Abstract
In this report, we present the identification of the main polypeptides that are extracted from purified cell walls of a Saccharomyces cerevisiae mnn1 mnn9 strain by reducing agents. Treatment of the purified cell walls of this strain with β-mercaptoethanol releases several mannoproteins, of which three, with apparent sizes of 120, 45, and 40 kDa, are the most abundant. Analysis of the amino-terminal sequences revealed that the 120-kDa mannoprotein is Bar1p, the protease involved in the so-called barrier activity in yeast cells, and that the 45- and 40-kDa mannoproteins are the Kex2-unprocessed and Kex2-processed forms of the gene product of open reading frame (ORF) YJL158c, an ORF that belongs to the PIR (protein with internal repeats) family of genes, composed thus far of PIR1, PIR2/HSP150, and PIR3. Accordingly we have named this gene PIR4, and Pir4 denotes the 40-kDa Kex2-processed form of the mannoprotein. We have characterized Pir4 and have shown the feasibility of using it as a fusion partner for the targeting of recombinant proteins to the cell wall.
The cell wall of Saccharomyces cerevisiae represents some 30% of the total weight of the cell and is made up of β-glucans, mannose-containing glycoproteins (mannoproteins), and small amounts of chitin (9, 15). The mannoproteins can be divided into three groups according to the linkages that bind them to the structure of the cell wall: (i) noncovalently bound, (ii) covalently bound to the structural glucan, and (iii) disulfide bound to other proteins that are themselves covalently bound to the structural glucan of the cell wall (8). Our work has focused on the disulfide-bound mannoproteins, probably the least well known of the three groups mentioned above. Previous work (25) showed that treatment of whole yeast cells with a reducing agent releases four mannoproteins, with molecular masses of 38, 49, 68, and 88 kDa, and a highly polydisperse high-molecular-weight material. Additionally, in the case of cells of sexual mating type a previously treated with α-factor, treatment with a reducing agent releases an O-glycosylated 22-kDa mannoprotein (25). This 22-kDa mannoprotein is one of the subunits of the a-agglutinin and is coded by the AGA2 gene (6).
Extraction of whole cells with reducing agents can cause the release of proteins that are not part of the cell wall, and for this reason we chose to use purified cell walls, previously extracted with hot sodium dodecyl sulfate (SDS), as starting material for the β-mercaptoethanol extraction. Also, extraction of the wild-type strain releases mannoproteins that are polydisperse when run in SDS-polyacrylamide gel electrophoresis (PAGE). This polydispersity is due to the fact that some of them are highly glycosylated proteins and thus very difficult to characterize. To minimize this problem, we used both the wild-type strain and an mnn1 mnn9 strain deficient in glycosylation (2, 11). The results presented in this work consist of the characterization of two mannoproteins released from the purified cell walls of the mnn1 mnn9 strain by β-mercaptoethanol.
MATERIALS AND METHODS
Strains and media.
Escherichia coli DH5α was used for the propagation of plasmids; it was grown in Luria broth supplemented with 100 μg of ampicillin per ml when necessary. The standard S. cerevisiae strains X2180-1A (MATa SUC2 mal mel gal2 cup1) and BMA64-1A (MATa ade2-1 can1-100 ura3-1 leu2-3,112 trp1-D 2 his3-11) were used. All strains except the mnn1 mnn9 mutant were provided by the Spanish Type Culture Collection; the mutant strain was provided by Luis Miguel Hernandez (Universidad de Extremadura, Badajoz, Spain). Yeast strains were grown in YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose) or synthetic minimal medium SD (0.7% yeast nitrogen base without amino acids, 2% glucose, and amino acids as required).
Reagents.
Agar, yeast extract, peptone, and yeast nitrogen base were purchased from Difco Laboratories (Detroit, Mich.); phenylmethylsulfonyl fluoride (PMSF) was from Boehringer Mannheim; DNA restriction and modification enzymes were from Boehringer Mannheim, New England Biolabs Inc. (Beverly, Mass.), and Amersham-Pharmacia (Amersham, United Kingdom). The usual chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) and from Panreac (Barcelona, Spain). Electrophoresis reagents were from Bio-Rad Laboratories. Nitrocellulose membranes and the enhanced chemiluminescence reagents for developing Western immunoblots were from Amersham. Goat anti-rabbit immunoglobulin G (IgG)-peroxidase, and fluorescein-isothiocyanate-conjugated goat anti-rabbit IgG for immunofluorescence labeling, were from Bio-Rad.
Isolation of cell wall mannoproteins.
Cell walls from S. cerevisiae were purified and extracted with β-mercaptoethanol as follows. Cells in the early logarithmic phase were harvested and washed twice in Tris-HCl (10 mM, pH 7.4), 1 mM in PMSF (buffer A). The harvested biomass was resuspended in buffer A in a proportion of 2 ml per g (wet weight), glass beads (0.45 mm in diameter) were added to 50% of the final volume, and the cells were broken by shaking four times for 30 s, with 1-min intervals, in a CO2 refrigerated MSK homogenizer (Braun Melsungen AG, Melsungen, Germany). Breakage was confirmed by phase-contrast microscopy, and the walls were washed six to eight times in buffer A. Removal of noncovalently bound proteins was achieved by boiling the walls in buffer A containing 2% SDS (10 ml per g [wet weight] of walls) for 10 min, followed by six to eight washes in buffer A. The purified cell walls were finally resuspended in 10 mM ammonium acetate buffer (pH 6.3) containing 2% (vol/vol) β-mercaptoethanol (5 ml per g [wet weight] of walls) and incubated for 3 h at 30°C in an orbital incubator at 200 rpm. The extract was separated from the cell walls by centrifugation and concentrated by lyophilization.
Preparation of the polyclonal antibody against the β-mercaptoethanol extract from the mnn1 mnn9 strain.
Approximately 100 μg of the β-mercaptoethanol extract from the cell walls of the S. cerevisiae mnn1 mnn9 strain was resuspended in 1 ml of 0.9% NaCl in 10 mM phosphate-buffered saline, pH 7.2 (PBS), and mixed with an identical volume of complete Freund’s adjuvant (Difco). This emulsion was injected subcutaneously into the back of a 2-kg female New Zealand White rabbit. Boosts containing the same amount of protein in PBS mixed with incomplete Freund’s adjuvant were given at intervals of 6 weeks. Fifteen days after the second boost, the rabbit was exsanguinated; the blood was left to coagulate, the serum components were separated by centrifugation and kept in aliquots of 250 μl at −20°C. Monospecific antibodies against the Pir4 polypeptide were immunopurified as follows. A volume of 100-300 μl of the serum was laid on the antigenic side of a strip of nitrocellulose containing the band corresponding to Pir4, previously separated by SDS-PAGE and transferred to nitrocellulose by Western blotting, and incubated with gentle rocking for 2 h. After removal of the serum, the strip was washed three times in Tris-buffered saline containing 0.05% Tween 20 (TBST), and the immunoabsorbed antibodies were eluted by laying 200 to 400 μl of 0.2 glycine (pH 2.4) on the nitrocellulose strip and incubating the strip for 15 min. Finally, this solution, containing the eluted immunopurified antibodies, was recovered, and the pH was neutralized by adding an identical volume of cold 100 mM Tris base. The affinity of the antibody was tested by Western blotting.
SDS-polyacrylamide gels and Western blot analysis.
Proteins were separated by SDS-PAGE by the method of Laemmli (16) in 10 or 12% polyacrylamide gels. The proteins separated by SDS-PAGE were either stained with Coomassie brilliant blue or transferred onto Hybond-C nitrocellulose membranes as described by Towbin et al. (34) and Burnette (5). Membranes were blocked overnight in TBST–5% nonfat milk. The blocked membranes were washed three times in TBST and incubated for 1 h in TBST containing the antibody at a dilution of 1:5,000. After three washes in TBST, the membranes were incubated for 20 min in TBST containing goat anti-rabbit IgG-peroxidase at a dilution of 1:12,000 and washed in TBST. Finally, antibody binding was visualized on X-ray film by the enhanced chemiluminescence method (Amersham).
Amino-terminal sequencing of polypeptides.
Proteins separated by SDS-PAGE were transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore) as instructed by the manufacturer; the membranes were then stained with Coomassie brilliant blue, and the individual protein bands were excised. The amino-terminal sequences of the proteins isolated in this way were determined by Edman degradation in a Millipore ProSequencer 6600 automatic sequencer.
Immunofluorescence microscopy.
A small volume of exponentially growing culture was harvested and washed twice in PBS. The cells were resuspended in 50 μl of PBS containing the monospecific antibody immunopurified against Pir4 diluted 1:25 and incubated for 1 h at 37°C. After incubation, the cells were washed three times in PBS, resuspended in 50 μl of PBS containing fluorescein isothiocyanate-conjugated goat anti-rabbit IgG diluted 1:20, and incubated at 37°C for a further 2 h. After washing out the unreacted antibody, cells were mounted on glass slides and examined with a Zeiss Photomicroscope III.
Transformation of strains, DNA isolation, and sequencing.
Basic DNA manipulation and transformation in E. coli were performed as described by Sambrook et al. (28). Yeast transformation was carried out by the lithium acetate method (12). Plasmid DNA from E. coli was prepared by using a Flexi-Prep kit (Pharmacia), and DNA fragments were purified from agarose gels by using a Sephaglass Band-Prep kit, also from Pharmacia. Sequencing was performed by using Amplytaq polymerase with a Dye Terminator kit (Perkin-Elmer) in an Applied Biosystems 373A automatic sequencer.
Construction of the deletion cassette and confirmation of the deletion mutant by PCR.
Replacement of the genomic copy of PIR4 by the deletion cassette was performed by the one-step transplacement method (26). A fragment of 882 bp containing the PIR4 open reading frame (ORF) was generated by PCR using Taq DNA polymerase and the oligonucleotides ATGCAATTCAAAAACGTCGCCCCAG (starting at the ATG of the ORF) and GTGTATATTAAAGCTGCATGTG (located 195 bp downstream from the TAA) as primers. This 882-bp fragment was subcloned in pGEMT (Promega) to give plasmid pIMDB1. This plasmid was then digested with BglII and EcoRI, and the 85-bp fragment released was substituted by the KanMX4 marker digested from pFA6 (37) also with BglII and EcoRI to give plasmid pIMDB2. Finally, a 2.1-kbp disruption cassette comprising most of the PIR4 ORF interrupted by the KanMX4 marker was released from pIMDB2 by digestion with SalI and purified, and approximately 1 μg was transformed into strains BMA64A and FY1679. To confirm the replacement in the PIR4 locus, stable Geneticin-resistant transformants of the viable haploid form were analyzed by PCR using the oligonucleotides GCATTCCATACGATTTCCACGG (located 351 bp upstream from the ATG) and GTGTATATTAAAGGCTGCATGTG (located 195 bp downstream from the TAA) as primers. The sequence of these oligonucleotides and the length of the predicted PCR product were derived from the yeast genome sequence.
Generation by PCR of the ORF and its regulatory sequences and subcloning in YEplac112.
The PIR4 ORF was generated by PCR using the oligonucleotides GCATTCCATACGATTTCCACGG (located 351 bp upstream from the ATG) and GTGTATATTAAAGGCTGCATGTG (located 195 bp downstream from the TAA) as primers. A DNA polymerase with 3′-5′ proofreading activity (Vent polymerase; New England Biolabs) was used to improve fidelity. A 1,234-bp fragment that included the complete PIR4 ORF, 351 bp upstream from the ATG and 195 bp downstream from the stop codon, presumably containing the promoter and terminator sequences of PIR4, was obtained. This blunt-ended fragment was ligated to SmaI-digested YEplac 112 (10) to give rise to plasmid pIMDB3, which was transformed into the wild-type strain and the disruptant strain obtained as described above, and the resulting strains were then tested for Pir4 overexpression.
RESULTS AND DISCUSSION
Identification of the three main mannoproteins extracted from purified cell walls of the S. cerevisiae mnn1 mnn9 strain by treatment with β-mercaptoethanol.
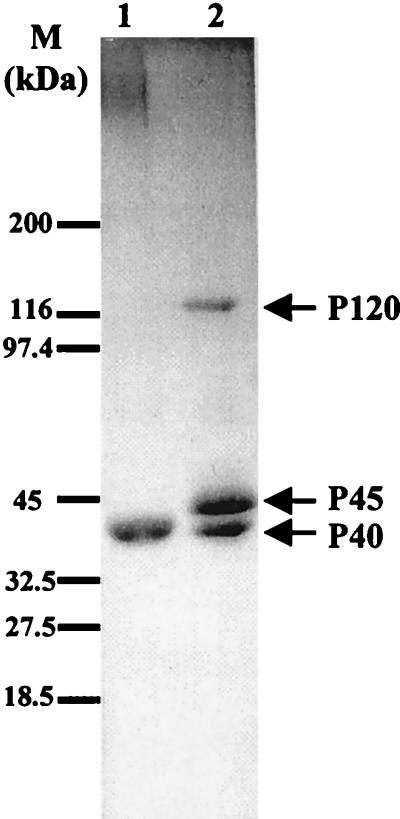
SDS-PAGE analysis of the β-mercaptoethanol-extracted material from the purified cell walls of the wild-type strain showed the presence of a 40-kDa (P40) band (Fig. 1, lane 1). Analysis of the same material extracted from the mnn1 mnn9 strain showed the presence of the P40 band and of two additional bands of 45 (P45) and 120 (P120) kDa (lane 2). To identify the three bands isolated in the extracts, we sequenced their amino termini and compared the sequences obtained with those in the EMBL data bank.
FIG. 1.
SDS-PAGE (Coomassie blue-stained gel) of β-mercaptoethanol extracts from purified cell walls of S. cerevisiae wild-type (lane 1) and mnn1 mnn9 mutant (lane 2) strains. M, size markers.
The amino-terminal sequence determined for P120 (LTNDGTGXLXFLLQHE) was, except for the amino acids that could not be determined (X), identical to that encoded by the BAR1 gene (18), a protease produced by mating type a S. cerevisiae cells that, by degrading α-factor, is responsible for the so-called barrier activity that antagonizes α-factor activity. To the best of our knowledge, this is the first report of the association of the BAR1 gene product to the cell wall through disulfide bridges, since it had thus far been considered to be secreted into the medium (1, 18, 19). The retention of Bar1p in the cell wall may avoid the activity dilution effect associated to its secretion; we cannot, however, discard the possibility that only a small part of Bar1p is retained or that this retention is merely transitory. MacKay et al. (18) found 95% of the Bar1p activity in the culture medium. We have not performed activity assays, but it is reasonable to assume that the cell wall-associated protein is active since it has been reported that Bar1p is active in the early stages of the secretory pathway (1).
The amino-terminal sequences determined for both P45 (EGYTPGEPWSTLTPTGSISXGSSEYT) and P40 (DVISQIGDGQVQATSAATAQATDEQ) were, except for the amino acids that could not be determined (X), identical to different parts of the sequence coded by ORF YJL158c. This ORF codes for 227 amino acids of which 54 are serine or threonine and 6 are cysteine; it contains also a putative signal peptide whose cleavage site matches the amino-terminal sequence of the P45 polypeptide as well as a putative cleavage site for the Kex2 protease that matches the amino-terminal sequence of the P40 polypeptide. These data suggest that both P40 and P45 polypeptides are encoded by ORF YJL158c, P40 corresponding to the mature gene product as processed by Kex2, a hypothesis that was confirmed by analyzing the β-mercaptoethanol extracts of the cell walls of a kex2-deficient strain (data not shown). YJL158c is clearly related to the PIR (protein with internal repeats) family of genes (27, 33): it contains the ISQIGDGQVQA repeat motif, albeit only once, and its homology with PIR2 is 55% overall; homology rises to 81% if only the last 80 carboxy-terminal amino acids are considered. Accordingly we propose the name PIR4 for this ORF and Pir4 for the Kex2-processed, β-mercaptoethanol-extractable P40 polypeptide encoded by it.
The protein encoded by YJL158c has been also described as a covalently linked cell wall protein (Ccw5p/Ccw11p) of 37 kDa that can be extracted by either laminarinase or mild alkali treatment (21). These results are, in principle, not easy to reconcile with our own. We base our conclusion regarding the way in which the gene product of YJL158c is bound to the cell wall on our results; a protein that is extracted by the simple action of reducing agents cannot be attached to the cell wall through any kind of covalent linkages apart from, obviously, disulfide bridges. This conclusion is reinforced by the fact that Ccw5p/Ccw11p has recently been identified as Scw8p by Cappellaro et al. (7), who describe it as dithiothreitol extractable. However, we cannot discard the possibility that the hypothetical cell wall protein to which Pir4 is bound through disulfide bridges could itself be bound to the structure of the cell wall through the type of linkages described by Mrsa et al. (21), or that part of Pir4 is bound directly to the structure of the cell wall through such linkages. In this context, Kapteyn et al. (13) have recently shown that part of Pir2/Hsp150, previously described as a secreted protein (27), remains attached to the β-1,3 glucan of the cell wall through the linkage described by Mrsa et al. (21); moreover, we have also detected a small amount of Pir2/Hsp150 in our β-mercaptoethanol extracts (data not shown). A possible hypothesis explaining all of these seemingly contradictory results is that the association of the Pir proteins to the wall structure through disulfide bridges would, in some cases, be an intermediate and transient stage before secretion, in others an intermediate stage before linkage to β-1,3 glucan, and in some others the final localization of the mature protein, each of these three possible localizations being, to some degree, interchangeable for each individual Pir cell wall protein. This hypothesis would explain why some Pir proteins are extracted from the cell wall, either with weak alkali or reducing agents, and at the same time secreted into the medium.
Disruption and overexpression of PIR4 do not affect the phenotype of the cells.
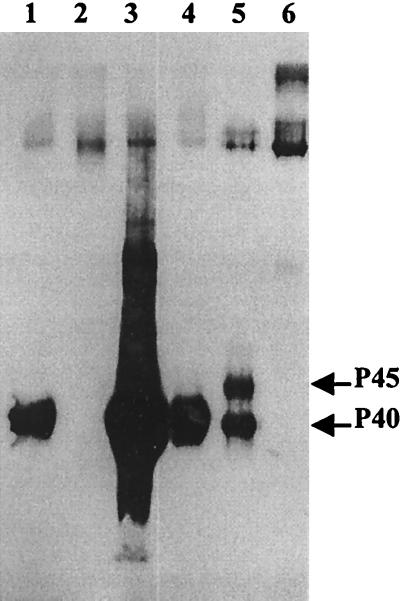
The possible role of Pir4 remains unclear. It belongs to the Pir family of proteins, which includes the heat shock-inducible Hsp150 (27), but it is not heat shock inducible (reference 21 and our results), and does not seem to be involved in the recovery of the cells following heat shock. YJL158c/PIR4 has also been named CIS3, as it is a multicopy suppressor of cik1 (20), a mutation that affects microtubule-based processes, causing a delay at mitosis with formation of a large bud and lysis at 37°C. However, the multicopy suppression effect of YJL158c could simply consist in a limitation of the lysis effect through a reinforcement of the surface of the growing bud (19a). To investigate the possible role of Pir4 in the cell wall, we first disrupted the PIR4 gene and then overexpressed it in S. cerevisiae cells. The construction made to test the effect of overexpression on the phenotype was based on the PIR4 gene amplified by PCR, subcloned in YEplac112 (10), and transformed in S. cerevisiae cells. Disruption of the gene was performed with a disruption cassette that included the KanMX4 gene as a marker (38). Analysis of the extracts of the strains revealed that both the disrupted wild-type (Fig. 2, lane 2) and mnn1 mnn9 (lane 6) strains lacked the P40 and the P40 and P45 polypeptides respectively, while the extract of the wild-type strain transformed with a multicopy plasmid harboring the PIR4 gene showed a corresponding increase in the level of the P40 polypeptide (lane 3). These results confirm that the PIR4 gene encodes both P40 and P45 and that the disruption and overexpression constructions were correct. However, neither disruption of the gene nor its overexpression affected the viability or morphology of the cells, their ability to recover from heat shock (tested as described in reference 24), their mating efficiency (tested as described in reference 31), their sensitivity to the killer toxin (tested as described in reference 4), or their sensitivity to calcofluor white, Congo red, or Zymolyase (tested as described in reference 36), agents that interfere with the synthesis of the cell wall or degrade it (data not shown); consequently, the possible role of Pir4 in the structure or in the synthesis of the cell wall remains unsolved.
FIG. 2.
Western immunoblot of β-mercaptoethanol extracts from the wild-type strain (lanes 1 and 4), disrupted PIR4::KanMX4 strain (lane 2), wild-type strain transformed with a multicopy plasmid containing PIR4 (lane 3), mnn1 mnn9 strain (lane 5), and mnn1 mnn9 strain containing the PIR4::KanMX4 disruption (lane 6), probed with a polyclonal antibody raised against the β-mercaptoethanol extract from the mnn1 mnn9 strain.
Pir4 is localized predominantly on the surface of growing buds.
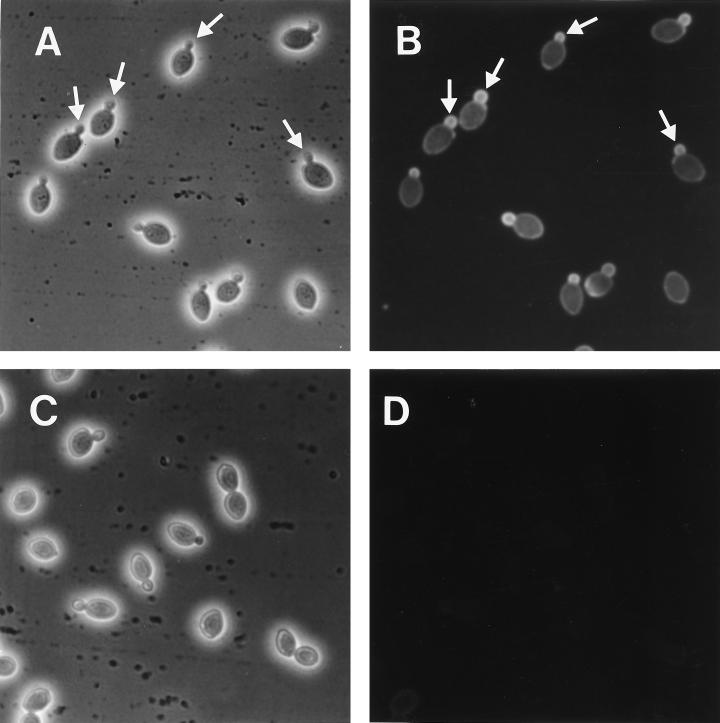
As an alternative approach to studying the role of Pir4, we examined the possible localization of Pir4 on the cell surface by using a polyclonal antibody, immunopurified against Pir4, in an indirect immunofluorescence technique. The results (Fig. 3) show that Pir4 is localized in the growing buds and not in the older mother cells. This result, together with the recently published results of Spellman et al. (32), who, by using a microarray hybridization technique, identify PIR4 as a cell cycle-regulated gene that is expressed during the G2 phase, suggest a role of Pir4 in the synthesis of the newly formed wall of the growing bud, albeit a nonessential one, since disruption of the PIR4 gene does not affect the viability of the cells.
FIG. 3.
Indirect immunofluorescence, using a polyclonal antibody immunopurified against Pir4, on S. cerevisiae cells. (A and C) Phase contrast; (B and D) fluorescence. (A and B) Wild-type cells; (C and D) cells from the disrupted PIR4::KanMX4 strain. Arrows point to the labeled growing buds.
Pir4 can be used as a carrier for the targeting of recombinant proteins to the cell wall.
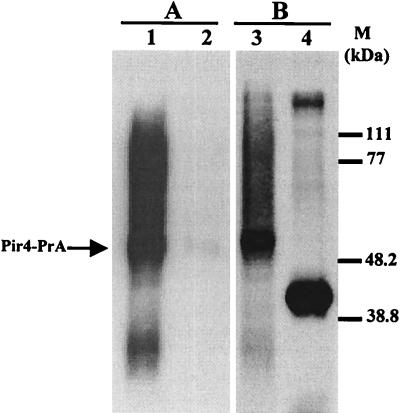
Targeting of recombinant proteins to the cell wall of S. cerevisiae can be used, among other things, for the immobilization of enzymes on the surface of yeast cells, for the inclusion of proteins that would facilitate the immobilization of the yeast cells to some solid matrix, or for the modification of the agglutinability properties of the cells. To date, glycosylphosphatidylinositol-anchored cell wall proteins have been used for the targeting of different enzymes to the surface of yeast cells (22, 23, 29, 30, 35, 37), and a disulfide-bound protein, the ligand/recognition peptide of the a-agglutinin Aga2p, has been used as a carrier to present a library of antibody fragments on the cell surface (3, 14). The feasibility of using Pir4 as a carrier for the targeting of recombinant proteins to the cell wall was tested by creating a fusion between the PIR4 gene and a portion of the gene coding for Staphylococcus aureus protein A (17). Staphylococcal protein A can be easily detected due to its reactivity with the Fc fraction of the IgGs of many species. The fusion was made by subcloning a PCR generated fragment containing two Fc binding domains of protein A flanked by BglII sites in the naturally occurring BglII site in PIR4 previously subcloned in YEplac112. As shown in Fig. 4, the resulting recombinant fusion protein was correctly targeted to the cell wall and could be released from this structure by extraction with β-mercaptoethanol, demonstrating that the insertion of a recombinant protein of moderate size in the amino acid sequence of Pir4 does not affect its ability to be correctly retained in the cell wall, consequently opening the way for its use as a fusion partner for the targeting of other proteins of interest to the yeast cell wall.
FIG. 4.
Western immunoblot of the β-mercaptoethanol extracts of the wild-type strain (lanes 2 and 4) and disrupted PIR4::KanMX4 strain harboring a PIR4-protein A gene fusion in a multicopy plasmid (lane 1 and 3), probed with a polyclonal antibody raised against β-mercaptoethanol extracts from the mnn1 mnn9 strain (B) and with serum of a nonimmunized rabbit (A). M, size markers.
ACKNOWLEDGMENTS
We thank Frans Klis (Department of Molecular Cell Biology, BioCentrum Amsterdam, University of Amsterdam, Amsterdam, The Netherlands) for critical reading of the manuscript.
This work was supported by grant PM96-0019 from the Secretaría de Estado de Universidades, Investigación y Desarrollo. I.M. was supported by a fellowship from the Ministère de l’Education Nationale du Maroc. L.J. was supported by a fellowship from Colegio Mayor La Coma.
REFERENCES
- 1.Ballensiefen W, Schmitt H D. Periplasmic Bar1 protease of Saccharomyces cerevisiae is active before reaching its extracellular destination. Eur J Biochem. 1997;247:142–147. doi: 10.1111/j.1432-1033.1997.00142.x. [DOI] [PubMed] [Google Scholar]
- 2.Ballou C E. Isolation, characterization and properties of Saccharomyces cerevisiae mnn mutants with non-conditional glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- 3.Boder E T, Wittrup K D. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 4.Brown J L, Roemer T, Lussier M, Sdicu A M, Bussey H. The K1 killer toxin: molecular and genetic applications to secretion and cell surface assembly. In: Johnston J R, editor. Molecular genetics of yeast: a practical approach. Oxford, United Kingdom: IRL Press, Oxford University Press; 1994. pp. 217–231. [Google Scholar]
- 5.Burnette W N. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 6.Cappellaro C, Hauser K, Mrsa V, Watzele M, Watzele G, Gruber C, Tanner W. Saccharomyces cerevisiae a-agglutinin and α-agglutinin. Characterization of their molecular interaction. EMBO J. 1991;10:4081–4088. doi: 10.1002/j.1460-2075.1991.tb04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappellaro C, Mrsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Nobel H, Lipke P N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 1994;4:42–46. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 9.Fleet G H. Cell walls. In: Rose A H, Harrison J S, editors. The yeasts. 2nd ed. 14. Yeast organelles. London, United Kingdom: Academic Press; 1991. pp. 199–277. [Google Scholar]
- 10.Gietz R, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez L M, Ballou L, Alvarado E, Gillece-Castro B L, Burlingame A L, Ballou C E. A new Saccharomyces cerevisiae mnn mutant N-linked oligosaccharide structure. J Biol Chem. 1989;246:11846–11856. [PubMed] [Google Scholar]
- 12.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapteyn J C, Van Egmond P, Slevi E, Van Den Ende H, Makarow M, Klis F M. The contribution of the O-glycosylated protein Pir2/Hsp150 to the construction of the yeast cell wall in wild-type cells and β 1,6-glucan-deficient mutants. Mol Microbiol. 1999;31:1835–1844. doi: 10.1046/j.1365-2958.1999.01320.x. [DOI] [PubMed] [Google Scholar]
- 14.Kieke M C, Cho B K, Boder E T, Kranz D M, Wittrup K D. Isolation of anti-T cell receptor scFv mutants by yeast surface display. Protein Eng. 1997;10:1303–1310. doi: 10.1093/protein/10.11.1303. [DOI] [PubMed] [Google Scholar]
- 15.Klis F M. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lowenadler B, Nilsson B, Abrahmsen L, Moks T, Ljungqvist L, Holmgren E, Paleus S, Josephson S, Philipson L, Uhlen M. Production of specific antibodies against protein A fusions. EMBO J. 1986;5:2393–2398. doi: 10.1002/j.1460-2075.1986.tb04509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKay V L, Welch S, Insley M Y, Manney T R, Holly J, Saari G C, Parker M L. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci USA. 1988;85:55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacKay V L, Armstrong J, Yip C, Welch S, Walker K, Osborn S, Sheppard P, Forstrom J. Characterization of the Bar proteinase, an extracellular enzyme from the yeast Saccharomyces cerevisiae. Adv Exp Med Biol. 1991;306:161–172. doi: 10.1007/978-1-4684-6012-4_21. [DOI] [PubMed] [Google Scholar]
- 19a.Manning, B. Personal communication.
- 20.Manning B D, Padmanabha R, Snider M. The Rho-GEF Rom2 localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mrsa V, Seidl T, Gentzsch M, Tanner W. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1145–1154. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Murai T, Ueda M, Yamamura M, Atomi H, Shibasaki Y, Kamasawa N, Osumi M, Amachi T, Tanaka A. Construction of a starch-utilizing yeast by cell surface engineering. Appl Environ Microbiol. 1997;63:1362–1366. doi: 10.1128/aem.63.4.1362-1366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murai T, Ueda M, Atomi H, Shibasaki Y, Kamasawa N, Osumi M, Kawaguchi T, Arai M, Tanaka A. Genetic immobilization of cellulase on the cell surface of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;48:499–503. doi: 10.1007/s002530051086. [DOI] [PubMed] [Google Scholar]
- 24.Navarro-García F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlean P, Ammer H, Watzele M, Tanner W. Synthesis of an O-glycosylated cell surface protein induce by alpha factor. Proc Natl Acad Sci USA. 1986;83:6263–6266. doi: 10.1073/pnas.83.17.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 27.Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc Natl Acad Sci USA. 1992;89:3671–3675. doi: 10.1073/pnas.89.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schreuder M P, Brekelmans S, Van den Ende H, Klis F M. Targeting of a heterologous protein to the cell wall of Saccharomyces cerevisiae. Yeast. 1993;9:399–409. doi: 10.1002/yea.320090410. [DOI] [PubMed] [Google Scholar]
- 30.Schreuder M P, Mooren A T A, Toschka H Y, Verrips C T, Klis F M. Immobilizing enzymes on the surface of yeast cells. Trends Biotechnol. 1996;14:115–120. doi: 10.1016/0167-7799(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 31.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 32.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P Q, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toh-e A, Yasunaga S, Nisogi H, Tanaka K, Oguchi T, Matsui Y. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast. 1993;9:481–494. doi: 10.1002/yea.320090504. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Berkel M A A, Caro L H P, Montijn R C, Klis F M. Glucosylation of chimeric proteins in the cell wall of Saccharomyces cerevisiae. FEBS Lett. 1994;349:135–138. doi: 10.1016/0014-5793(94)00631-8. [DOI] [PubMed] [Google Scholar]
- 36.Van der Vaart J M, Caro H P, Chai Man J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Vaart J M, te Biesebeke R, Chapman J W, Toschka H Y, Klis F M, Verrips T. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl Environ Microbiol. 1997;63:615–620. doi: 10.1128/aem.63.2.615-620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wach A, Brachar A, Pohlmann R, Philipsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]