Besides, all fruits from determinate cultivars usually ripen in a short period from simultaneous flowering, which is beneficial for facilitating mechanical harvest.
In this study, we have successfully regenerated plants from cultivated tomato transfected protoplasts within four months after transfection. Furthermore, the regenerated plants have a high editing rate when targeting both SP and SP5G genes simultaneously. Hence, we have improved the process of tomato protoplast isolation and solved the challenge of shoot regeneration from RNP-transfected protoplasts using CRISPR/Cas9 technology.
Materials and methods
Plant material and in vitro culture conditions
Seeds of tomato (S. lycopersicum) cultivars (cvs) Red Setter, Ailsa Craig, M82 and Moneymaker were used in this study. Seeds were surface sterilized by washing with 70% ethanol for 5 min, followed by 15% (w/v) calcium hypochlorite (CaCl2O2) for 3 min and then rinsed 5 times with sterile distilled water. Sterilized seeds were placed in Plante Containers (Sakata Ornamentals Europe A/S, Denmark) with germination medium containing 0.2 mg/L Indole Acetic acid (IAA), 15 g/L sucrose, 8 g/L phyto agar, and half-strength Murashige & Skoog (MS) with vitamins (Duchefa Biochemie M0222, Haarlem, Netherlands) (2.2 g/L) with additional 0.2 mg/L Thiamine and 50 mg/L Myo-Inositol at pH 5.9.
In vitro culture mentioned in this study was carried out in a controlled chamber at a temperature of 24 °C/18 °C (light/dark), under a photoperiod of 16 h at 120–140 μE m−2 s−1 light and 8 h dark.
Protoplast isolation, transfection and callus induction in liquid medium
Protoplast isolation, transfection and early callus induction in liquid medium were done as previously described by Nicolia et al. (2021b) with some modifications to improve the yield of isolated protoplasts and regeneration. The components of Medium C, E, F, wash solution, PEG solution, alginate solution and transient expression solution mentioned below can be found in Nicolia et al. (2021b).
In brief, the modifications in protoplast isolation were as follows: preconditioning treatment of in vitro cultured seedlings prior to protoplast isolation was done by placing the Plante Containers in a fridge (4 °C) in darkness one day before isolation. Cotyledons and first true leaves at different ages (14, 17 and 21 d) were used for protoplast isolation. The leaf tissues were sliced and treated with enzyme solution (medium C) at different temperatures (15 and 25 °C) and different time durations of enzyme digestion (14 and 16 h). Protoplasts were collected after centrifugation and the protoplast yield was quantified immediately after isolation by a hemocytometer (FuchsRosenthal 0.2 mm chamber, Horsham, UK) under microscope. The optimization of protoplast isolation was carried out with two cvs (Red Setter and Ailsa Craig) and optimized conditions were confirmed in all four cvs.
Freshly isolated protoplasts were transfected via PEG mediated delivery of RNPs. For each transfection, two different RNP complexes were assembled by mixing two 0.1 nmol synthetic sgRNAs (Synthego) with 10 µg TrueCut™ Cas9 v.2 (Thermo Fisher, Waltham, USA) in a 15 ml tube at room temperature for 15 min. The sgRNA was used as one synthetically produced component including the 20 bp target and an 80-mer SpCas9 scaffold from the suppliers’ standard products. Then, 100 µl of protoplast suspension (1.0 × 106 protoplasts/ml) was added to the same tube and gently mixed before and after adding 120 µl 25% (w/v) PEG solution. The transfection was stopped after 3 min by 5 ml wash solution. Two control experiments, one with and one without PEG solution were also conducted. For estimation of transfection efficiency, protoplasts were transfected by replacing RNP with 20 µg plasmid vector expressing Green Fluorescent Protein (GFP) (pCW498-35S-GFiP-OcsT) and incubated with transient expression solution at room temperature in darkness. After 24 h, the expression of GFP signal was detected under a confocal microscope (Zeiss LSM 880 Airyscan confocal laser scanning microscope, Oberkochen, Germany).
After transfection, protoplasts were embedded in alginate and incubated in Medium E at 25 °C in darkness for 5 d where after the light was gradually increased by replacing the aluminum foil with a white paper sheet under the light intensity at ca. 10 μE m−2 s−1. After two weeks, Medium E was replaced by Medium F and calli were exposed to full light with fresh Medium F changed every week.
Shoot and root regeneration on solid medium
After two weeks of incubation in Medium F, calli of 1–3 mm in size were released from the alginate using forceps and transferred directly to solid media for further shoot regeneration. Different solid media were designed to test their respective potential for tomato protoplast shoot regeneration. The composition of each medium is listed in Supp. Table 1. Solid shoot media were renewed every two weeks until shoots were regenerated. The number of regenerated shoots was evaluated continuously on different regeneration media until six months after protoplast isolation.
Individual regenerated shoots were excised from calli and moved to root regeneration medium containing 4.405 g/L MS medium with vitamins, 30 g/L sucrose and 6 g/L phyto agar at pH 5.8 in Plante Containers. Regenerated plants with roots were moved to soil for subsequent seed production and phenotypic observation.
Identification of SP and SP5G genes and sgRNA design
Genomic DNA was extracted from tomato leaf tissue of the four tomato cultivars using the GeneJet Plant Genomic DNA Purification Mini Kit (Thermo Fisher Scientific, Waltham USA) for amplification of the target regions in SP and SP5G genes. For each gene, two pairs of primers were designed based on the sequence of Solyc06g074350 (SP) and Solyc05g053850 (SP5G) (https://solgenomics.net/). Amplification of the target regions was conducted in a total reaction of 10 µl containing 5X Phusion HF buffer, 0.2 mM dNTPS, 0.15 µM primers, 0.02 U/µl Phusion DNA polymerase and 1 µl of extracted gDNA. PCR was conducted as follows: 98 ℃ for 1 min, 35 cycles of 98 °C 10 s, 59 °C 15 s, 72 °C 15 s and a final extension of 72 °C for 10 min. PCR products were cloned using the CloneJET PCR cloning Kit (Thermo Fisher Scientific, Waltham USA) and six random clones from each amplicon were selected for Sanger Sequencing (Eurofins). sgRNAs were designed to target all alleles in the four tomato cvs. according to the Sanger Sequencing using CRISPR RGEN Cas-Designer (Bae et al. 2014) and CRISPOR (Concordet and Haeussler 2018). All primers and sgRNAs used in this study are summarized in Supp. Table 2.
Genotyping of SP and SP5G mutants
Initial screening of mutations was performed with High Resolution Fragment Analysis (HRFA) according to Andersson et al. (2017). Genomic DNA was extracted from single young leaf tissue from each in vitro regenerated plant using GeneJet Plant Genomic DNA Purification Mini Kit. Multiplexing PCR was applied to amplify the regions covering target sites of both SP and SP5G genes simultaneously with forward primers labeled with FAM and HEX fluorescent dye (Thermo Fisher Scientific, Waltham USA), respectively. Labeled amplicons were analyzed in a 3500 Genetic Analyzer (Applied Biosystems) and the size of fragments was determined with GeneMarker Software (SoftGenetics, Pennsylvania, USA) compared with the size of wild type amplicons. Sanger Sequencing was conducted for further characterization of mutations using unlabeled primers.
Statistical analysis
To evaluate the effect of different media on shoot regeneration rate, the number of shoots on each medium was recorded until six months after protoplast isolation. On the representative media (Medium TRS-a, b and c), the mean regeneration rate of each treatment was calculated with three replicates on individual culture dishes, containing ca. 40–60 calli per replicate. For other shoot regeneration media, the regeneration rate of each treatment was calculated based on one replicate containing 50 calli. Data were analyzed with one-way ANOVA and Tukey’s test using software IBM SPSS Statistics version 27.
Results
Improvement on protoplast isolation
To improve yield, viability and regenerability of isolated protoplasts, the process was optimized in this study based on a previously published method (Nicolia et al. 2021a). The key steps of protoplast isolation are shown in Fig. 1a–d. The yield of protoplasts with initial isolation conditions (14 d seedlings, enzyme treatment: 15 °C for 17 h) from the four cultivars is shown in Fig. 2 and the optimization efforts were made on two cvs, Red Setter and Ailsa Craig, using variables, such as different seedling age, enzyme digestion temperature and duration, and preconditioning treatment.
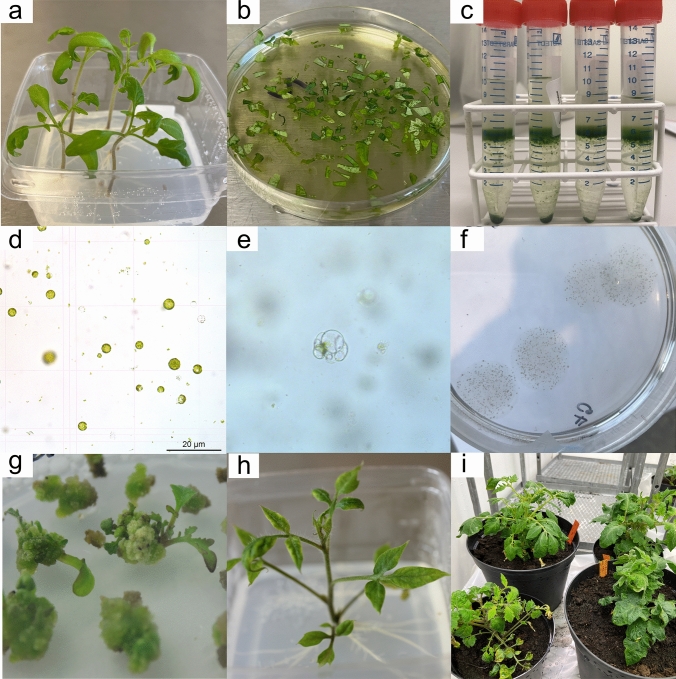
Fig. 1.
Protoplast isolation and regeneration from tomato (S. lycopersicum) cv. Red Setter. a Cotyledons and first true leaves from 21-d-old in vitro seedlings used for protoplast isolation. b Sliced cotyledons and first true leaves incubated in enzyme solution after 16 h under 25 °C before protoplast purification. c Dark green bands containing released intact protoplasts appeared at the interface of sucrose solution and wash solution after centrifugation. d Freshly isolated green protoplasts under microscope. e Cell division 5 d after protoplast isolation. f Callus formation derived from protoplasts embedded in alginate after 12 d from protoplast isolation. g Calli released from alginate and cultured on solid shoot regeneration Medium TSR-b with first regenerated shoots observed three months after protoplast isolation. h A regenerated plant with well-developed roots on root regeneration medium three months after protoplast isolation. i Regenerated plants moved to soil in biotron four months after protoplast isolation
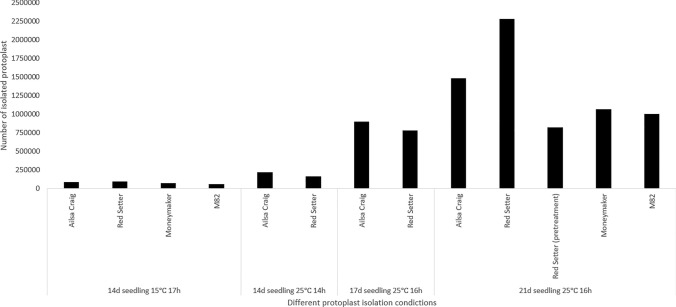
Fig. 2.
Comparison of effects of different protoplast isolation conditions on the protoplast yield from four different tomato cultivars (Red Setter, Ailsa Craig, M82 and Moneymaker). The number of isolated protoplasts was calculated from the extraction and sampling of 1 g seedlings (results are normalized)
We found that the yield of extracted protoplasts was improved using older seedlings, higher enzyme treatment temperature and longer incubation time. The number of isolated protoplasts from seedlings at the age of 21 d with the enzyme digestion at 25 °C for 16 h was 15–25 times higher than when using 14-d-old seedlings with the enzyme treatment at 15 °C for 17 h in all four tested cultivars, where a thick dark green band was formed after purification using sucrose density gradient centrifugation (Fig. 1c). An extra pretreatment step of in vitro seedlings (cv. Red Setter) before protoplast isolation did not increase the yield of protoplasts, as shown in Fig. 2.
To confirm our findings for optimized protoplast isolation, we tested two additional tomato cvs, M82 and Moneymaker, under the optimized protoplast isolation conditions. The results illustrated that the yield of protoplasts obtained from cvs M82 and Moneymaker was also improved using the new protoplast isolation conditions.
Cell division and callus formation from RNP-transfected protoplasts (week 1–4)
Freshly isolated protoplasts (Fig. 1d) were used for PEG transfection with RNP complexes or a vector harboring GFP. Expression of GFP was observed under microscope after 24 h incubation at room temperature and the estimated transfection efficiency was 30–50% (Supp. Figure 1a). RNP-transfected protoplasts were embedded in alginate and incubated at 25 °C. After 4–5-d incubation in the dark, initial cell division was observed under microscope (Fig. 1e). Light was gradually increased, and the mini-calli were usually visible to the naked eye 2 weeks after transfection (Fig. 1f).
Shoot regeneration on different solid media (week 4–10) and root formation (week 9–12)
To find the optimal solid tomato shoot regeneration (TSR) medium, different media compositions were assessed. Calli were released from alginate when the size reached 1–3 mm (usually 5 weeks after transfection) and moved to the various solid media for assessment of shoot regeneration. Media were designed to study the effect of different combinations or concentrations of plant hormones, different gelling agents and different carbon sources on shoot regeneration. The results of shoot regeneration rate from non-treated and treated protoplasts of the cv. Red Setter on three different shoot regeneration media, are summarized in Table 1 (for results with all tested media see Supp. Table 3). Shoot primordia were observed 2–12 weeks after moving to different shoot regeneration media. The highest shoot regeneration rate (from RNP-transfected protoplasts) was on Medium TSR-a (31.4%) and Medium TSR-b (21.3%), without significant difference. While there was no significant difference among the three treatments (p = 0.844), differences for the various media (p = 0.007) and the interaction between treatments and media were both significant (p < 0.001).
Table 1.
Shoot regeneration rate1 (%) on three media, TSR a-c (cv. Red Setter)
| Treatment | Regeneration rate (%) on TSR Media2 | ||
|---|---|---|---|
| TSR-a | TSR-b | TSR-c | |
| Protoplasts + PEG + RNPs | 31.4 a | 21.3ab | 18.4b |
| Protoplasts + PEG | 19.3a | 30.3a | 25.4a |
| Protoplasts | 8.1b | 44.6a | 24.5ab |
1The calculation of regeneration rate is described in detail in material and methods. Values in a row followed by the same letters were not statistically different at p = 0.05 (n = 3)
2TSR Medium a, b and c are different media for shoot regeneration and the components of each medium are shown in Supp. Table 3
When the shoots reached a length of 1–2 cm and at least two leaves had developed, they were excised from the calli (the calli was discarded after picking one shoot) and transferred to root regeneration medium. Usually well-developed roots were formed within two weeks.
Interestingly, there was morphological difference among shoots regenerated on different shoot regeneration media using cultivar Red Setter (Supp. Figure 1), which had an effect on root formation. For example, the shoots from Medium TSR-b (Fig. 1g) were green and healthy without any evident defect and usually produced well and fast developed roots (Fig. 1h) with normal rooting and acclimation in pots (Fig. 1i). On the other hand, the shoots regenerated from Medium TSR-a were curved, less green, with a grass-like shape (Supp. Figure 1b). When moved to root regeneration medium, roots developed more slowly and shoots even failed to survive. Contrasting with this, the shoots on Medium TSR-c (Supp. Figure 1c) were thicker, and faced the same rooting issue as using Medium TSR-a.
SP and SP5G allele sequencing and sgRNA design
For the determination of SP and SP5G allele gene sequences, we designed two different primer pairs for PCR amplification covering the exon 1 region of all four used cultivars (Fig. 3a). Sequence results showed that in the amplified region, they were identical to the public tomato reference genomic sequences, except for the SP gene, where one single-nucleotide polymorphism (SNP) was identified among the four cultivars (Supp. Figure 2).
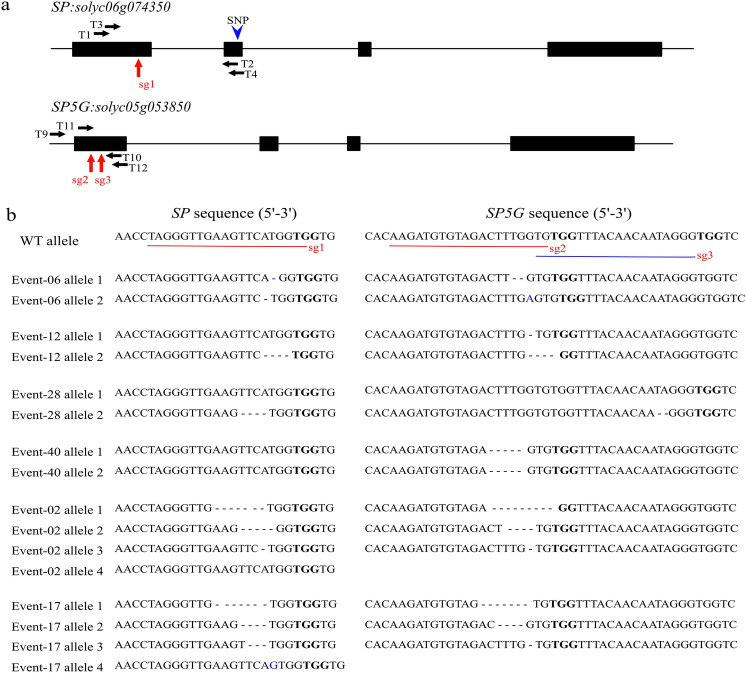
Fig. 3.
DNA-free CRISPR/Cas9 mediated genome editing in tomato multiplexing of SP and SP5G genes. a Structure of SP and SP5G genes. Exons are indicated in black boxes. Primers used for genotyping and sequencing are noted with black arrows. For each gene, sgRNAs (red arrows) were designed, all targeting exon 1. Only one SNP (blue arrow) was found within the amplification region of the SP gene. b Genotyping of first-generation events (M0) by Sanger Sequencing. The DNA sequence of each allele was aligned to wild type (WT) allele and deletions are shown with hyphens and insertions marked with blue color. Protospacer Adjacent Motif (PAM) is shown in bold
One sgRNA (sgRNA1) for SP and two sgRNAs for SP5G (sgRNA2 and sgRNA3) were designed (Fig. 3a) and used for multiplexing of the targets in two different combinations, sgRNA1 + 2 and sgRNA1 + 3.
Identification of mutants after multiplexed targeting of SP and SP5G
In total we analyzed 110 regenerated shoots (events, M0 plants) by HRFA analysis (for HRFA results on all mutants see Supp. Table 4), where SP and SP5G genes were targeted simultaneously by either sgRNA1 + 2 or sgRNA1 + 3. Among all 110 events, 66 (60.0%) were identified with mutations (indels found in at least one allele) in either SP or SP5G genes (Table 2). Of mutated events, 10 (9.1%) and 21 (19.1%) events were edited only in SP and SP5G, respectively, while the remaining were mutated in both genes. Furthermore, 34 (30.9%) events were found to be potentially chimeric from the observation that more than two allelic variants of either gene were detected during HRFA analysis. We selected 20 events for genotyping by Sanger Sequencing and the results were in line with the indel sizes identified with HRFA analysis, as can be seen in Fig. 3b and Supp. Table 4. We selected 14 representative regenerated M0 plants and five unedited regenerated plants and grew them in the biotron for further phenotypical assessment (Supp. Figure 3).
Table 2.
Mutation rate1 of regenerated events (M0) from transfected protoplasts, tomato cv. Red Setter
| total # of events analyzed | # of events with mutation2 | # of events with mutation only in SP | # of events with mutation only in SP5G | # of events with mutation in both SP and SP5G | # of events possibly chimeric3 |
|---|---|---|---|---|---|
| 110 | 66 (60.0%) | 10 (9.1%) | 21 (19.1%) | 35 (31.8%) | 34 (30.9%) |
1Mutations (indels) were determined on a single leaf from 110 regenerated plants by HRFA analysis where both SP and SP5G genes were targeted simultaneously
2Mutations in at least one allele in either SP or SP5G genes. The results of HRFA analysis of all 66 mutated events are shown in Supp. Table 4
3More than two allelic variants for either SP or SP5G detected in an event
Discussion
Genome editing has become a complementary method to traditional breeding of many crops including tomato and the CRISPR/Cas9 technology is the most utilized tool in recent years. Tomato, as an important horticultural crop with high commercial value, has already been well studied genetically, which makes the application of modern molecular breeding possible (Foolad 2007). Currently, there is no DNA-free genome editing method established for cultivated tomato, which is an important drawback when utilizing this important technology for breeding or in research. A DNA-free genome editing method requires efficient and reproducible shoot regeneration from single cells, which is still a challenge and can be highly genotype dependent (Peres et al. 2001). A recent study reported protoplast regeneration via a DNA-free method on wild tomato, which is the closest study so far to cultivated tomato (Lin et al. 2022). Based on a previously published protocol for cultivated tomato protoplast genome editing via RNP-based CRISPR/Cas9 (Nicolia et al. 2021a), we have improved the protoplast isolation process and solved the challenge of shoot regeneration from RNP-transfected protoplasts.
The yield and quality of isolated protoplasts further affect shoot regeneration. Here we have optimized the process of protoplast isolation based on seedling age and enzyme digestion temperature and duration. With the optimized conditions (21 d seedlings, enzyme treatment: 25 °C for 16 h), we successfully increased the yield of protoplasts to 1.0–2.2 × 106 per gram of leaf materials, which were comparable results to previous reports on tomato (Morgan and Cocking 1982; Niedz et al. 1985; Tan et al. 1987). We also found in this study that preconditioning treatment of donor plants under 4 °C prior to protoplast isolation had a negative effect on protoplast yield. By contrast, Tan et al. (1987) got the opposite result from preconditioning treatment, where they found that cold treatment increased the stability of protoplasts and thus yielded more viable protoplasts.
The formation of callus from transfected protoplasts is achieved by stimulation of cell wall development and cell divisions. Moreover, there are many factors that can affect the success of shoot regeneration, such as osmotic pressure, different types and concentration of hormones, carbon sources and gelling reagents. In this study, shoot regeneration was analyzed on ten different shoot regeneration media, but with extra focus on three of them. Cytokinins, such as zeatin and 6-Benzylaminopurine (6-BAP), are involved in early cell division as well as initiation and elongation of shoots, and are widely used in protoplast-derived shoot regeneration in many plant species. Previous studies showed that zeatin was necessary for tomato shoot regeneration (Morgan and Cocking 1982) and we found that the 0.1 mg/L IAA and 0.75 mg/L zeatin was the most suitable combination for shoot regeneration in all ten media tested with cv. Red Setter. On the other hand, when the calli were cultured on 6-BAP-based media together with IAA or 1-Naphthaleneacetic acid (NAA) (Medium TSR-d and TSR-e) (Supp. Figure 1d, e), the browning of calli seemed to accelerate or smaller calli were generated and no shoots were regenerated after six months. Gibberellins such as Gibberellic acid (GA3) have been proven to be beneficial for stimulating shoot elongation (Niedz et al. 1985). Shahin (1985) observed higher regeneration rate when using both zeatin and GA3 compared with using zeatin alone. In contrast, we found that when GA3 was added in early shoot induction process, it had an adverse effect on shoot morphology which was curved, thin and grass-like as observed from most of the shoots regenerated from Medium TSR-a, TSR-f and TSR-g (Supp. Figure 1b, f and g) where GA3 concentration varied from 0.34 mg/L to 1 mg/L. Auxins are also an essential component in shoot regeneration medium, such as the frequently used IAA and NAA. We found that when IAA was replaced by NAA, the calli on Medium TSR-h were inflated, less green and not able to generate shoots (Supp Fig. 1h), which did not concur with the conclusions from Niedz et al. (1985). We did not find an obvious difference between two carbon sources and gelling agent in this study.
In this study, four different tomato cvs Red Setter, Ailsa Craig, M82 and Moneymaker were used to study protoplast regeneration from RNP-transfected protoplasts. Cultivar Red Setter was superior to other cvs with a regeneration rate up to 31.4% from RNP-transfected protoplasts. Five shoots were obtained from 200 RNP-transfected protoplast-derived calli from cv. M82, with a high mutation rate (80%), although all four mutant regenerated events were chimeras (Supp. Figure 4). On the contrary, plating efficiency was low on both cvs Ailsa Craig and Moneymaker and all attempts to regenerate shoots from RNP-transfected protoplasts failed, indicating that a further adaptation of the protocol will be required for these cultivars. In this study, we clearly observed genotype differences among the four tested cultivars, which was in line with previous reports where variable regeneration rate among cultivars was found (Morgan and Cocking 1982; Niedz et al. 1985; Tan et al. 1987).
The mutations identified in M0 events were a mix of bi-allelic, mono-allelic and chimeric with small deletions or insertions at the target site with an editing efficiency up to 60% in considering at least one allele mutated in either SP or SP5G genes and 31.8% considering on both genes simultaneously targeted. Previously, 30 and 90% of protoplast-derived calli were found to be mutated in at least one allele of CCD7 or CCD8 genes, respectively, after multiplex RNP delivery (Nicolia et al. 2021a). Such results indicate that the CRISPR/Cas9 system using RNP-transfected protoplasts can be highly efficient to generate desired mutations in cultivated tomato, without any stable integration of foreign DNA. Brooks et al. (2014) were the first to report the successful application of the CRISPR/Cas9 system on tomato via stable Agrobacterium tumefaciens-mediated transformation with an editing efficiency of 48% on T0 plants with two sgRNAs targeting at the same gene. More studies using CRISPR/Cas9 via Agrobacterium tumefaciens-mediated transformation have been published in recent years and some of these studies had a very high editing efficiency of up to 100% of the transgenic shoots (Ito et al. 2015; Ueta et al. 2017; Dahan-Meir et al. 2018). With the latter method, however, comes the use of antibiotic selection as well as either selfing or backcrossing to remove T-DNA insertions if a transgene-free plant is desirable.
We observed a higher rate of potential chimeras in tomato than for example, potato, using RNP complexes and similar protoplast density (Andersson et al. 2018). This might be because the ratio between RNP complexes and protoplasts was not optimal in our study, as Sidorov et al. (2021) also observed high number of chimeras (33.3%) on regenerated calli from RNP-transfected protoplasts in canola (Brassica napus L.). It might be possible to address it by testing higher concentrations of RNPs as the efficiency of RNP is dose dependent (Zhang et al. 2022). Another possible reason might be endopolyploidy according to a report by Smulders et al. (1994) on varying ploidy level in different tomato somatic tissues. A high chimeras using CRISPR/Cas9 on tomato was also identified in earlier studies on Agrobacterium tumefaciens-mediated transformation (Brooks et al. 2014; Dahan-Meir et al. 2018). However, a high frequency of chimeric events is less important in tomato than in clonally propagated crops, due to sexual generation and the possibility of selecting and producing homozygous mutations in the next generation. Moreover, the use of protoplasts allows to scale-up the experiments of mutagenesis easily, so that among a high number of regenerated mutant M0 plants those indicating chimerism can be discarded.
Our findings illustrate that the challenge of shoot regeneration from transfected protoplasts of cultivated tomato has been overcome and we have successfully obtained regenerated plants from non-treated protoplasts from all four studied cultivars, as well as regenerated plants with induced mutations in two cultivars (Red Setter and M82) via DNA-free CRISPR/Cas9. The availability of this reported method in the determinate cvs Red Setter or M82 provides opportunities for important research and breeding efforts oriented toward tomato field cultivation and industrial processing, such as improving the fruit quality (e.g., flavor, sugar content, acidity) and plant resistance to biotic (e.g., soil born, virus, parasitic plants) and abiotic stress (e.g., water deficiency, salinity).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Mirela Beganovic and Marina Kuzmenkova for technical support in tissue culture; Martin Friberg for help with collecting seeds in biotron; Sjur Sandgrind for help with the confocal microscope; Rui Guan for suggestions in shoot regeneration; and Yuzhou Lan for support in Statistical analysis.
Author contributions
YL, MA, TC, PH and AN conceived the project and contributed to the study conception. YL designed and conducted the experiment and wrote the first draft of the manuscript and all authors commented on previous versions of the manuscript. AG contributed with design of shoot regeneration Medium TSR-b. All authors read and approved the manuscript.
Funding
Open access funding provided by Swedish University of Agricultural Sciences. This study is supported by a grant from Nilsson-Ehle-donationerna.
Data availability
All the data in this study are included in this manuscript and supplementary data file.
Declarations
Conflict of interest
The authors declare non-financial interests that are directly or indirectly related to the work submitted for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andersson M, Turesson H, Nicolia A, et al. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017;36:117–128. doi: 10.1007/s00299-016-2062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Turesson H, Olsson N, et al. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant. 2018;164:378–384. doi: 10.1111/ppl.12731. [DOI] [PubMed] [Google Scholar]
- Bae S, Kweon J, Kim HS, Kim J. Microhomology-based choice of Cas9 nuclease target sites. Nat Publ Gr. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lipppman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel-Goren L, Liu YS, Lifschitz E, Zamir D. The self-pruning gene family in tomato. Plant Mol Biol. 2003;52:1215–1222. doi: 10.1023/B:PLAN.0000004333.96451.11. [DOI] [PubMed] [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2016;15:257–268. doi: 10.1111/pbi.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, et al. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Concordet J, Haeussler M. CRISPOR : intuitive guide selection for CRISPR / Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:242–245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, et al. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018;95:5–16. doi: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- Foolad MR. (2007) Genome mapping and molecular breeding of tomato. Int J Plant Genomics. 2007;1:52. doi: 10.1155/2007/64358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C. Genome engineering for crop improvement and future agriculture. Cell. 2021;184:1621–1635. doi: 10.1016/j.cell.2021.01.005. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, et al. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467:76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:1–12. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CT, Heo J, Lemmon ZH, et al. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat Biotechnol. 2020;38:182–188. doi: 10.1038/s41587-019-0361-2. [DOI] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, et al. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol. 2018 doi: 10.1038/nbt.4273. [DOI] [PubMed] [Google Scholar]
- Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41:63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:1–5. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-S, Hsu C-T, Yuan Y-H, et al. DNA-free CRISPR-Cas9 gene editing of wild tetraploid tomato Solanum peruvianum using protoplast regeneration. Plant Physiol. 2022 doi: 10.1093/plphys/kiac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A, Cocking EC. Plant Regeneration from protoplasts of Lycopersicon esculentum Mill. Z Pflanzenphysiol. 1982;106:97–104. doi: 10.1016/s0044-328x(82)80071-8. [DOI] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, et al. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- Nicolia A, Andersson M, Hofvander P, et al. Tomato protoplasts as cell target for ribonucleoprotein ( RNP )- mediated multiplexed genome editing. Plant Cell Tissue Organ Cult. 2021;144:463–467. doi: 10.1007/s11240-020-01954-8. [DOI] [Google Scholar]
- Nicolia A, Fält A-S, Hofvander P, Andersson M. Protoplast-Based Method for Genome Editing in Tetraploid Potato Methods in Molecular Biology. New york: Springer; 2021. pp. 177–186. [DOI] [PubMed] [Google Scholar]
- Niedz RP, Rutter SM, Handley LW, Sink KC. Plant regeneration from leaf protoplasts of six tomato cultivars. Plant Sci. 1985;39:199–204. doi: 10.1016/0168-9452(85)90175-X. [DOI] [Google Scholar]
- Pan C, Ye L, Qin L, et al. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep. 2016;6:1–10. doi: 10.1038/srep24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres LEP, Morgante PG, Vecchi C, et al. Shoot regeneration capacity from roots and transgenic hairy roots of tomato cultivars and wild related species. Plant Cell Tissue Organ Cult. 2001;65:37–44. doi: 10.1023/A:1010631731559. [DOI] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Sakata Y, NISHIO T, TAKAYANAGI K, Plant regeneration from mesophyll protoplasts of tomato cv. Ponderosa J Jpn Soc Hortic Sci. 1987;56:334–338. doi: 10.2503/jjshs.56.334. [DOI] [Google Scholar]
- Shahin EA. Totipotency of tomato protoplasts. Theor Appl Genet. 1985;69:235–240. doi: 10.1007/BF00662431. [DOI] [PubMed] [Google Scholar]
- Sidorov V, Wang D, Nagy ED, et al. Heritable DNA-free genome editing of canola (Brassica napus L) using PEG-mediated transfection of isolated protoplasts. Vitr Cell Dev Biol Plant. 2021 doi: 10.1007/s11627-021-10236-7. [DOI] [Google Scholar]
- Smulders MJM, Rus-Kortekaas W, Gilissen LJW. Development of polysomaty during differentiation in diploid and tetraploid tomato (Lycopersicon esculentum) plants. Plant Sci. 1994;97:53–60. doi: 10.1016/0168-9452(94)90107-4. [DOI] [Google Scholar]
- Soyk S, Müller NA, Park SJ, et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49:162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, et al. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:1–7. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MMC, Rietveld EM, et al. Regeneration of leaf mesophyll protoplasts of tomato cultivars (L. esculentum): factors important for efficient protoplast culture and plant regeneration. Plant Cell Rep. 1987;6:172–175. doi: 10.1007/BF00268470. [DOI] [PubMed] [Google Scholar]
- Ueta R, Abe C, Watanabe T, et al. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep. 2017;7:1–8. doi: 10.1038/s41598-017-00501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-Guided genome editing for target gene mutations in wheat. G3 Genes. Genomes Genet. 2013;3:2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang S, Wang W, et al. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015;34:1473–1476. doi: 10.1007/s00299-015-1816-7. [DOI] [PubMed] [Google Scholar]
- Woo JW, Kim J, Il KS, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1165. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, et al. High-Efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant. 2015;8:1820–1823. doi: 10.1016/j.molp.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:1–8. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Fang H, et al. Highly efficient genome editing in plant protoplasts by ribonucleoprotein delivery of CRISPR-Cas12a nucleases. Front Genome Ed. 2022;4:1–11. doi: 10.3389/fgeed.2022.780238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, et al. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data in this study are included in this manuscript and supplementary data file.