Abstract
The gene encoding OprF, a major outer membrane protein in Pseudomonas species (formerly known as type 1 pseudomonads), was thought to be constitutively transcribed from a single sigma 70 promoter immediately upstream of the gene. We now report the identification of a novel putative ECF (extracytoplasmic function) sigma factor gene, sigX, located immediately upstream of oprF in both Pseudomonas aeruginosa PAO1 and Pseudomonas fluorescens OE 28.3 and show that disruption of this gene significantly reduces OprF expression. In P. aeruginosa, Northern analysis demonstrated that this reduction was a result of an effect on transcription of monocistronic oprF combined with a polar effect due to termination of a transcript containing sigX and oprF. Comparison of sigX-disrupted and wild-type cell transcripts by primer extension indicated that monocistronic transcription of oprF occurs from two overlapping promoters, one that is SigX-dependent and resembles ECF sigma factor promoters in its minus-35 region and another promoter that is independent of SigX and is analogous to the sigma 70-type promoter previously reported. Complementation of the P. aeruginosa sigX-disrupted mutant with plasmid-encoded OprF did not resolve the phenotypes associated with this mutant, which included a markedly reduced logarithmic-phase growth rate in rich medium (compared to that in minimal medium), further reduction of the growth rate in a low-osmolarity environment, secretion of an unidentified pigment, and increased sensitivity to the antibiotic imipenem. This indicates that SigX is involved in the regulation of other genes in P. aeruginosa. Disruption of the sigX gene in P. fluorescens also had an effect on the logarithmic-phase growth rate in rich medium. A conserved sigX gene was also identified in a Pseudomonas syringae isolate and six P. aeruginosa clinical isolates. Collectively, these data indicate that an ECF sigma factor plays a role in the regulation and expression of OprF and also affects other genes.
OprF, a major outer membrane protein in type I Pseudomonas spp., is a nonspecific porin that plays a role in the maintenance of cell shape and is required for growth in a low-osmolarity environment (8, 10, 19, 29). Clinically derived mutants of the opportunistic human pathogen Pseudomonas aeruginosa which are multiply antibiotic resistant (MAR) and are deficient in the major outer membrane protein OprF have been isolated (20). Sequencing of the oprF gene in such a clinical isolate has shown that the oprF gene and promoter are intact, suggesting that a possible regulatory mutation is involved (21, 21a). In plant root-colonizing Pseudomonas fluorescens, regulation of OprF is of interest because of the in vitro-demonstrated role of OprF in adhesion to plant roots (4).
Previous transcriptional analysis of P. aeruginosa oprF by primer extension, S1 nuclease mapping, and Northern blot analysis indicated that there was a single transcriptional start site 57 bp upstream of oprF (5). A putative rho-independent transcription terminator was identified immediately downstream of oprF, followed by a gene transcribed in a convergent direction. The promoter upstream of oprF was found to share similarity with other sigma 70-type promoters (5), and changes in OprF expression were not observed under a variety of growth conditions. Therefore, oprF was thought to be constitutively transcribed as a monocistronic unit, and thus, studies of its regulation, or upstream genes, were not pursued further.
However, we now report the identification of sigX, a new putative ECF (extracytoplasmic function) sigma factor gene located upstream of oprF in both P. aeruginosa PAO1 and P. fluorescens OE 28.3, and show that SigX plays a role in OprF expression. Characterization of sigX-disrupted mutants, and the mutant complemented with oprF on a plasmid, demonstrated that this probable sigma factor also affects the expression of other genes in P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used are listed in Table 1. Strains were grown in Mueller-Hinton, brain heart infusion, chocolate agar, or Luria Bertani (LB) medium (1% tryptone, 0.5% yeast extract, pH 7.0; Difco). BM2 medium (9) containing succinate as a carbon source was used as a minimal medium for P. aeruginosa, and either BM2 or M9 medium (containing glucose as a carbon source for M9 [15]) was used for P. fluorescens. Varying concentrations of NaCl were added to LB or minimal medium (no-salt, low-salt, normal-salt, high-salt, and very-high-salt medium represented additions of 0, 8, 85, 200, and 500 mM NaCl, respectively). In some experiments, 171 mM NaCl was added. Antibiotics were used when necessary at the following concentrations unless otherwise indicated: streptomycin, 500 μg/ml; ampicillin, 100 μg/ml; carbenicillin, 300 μg/ml; chloramphenicol, 25 μg/ml; kanamycin (Km), 50 μg/ml; gentamycin (Gm), 12 μg/ml; and spectinomycin, 50 μg/ml. For growth curve studies, frozen aliquots of cells were first grown overnight on agar containing appropriate antibiotics for maintenance of the strain genotype, and then the cells were grown overnight in the appropriate antibiotic-free broth medium. This overnight culture was then diluted 1:100 into 20 ml of fresh medium in a 250-ml sidearm flask. Growth of the cells in this 20-ml culture was then monitored by determining the optical density either with a Klett spectrophotometer with a red filter or by determination of absorbance at 610 nm with a standard UV and visual spectrophotometer. The culture was subjected to constant shaking to provide aeration and incubated at 37°C for P. aeruginosa or 30°C for P. fluorescens unless otherwise described. Some growth curves of P. fluorescens were determined with a Bioscreen growth analyzer (Labsystems, Helsinki, Finland) with constant shaking at 30°C and absorbance measurements at 600 nm.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Description | Reference |
|---|---|---|
| pSUP202 | pBR325 containing RP4 Mob site; Cmr Tcr Apr | 24 |
| pUC4K | pUC4 with aminoglycoside 3′-phosphotransferase gene from Tn903 (Kmr) | Pharmacia Biotech |
| λFAJ2001 | λEMBL3 with Sau3AI fragment carrying P. fluorescens OE 28.3 oprF gene | 1 |
| pFAJ2001 | pCL1921 with 3.3-kb SmaI fragment of λFAJ2001 carrying oprF gene in SmaI site | 1 |
| pCL1821 | Low-copy-number E. coli plasmid; Spr | 12 |
| pFAJ2052 | pCL1921 with 3.3-kb SmaI fragment of λFAJ2001 carrying oprF gene in HindIII site | This work |
| pFAJ2073 | pFAJ2052 with 1.26-kb HindIII fragment of pUC4K conferring Kmr inserted in NruI site of oprF | This work |
| pFAJ2082 | pSUP202 with BamHI-HindIII fragment of pFAJ2073 containing disrupted oprF | This work |
| pFAJ2425 | pFAJ2052 with 1.26-kb HindII fragment of pUC4K inserted in Asp700 site of sigX | This work |
| pFAJ2431 | pSUP202 with BamHI-HindIII fragment of pFAJ2425 containing disrupted sigX | This work |
| pFAJ2432 | pUC19 with PCR fragment containing P. fluorescens M114 sigX gene in BamHI site | This work |
| pFAJ2448 | pUC18 with PCR fragment containing P. syringae pv. syringae sigX gene, in BamHI site | This work |
| pFAJ2511 | pUC18 with 1.5-kb SacI fragment of λFAJ2001 (upstream of sigX) | This work |
| pFAJ2672 | pUC18 with 4.8-kb XhoI fragment of λFAJ2001 (upstream of sigX) | This work |
| pRW5 | pUCP19 + 1.47-kb HindIII-EcoRI fragment coding for P. aeruginosa H103 OprF | 27a |
| pEX100t | pUC19-based gene replacement vector with a counterselectable sacB marker | 23 |
| pX1918GT | Contains xylE-Gmr cassette (SmaI fragment) for construction of gene disruptions and reporter gene fusions | 23 |
| pWW2300 | pUC19 + 1.3-kb PstI-SalI insert (contains 3′ end of cmpX, sigX, and the 5′ end of oprF from P. aeruginosa H103) | 28 |
| pWW1701 | pUC19 + 1.7-kb SmaI insert (contains cmpX, sigX, and the 5′ end of oprF from P. aeruginosa H103) | 28 |
| pWW1901 | pUC19 + 1.9-kb SalI insert (contains cmaX, crfX, cmpX, sigX, and the 5′ end of oprF from P. aeruginosa H103) | 28 |
| pFB2e1 | xylE-Gmr cassette (SmaI fragment) inserted into the EcoRV site within P. aeruginosa sigX in pWW1701 | This work |
| pFB2e1a3 | 3.4-kb FspI-SmaI fragment of pFB2e1 (xylE-Gmr cassette in SigX) cloned into Sma I site of pEX100t | This work |
| E. coli | ||
| CL83 | recA mutant (recA56) derivative of JM83 | 12 |
| DH5α | Strain used for all standard cloning experiments | |
| S17-1 | Mobilizing strain for RP4 Mob-containing plasmids | 24 |
| P. aeruginosa | ||
| H103 | PAO1 prototroph; wild-type reference strain | 9 |
| H636 | oprF::Strr mutant of P. aeruginosa H103 | 29 |
| H814 | sigX::Gmr mutant of P. aeruginosa H103 | This work |
| H845 | P. aeruginosa H814 + pRW5 (sigX::Gmr mutant with OprF expressed from a plasmid) | This work |
| P. fluorescens | ||
| FAJ2026 | oprF mutant of P. fluorescens OE 28.3 | This work |
| FAJ2030 | sigX mutant of P. fluorescens OE 28.3 | This work |
| M114 | Isolated from sugarbeet rhizosphere | 7 |
| OE 28.3 | Isolated from wheat rhizosphere | 4 |
| P. syringae LMG 1247T | Isolated from Syringa vulgaris; pathovar reference strain |
General DNA procedures and sequencing.
Most common DNA procedures were performed as described by Sambrook et al. (22). For PCR, either Vent or Taq DNA polymerase (Fisher Scientific) was used under the standard conditions suggested by the manufacturer, unless otherwise described. Most PCR experiments were performed with an MJ Research thermal cycler or a Trio-thermoblock PCR apparatus (Biometra) with the following thermal profile repeated 30 times: 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. DNA was sequenced (both strands) with either an ABI 373A automated sequencing system (Perkin-Elmer, Norwalk, Conn.) or an ALF sequencer (Pharmacia) according to the manufacturer’s instructions, and oligonucleotides were synthesized on an ABI 392 DNA-RNA synthesizer as described by the manufacturer. For primer extension experiments, sequencing was performed with the fmol sequencing kit (Promega) as described by the manufacturer with [γ-32P]ATP end-labeled primer (see “RNA analysis” below). For P. fluorescens OE 28.3, the complete sequences of the inserts in plasmids pFAJ2001, pFAJ2511, and pFAJ2672 (described in Table 1) were determined, resulting in a total of 5.6 kb of sequence determined upstream of oprF. For P. aeruginosa, the complete sequences of the inserts in pWW1901, pWW1701, and pWW2300 were determined (providing a total of 3,020 bp of sequence information upstream of oprF) and additional sequence information (to a total of 5,519 bp of sequence upstream of oprF) was obtained from the Pseudomonas Genome Project sequence data (20a). The sigX genes of P. syringae pv. syringae LMG 1247 and of P. fluorescens M114 were also amplified by PCR with, as a forward primer, 5′-ATAGGATCAAGGAGGACTTGTTATGAATAAAGCCCAAACG-3′ (italics indicate the added BamHI tag and ribosome binding site), and with ATAGGATCCGCACTAAGTTTCAGTCTCGCC-3′ as a reverse primer. Note that the possible TTG start codon in the forward primer was replaced with ATG (represented in boldface), to assist in prospective expression studies in Escherichia coli. The resulting BamHI-digested PCR amplicons obtained from P. syringae pv. syringae LMG 1247 and P. fluorescens M114 were cloned for sequence analysis in pUC18 and pUC19 to generate pFAJ2448 and pFAJ2432, respectively. The sequence of sigX was also determined from six P. aeruginosa clinical isolates, H246, H344, H397, H411, H580, and H813, by direct sequencing of PCR amplicons of the sigX gene obtained with primers FL37 (5′-GGCCAACCGTCTACTGCTCG-3′) and FL9 (5′-TTGTCCAACAATCAGCCGCA-3′), which flank the gene.
RNA analysis.
For RNA analysis, cells were grown in high-salt LB medium as described above to early log phase, mid-log phase, or late log phase or were grown overnight (stationary phase). Total P. aeruginosa RNA was extracted from the cells with the protocol and materials supplied in the RNeasy RNA isolation kit from Qiagen (Hilden, Germany). All RNA preparations were treated with RNase-free DNase and repurified by the Qiagen protocol before electrophoresis, reverse transcription (RT)-PCR, or primer extension experiments. Five micrograms of this RNA was subjected to electrophoresis in 1% formaldehyde agarose gels as described by Sambrook et al. (22). Total RNA from P. fluorescens was prepared according to the method of Nagy et al. (18). For Northern analysis, RNA was transferred to positively charged nylon membranes (Boehringer Mannheim), and hybridization and probe preparation were performed with the AlkPhos direct DNA labeling and detection system from Amersham. This method for direct labeling of the probe bypasses the need for antibody detection and is reported to be quantitative by the manufacturer. A probe containing oprF sequences was PCR amplified from the plasmid pRW5, which contains only 60 bp of sequence upstream of oprF (plus a mutated promoter region) and so provided a good template for preparation of an oprF probe that was free of any sigX sequences. The sigX probe was prepared by PCR amplification from pWW2300 using one set of primers within the gene, followed by a second round of PCR using the resulting amplicon as a template and primers internal to the first-round PCR primers. This also produced a probe that, according to control experiments, was not contaminated with upstream (cmpX) or downstream (oprF) gene sequences. For RT-PCR experiments, RNA was first reverse transcribed with specific primers and Moloney murine leukemia virus reverse transcriptase (Promega or Roche, Basel, Switzerland) or avian myeloblastosis virus reverse transcriptase (Promega) according to the manufacturer’s instructions. The resulting cDNA was then subjected to amplification by PCR with a second set of internal primers. For primer extension experiments, 5 μg of RNA was reverse transcribed with SuperscriptII (Life Technologies) according to the manufacturer’s instructions with the following end-labeled primer which is complementary to the reverse strand of the 5′ end of oprF: 5′-CAACCAGCGAGCCGATGACA-3′. The primer was end-labeled with Redivue [γ-32P]ATP (Amersham). A corresponding sequencing reaction was performed as described above with the same end-labeled primer. RNA for the primer extension experiments was obtained from mid-log-phase cells that had been grown on high-salt LB medium as described above.
Protein procedures.
Outer membranes were prepared by the one-step sucrose gradient method of Hancock and Carey (9). Cell envelopes were prepared by subjecting the cells (resuspended in 10 mM Tris-HCl, 5 mM MgSO4, pH 7.5) to breakage in a French pressure cell (twice at 15,000 lb/in2), followed by centrifugation of the lysate at low speed (1,200 × g) to remove unbroken cells and debris. The supernatant was then subjected to one high-speed centrifugation at 151,000 × g, and the pellet, containing cell envelope proteins, was resuspended in water. The proteins were solubilized in solubilization buffer at 100°C for 10 min and then separated by electrophoresis in 12.5% polyacrylamide gels as previously described (9). Two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell extracts of P. fluorescens was carried out as described previously (2, 3). The gels were stained with Coomassie brilliant blue R250 (Bio-Rad) for visualization of proteins. Western immunoblot analysis for the detection of OprF proteins was performed as described previously, using the monoclonal antibody MA7-1 for P. aeruginosa (17) or MA28B9 for P. fluorescens (2). Densitometric analysis of both protein gels and Northern blots was performed with small amounts of sample analyzed with the AlphaImager 1200 documentation and analysis system (Alpha Innotech Corporation). To roughly confirm the densitometric analysis, different amounts of sample loaded on a gel were compared with each other to determine the amounts required to obtain the same band intensity.
Construction of gene disruptions.
For the construction of a P. fluorescens OprF-deficient mutant, the 3.3-kb SmaI fragment carrying the P. fluorescens OE 28.3 oprF gene (1) was cloned into the HindIII site of pCL1921 (12) to generate pFAJ2052. The Tn903 kanamycin resistance gene, obtained as a 1.26-kb HindIII fragment from pUC4K (Pharmacia Biotech), was then inserted in the NruI site of the oprF gene in pFAJ2052. From this construct, pFAJ2073, with the aminoglycoside 3′-phosphotransferase gene in the opposite orientation to oprF, the interrupted oprF gene was recovered as the BamHI-HindIII fragment and cloned into pSUP202 (24). The resulting plasmid, pFAJ2082, was transferred from E. coli S17-1 to P. fluorescens OE 28.3 by biparental conjugation. Among the kanamycin-resistant Pseudomonas transconjugants, a strain (FAJ2026) sensitive to chloramphenicol was selected, since this antibiotic resistance pattern would correspond to a double crossover, resulting in loss of the vector-associated chloramphenicol resistance. The anticipated genomic rearrangement interrupting the oprF gene in strain FAJ2026 was confirmed by Southern blot analysis (using vector-, kanamycin cassette-, and oprF-specific probes). No OprF protein was detectable in mutant FAJ2026 by Western blotting with the OprF-specific monoclonal antibody MA28B-9 (2).
For the creation of the P. fluorescens sigX disruption, the Tn903 kanamycin resistance gene was inserted in an opposite orientation into pFAJ2052, which was linearized at an Asp700 site within sigX by partial digestion. A BamHI (complete) and HindIII (partial) digest of this clone was used to obtain a fragment containing the disrupted sigX gene, which was cloned into BamHI-HindIII-digested pSUP202. This plasmid, pFAJ2431, was mobilized from E. coli S17-1 into P. fluorescens OE 28.3, and a putative sigX mutant resulting from double homologous recombination (FAJ2030) was selected as described above for the oprF::Kmr mutant FAJ2026.
The P. aeruginosa sigX disruption was constructed with a 2.4-kb xylE-gentamicin resistance cassette from pX1918GT, which was cloned into a unique EcoRV site within sigX present in pWW1701. A 3.4-kb FspI-SmaI fragment from the resulting plasmid, pFB2e1, was cloned into the SmaI site of pEX100t (23), producing pFB2e1a3. This plasmid, which encodes a counterselectable sacB marker, was transformed into E. coli S17-1 for mobilization into P. aeruginosa H103. After conjugation, colonies which grew on BM2 minimal medium containing 78 mM salt, 4 μg of gentamicin/ml, and 150 μg of carbenicillin/ml were plated onto LB medium containing 5% sucrose and 4 μg of gentamicin/ml. Sucrose-resistant colonies were screened for sensitivity to 300 μg of carbenicillin/ml, indicating a double-crossover event had occurred. Each colony was confirmed to be a sigX::Gmr mutant by its gentamicin resistance and production of a yellow color when exposed to catechol (indicating xylE function) by PCR with primers yielding appropriately sized fragments containing both sigX and the gentamicin cassette sequences and by Southern blots probed with the gentamicin cassette. Four separately obtained cultures were subjected to preliminary phenotypic analysis, and all had identical phenotypes under the conditions observed (OprF expression levels and growth rate in low-salt and high-salt media). One such culture was given the strain name H814.
For mobilization of the pRW5 plasmid into strain H814, the cells were electroporated as described by Farinha and Kropinski (6), except that high-salt medium containing 50 mM MgCl2 was used for the recovery period after electroporation.
MIC and other phenotypic tests.
MICs were determined as previously described (29) for P. aeruginosa H103 (wild type), P. aeruginosa H814 (sigX::Gmr), and P. aeruginosa H636 (oprF::Strr) by using the following compounds: tetracycline, chloramphenicol, ciprofloxacin, nalidixic acid, enoxacin, carbenicillin, ceftazidime, cefpirome, gentamicin, imipenem, rifampin, polymyxin B, erythromycin, SDS, HgCl2, ZnSO4, CuCl2, Co(NO3)2, K2CrO4, Ni(NO3)2, and AgNO3. The Biolog GN Microplate (Biolog Inc., Hayward, Calif.), and API20 NE (BioMérieux, Marcy l’Étoile, France) metabolic tests were conducted as described by the manufacturers. Uptake of the hydrophobic probe 1-N-phenyl-1-naphthylamine was examined with a fluorometer as described previously (13).
Nucleotide sequence accession numbers.
The accession numbers for the sequences reported in this paper are AF027290, AF115334, AF115335, and AF115338.
RESULTS
Upstream of oprF is a putative ECF sigma factor gene, sigX.
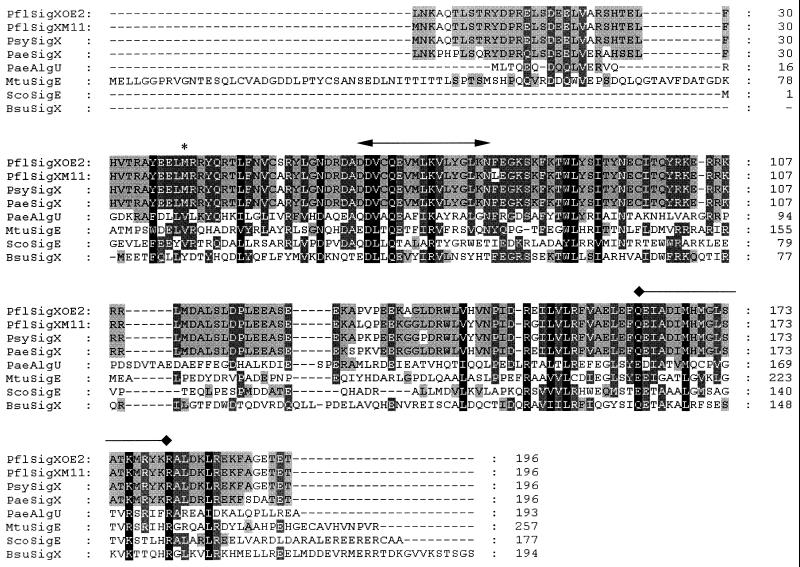
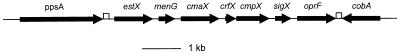
DNA sequencing upstream of the oprF gene in P. fluorescens OE 28.3 and P. aeruginosa H103 revealed the existence of an open reading frame (ORF) 108 bp upstream of oprF that shared significant similarity with sigma factor genes of the ECF family (approximately 40 to 45% amino acid similarity; BLASTP Expect values of up to 8e-12). This similarity was most pronounced in regions known to be highly conserved among well-studied ECF sigma factors and included putative helix-turn-helix and RNA polymerase-binding domains (Fig. 1). This ORF was named sigX in both P. fluorescens and P. aeruginosa, since the high degree of similarity between these two putative genes (94% amino acid similarity) and the conserved gene order surrounding the genes (Fig. 2) suggests that these genes are orthologs. Of note, sigX, and most of the sigma factor genes it closely resembled, did not have a downstream (or upstream) regulatory gene, as is seen for some ECF sigma factors (14). Interestingly, the sigX genes were most similar to known or putative ECF sigma factor genes from gram-positive rather than gram-negative bacteria.
FIG. 1.
Alignment of the deduced amino acid sequences from the sigX genes from P. aeruginosa H103 (PaeSigX), P. fluorescens OE 28.3 (PflSigXOE2), P. fluorescens M114 (PflSigXM11), and P. syringae pv. syringae LMG 1247 (PsySigX), as well as four other selected ECF sigma factors that have had their sigma factor function confirmed (P. aeruginosa AlgU [PaeAlgU], M. tuberculosis SigE [MtuSigE], Streptomyces coelicolor SigE [ScoSigE], and Bacillus subtilis SigX [BsuSigX]). Note that for P. fluorescens M114 and P. syringae pv. syringae LMG 1247, the N-terminal sequence MNKAQT and the C-terminal sequence GETET correspond to parts of the primers used to amplify the genes and so do not necessarily reflect actual deduced SigX sequence for these organisms. The asterisk marks a possible alternate ATG start site for sigX. The bar with arrowheads marks the proposed location of the polymerase core binding region, and the bar with diamond ends is above the proposed helix-turn-helix motif region. Residues conserved in 50 to 74% of the shown sequences are shaded in grey, residues conserved in 75 to 99% of the sequences are shaded in dark grey, and residues conserved in 100% of the sequences are shaded in black.
FIG. 2.
Schematic diagram of the organization of genes surrounding sigX and oprF in P. aeruginosa H103 and P. fluorescens OE 28.3. The same gene organization is observed in both species, and a combination of PCR and sequence data suggests that cmpX (putative cytoplasmic membrane protein X), sigX, and oprF are also conserved in size and gene order in P. syringae pv. syringae (data not shown). ORFs are shown as thick arrows, and the open boxes mark the locations of putative rho-independent transcription terminators. The sequence of the cobA gene in P. fluorescens has been previously reported (accession no. U09566), but the P. aeruginosa cobA gene was deduced from the Pseudomonas Genome Project sequence (20a). All other genes are putative, and similarity of cmaX, menG, estX, and ppsA was observed to an unidentified ORF from E. coli (accession no. AE000232), the E. coli S-adenosyl-methione-2-demethyl menaquinone methyl transferase gene menG, human gastric lipase, and phosphoenol pyruvate synthase, respectively. The diagram is not drawn accurately to scale.
Based on sequence similarity with certain other ECF sigma factor genes (e.g., the genes for P. aeruginosa AlgU and Mycobacterium tuberculosis SigE in Fig. 1), the start site for sigX in both organisms was designated as a TTG codon, 585 bp upstream of the gene’s stop codon. However, upstream of this start codon is a very weak ribosome binding site. Another very likely start codon is an in-frame ATG that is 468 bp upstream of the stop codon and has a better ribosome binding site upstream that is well conserved in both Pseudomonas species.
We were also able to PCR amplify a similar sigX sequence from upstream of oprF in P. syringae pv. syringae, P. fluorescens M114 (Fig. 1), and six clinical isolates of P. aeruginosa. SigX was highly conserved in all of these pseudomonads. The six P. aeruginosa clinical isolates (H246, H344, H397, H411, H580, and H813) had identical sigX sequences, except for H411, which had a single silent T-to-C transition mutation 369 bp upstream of the proposed stop codon for sigX.
Transcriptional linkage and analysis of other genes upstream of oprF.
Other probable genes in the same orientation as sigX-oprF that were similar in sequence and gene order in both P. aeruginosa H103 and P. fluorescens OE 28.3 were identified by sequencing the region upstream of sigX. A schematic diagram of this region is shown in Fig. 2, with the genes described in the respective GenBank sequence submissions.
To examine possible transcriptional linkage among these genes, RT-PCR experiments were performed with both P. aeruginosa and P. fluorescens with primers that flanked the intergenic sequences within this region (data not shown). A primer complementary to a sigX sequence and a primer complementary to the 5′ end of oprF were able to successfully amplify a PCR product of the expected size from reverse-transcribed total RNA isolated from log-phase wild-type cells. Primers flanking the intergenic region between cmpX and sigX and primers coupling cmaX-crfX, crfX-cmpX, menG-cmaX, and estX-menG were also able to produce a PCR amplicon. No PCR amplicon or, in one instance, a very faint PCR amplicon was obtained for primers coupling ppsA and estX in P. aeruginosa (this region contained a putative transcription terminator in P. aeruginosa). These results are consistent with the concept that there is some degree of transcriptional linkage between neighboring genes in this region from estX to oprF.
Disruption of sigX reduces OprF expression: protein and transcriptional analysis.
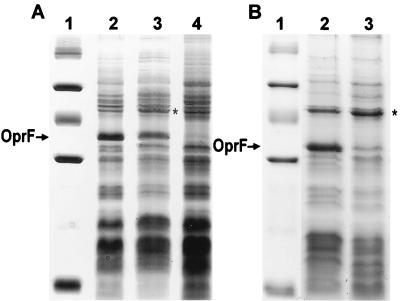
In both P. fluorescens OE 28.3 and P. aeruginosa H103, we constructed disruptions of sigX. These disruptions also terminated any transcript produced from a promoter upstream of sigX, since rho-independent transcription terminators flank the inserted antibiotic resistance cassettes. Examination of outer membrane protein preparations, cell envelope preparations (Fig. 3), and whole-cell lysates revealed that there was an estimated 2- to 10-fold reduction in the levels of OprF as a result of this disruption. This was confirmed by a Western immunoblot probed with an anti-OprF monoclonal antibody (data not shown). A slight increase in another unidentified outer membrane protein (approximately 47 kDa) was also observed for the sigX-disrupted mutant versus wild type for both species (Fig. 3) (also observed in outer membrane preparations). This change was not observed in the OprF-deficient mutant for each species. Comparative two-dimensional SDS-PAGE analysis of whole-cell proteins of the sigX::Kmr mutant and wild-type strains of P. fluorescens did not reveal other quantitative changes or qualitative changes (data not shown).
FIG. 3.
Coomassie blue-stained SDS-PAGE gel of cell envelope preparations of P. aeruginosa (A) and P. fluorescens (B) cells. (A) Lane 1, molecular weight marker (103, from top to bottom) 94, 67, 43, 30, 20.1, and 14.4; lane 2, P. aeruginosa H103 (wild type); lane 3, P. aeruginosa H814 (sigX disrupted); lane 4, P. aeruginosa H636 (oprF disrupted). (B) Lane 1, molecular weight marker (same as for panel A); lane 2, P. fluorescens OE 28.3 (wild type); lane 3, P. fluorescens FAJ2030 (sigX disrupted). The position of the band corresponding to OprF is indicated with an arrow next to both gels. Note that OprF in P. fluorescens OE 28.3 does not contain the “disulfide bridge” region (a 23-amino-acid insertion containing two disulfide bonds) found in OprF proteins in some pseudomonad strains, including P. aeruginosa H103, and so migrates further when subjected to SDS-PAGE. Note also that the cell envelope preparations shown were picked to indicate the variability in the level of reduction of OprF. The P. aeruginosa sigX mutant would frequently have a more marked reduction in OprF expression than is observed here. An asterisk marks the location of a 47-kDa protein that is increased in expression in the sigX mutant.
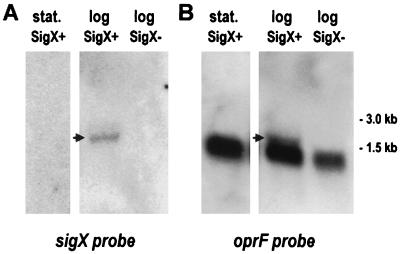
To determine whether the reduction in OprF in the sigX-disrupted mutant was due to a polar effect (because of cotranscription of sigX-oprF) or to a role of SigX in the transcription of monocistronic oprF, Northern blots of total RNA from P. aeruginosa cells grown to different stages of growth were probed with PCR-amplified DNA fragments containing either sigX or oprF sequences. The probes were prepared (see Materials and Methods) to ensure that they were free of any contaminating flanking sequences. For cells grown overnight (stationary phase) in LB medium with 200 mM salt, a 1.2-kb transcript predominated with the oprF probe, corresponding to monocistronic transcription of oprF (Fig. 4), though faint transcripts of sizes possibly corresponding to cmpX-sigX-oprF and to sigX-oprF were visible upon overexposure of the Northern blot. However, cells grown to the early logarithmic or mid-logarithmic stages of growth clearly contained the predominant 1.2-kb transcript plus an additional minor transcript that hybridized with both the sigX and oprF probes, indicating cotranscription of these genes and confirming their linkage by RT-PCR (Fig. 4). The size of this putative sigX-oprF transcript seemed too small to accommodate the size of sigX predicted from the TTG start site mentioned above, consistent with the proposed alternate ATG start site for the gene; however, the size was not accurately determined. For similarly probed blots containing RNA from the sigX-disrupted mutant, the sigX-oprF transcript was not observed and there was a reproducible, marked reduction in the amount of the 1.2-kb monocistronic oprF transcript (Fig. 4). The lack of sigX-oprF transcript was also confirmed by RT-PCR (with the same primers used above to show linkage between these genes in the wild-type strain). Densitometric analysis of these data (as well as comparisons of different amounts loaded on a gel) suggests that these reductions in sigX-oprF and moncistronic oprF transcript levels could account for the reduction in OprF expression observed. These data therefore suggest that the reduction of OprF expression in the sigX mutant is due to an effect of SigX on transcription of monocistronic oprF (either directly or indirectly) combined with a more minor polar effect of the knockout on the downstream oprF gene.
FIG. 4.
Northern blots of P. aeruginosa wild-type and (SigX+) and sigX mutant (SigX−) RNA obtained from log-phase (log) and stationary-phase (stat.) cells. The RNA was probed with sigX (A) and oprF (B) sequences. The arrows mark the bands corresponding to a proposed sigX-oprF transcript. The thicker bands observed in the blots probed with oprF correspond to the monocistronic oprF transcripts.
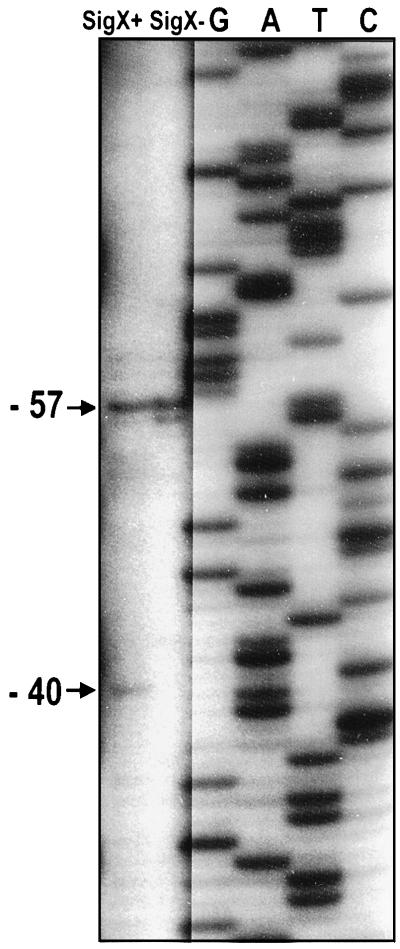
The effect of SigX on transcription of monocistronic oprF was further examined through primer extension analysis. Experiments were performed with a primer complementary to the reverse strand of the 5′ end of oprF and with P. aeruginosa RNA extracted from wild-type and sigX mutant cells grown in LB medium with 200 mM salt to mid-logarithmic stages of growth. A major primer extension product (Fig. 5), of equal intensity for both wild-type and sigX mutant cells, was observed that corresponded to the sigma 70 transcriptional start site previously reported by Duchêne et al. (5) 57 bp upstream of oprF. However, an additional primer extension product was also observed 40 bp upstream of oprF that was present for wild-type cells but absent from the sigX mutant (Fig. 5). These results, therefore, suggest that there are two overlapping promoters upstream of oprF (Fig. 6), one which is SigX independent and one which is SigX dependent. Upstream of the SigX-dependent start site in both the P. aeruginosa and P. fluorescens sequences were possible −35 regions (GAAGTT in P. aeruginosa and CAAGTT in P. fluorescens) which shared notable similarity to consensus −35 sequences for other ECF sigma factors (11, 16) (Fig. 6). The −10 region is not normally conserved among different ECF sigma-type promoters and in this case was proposed to be GTTGTG and GTTGTC in P. aeruginosa and P. fluorescens, respectively (Fig. 6).
FIG. 5.
Primer extension analysis of transcriptional start sites immediately upstream of oprF in wild-type (H103) (SigX+) and sigX mutant (H814) (SigX−) P. aeruginosa. Cells were harvested from high-salt LB broth at mid-log phase. The band corresponding to a SigX-dependent transcriptional start site is marked −40, indicating the start site’s position upstream of the initiation codon of oprF. The band corresponding to the SigX-independent start site is at location −57 from the start of oprF. The cutting and pasting for this figure was necessitated by the fact that it required different exposure times to optimally visualize the sequencing ladder (G, A, T, C) and primer extension products.
FIG. 6.
Alignment of P. aeruginosa H103 (P. aerug) and P. fluorescens OE 28.3 (P. fluor) sequence between sigX and oprF (boxed), showing the location of putative promoter sequences. Identical bases are indicated with a colon. The two transcriptional start sites, as determined from primer extension analysis (Fig. 5), are indicated with arrows, and the nucleotides (−35 and −10 sites) proposed to be associated with the SigX-dependent promoter and the SigX-independent promoter are marked with underlined italics and underlined boldface characters, respectively. The −35 site for the SigX-independent promoter proposed by Duchêne et al. (5) seems somewhat unlikely based on the lack of alignment between the P. aeruginosa and P. fluorescens sequences and the nonoptimal 20-nucleotide spacing from the −10 site, but it was not further investigated in this paper.
Growth rate of the sigX-disrupted mutant: evidence that SigX regulates genes other than oprF.
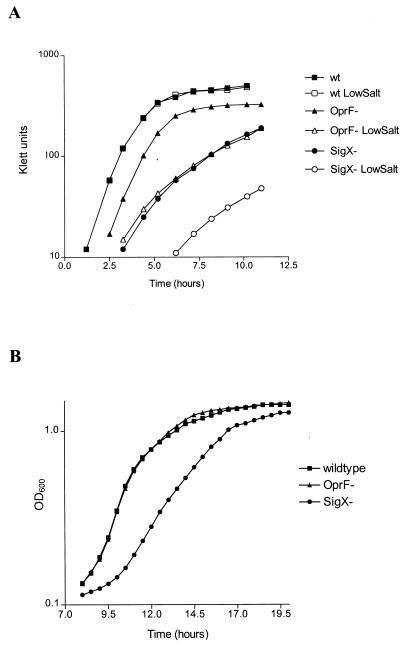
The most noticeable phenotype associated with the sigX disruption was a marked reduction in logarithmic-phase growth rate and/or an increase in lag phase for cultures of the mutant grown in various media (Fig. 7 and Table 2). For P. aeruginosa, this reduction in growth rate was particularly apparent under low-salt conditions (8 mM), and the mutant did not grow at all in medium containing no salt. OprF-deficient P. aeruginosa is also unable to grow in medium containing no salt (30); however, an oprF-disrupted strain grew with a shorter doubling time than the sigX knockout in LB medium containing low salt (8 mM [Table 2]). The inclusion of 200 mM salt in LB medium restored the growth of the OprF-deficient strain to near wild-type levels, but only partially reversed the growth defect of the sigX::Gmr mutant. When the sigX mutant was complemented with oprF on a plasmid (by using a cloned oprF gene with a mutated promoter, resulting in expression of relatively normal levels of OprF, as confirmed for the complemented sigX mutant), the growth rate was not restored to wild-type levels under any growth condition examined, including low-salt media (Table 2). This indicated that factors other than the reduced expression of OprF played a role in the inability of the sigX-disrupted mutant to grow like its parent wild type in low-salt medium.
FIG. 7.
Growth of P. aeruginosa (A) and P. fluorescens (B) SigX mutants (circles) and parent wild-type (squares) and OprF-deficient strains (triangles) in LB medium. Solid symbols denote cultures grown in high-salt (200 mM) media, and open symbols are used for cultures grown in low-salt (8 mM) media. OD600, optical density at 600 nm.
TABLE 2.
Influence of OprF and SigX on doubling times of P. aeruginosa and P. fluorescens grown in different liquid media with constant aeration
| Growth conditionsa | Doubling time (min)
|
||||||
|---|---|---|---|---|---|---|---|
|
P. aeruginosa
|
P. fluorescens
|
||||||
| H103 (wild type) | H636 (OprF−) | H814 (SigX−) | H845 (SigX− + OprF) | OE 28.3 (wild type) | FAJ2026 (OprF−) | FAJ2030 (SigX−) | |
| LB + 8 mM salt | 30 | 54 | 90 | 78 | ND | ND | ND |
| LB + 200 mM salt | 30 | 30 | 54 | 51 | 27 | 27 | 36 |
| LB + 200 mM salt at 22°C | 73 | 77 | 125 | ND | ND | ND | ND |
| LB + 500 mM salt | 36 | NDb | 60 | 54 | ND | ND | ND |
| LB + 500 mM salt + succinatec | 36 | ND | ND | 54 | ND | ND | ND |
| MMd + 200 mM salt | 42 | 51 | 63 | 54 | 38 | 42 | 53 |
| MM + 500 mM salt | 54 | 81 | 63 | 54 | 54 | ND | 55 |
| MM + 500 mM KCl | 54 | ND | 60 | 60 | ND | ND | ND |
| MM + 500 mM sucrose | 54 | ND | 57 | 54 | ND | ND | ND |
All P. aeruginosa cultures were grown at 37°C and P. fluorescens cultures were grown at 30°C unless otherwise noted.
ND, not done.
7 mM succinate.
MM, minimal medium (BM2).
A reduction in growth rate was observed regardless of whether the sigX mutant was grown in LB, Mueller Hinton, or brain heart infusion broth, and smaller colonies were observed for the mutant and the wild type when grown for the same length of time on chocolate agar, indicating that even very rich medium did not complement this slow-growth phenotype. In minimal medium (BM2), the differences in doubling time between the wild type and the sigX mutant were less pronounced but still apparent when the medium contained high salt (200 or 171 mM—both concentrations produced essentially identical results). With the addition of very high salt to the minimal medium (500 mM) the growth rate of the wild type and the sigX mutant became similar (Table 2). While there was slower growth of the wild type in this very-high-salt environment, it is notable that the growth rate of the sigX mutant did not correspondingly decrease, as it did when grown under other stress conditions, such as extremes of pH (data not shown) or low temperature (Table 2). A similar growth rate for the wild type and the sigX::Gmr mutant was also observed when the high salt in this minimal medium was replaced by high sucrose or KCl (both 500 mM [Table 2]). This suggests that it was not high salt, but rather high osmolarity, that allowed the mutant to grow at the same rate as the wild type. However, the mutant still grew more slowly than the wild type in LB medium containing 500 mM salt (Table 2), suggesting that osmolarity was not the only factor affecting growth of the sigX mutant in rich media. Addition of succinate to the LB medium containing 500 mM salt did not restore the sigX mutant to wild-type growth, showing that the differences between growth in LB and minimal media under these very-high-salt conditions were not solely due to differences in carbon sources that the bacterium could utilize in each medium.
For P. fluorescens, the growth rate differences between the wild type and the sigX mutant grown in rich medium (LB) were less pronounced than that observed for the corresponding P. aeruginosa strains; however, differences were still apparent (Table 2). A notable difference in lag phase was observed, with smaller changes in growth rate, suggesting that the sigX knockout may affect different genes (or possibly fewer genes) in P. fluorescens than in P. aeruginosa.
Other phenotypes resulting from the disruption of sigX in P. aeruginosa.
Some small but notable MIC differences for some antimicrobials and metals were observed for the P. aeruginosa sigX mutant versus its parent wild type. The sigX knockout was 4-fold more susceptible to imipenem, 2-fold more susceptible to polymyxin B, and 1.5- to 2-fold more resistant to chromate ions and the fluoroquinolone enoxacin (see Materials and Methods for a full list of antimicrobials and metals examined). A MAR phenotype was not seen for this sigX mutant, indicating that it does not play a direct role in determining the phenotype of MAR OprF-deficient clinical isolates (20).
Another phenotypic change involved the production of an unidentified bright-yellow pigment in the P. aeruginosa sigX mutant. When grown in LB, this pigment appeared as the culture reached an optical density at 610 nm of 0.7, while the wild type remained colorless at this stage of growth; after overnight growth, the mutant always had a more intense yellow color. This pigment, which was secreted into the culture supernatant, was not fluorescent and, in aqueous solution at pH 7.5 to 8.0, had an absorption maximum of 380 nm. However, we are uncertain of the significance of the pigment, since alterations in the levels of Pseudomonas pigments are commonly associated with different growth conditions and mutations (27).
The magnitude of differences in growth rate between the P. aeruginosa wild type and the sigX mutant remained similar during growth at low or high temperatures (8, 15, 22, 42, and 45°C), at low or high pH (pHs 3, 4, 5, 9, 10, 11, and 12), or under anaerobic conditions. Similarly, no significant differences were observed for uptake of the hydrophobic probe N-phenyl-1-naphthylamine (13), which is often used to measure changes in outer membrane permeability to hydrophobic compounds and/or efflux changes. The sigX mutant was motile and had metabolic profiles similar to those of the wild type, according to Biolog GN Microplate and API20 NE strip tests. When examined under a microscope at ×100, the cells did not appear to be different from the wild type (while OprF-deficient cells are noticeably shorter [30]).
DISCUSSION
Our results indicate that OprF in P. aeruginosa is not just expressed constitutively from a sigma 70 promoter but rather the sigX gene product, a probable ECF sigma factor, plays a role in its expression. Transcriptional linkage of sigX and oprF was detected, and there appear to be two overlapping promoters immediately upstream of oprF, one independent of SigX and resembling the sigma 70 consensus sequence and another that is SigX dependent and resembles an ECF sigma promoter. This SigX-dependent transcription was not previously identified in the study by Duchêne et al. (5); however, their results are consistent with our results obtained for late-log- or stationary-phase cells, when sigX is not well expressed. The transcript initiating from the sigma 70-like promoter is still a significant source of oprF transcript; however, clearly whatever conditions effect sigX transcription may also have an impact on OprF expression.
OprF, and its deficiency in some clinical isolates of P. aeruginosa, has been previously correlated with significant antibiotic resistance; however, the level of antibiotic resistance in both an oprF knockout (29) and the sigX knockout described here is modest and does not match the resistance observed in an OprF-deficient clinical isolate (20). Since OprF is apparently subject to more complex regulation than was previously thought, it is possible that the MAR- and OprF-deficient clinical phenotype is due to a regulatory mutation that affects both OprF and MAR determinants. The one clinical isolate studied in detail to date is notable for its frequent reversion to normal antimicrobial sensitivity and an OprF+ phenotype in a single step, and it contains no significant mutations in the oprF gene or in the upstream promoter region (20, 21, 21a). However, our studies demonstrate that this regulatory mutation would not likely be within the sigX gene, since sigX disruption did not result in complete OprF deficiency and the MAR phenotype was not observed for the sigX mutant. Though a MAR phenotype was not observed, the increased imipenem sensitivity of the P. aeruginosa sigX mutant is interesting, since there was increased expression of a 47-kDa protein in the sigX mutant that is approximately the size of OprD, an outer membrane protein known to be involved in uptake of imipenem.
Other genes subject to control by SigX have not yet been identified; however, phenotypic analysis of the two sigX knockout mutants, and the P. aeruginosa sigX mutant complemented with oprF on a plasmid, clearly indicated that other genes require SigX for expression. The sigX mutant showed a marked inability to grow in low-osmolarity medium, even more so than an OprF-deficient mutant, and was only able to grow at the same rate as the wild type in a very-high-osmolarity environment. This would suggest that SigX is involved, directly or indirectly, in growth and/or survival in low-osmolarity environments. SigX was clearly required for optimal growth in a low-osmolarity environment, a notable factor for both P. aeruginosa and P. fluorescens, given that their niches include water. SigX in P. aeruginosa also seemed to be required for utilization of nutrients in rich medium such as LB medium), since growth of the mutant was always markedly slower than that of the wild type in rich media, even under the very-high-salt conditions in which the wild type and mutant grew at similar rates in minimal medium.
In conclusion, we have presented evidence that a probable ECF sigma factor plays a role in OprF expression and that this putative sigma factor also influences other genes. These results shed new light on the nature of the regulation and expression of OprF, which could impact on our understanding and study of this protein’s involvement in rhizosphere colonization by P. fluorescens and its relationship with clinical antibiotic resistance in P. aeruginosa.
ACKNOWLEDGMENTS
This work was funded in part by the Canadian Cystic Fibrosis Foundation (CCFF), the Medical Research Council (MRC) of Canada, and the Biotechnology Program of the European Union (BIO2-CT93-0196). R.E.W.H. is a recipient of the MRC Distinguished Scientist Award. F.S.L.B. is the recipient of a fellowship from the CCFF. R.D. is a Senior Research Associate with the Fund for Scientific Research (Flanders).
We acknowledge Colin Crist for studies of the conservation of the genes in P. syringae, Rick Heffernan for help with phenotypic analysis of the SigX mutant in P. aeruginosa, Alex Beysen for constructing P. fluorescens FAJ2026, and Margaret Pope for aid with primer extension experiments with P. aeruginosa.
REFERENCES
- 1.De Mot R, Proost P, Van Damme J, Vanderleyden J. Homology of the root adhesin of Pseudomonas fluorescens OE 28.3 with porin F of P. aeruginosa and P. syringae. Mol Gen Genet. 1992;231:489–493. doi: 10.1007/BF00292721. [DOI] [PubMed] [Google Scholar]
- 2.De Mot R, Schoofs G, Roelandt A, Declerck P, Proost P, Van Damme J, Vanderleyden J. Molecular characterization of the major outer membrane protein OprF from plant root-colonizing Pseudomonas fluorescens. Microbiology. 1994;140:1377–1387. doi: 10.1099/00221287-140-6-1377. [DOI] [PubMed] [Google Scholar]
- 3.De Mot R, Vanderleyden J. Application of two-dimensional protein analysis for strain fingerprinting and mutant analysis of Azospirillum species. Can J Microbiol. 1989;35:960–967. [Google Scholar]
- 4.De Mot R, Vanderleyden J. Purification of a root-adhesive outer membrane protein of root-colonizing Pseudomonas fluorescens. FEMS Microbiol Lett. 1991;81:323–328. [Google Scholar]
- 5.Duchêne M, Schweizer A, Lottspeich F, Krauss G, Marget M, Vogel K, von Specht B-U, Domdey H. Sequence and transcriptional start site of the Pseudomonas aeruginosa outer membrane porin protein F gene. J Bacteriol. 1988;170:155–162. doi: 10.1128/jb.170.1.155-162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farinha M A, Kropinski A M. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;70:221–226. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- 7.Fenton A M, Stephens P M, Crowley J, O’Callaghan M, O’Gara F. Exploitation of gene(s) involved in 2,4-diacteylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh N, Wakebe H, Yoshihara E. Role of protein F in maintaining structural integrity of the Pseudomonas aeruginosa outer membrane. J Bacteriol. 1989;171:983–990. doi: 10.1128/jb.171.2.983-990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W, Irwin R T, Costerton J W, Carey A M. The outer membrane of Pseudomonas aeruginosa: peptidoglycan associated proteins. J Bacteriol. 1981;145:628–631. doi: 10.1128/jb.145.1.628-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Helmann J D. Identification of target promoters for the Bacillus subtilis ςX factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- 12.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh B, Grant C, Hancock R E W. Use of the fluorescent probe 1-N-phenylnapthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller H. Practical aspects of preparing phage and plasmid DNA: growth, maintenance, and storage of bacteria and bacteriophage. Methods Enzymol. 1987;152:145–170. doi: 10.1016/0076-6879(87)52016-x. [DOI] [PubMed] [Google Scholar]
- 16.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 17.Mutharia L M, Hancock R E W. Surface localization of Pseudomonas aeruginosa outer membrane porin protein using monoclonal antibodies. Infect Immun. 1983;42:1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy I, Schoofs G, De Schrijver A, Vanderleyden J, De Mot R. New method for RNA isolation from actinomycetes: application to the transcriptional analysis of the alcohol oxidoreductase gene thcE in Rhodococcus and Mycobacterium. Lett Appl Microbiol. 1997;25:75–79. doi: 10.1046/j.1472-765x.1997.00179.x. [DOI] [PubMed] [Google Scholar]
- 19.Nicas T I, Hancock R E W. Pseudomonas aeruginosa outer membrane permeability: isolation of a porin F-deficient mutant. J Bacteriol. 1983;153:281–285. doi: 10.1128/jb.153.1.281-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piddock L J, Hall M C, Bellido F, Bains M, Hancock R E W. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1057–1061. doi: 10.1128/aac.36.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Pseudomonas Genome Project. 15 March 1999, posting date. Sequence data. [Online.] http://www.pseudomonas.com. [20 June 1999, last date accessed.]
- 21.Pumbwe L, Everett M J, Hancock R E W, Piddock L J. Role of gyrA mutation and loss of OprF in the multiple antibiotic resistance phenotype of Pseudomonas aeruginosa G49. FEMS Microbiol Lett. 1996;143:25–28. doi: 10.1111/j.1574-6968.1996.tb08456.x. [DOI] [PubMed] [Google Scholar]
- 21a.Pumbwe, L., et al. Unpublished data.
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 24.Simon R, O’Connell M, Labes M, Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other Gram-negative strains. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 25.Smith R L, Gottlieb E, Kucharski L M, Maguire M E. Functional similarity between archaeal and bacterial CorA magnesium transporters. J Bacteriol. 1998;180:2788–2791. doi: 10.1128/jb.180.10.2788-2791.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith R L, Banks J L, Snavely M D, Maguire M E. Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J Biol Chem. 1993;268:14071–14080. [PubMed] [Google Scholar]
- 27.Turner J M, Messenger A J. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 27a.Wong, R. S. Unpublished results.
- 28.Woodruff W A. Ph.D. thesis. Vancouver, British Columbia, Canada: University of British Columbia; 1989. [Google Scholar]
- 29.Woodruff W A, Hancock R E W. Construction and characterization of Pseudomonas aeruginosa porin protein F-deficient mutants after in vivo and in vitro mutagenesis of the cloned protein F gene in Escherichia coli. J Bacteriol. 1988;170:2592–2598. doi: 10.1128/jb.170.6.2592-2598.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodruff W A, Hancock R E. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171:3304–3309. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]