Abstract
Chlamydia was the only genus in the order Chlamydiales until the recent characterization of Simkania negevensis ZT and Parachlamydia acanthamoebae strains. The present study of Chlamydiales 23S ribosomal DNA (rDNA) focuses on a naturally occurring group I intron in the I-CpaI target site of 23S rDNA from S. negevensis. The intron, SnLSU · 1, belonged to the IB4 structural subgroup and was most closely related to large ribosomal subunit introns that express single-motif, LAGLIDADG endonucleases in chloroplasts of algae and in mitochondria of amoebae. RT-PCR and electrophoresis of in vivo rRNA indicated that the intron was not spliced out of the 23S rRNA. The unspliced 658-nt intron is the first group I intron to be found in bacterial rDNA or rRNA, and it may delay the S. negevensis developmental replication cycle by affecting ribosomal function.
Group I introns are mobile genetic elements that have not previously been found in bacterial ribosomal DNA (rDNA), despite the fact that nearly all bacterial 23S rDNAs have conserved target sequences for the intron-encoded homing endonucleases I-CeuI (1, 35) and I-CpaI (1, 49, 50). Upon intron entry into cells, homing endonucleases are expressed and mediate intron insertion into host DNA by cleaving intronless target sites (5). Group I 23S rDNA introns are widespread in algal chloroplasts (49), which are thought to be derived from bacterial ancestors (21). The 23S rDNA group I introns are also found in the apparently bacterially derived mitochondria (32) of an amoeba and other lower eukaryotes (22, 24, 36). They are present in the nuclear rDNA of lower eukaryotes and archaea (22, 24, 37, 38). It is not known why bacteria lack rDNA introns.
Some bacterial ribosomal genes encode intervening segments (IVSs) that are approximately the size of small introns. These are excised from the rRNA by ribonucleases, leaving fragmented but functional rRNA. IVSs are found in highly variable regions, not in functionally essential domains (23). In contrast, group I introns are located in functionally vital loci and must be removed from transcripts by autocatalytic splicing or by splicing that is facilitated by maturase protein (7, 12, 39). Splicing occurs coordinately with ligation of the RNA exons (12, 39). The intron transcript folds to form a catalytic core for carrying out the splicing and ligation. The core structure can be predicted by RNA folding analysis (24), and 10 complementary domains with specific roles in core formation, P1 to P10, have been deduced by sequence similarity, covariance of distant positions, and stereochemical modeling (39). Neither sequence nor folding analysis, however, predicts whether group I splicing will be autocatalytic or maturase facilitated. Autocatalysis can be tested by an in vitro assay (26). Maturases must be encoded by the introns themselves or supplied endogenously or exogenously.
Although the 16S small ribosomal subunit (SSU) genes of species belonging to the bacterial order Chlamydiales are well characterized, chlamydial 23S rRNA large ribosomal subunit (LSU) genes have undergone only partial and limited scrutiny (17, 47). Chlamydiae are obligately intracellular bacteria that replicate only within endocytic vacuoles of eukaryotic cells. Four families of chlamydiae are known to parasitize vertebrates or have been associated with vertebrates (8, 17, 18, 29, 34, 40, 45), and those strains belonging to Parachlamydia acanthamoebae also live in amoebae (2, 3). In a comprehensive analysis of chlamydial 23S rRNA, a group I intron was identified in Simkania negevensis ZT, a Chlamydiales strain for which the natural eukaryotic host is not known. This is the first bacterium that has been found to have a 23S rRNA group I intron. The intron is characterized in this study.
MATERIALS AND METHODS
Bacteria and cell culture.
S. negevensis ZT (ATCC VR-1471) (18, 28) was grown at 37°C in confluent monolayers of cultured Vero cells (ATCC CCL81; American Type Culture Collection, Rockville, Md.) in RPMI containing 15% fetal bovine serum, 1% glucose, 10 μg of ampicillin/ml, 100 μg of gentamicin/ml, 160 μg of vancomycin/ml, and 1 μg of cycloheximide/ml. Chlamydia trachomatis L2/434/BU was similarly grown in Vero cells, but without ampicillin.
Sequence analyses.
Double-stranded sequence data for Chlamydiaceae 23S rRNA genes and for the S. negevensis 23S rRNA gene were obtained by direct PCR product sequencing at the Iowa State University DNA Sequencing and Synthesis Facility, Ames. Sequences were assembled with Sequencher data analysis software (Gene Codes, Ann Arbor, Mich.).
Sequence analysis programs, described elsewhere (20), were used to compare 23S rDNA sequences of Simkania, Parachlamydia, and Chlamydiaceae spp. to identify the SnLSU · 1 insertion site and to identify an open reading frame (ORF). These programs were also used to identify a hairpin structure at position 1931 and the probable start and stop sites of the S. negevensis 23S rRNA gene. The programs used included Reformat, Assemble, PileUp, LineUp, FoldRNA, and Squiggles. The homology of the intron and intron ORF with other genes was determined by BLASTN, BLASTP, and TFASTA searching of the GenBank, PIR, and SWISSPROT databases.
ORF analysis.
Endonuclease homologs were aligned with I-CreI by using alignments by Turmel et al. (51) and structural analysis by Heath et al. (25). Phylogenetic analysis of the EndA gene sequence from SnLSU · 1 was carried out using the tree bisection reconnection option of the maximum parsimony routine of PAUP version 3.1 (48). Input order was randomized 10 times, and 1,000 bootstrap replicates were run to provide statistical support for branching order (19). A saturation plot of the phyletic distance versus the percent pairwise distance between isolates was constructed to determine whether homoplasy interfered with the analysis (52).
Intron structure.
Intron secondary structure was inferred with the comparative sequence analysis program AE2, and the secondary structure diagrams were drawn with the program XRNA (16). The SnLSU · 1 structure diagram was altered by hand to enhance readability.
RNA and DNA preparation.
RNA and DNA were prepared from S. negevensis and from two negative controls, C. trachomatis and uninfected Vero cells. Numerous S. negevensis RNA preparations from replicating reticulate bodies (RBs) and also from metabolically inactive elementary bodies (EBs) were harvested at many time points in the 2 to 12 days postinfection. Standard methods were used to isolate chlamydiae (11): infected preparations were removed from flasks by using glass beads, mildly sonicated, and centrifuged in Renografin gradients (Solvay Animal Health Inc., Mendota Heights, Minn.). C. trachomatis was harvested 2 to 3 days postinfection. EB and RB preparations were standardized at 1 μg/μl of protein before extraction of nucleic acids. RNA and DNA were separately extracted with the TriReagent RNA-DNA-protein isolation kit TR-118 (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer’s recommendations. RNA was also extracted by a second method, using the SV total RNA isolation system Z3101 and DNase (Promega, Madison, Wis.).
RNA analysis and in vitro autocatalysis.
RNAs prepared with TriReagent from S. negevensis RBs, C. trachomatis RBs, and Vero cells were electrophoresed under denaturing conditions with formaldehyde in 1.5% agarose gels (46). C. trachomatis RNA provided an intron-negative control. Vero cell RNA provided a host cell control. Each loaded sample contained 10 μmol of ethidium bromide. Electrophoresis of RNA from different preparations was repeated several times, with consistent results each time. RNA markers (G3191; Promega) were used.
In vitro autocatalytic intron splicing was attempted with RNAs prepared with TriReagent from S. negevensis RBs, under conditions that have previously been used for in vitro autocatalysis and splicing (26). Equal volumes of S. negevensis RNA in water and 200 mM MgCl2 were separately incubated at 95°C for 2 min and then were mixed together and allowed to cool to 37°C. The preparation was brought to a final concentration of 60 mM MgCl2, 100 mM GTP, 100 mM KCl, and 50 mM Tris, pH 7.5 and was incubated for 1 h at 37°C. Both treated and untreated RNAs were used as templates for reverse transcription (RT)-PCR.
RT-PCR, PCR, and electrophoresis.
RT-PCR with Tth DNA polymerase was performed in 1× RT-PCR buffer (Boehringer Mannheim, Indianapolis, Ind.) according to the instructions of the manufacturer. Amplification of the intron from 23S rRNA or ribosomal DNA (rDNA) was done with primer AF (5′ CACAGGTAGGCATGATGA 3′), which matched the 23S rDNA 319 bases upstream of the intron, and primer BR (5′ CTAGCTGCGGGTAAACG 3′), which complemented the 23S rDNA 122 bases downstream of the intron. The primers perfectly matched the S. negevensis rRNA, but primer AF had three mismatches with C. trachomatis and primer BR had five mismatches with C. trachomatis. RT was carried out in 1 cycle of 30 min at 60°C and 60 s at 94°C followed by PCR cycling: 10 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C; 20 cycles of 30 s at 94°C, 35 s at 55°C, and 100 s at 72°C; and finally a 7-min cycle at 72°C. RT-PCR amplification of 338 nucleotides (nt) of the SnLSU · 1 intron alone was carried out under these conditions, using primers INTF (5′ TTAGATGCACAATGGATAGTTGGA 3′) and INTR (5′ CCATCAGCGCTCATGTGCTCA 3′).
PCR amplification was performed with Taq DNA polymerase (Takara Shuzo Co., Ltd., Kyoto, Japan) according to instructions of the manufacturer. The PCR amplification conditions were 1 cycle for 6 min at 94°C; 30 cycles of 60 s at 94°C, 60 s at 53°C, and 60 s at 73°C; and one cycle of 60 s at 94°C, 60 s at 53°C, and 10 min at 73°C. PCR and RT-PCR products were electrophoresed on 1% agarose gels in Tris acetate-EDTA buffer and stained with ethidium bromide. Electrophoresis of products obtained from different preparations was repeated many times, with consistent results each time. The AmpliSize DNA size standard (170-8200; Bio-Rad, Hercules, Calif.) was used.
Nucleotide sequence accession number.
The GenBank accession number of the S. negevensis ribosomal operon, including EndA, is U68460.
RESULTS AND DISCUSSION
Sequence and ORF analysis.
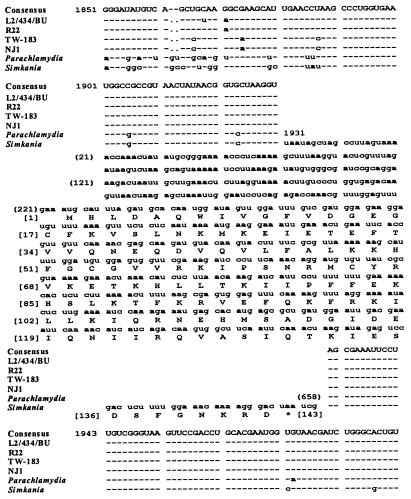
A 23S rDNA group I intron in S. negevensis Z was initially identified in a sequence survey of 23S rDNAs from species belonging to the order Chlamydiales. In this survey, all 10 Chlamydiaceae strains examined were found to have a base change at position 1923 (Escherichia coli numbering [44]) (Fig. 1) compared to nearly all other bacteria, including Simkania and Parachlamydia. This is the target intron insertion site for the homing endonuclease I-CeuI (5). I-CeuI is encoded by group I intron CeLSU · 5 in the 23S rRNA in chloroplasts of Chlamydomonas eugametos algae. Comparably positioned I-CeuI introns are found in eight other species of algae (51). The base difference in the Chlamydiaceae strains would make them naturally resistant to cleavage by I-CeuI (38).
FIG. 1.
23S rRNA, bases 1851 to 1992, from several Chlamydiales strains (E. coli numbering). In S. negevensis ZT, a group I intron was located between positions 1930 and 1931. The predicted intron-encoded endonuclease, EndA, is shown. The full-length 23S rRNA gene of Chlamydiaceae strains L2/434/BU, R22, MoPn, TW-183, 6BC, NJ1, FP Baker, EBA, IPA, and GPIC were sequenced.
S. negevensis ZT, which belongs to family II, the Simkaniaceae (18), in the order Chlamydiales, had a 658-base insertion at position 1931 in the 23S rDNA (Fig. 1) compared to other bacteria. Position 1931 is the target site for homing endonuclease I-CpaI (5), which is encoded by the 23S rRNA group I intron CpLSU · 2 in chloroplasts of Chlamydomonas pallidostigmatica algae (50). Comparably positioned introns are found in other species of algae (24) and in the mitochondria of Acanthamoeba castellanii (36). The S. negevensis insertion contained a 432-bp ORF (Fig. 1). A low-molecular-weight [35S]methionine-labeled product could be produced by in vitro transcription-translation of a PCR product that contained the full-length insert (not shown). Because it was not known if the product of the S. negevensis ORF was an active endonuclease, it could not be given a formal name and was designated EndA (5, 9, 30, 31).
EndA had 43% deduced amino acid identity with I-CpaI. EndA had 41% identity with YMF46, the I-CpaI homolog encoded by AcLSU · m1 in position 1931 of A. castellanii mitochondrial 23S rDNA. These homologs are both single-motif LAGLIDADG homing endonuclease sequences. Comparison of EndA with several LAGLIDADG endonucleases, including I-CreI, which targets 23S rRNA position 2593 and for which the crystal structure is known, showed conservation of many structural features (Fig. 2). EndA had an apparent LAGLIDADG catalytic domain, a conserved glutamine in position 47 which is associated with Mg2+ binding, functionally similar amino acids in four antiparallel β-sheet DNA-binding domains, conservation within the four overlying α-helical segments, functionally similar amino acids in the turn segments, and conserved sequence deletions. A six-residue deletion in position 32 of EndA, in the turn segment linking β1 and β2 DNA-binding domains, was located well away from the α1 catalytic site. EndA could thus be predicted to have possible LAGLIDADG endonuclease activity and functional similarities with I-CpaI and YMF46 for recognition and cleavage of the 1931 target.
FIG. 2.
Structural and sequence comparison of single-motif LAGLIDADG homing endonucleases. The proteins were aligned with I-CreI, for which the secondary structure is known (25), with alignments by Turmel et al. (51). I-CreI numbering has been used. The LAGLIDADG catalytic domain is underlined; boldface, β-pleated sheet (binds the DNA); italics, α helices overlying the β-sheet; T, turn structure; •, stop codon; *, acidic residue required for catalytic activity of I-CeuI; Mg++, magnesium binding required for catalytic activity of I-CeuI; †, possible substrate recognition site in I-CeuI. GenBank accession numbers: I-CpaI, L36830; YMF46, U12386 and U03732 (the AcLSU · ml ORF in both A. castellanii SGC6 and A. castellanii Neff mitochondria) (10); CmeLSU · 1 is from reference 51; I-CeuI, Z17234 (a partial sequence is shown, beginning with residue 47); PaND3 · 1, X14485 (the first half of the double-motif endonuclease in the ND3 gene of the mitochondrion in the fungus Podospora anserina is shown); SsSSU · 1, U07553 (the first half of the double-motif endonuclease in the SSU rRNA of the mitochondrion in the fungus Sclerotinia sclerotiorum is shown); I-CreI sequence and structure are according to reference 25, but viewed as looking down on homodimers bound to the DNA. β-Sheets are in direct contact with the DNA, and α-helices form the catalytic domain and overlying structure.
Both single-motif and bifunctional LAGLIDADG endonucleases have been identified in the LSU rDNA. I-CeuI, I-CpaI, YMF46, I-CreI, and EndA are single-motif endonuclease sequences, each having only one LAGLIDADG domain (5). Single-motif LAGLIDADG endonucleases are expressed only by LSU introns (Table 1). It is thought that the single motif may have been ancestral to nucleases with more complex LAGLIDADG motifs (5).
TABLE 1.
Introns encoding single-motif, LAGLIDADG endonucleases
| LSU target | Intron | ORF | Host organism | Splicing subgroup |
|---|---|---|---|---|
| 1923 | CeLSu · 5 | I-CeuI | Algal chloroplast | IB4 |
| 1931 | AcLSU · m1 | YMF46a | Amoeba mitochondrion | IB4 |
| 1931 | CmeLSU · 1 | a | Algal chloroplast | IA1 |
| 1931 | CpLSU · 2 | I-CpaI | Algal chloroplast | IB4 |
| 1931 | SnLSu · 1 | EndAa | S. negevensis ZT | IB4 |
| 1951 | AcLSU · m2 | a | Amoeba mitochondrion | IA3 |
| 2593 | CrLSU · 1 | I-CreI | Algal chloroplast | IA3 |
Endonuclease activity has not been demonstrated in vitro.
Ruling out an IVS.
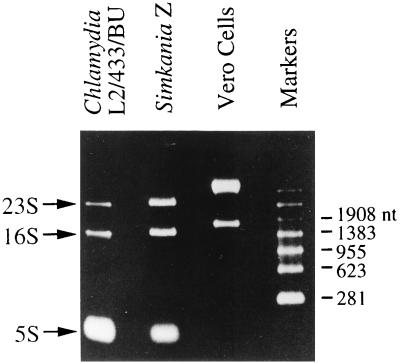
To examine whether EndA was encoded by a group I intron, it was first essential to determine whether the 658-bp insert could be an IVS. IVSs are common in bacterial rDNA and are easily identified because they are typically located in nonvital loci (23) and are excised from rRNA in vivo without ligation, producing fragmented rRNA. To rule out IVS excision and RNA fragmentation, in vivo S. negevensis rRNA was isolated and subjected to denaturing electrophoresis (Fig. 3). In the event of IVS excision, the 3,600-nt S. negevensis 23S rRNA would be cleaved into 2,000-, 1,000-, and 658-nt fragments. When electrophoresis of S. negevensis rRNA was carried out, these sizes were not evident (Fig. 3). In both S. negevensis and C. trachomatis, only intact 23S, 16S, and 5S rRNAs were observed (Fig. 3). The S. negevensis rRNAs appeared to be slightly larger than the 1,558- and 2,981-nt C. trachomatis 16S and 23S rRNAs. There was no evidence of contamination of these rRNAs with rRNA from the Vero cell line that was the in vitro host cell for both S. negevensis and C. trachomatis. Because the 23S rRNA was not fragmented, it was concluded that the S. negevensis 23S rDNA insert was not excised without ligation in vivo and was therefore not a bacterial IVS.
FIG. 3.
Purified rRNA from C. trachomatis, S. negevensis, and host Vero cells. The sizes of Vero cell rRNAs were consistent with the known sizes of human 18S and 28S rRNAs (1,869 and 5,025 nt, respectively; GenBank accession no. X03205 and M11167).
RNA folding analysis.
Target and sequence analysis suggested that the S. negevensis 23S rDNA insertion was an I-CpaI-like intron (Fig. 1 and 2). Eleven different group I intron structural subgroups have been identified by target sequence, sequence homology, and RNA folding pattern (39). The S. negevensis 23S rDNA-insert sequence was examined by folding analysis to determine whether it belonged to any known subgroup. Identifiable P6′ and P7 domains were present that could form hydrogen bonds with portions of the 658-nt segment to produce P8, P3, and P9 domains (Fig. 4A). The 5′ end of the intron formed a hairpin and flawless splicing recognition site with the exon. Thus, the essential nucleotide sequence requirements for intron cleavage and transesterification were present in the S. negevensis insert. The predicted folding similarity of the S. negevensis insert, CpLSU · 2, and AcLSU · m1 (Fig. 4), indicated that all three belong to the IB4 structural subgroup of group I introns. Therefore, the S. negevensis segment was given an appropriate intron designation, SnLSU · 1. Folding analysis of SnLSU · 1 also indicated that the entire 3′ end of the IB4 splicing apparatus was the C-terminal 30% of the EndA gene (Fig. 4A). Such a large functional overlap has not previously been described in group I introns.
FIG. 4.
Folding analysis of the three position-1931 LSU group I introns belonging to structural subgroup IB4. The large arrows near the 5′ and 3′ ends of the introns indicate predicted splice sites. Single-motif endonuclease ORFs were located in the loops marked 485, 494, and 454 nt, but the SnLSU · 1 ORF extended out of the loop to the 3′ end of the intron (boldface).
Phyletic and functional relatedness.
The folding and splicing domains and the endonuclease-encoding domain of group I introns are generally regarded as having evolutionarily distinct origins, one deriving from ribozymes and the other from protein-coding genes (6, 39). The 5′ ribozyme domain of SnLSU · 1 had little homology with other sequences, including bacterial tRNA group I introns, which belong to the IC3 folding subgroup. Phyletic analysis of the deduced EndA protein showed that it was related to endonucleases in group I LSU introns (Fig. 5). EndA clustered with the algal chloroplast intron, I-CpaI. Algal chloroplasts are the largest currently known reservoir of 23S rRNA group I introns and so may at one time have provided the ancestral SnLSU · 1 intron. Because, to a significant extent, the endonuclease and ribozyme in S. negevensis are a single genetic element, an ancient duplication of an SnLSU · 1-like element in algae may have led to the separate and diverse group I ribozyme and coding elements in algae.
FIG. 5.
Phylogeny constructed by maximum parsimony analysis of the endonuclease DNA sequence in SnLSU · 1 and related endonuclease genes (48) (from Fig. 2). The percent confidence in each node was determined with 1,000 bootstrap replicates, and the consistency index was 0.90. The linearity of the saturation plot (inset) suggested that long branch attraction did not adversely affect the resolution of this phylogeny, despite these sequences being only distantly related to each other. +, points that compare S. negevensis to one of the other genes.
Absence of splicing.
The ribozyme and coding overlap raised the question of whether splicing and translation might conflict at the 3′ end of the intron. Splicing is either autocatalytic or maturase facilitated, but folding analysis does not predict which. Most IB4 introns have so far been shown not to be autocatalytic (14, 26) and autocatalysis of neither CpLSU · 2 nor AcLSU · m1 has been reported. S. negevensis SnLSU · 1 encoded only a single-motif endonuclease and so did not supply a maturase.
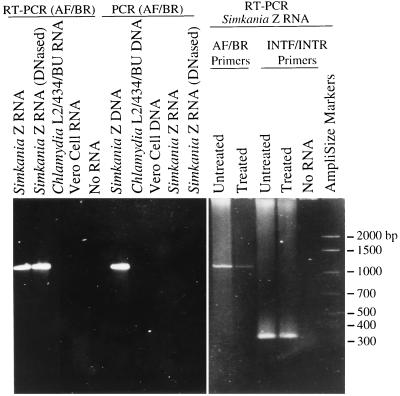
To determine whether SnLSU · 1 was spliced out and the exons ligated in vivo, RT-PCR of S. negevensis 23S rRNA was carried out with purified in vivo RNA (as seen in Fig. 3) and also with in vivo RNA that had first been subjected to in vitro autocatalytic conditions (26) (Fig. 6). RT-PCR primers AF and BR, which matched and complemented the 23S rRNA gene, would generate a 441-bp PCR product if the intron were removed and the exons ligated, a 1,099-bp product if the intron were not spliced out, both PCR product sizes if spliced and unspliced RNAs were present, or no product if the intron had been removed by excision without repair. RT-PCR of rRNA with primers AF and BR produced only 1,100-bp amplicons (Fig. 6). This result was reproducible whether the S. negevensis rRNA had been purified from early, middle, or late stages of the S. negevensis developmental cycle (not shown). DNA-dependent PCR amplification with rRNA preparations (after treatment of template with RNase or without prior RT) reproducibly yielded no PCR product (Fig. 6). There was no amplification of Vero cell or C. trachomatis template. RT-PCR of purified S. negevensis rRNA that was exposed to autocatalytic splicing conditions produced only 1,100-bp amplicons with primers AF and BR.
FIG. 6.
SnLSU · 1 amplification from S. negevensis, C. trachomatis, and Vero cell rRNAs. Amplification primers AF and BR were used for all lanes except the three INTF/INTR lanes. AF and BR would amplify a 441-bp PCR product from intronless rRNA and a 1,099-bp PCR product from intron-containing rRNA. INTF and INTR matched the intron, amplifying a 338-bp intron-only segment. All rRNA templates were used directly except for “treated” S. negevensis template, which was subjected to autocatalytic splicing conditions prior to amplification.
RT-PCR of purified S. negevensis rRNA with intron-specific primers, INTF and INTR, produced only 338-bp amplicons (Fig. 6). The ratio of detectable AF-BR to INTF-INTR product was reduced after exposure of S. negevensis RNA to autocatalytic conditions, indicating that limited nucleolytic attack had occurred under these conditions. This was consistent with either incomplete autocatalysis (cleavage at only one end of the intron) or with template degradation under the autocatalytic test conditions.
RT-PCR can amplify short amplicons in preference to long amplicons and is therefore sensitive to the presence of very small amounts of spliced rRNA. The amplicon sizes produced with RT-PCR from purified in vivo RNA and from RNA subjected to autocatalytic conditions indicated that the intron was not spliced out of the 23S rRNA. Autocatalytic splicing capability may have been lost through mutation, or the organism that donated the ancestral intron may have utilized an endogenous or exogenous maturase for splicing (13, 14, 26).
Ribosome function and prolonged developmental cycle.
In ribosomes, 23S rRNA base 1931 is located in domain IV at the interface between the SSU and the LSU, in close proximity to the highly conserved 23S peptidyl-transferase center (33). S. negevensis is a viable bacterium with functional SSU and LSU, despite the presence of SnLSU · 1 in position 1931 of the 23S rRNA. Three factors may contribute to the viability of S. negevensis. First, S. negevensis is an obligately intracellular bacterium and benefits from replicating in a nutrient-rich intracellular environment. Second, there is only one rRNA operon in the S. negevensis genome (27), and for survival, S. negevensis has had to unilaterally adapt to the presence of the intron. Third, in other organisms, it has been shown that point mutations in 23S rDNA sites 1926 or 1940 can make LSU defective for association with SSU (33). Experiments with recombinant E. coli have shown that 23S rRNA with a heterologous intron in position 1926 assembles into LSU but does not compete effectively for SSU, compared to endogenous LSU without introns (41). The dissociation constant for these SSU–LSU–unspliced-intron complexes is thus considerably higher than it is for SSU-LSU complexes that do not have introns. The site 1931 intron in S. negevensis 23S rRNA domain IV is located on the opposite side of the helical hairpin containing bases 1926 and 1940. If the ribonucleotides in positions 1926 and 1940 are occupied in the peptidyl-transferase center, the unspliced SnLSU · 1 intron in position 1931 may extend away from the active interface and out into the LSU. Yonath and Berkovitch-Yellin have, after all, observed that the ribosome is not a compact body but contains hollows, gaps, and tunnels (53). In addition, new evidence indicates that domain IV is less important than previously believed for peptide bond formation by 23S rRNA complexes (42).
Because of the proximity of the unspliced SnLSU · 1 RNA to the peptidyl-transferase center of the ribosome, it might be supposed that the intron would affect ribosome function in S. negevensis. As it turns out, S. negevensis has a uniquely prolonged developmental cycle of replication (12 to 14 days) compared to other Chlamydiales (27). C. trachomatis, for example, completes a cycle of replication by damaging or rupturing host cells just 2 or 3 days after infection. S. negevensis grows exponentially for 2 to 3 days and then enters a 7- to 10-day stationary phase. By light microscopy, changes in the appearance of an infected cell are dramatic as the cycle progresses. During exponential growth, the infected areas in the cytoplasm are tarry masses of tiny vacuoles. The vacuoles enlarge over time and become angular, but still appear to be empty. In the final 7 to 14 days the vacuoles fill with small flickering particles. The long stationary phase in the S. negevensis replication cycle might be caused by slowing of translation and growth, due to the intron. Other explanations, such as an unknown auxotrophy, must also be ruled out. It is not known whether the S. negevensis growth cycle would be quite so prolonged in the natural host. It is possible that the natural host cells encode maturases that are transported into S. negevensis to facilitate SnLSU · 1 splicing. The discovery of SnLSU · 1 may eventually make it possible to deduce what host cells S. negevensis grows in naturally, both by intron homology and by a reversion of the S. negevensis phenotype to rapid growth in that host.
Conclusions.
SnLSU · 1 is the first group I intron to be found in bacterial rDNA and the only group I intron that is not naturally spliced out of the rRNA. EndA, which is encoded by SnLSU · 1 and which may be a functional endonuclease, also encodes the 3′ splicing apparatus of the intron. EndA is closely related to endonucleases expressed by group I introns in chloroplasts and mitochondria. Because Simkania is an obligate intracellular bacterium, S. negevensis ZT may have acquired the rDNA intron by horizontal transfer within the eukaryotic environment. Interorganellar genetic transfer of SnLSU · 1 could occur among chlamydiae, chloroplasts, mitochondria, or other endosymbionts in amoebae. Such horizontal transfer would be consistent with the recent discovery of a rich mosaic of bacterial, chloroplast, plant, and other eukaryotic gene homologs in C. trachomatis, a relative of S. negevensis (47). The modified I-CeuI target sequence in Chlamydiaceae strains suggests that introns may at one time have affected this lineage. C. trachomatis, however, lacks genes for acquisition of exogenous DNA, making intron acquisition by these bacteria a dubious event (47). In comparison, the genome of Rickettsia prowazekii, a much more distantly related obligate intracellular bacterium, does not contain plant genes or introns (4).
An alternate explanation for the presence of SnLSU · 1 in S. negevensis rDNA and, indeed, for the origin of the Chlamydiales may be that Simkania, Chlamydia, and Parachlamydia are relics of the dawn of eukaryotic history: their common ancestor may have participated in the ancient chimeric events that led to the formation of the plant and animal lineages (23a). Such a proposal would be consistent with the current model for tRNA group I intron inheritance in cyanobacteria and other species of bacteria, including C. trachomatis (1, 15, 17, 43). Ancient parasitic intracellular bacteria such as chlamydiae could be viewed as vehicles that delivered packages of genetic material to cells.
Further characterization of the presence of SnLSU · 1 in the LSU will contribute to our understanding of ribosome structure and function. Experiments to mutagenize and delete a large portion of the intron can be predicted to produce a strain of Simkania that would behave more like other chlamydiae.
ACKNOWLEDGMENTS
We thank Robin R. Gutell for folding analysis, Thomas P. Hatch for in vitro transcription-translation of the Simkania ORF, Shirley M. Halling for providing sequence analysis software and computer facilities, Wolfgang Baehr for constructive critique of the manuscript, and Arthur A. Andersen for supporting this research.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Springer N, Schonhuber W, Ludwig W, Schmid E N, Muller K D, Michel R. Obligate intracellular bacterial parasites of ancanthamoebae related to Chlamydia spp. Appl Environ Microbiol. 1997;63:115–121. doi: 10.1128/aem.63.1.115-121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Scheiffer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 5.Belfort M, Roberts R J. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biniszkiewicz D, Cesnaviciene E, Shub D A. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birtles R J, Rowbotham T J, Storey C, Marrie T J, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–926. doi: 10.1016/s0140-6736(05)62701-8. [DOI] [PubMed] [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Burger G, Plante I, Lonergan K M, Gray M W. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J Mol Biol. 1995;245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- 11.Caldwell H D, Kromhout J, Schacter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cech T R. Structure and mechanism of the large catalytic RNAs: group I and group II introns and ribonuclease P. In: Gesteland R F, Atkins J F, editors. The RNA world. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 239–269. [Google Scholar]
- 13.Cech T R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 14.Cote M J, Turmel M. In vitro self-splicing reactions of chloroplast and mitochondrial group-I introns in Chlamydomonas eugametos and Chlamydomonas moewusii. Curr Genet. 1995;27:177–183. doi: 10.1007/BF00313432. [DOI] [PubMed] [Google Scholar]
- 15.Cousineau, B., and R. Cedergren. GenBank accession number U57090.
- 16.Damberger S H, Gutell R R. A comparative database of group I intron structures. Nucleic Acids Res. 1994;22:3508–3510. doi: 10.1093/nar/22.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett K D E, Andersen A A. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int J Syst Bacteriol. 1997;47:461–473. doi: 10.1099/00207713-47-2-461. [DOI] [PubMed] [Google Scholar]
- 18.Everett K D E, Bush R M, Andersen A A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 21.Giovannoni S J, Turner S, Olsen G J, Barns S, Lane D J, Pace N R. Evolutionary relationships among cyanobacteria and green chloroplasts. J Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray M W, Schnare M N. Evolution of rRNA gene organization. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 49–69. [Google Scholar]
- 23a.Gupta R S, Golding G B. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 24.Gutell, R. R. 13 March 1995, posting date. Group I intron database. [Online.] http://pundit.icmb.utexas.edu/RNA/GRPI/introns.html. [29 June 1999, last date accessed.]
- 25.Heath P J, Stephens K M, Monnat R J, Jr, Stoddard B L. The structure of I-CreI, a group I intron-coded homing endonuclease. Nat Struct Biol. 1997;4:468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 26.Hur M, Geese W J, Waring R B. Self-splicing activity of the mitochondrial group-I introns from Aspergillus nidulans and related introns from other species. Curr Genet. 1997;6:399–407. doi: 10.1007/s002940050294. [DOI] [PubMed] [Google Scholar]
- 27.Kahane S, Everett K D E, Kimmel N, Friedman M G. Simkania negevensis, strain ZT: growth, antigenic and genome characteristics. Int J Syst Bacteriol. 1999;49:815–820. doi: 10.1099/00207713-49-2-815. [DOI] [PubMed] [Google Scholar]
- 28.Kahane S, Gonen R, Sayada C, Elion J, Friedman M G. Description and partial characterization of a new chlamydia-like microorganism. FEMS Microbiol Lett. 1993;109:329–334. doi: 10.1016/0378-1097(93)90041-y. [DOI] [PubMed] [Google Scholar]
- 29.Kahane S, Greenberg D, Friedman M G, Haikin H, Dagan R. High prevalence of S. negevensis, a novel Chlamydia-like bacterium, in infants with acute broncholitis. J Infect Dis. 1998;177:1425–1427. doi: 10.1086/517830. [DOI] [PubMed] [Google Scholar]
- 30.Kleman-Leyer K, Armbruster D W, Daniels C J. Properties of H. volcantii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 31.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kryrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Goldek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 32.Lang B F, Burger G, O’Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 33.Leviev I, Levieva S, Garrett R A. Role for the highly conserved region of domain IV of 23S-like rRNA in subunit-subunit interactions at the peptidyl transferase centre. Nucleic Acids Res. 1995;23:1512–1517. doi: 10.1093/nar/23.9.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman D, Kahane S, Lieberman D, Friedman M G. Pneumonia with serological evidence of acute infection with the chlamydia-like microorganism “Z.”. Am J Respir Crit Care Med. 1997;156:578–582. doi: 10.1164/ajrccm.156.2.9608081. [DOI] [PubMed] [Google Scholar]
- 35.Liu S L, Hessel A, Sanderson K E. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonergan K M, Gray M W. The ribosomal RNA gene region in Acanthamoeba castellanii mitochondrial DNA. A case of evolutionary transfer of introns between mitochondria and plastids? J Mol Biol. 1994;239:476–499. doi: 10.1006/jmbi.1994.1390. [DOI] [PubMed] [Google Scholar]
- 37.Lykke-Andersen J, Garrett R A, Kjems J. Mapping metal ions at the catalytic centres of two intron-encoded endonucleases. EMBO J. 1997;16:3272–3281. doi: 10.1093/emboj/16.11.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall P, Lemieux C. The I-CeuI endonuclease recognizes a sequence of 19 base pairs and preferentially cleaves the coding strand of the Chlamydomonas moewusii chloroplast large subunit rRNA gene. Nucleic Acids Res. 1992;20:6401–6407. doi: 10.1093/nar/20.23.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 40.Michel R, Hauröder-Philippczyk B, Müller K-D, Weishaar L. Acanthamoeba from human nasal mucosa infected with an obligate intracellular parasite. Eur J Protisol. 1994;30:104–110. [Google Scholar]
- 41.Nikolcheva T, Woodson S A. Association of a group I intron with its splice junction in 50S ribosomes: implications for intron toxicity. RNA. 1997;3:1016–1027. [PMC free article] [PubMed] [Google Scholar]
- 42.Nitta I, Kamada Y, Noda H, Ueda T, Watanabe K. Reconstitution of peptide bond formation with Escherichia coli 23S ribosomal RNA domains. Science. 1998;281:666–669. doi: 10.1126/science.281.5377.666. [DOI] [PubMed] [Google Scholar]
- 43.Paquin B, Kathe S D, Nierzwicki-Bauer S A, Shub D A. Origin and evolution of group I introns in cyanobacterial tRNA genes. J Bacteriol. 1997;179:6798–6806. doi: 10.1128/jb.179.21.6798-6806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raué H A, Musters W, Rutgers C A, Van’t Riet J, Planta R J. rRNA: from structure to function. In: Hill W E, Moore P B, Dahlberg A, Schlessinger D, Garrett R A, Warner J R, editors. The ribosome-structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 217–235. [Google Scholar]
- 45.Rurangirwa F R, Dilbeck P M, Crawford T B, McGuire T C, McElwain T F. Analysis of the 16S rRNA gene of microorganism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int J Syst Bacteriol. 1999;49:577–581. doi: 10.1099/00207713-49-2-577. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 47.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 48.Swofford D L. PAUP: phylogenetic analysis using parsimony, version 3.1. Champaign, Ill: Illinois Natural History Survey; 1993. [Google Scholar]
- 49.Turmel M, Gutell R R, Mercier J-P, Otis C, Lemieux C. Analysis of the chloroplast large subunit ribosomal RNA gene from 17 Chlamydomonas taxa. J Mol Biol. 1993;232:446–467. doi: 10.1006/jmbi.1993.1402. [DOI] [PubMed] [Google Scholar]
- 50.Turmel M, Cote V, Otis C, Mercier J-P, Gray M W, Lonergan K M, Lemieux C. Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion) Mol Biol Evol. 1995;12:533–545. doi: 10.1093/oxfordjournals.molbev.a040234. [DOI] [PubMed] [Google Scholar]
- 51.Turmel M, Otis C, Cote V, Lemieux C. Evolutionarily conserved and functionally important residues in the I-CeuI homing endonuclease. Nucleic Acids Res. 1997;25:2610–2619. doi: 10.1093/nar/25.13.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vuillaumier S, Kaltenboeck B, Lecointre G, Lehn P, Denamur E. Phylogenetic analysis of cystic fibrosis transmembrane conductance regulator gene in mammalian species argues for the development of a rabbit model for cystic fibrosis. Mol Biol Evol. 1997;14:372–380. doi: 10.1093/oxfordjournals.molbev.a025773. [DOI] [PubMed] [Google Scholar]
- 53.Yonath A, Berkovitch-Yellin Z. Hollows, voids, gaps and tunnels in the ribosome. Curr Opin Struct Biol. 1993;3:175–181. [Google Scholar]