Abstract
Furin is an important mammalian proprotein convertase that catalyzes the proteolytic maturation of a variety of prohormones and proproteins in the secretory pathway. In the brain, the substrates of furin include the proproteins of growth factors, receptors and enzymes. Emerging evidence, such as reduced FURIN mRNA expression in the brains of Alzheimer’s disease patients or schizophrenia patients, has implicated a crucial role of furin in the pathophysiology of neurodegenerative and neuropsychiatric diseases. Currently, compared to cancer and infectious diseases, the aberrant expression of furin and its pharmaceutical potentials in neurological diseases remain poorly understood. In this article, we provide an overview on the physiological roles of furin and its substrates in the brain, summarize the deregulation of furin expression and its effects in neurodegenerative and neuropsychiatric disorders, and discuss the implications and current approaches that target furin for therapeutic interventions. This review may expedite future studies to clarify the molecular mechanisms of furin deregulation and involvement in the pathogenesis of neurodegenerative and neuropsychiatric diseases, and to develop new diagnosis and treatment strategies for these diseases.
Keywords: Furin, Proteolytic cleavage, Neurodegenerative disease, Neuropsychiatric disease, Brain-derived neurotrophic factor
Introduction
Furin is the first proprotein convertase (PC) found in mammals in 1990 [1]. It catalyzes the proteolytic maturation of large numbers of prohormones and proproteins in the secretory pathway compartments [1–3]. The substrates of furin include hormones, cytokines, growth factors and enzymes, which play important roles in cell proliferation, anti-apoptosis, immunity and inflammation [1]. Furin also participates in the proteolytic processing of proteins in viruses and bacteria [4], such as the maturation of SARS-CoV-2 spike protein [5–7]. Thus, aberrant activity of furin has been found to be associated with a strikingly diverse range of pathological events, including cancer, cardiovascular disorders, infectious diseases and neurological diseases [4, 8–10]. Among these disorders, the role of furin in neurological diseases is the most poorly understood.
In the brain, the proprotein substrates cleaved by furin in vivo include precursors of growth factors such as brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) [11, 12], α- and β-secretases [13, 14], multiple matrix metalloproteases (MMPs) [15, 16], and other enzymes and receptors [1, 17, 18]. Since these substrates play vital roles in neuronal survival, axon growth, dendritic development, synaptogenesis, neurodegeneration and inflammation [19–22], a stable activity of furin is crucial for maintaining the homeostasis of the central nervous system.
A growing body of evidence has suggested that alterations of furin expression and abnormal cleavage of its substrates contribute to the pathophysiological mechanisms of neurodegenerative and neuropsychiatric diseases. Reduced expression of FURIN mRNA has been found in the brains of Alzheimer’s disease (AD) patients [13], and decreased protein levels of furin are found in the cortex of AD mice [23]. The FURIN mRNA expression is decreased in the prefrontal cortex of schizophrenia (SCZ) patients [24], whereas increased protein levels of furin are found in the temporal cortex of epilepsy patients [25]. Moreover, studies have also shown that increasing furin expression in the mouse brain enhances BDNF maturation and promotes dendritic spine density and memory in transgenic mice [26], and that inhibiting furin expression reduces the spontaneous rhythmic electrical activity of cerebral neurons and suppresses epileptic seizure activity in epileptic mice [25]. These findings indicate the involvement of furin dysregulation in these neurological disorders, leading to increased interest in furin as a potential biomarker for diagnosis of or as a therapeutic target for treatment of neurological disorders.
In this review, we present an overview on the physiological roles of furin in the brain and deregulations of furin expression and its substrates in neurodegenerative and neuropsychiatric disorders, such as AD, Parkinson’s disease (PD), epilepsy, cerebral ischemia, SCZ and depression. We further discuss the implications of these findings and current approaches that target furin for therapeutic interventions.
Overview of furin
Gene structure and transcription of FURIN
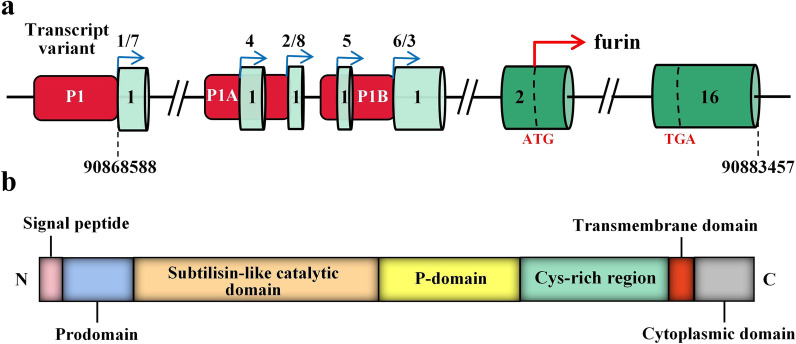
Furin was identified in 1990 as the first mammalian PC that catalyzes the proteolytic maturation of prohormones and proproteins of neurotrophic factors, receptors and enzymes, serum proteins and pathogen molecules [1–3]. The human FURIN gene is located at chromosome 15q26.1, an open reading frame upstream of the fes/fps proto-oncogene [27]. It has attracted more attention after being discovered as the first mammalian homologue of yeast Kex2 [4, 28, 29]. As shown in Fig. 1a, the human FURIN gene consists of 16 exons and encodes eight different transcript variants driven by three known promoters, P1, P1A and P1B [30, 31]. The respective transcripts differ only in the first untranslated exon and therefore generate identical furin precursor proteins [30, 32]. While the P1A and P1B promoters resemble those of constitutively expressed housekeeping genes, the P1 promoter is predicted to bind to many different transcription factors, including hypoxia-inducible factor-1 (HIF-1), C/EBPβ, and CREB (cAMP-responsive element binding protein) [33–36].
Fig. 1.
Human FURIN gene and furin protein structures. a The human FURIN gene consists of 16 exons and encodes eight different transcript variants driven by three known promoters, P1, P1A and P1B. Exons are shown as green boxes and introns are shown as lines. The red boxes indicate the three promoter regions. The blue arrows indicate the positions where different transcripts start. The red arrow indicates the translational start, and the start codon (ATG) and stop codon (TGA) are marked with dotted lines. b Furin protein contains an N-terminal signal peptide, a prodomain, a subtilisin-like catalytic domain, a middle P-domain, a cysteine-rich region, a transmembrane helix domain and a C-terminal cytoplasmic domain
Several intracellular and extracellular factors have been reported to regulate FURIN expression at the transcriptional level. Hypoxia remarkably increases the expression of FURIN mRNA via stabilizing HIF-1 and enhancing its binding to hypoxia-responsive element site at the P1 promoter [37]. Iron deficiency also upregulates FURIN transcription through stabilization of HIF-1α [35], whereas iron overload inhibits furin expression in a non-HIF-1α-dependent manner [35]. Transforming growth factor beta1 (TGFβ1) can induce transactivation of the FURIN P1 promoter through binding to Sma- and Mad-related protein 2 (SMAD2) and SMAD4 in complex with other DNA-binding partners, creating a constitutive activation/regulation positive feedback loop between TGFβ1 and furin [38]. Furthermore, extracellular regulated protein kinase 1 has been found to mediate the TGFβ–furin feedback loop in glioma-initiating cells [39]. In addition, bone morphogenetic protein 2 increases the transcription and translation of furin in human granulosa lutein cells by the activin receptor-like kinase (ALK)2/ALK3-SMAD4 signaling pathway [40].
Protein structure and expression of furin
Furin is a type I transmembrane protein and belongs to the subtilisin-like convertase family [1]. It is a calcium-dependent endoserine protease [8]. Furin protein is composed of a signal peptide, a prodomain, a subtilisin-like catalytic domain, a middle P-domain, a cysteine-rich region, a transmembrane helix domain and a cytoplasmic domain (Fig. 1b) [41]. The large extracellular region of furin has an overall homology with the same region of other members of the PC family [1]. The signal peptide directs translocation of the ~ 104-kDa pro-enzyme into the endoplasmic reticulum (ER), where the first cleavage in the inhibitory prodomain takes place via autocatalytic cleavage by the catalytic domain [42–44]. The second cut in the prodomain is made during trafficking of the propeptide-furin complex within the mildly acidic trans-Golgi network/endosomal system, which yields the active ~ 81-kDa mature enzyme. Furin circulates between the trans-Golgi network, cell surface and endosomes, in a tightly regulated manner, to catalyze various proproteins in different cellular components [43, 45, 46].
Furin is ubiquitously expressed in vertebrates and many invertebrates [9, 47, 48]. However, its mRNA and protein levels vary depending on the tissue and cell type [49–53]. FURIN has been found at high mRNA levels in the salivary gland, placenta, liver and bone marrow, and high protein levels in the brain, salivary gland, pancreas, kidney and placenta [49–53]. However, almost no expression is detected in skin, muscle and adipose tissues [49, 50], although substrates of furin have been identified in human adipose tissues [54]. In normal single cells, high expression of FURIN mRNA is identified in hepatocytes, exocrine glandular cells, pancreatic endocrine cells and syncytiotrophoblasts [50, 53]. This tissue- and cell-specific expression pattern of furin infers the different functions of furin in different tissues and organ systems.
Function of furin
Furin cleaves proproteins at the consensus site of Arg–X–Lys/Arg–Arg or Arg–X–X–Arg (X refers to any amino acid) [55, 56], and the cut is positioned after the carboxyl-terminal Arg residue [56]. The substrates cleaved by furin include a variety of precursor proteins within the secretory pathway, including hormones, growth factors and their receptors, neuropeptides, enzymes, adhesion molecules, metalloproteinases, bacterial toxins and viral glycoproteins [8, 29, 33]. As these molecules participate in many important cellular events, mouse embryos lacking Furin will die between days 10.5 and 11.5, with notable defects in ventral closure and axial rotation [57]. Deregulations of furin expression are found in diverse pathological conditions, including cancer, diabetes, cardiovascular disorders, inflammation and neurological diseases [10, 58–62].
Furin and its substrates in the brain
Furin expression in the brain
Brain is one of the organs that show the highest level of furin protein [50], particularly in the cerebral cortex, hippocampus and cerebellum, where the furin level is as high as that in the salivary gland [50]. Moreover, it has been reported that in the brains of epilepsy patients and epileptic mice, furin is predominantly expressed in neurons in the cortex and hippocampus, but barely in glial cells [25]. Double immunofluorescence staining showed a neuron-specific pattern of furin expression in the hippocampal CA3 and dentate gyrus (DG) regions in wild-type mice [63]. The neuron-specific expression may be related to the essential functions of furin in neurons. In addition, it has been reported that furin expression in glial cells may be increased in some pathological conditions as shown by the increase of furin expression in cultured rat astrocytes exposed to oxygen–glucose deprivation [64].
Substrates cleaved by furin in the brain
In the brain, the substrates proteolytically cleaved by furin include growth factors such as BDNF and NGF, proteases such as multiple MMPs, a disintegrin and metalloproteases (ADAMs) and beta-site APP cleaving enzyme 1 (BACE1), and receptors such as Notch receptor, low-density lipoprotein receptor-related protein 1 (LRP1), G protein-coupled receptor (GPR37) sortilin, integral membrane protein 2B (BRI2) and Ac45. Furin and its substrates potentially play important roles in diverse biological processes in the brain, including neuronal survival, differentiation, axonal outgrowth, dendritic development, synaptogenesis, inflammation and neurodegeneration (Fig. 2).
Fig. 2.
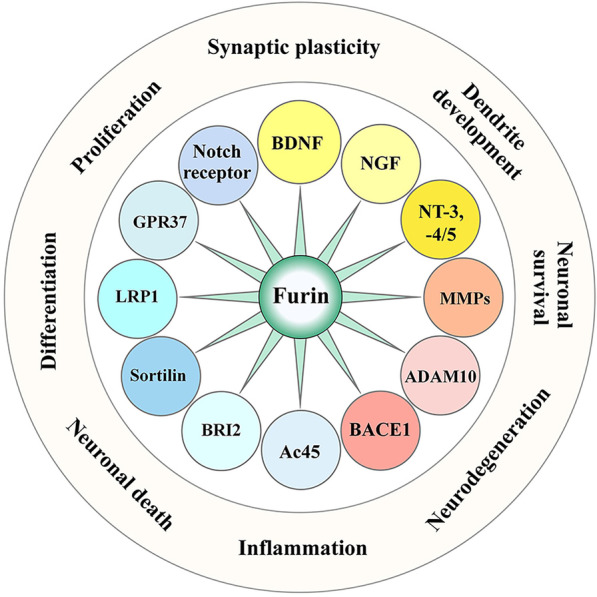
Activities mediated by furin and its substrates in the brain. The substrates of furin include growth factors such as BDNF and NGF, proteases such as MMPs, ADAMs and BACE1, and receptors such as Notch, LRP1, GPR37, sortilin, BRI2 and Ac45. They participate in diverse biological processes in the brain, including neuronal survival and death, proliferation and differentiation, dendritic development, synaptic plasticity, inflammation and neurodegeneration
BDNF
BDNF is a member of the neurotrophin family, which is widely distributed and extensively expressed in the brain [65–67]. BDNF is synthesized as pre-proBDNF and folded in the ER [68]. The pre-proBDNF harbors a signal peptide, a pro-domain and a mature domain [69], and is transported to the Golgi apparatus where it is converted into a full-length proBDNF (~ 32 kDa) after removal of the signal peptide [59]. The proBDNF is then cleaved by the protease furin to release the pro-domain and generate the biologically active ~ 14-kDa mature BDNF (mBDNF) [70]. The proBDNF can also be secreted into the extracellular space and then catalyzed by the extracellular proteases, such as MMPs [71, 72]. Furin is found to have higher efficiency than other PCs in cleavage of proBDNF in cultured rat astrocytes, and its aberrant activity leads to a significant change in mBDNF expression [64]. In terms of function, mBDNF binds to the tropomyosin-related receptor kinase B (TrkB) [73] and triggers downstream intracellular signaling pathways, including the phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt), the phospholipase C-γ/calcium-dependent protein kinase (PLCγ/CaMK), and the mitogen-activated protein kinase (MAPK)/ERK pathways [22, 74–76]. These signaling pathways mediate transcription of genes essential for neuronal survival, differentiation, axonal outgrowth, dendritic spine development, hippocampal long-term potentiation (LTP) and synaptic plasticity [22, 49, 74–76]. In contrast, proBDNF binds to p75 neurotrophic receptor (p75NTR) and induces apoptosis, spine shrinkage and long-term depression facilitation [77, 78]. Therefore, imbalances between proBDNF and mBDNF are involved in pathophysiological mechanisms of neurodegenerative diseases, as well as neuropsychiatric diseases [22, 73, 76, 79–81].
NGF
NGF is the first identified member of the neurotrophin family [82]. Like other proneurotrophins, the ~ 30-kDa proNGF is synthesized in the ER [83]. Its pro-domain is cleaved mainly intracellularly in the trans-Golgi network by furin, rather than in secretory granules by other PCs [84, 85], releasing the mature NGF (mNGF, ~ 17 kDa) [86, 87]. Similar to BDNF, proNGF and mNGF also differ significantly in receptor interaction properties and bioactivity. The mNGF binds to tropomyosin-related receptor kinase A (TrkA) and promotes cell survival, differentiation, growth and maintenance of specific types of neurons [88–90], whereas the proNGF binds to p75NTR with a high affinity and mediates neuronal cell death [91–93]. The balance between proNGF and mNGF levels is a key determinant of homeostasis in the brain, and disruption of the balance is associated with diseases such as epilepsy, AD, and ischemic stroke [94–96].
Other neurotrophins
The third type of growth factors of the neurotrophin family includes neurotrophin-3 (NT-3) and neurotrophin-4/5 (NT-4/5) [97, 98]. They are also synthesized as ~ 31–35-kDa precursors, and in turn proteolytically cleaved to release biologically active mature neurotrophins (~ 13–21 kDa) [84]. Similarly, intracellular cleavage of proneurotrophins is accomplished by furin [99]. The mature neurotrophins then bind to their corresponding receptors, the Trk family of receptor tyrosine kinases, and regulate neuronal survival and synaptic plasticity [100, 101]. Aberrant expressions of NT-3 and NT-4/5 participate in pathophysiological conditions including motor dysfunction, cognitive decline, stroke, and SCZ [102–107].
MMPs
MMPs are a family of zinc-dependent metalloproteases [108], with many members being reported to be expressed in the brain, such as MMP-1, MMP-2, MMP-3, MMP-7, MMP-9, MMP-14, and MMP-24 [108]. MMP-1 is expressed in both glia and neurons in the cortex, hippocampus and cerebellum [108, 109]; MMP-2 is mainly expressed in astrocytes [110]; MMP-3 is expressed in glia and neurons in the cerebellum, striatum and hippocampus [111]; and MMP-9 is mainly expressed in neurons in the cerebral cortex, hippocampus and cerebellum [112, 113]. Typically, MMPs consist of a signal peptide, a propeptide sequence, a catalytic metalloproteinase domain with zinc, a hinge region, and a hemopexin domain [114]. The signal peptide is removed in ER [115], and the propeptide is cut off by furin or other PCs at a furin-like recognition motif [116–118]. The MMPs are thus activated inside the cell before secretion or exposure to cell surface [119]. MMP-1 is shown to enhance the proliferation and neuronal differentiation of adult hippocampal neural progenitor cells via activating protease activated receptor 1 and subsequently increasing the cytoplasmic Ca2+ concentration [120, 121]. MMP-2 regulates astrocyte motility in connection with the actin cytoskeleton and integrins [122]. MMP-3 has a very broad range of substrates in the brain [123], and is upregulated in many pathological conditions, inducing neuroinflammation and apoptosis [124]. MMP-9 is specifically shown to regulate synaptic plasticity in the hippocampus by gain- and loss-of-function studies in vitro [125, 126]. Altered concentrations of MMP-3 and MMP-9 have been found in AD patients, indicating their involvement in AD pathophysiology [127]. MMP-1, MMP-2, MMP-9 and MMP-14 can cleave recombinant α-synuclein [128, 129]. Elevated levels of MMP-2 and MMP-3 have been identified in dopaminergic (DA) neurons in the substantia nigra in PD patients and animal models [129–131].
ADAM10
ADAMs are another major family of zinc-dependent metalloproteases involved in limited proteolysis and shedding [132]. In the brain, ADAM10 is mainly expressed in neurons [133], and is involved in the proteolytic processing of a variety of cell surface receptors and signaling molecules [134]. ADAM10 is synthesized in the ER as an inactive zymogen with a structure comprising a prodomain, a zinc-binding metalloprotease domain, a disintegrin domain, a cysteine-rich domain, a transmembrane domain and a C-terminal domain [133]. Furin cleaves the ~ 90 kDa pro-ADAM10, yielding a full-length active ADAM10 (∼65 kDa) [135], and after C-terminal shedding, a soluble ∼55-kDa ADAM10 is released [136]. ADAM10 has α-secretase activity [137]. It cleaves amyloid-β precursor protein (APP) to generate the soluble αAPP fragment (sAPPα) rather than the neurotoxic amyloid-β (Aβ), playing a protective role in AD [138].
BACE1
BACE1 is the major β-secretase that cleaves APP to generate Aβ [139]. BACE1 is a transmembrane aspartic protease, structurally similar to the pepsin family [140], containing two active catalytic site motifs in the luminal domain [141]. Like other aspartic proteases, BACE1 is synthesized as a precursor protein containing a N-terminal propeptide domain that is removed during maturation of the enzyme [142]. Furin or a furin-like PC is responsible for cleaving the BACE1 proprotein to yield the mature enzyme with the highest β-secretase activity [143]. Like APP, BACE1 is highly expressed in the brain [144]. Significant increases of BACE1 enzymatic activity and protein concentration have been detected in brain tissues, cerebrospinal fluid (CSF) and serum of AD patients and subjects with mild cognitive impairment [145–147]. BACE1 inhibitors have demonstrated therapeutic effects in preventing the initial cleaving events of APP in AD animal models [148–153].
Notch receptor
The Notch gene family encodes transmembrane receptors of ~ 300 kDa that are involved in cell-fate determination in vertebrates and invertebrates [154, 155]. The proteolytic processing of Notch receptor precursor is an essential step in the formation of biologically active Notch receptors. The constitutive processing of murine Notch1 requires a furin-like convertase, and mutations in the furin-cleavage site completely abolishes the proteolysis of the Notch1 receptor [155]. In the developing brain, activation of Notch receptors upon ligand binding is involved in the preservation of neural progenitors and inhibition of neurogenesis [156, 157]. In the adult brain, Notch signaling influences neuronal apoptosis, microglial activation and synaptic plasticity [158–161]. Deregulations of Notch signaling are involved in AD, depression, epilepsy, and stroke [159–163].
LRP1
LRP1 is a multifunctional receptor that belongs to the low-density lipoprotein receptor family [164]. It is synthesized as a ~ 600-kDa precursor, which is cleaved by furin in the trans-Golgi network and transported to the cell surface as a mature form consisting of α-chain and β-chain [8]. The mature LRP1 is further processed by other enzymes, such as MMPs and γ-secretase, to release the intracellular domain (ICD) [8]. LRP1 is highly expressed in neurons and glia of the brain, and functions to regulate proteinase activity, cytokine activity and cholesterol metabolism [165, 166]. The ligands for LRP1 include Aβ, ApoE and activated α2-macroglobulin [167]. In addition to controlling ligand metabolism, LRP1 can also regulate signaling pathways by coupling with other cell surface receptors or proteins, such as the N-methyl-D-aspartate (NMDA) receptors [168, 169]. The ICD of LRP1 can be transported into the nucleus, where it contributes to transcriptional regulation of target genes, including interferon-γ [170]. Accumulating evidence from preclinical and animal studies indicates that LRP1 is involved in AD pathogenesis not only by regulating the metabolisms of Aβ and ApoE, but also by influencing synaptic plasticity and inflammation through Aβ-independent pathways [171]. LRP1 is detected at an abundant level in post-synaptic sites of neurons, and it interacts with several synaptic proteins, including postsynaptic density protein 95, NMDA receptor and GluA1 [169, 171–173]. Deletion of LRP1 in neurons has been shown to affect lipid metabolism, leptin signaling, glucose metabolism, insulin signaling and anti-apoptotic signaling, resulting in neuroinflammation, motor dysfunction, and cognitive decline in mice [171, 172, 174, 175]. In addition, LRP1 is also found to modulate stem cell proliferation and survival, astroglial differentiation [176, 177], and oligodendrocyte progenitor cell differentiation [178].
GPR37
GPR37 is an orphan G-protein-coupled receptor that is widespread in several brain regions, including cerebral cortex, hippocampus, hypothalamus, midbrain and cerebellum [51]. It has a long extracellular N-terminal ectodomain which is recently demonstrated to be processed by both ADAM10 and furin [179]. The unfolded form of GPR37 is a substrate of parkin, and its intracellular retention leads to ER stress and DA neuronal death, linking to PD [180–182]. GPR37 is also involved in the DA signaling pathway by interacting with the dopamine transporter in mouse striatal presynaptic membranes, thereby modulating dopamine uptake [183]. In addition, GPR37 interacts with adenosine A2A receptors in the hippocampus, localized at the extrasynaptic plasma membrane of dendritic spines, dendritic shafts and axon terminals, regulating adenosinergic signaling [184]. GPR37 is also found in astrocytes and oligodendrocytes, and is demonstrated as a negative regulator of oligodendrocyte differentiation and myelination [185, 186]. Overexpression of GPR37 leads to profound neurodegeneration in animal models, selectively for DA neurons [187], while GPR37-knockout mice also show decreased dopamine levels in the striatum and specific motor deficits [188, 189]. GPR37 knockout also triggers non-motor behavioral phenotypes, such as anxiety and depression-like behaviors, in an age- and gender-dependent manner [190, 191].
Sortilin
Sortilin is a type I transmembrane protein that functions as an endocytosis receptor and plays a role in protein sorting and cell signaling [192]. Sortilin is synthesized as an inactive precursor protein, which is cleaved by furin to remove the N-terminal propeptide [193]. The resulting mature protein can be further processed by other proteases to shed its extracellular domain from the cell surface [193]. Sortilin is generally trafficked via the trans-Golgi network, endosomes and plasma membrane, binding to different proteins and directing them to the secretory pathway or for degradation [193]. Sortilin has been reported to function as a neuronal receptor for APP and its cleavage products sAPPα and Aβ [194, 195]. The ICD of sortilin interacts with APP and regulates its lysosomal and lipid raft trafficking [194]. Sortilin also binds to oligomerized Aβ, inducing endocytosis of Aβ and triggering apoptosis [195]. In addition, sortilin is found to be an essential component for transmitting pro-neurotrophin-dependent death signals from p75NTR, thereby playing roles in neuronal apoptosis, aging and brain injury [93, 196, 197]. On the other hand, sortilin has also been found to associate with TrkB receptors, which promotes cell survival [198]. Therefore, sortilin acts as a molecular switch from apoptotic response by interacting with p75NTR to neurotrophic effects via binding to TrkB receptors in neurons. Aberrant activity of sortilin has been found to be associated with the pathogenesis of AD and depression [193, 199, 200].
BRI2
BRI2 is a type II transmembrane protein of 266 amino acids, containing an extracellular region, a transmembrane region and a cytoplasmic region [201, 202]. During maturation, the ~ 4-kDa C-terminal propeptide of BRI2 is cleaved by furin at the trans-Golgi compartment, generating the membrane-bound form of mature BRI2 (mBRI2) [203, 204]. The mBRI2 contains an evolutionarily conserved BRICHOS domain that is found to act as a chaperone, facilitating proper folding of BRI2 and preventing Aβ formation [205, 206]. In the human brain, BRI2 is intensively expressed in cortical and hippocampal pyramidal neurons [207]. The BRICHOS domain of BRI2 interacts with APP and inhibits its processing, delaying fibrillation of Aβ [206–209]. Mutations in BRI2 and aberrant BRI2 expression have been reported to be associated with familial British dementia and involved in AD pathogenesis [210–212].
Ac45
Ac45, an accessory subunit of the vacuolar-type ATPase (V-ATPase) proton pump, is a type I transmembrane protein that is encoded by ATP6AP1 in humans [213–215]. Furin catalyzes the processing of Ac45 precursor protein to generate mature Ac45 [216, 217]. Furin-knockout β-cells show impaired cleavage of Ac45 [217]. Ac45 is ubiquitously expressed with the highest levels in neuronal and neuroendocrine cells and osteoclasts [218–220], and may be required for proper synaptic vesicle acidification and neurotransmitter release [221]. Ac45-deficient patients not only have immunodeficiency, but also display a spectrum of neurocognitive abnormalities [222]. These indicate that dysfunction of Ac45 can be potentially involved in neurological disorders such as AD and epilepsy.
Furin in neurodegenerative and neuropsychiatric diseases
So far, many studies have demonstrated associations of deregulation of furin expression with the pathophysiology of several neurodegenerative and neuropsychiatric diseases, as well as with alterations of substrates of furin in these diseases (Table 1).
Table 1.
Changes in the expression of furin and its substrates in neurodegenerative and neuropsychiatric diseases and the implications
| Disease | Patients/animal models | Furin expression | Expression of proteins processed by furin | Implications | References |
|---|---|---|---|---|---|
| AD | AD patients | FURIN mRNA (brain) ↓ | Furin reduction may be closely related to the mechanisms that lead to Aβ production in AD | [13] | |
| AD | Tg2576 mice | Furin mRNA (cortex) ↓ | Furin reduction downregulates α-secretase activity of ADAM10 and TACE, thereby enhancing Aβ production | [13] | |
| AD | APP-C105 mice | Furin (cortex) ↓ | ADAM10 (cortex) ↓ | Excess iron induces disruption of furin activity, which in turn reduces α-secretase-dependent APP processing | [23] |
| AD | AD patients | Furin (plasma) ↓ | Increased plasma iron concentration in AD downregulates furin level, impairing the ability of α-secretases to produce sAPPα, resulting in increased Aβ | [237] | |
| AD | AD patients |
BDNF mRNA (hippocampus) ↓; BDNF (hippocampus) ↓ |
Deficiency of BDNF may contribute to the progressive atrophy of neurons in AD | [240] [241] | |
| AD | AD patients | BDNF mRNA (cortex) ↓; mBDNF (cortex) ↓; mBDNF/proBDNF (cortex) ↓ | Imbalanced proBDNF and mBDNF play a role in synaptic loss and cellular dysfunction, leading to cognitive impairment in AD | [241] [242] [243] [244] [245] [246] | |
| AD | Tg2576 mice | mBDNF (hippocampus) ↓; mBDNF/proBDNF (hippocampus) ↓ | Abnormal cleavage of BDNF may be involved in AD-related traits triggered by excessive Aβ pathology | [247] | |
| AD | 5 × FAD mice | BDNF (hippocampus) ↓ | BDNF expression is reduced in 5 × FAD mice at the age of 3 and 7 months, contributing to the impairment of synaptic plasticity and memory | [248] [249] | |
| AD | AD patients | proNGF (cortex) ↑ | Decreased processing of proNGF to mNGF may be associated with AD pathology | [94] [250] | |
| AD | AD patients | proNGF (hippocampus) ↑ | Alterations in the hippocampal NGF signaling pathway in AD favor proNGF-mediated proapoptotic pathways | [251] | |
| AD | AD patients | Notch1 (hippocampus) ↑ | Notch1 is increased in AD and Pick’s disease, where abnormal tau aggregates are present, indicating a possible relationship between tau aggregation and Notch1 expression | [252] | |
| AD | AD patients | MMP-1 (cortex) ↑ | Enhanced MMP-1 activity in AD may contribute to the BBB dysfunction seen in AD | [255] | |
| AD | AD patients |
BACE1 mRNA (cortex) ↑ BACE1 (cortex) ↑ |
Increased BACE1 activity is correlated with Aβ level in AD | [253, 254] | |
| AD | AD patients | BACE1 (CSF) ↑ | Increased BACE1 in CSF is a predictor of mild cognitive impairment | [146] | |
| AD | 5× FAD mice |
MMP-2 (hippocampus) ↑; MMP-9 (hippocampus) ↑; MMP-14 (hippocampus) ↑ |
Different MMPs involved in APP/Aβ metabolism are differentially regulated in a spatio-temporal manner in the 5× FAD murine model of AD | [256] | |
| AD | AD patients | Sortilin (cortex) ↑ | Sortilin functions as a modulator of BACE1 retrograde trafficking and promotes the generation of Aβ | [199] | |
| AD | AD patients |
Sortilin (hippocampus) ↑; ProBDNF (hippocampus) ↑; ProBDNF/BDNF (CSF) ↑ |
ProBDNF-p75/sortilin signaling is an important contributor to the pathogenesis of AD, causing an increase of cell death and impairment of neuronal differentiation | [257] | |
| AD | AD patients |
LRP1 mRNA (cortex) ↑; LRP1 (brain) ↑ |
LRP1 expression may be upregulated in glial cells due to the neuroinflammation in AD | [258] [259] | |
| AD | AD patients | LRP1 (cortex) ↓ | LRP1 pathway may modulate Aβ deposition and AD susceptibility by regulating the removal of soluble Aβ | [260] | |
| AD | APP23 mice |
LRP1 (cortex) ↑; LRP1 (cortical blood vessels) ↓ |
LRP1 increase in the cortex and decrease in vascular endothelial cells may account for an imbalance of Aβ efflux and influx across the BBB in AD mice | [261] | |
| AD | AD patients | BRI2-BRICHOS (hippocampus) ↑; BRI2-APP (hippocampus) ↓ | Aberrant processing of BRI2 may promote its deposition and affect its function in halting Aβ aggregation | [212] | |
| PD | LRRK2-overexpressing Drosophila | Furin 1 (DA neurons) ↑ | LRRK2 enhances furin 1 translation in DA neurons, mediating neurotoxicity in the fly model of PD | [265] | |
| PD | Paraquat-treated Drosophila | Furin 1 (DA neurons) ↑ | Furin 1 may initiate a cellular program that is central to the process of neurodegeneration | [265] | |
| PD | PD patients | BDNF (CSF) ↑ | Altered BDNF level could be involved in the pathophysiology of PD | [267] | |
| PD | PD patients | BDNF (serum) ↓ | Lower serum levels of BDNF at early stage may be associated with the pathogenesis of PD | [268] [269] | |
| PD | PD patients | MMP-2 (substantia nigra) ↓ | Region-specific alterations of MMPs may contribute to the pathogenesis of PD | [131] | |
| PD | 6-Hydroxydopamine-treated rats | MMP-3 (substantia nigra) ↑ | Activation of MMP-3 processes the secreted α-synuclein in PD | [129] | |
| PD | PD patients | MMP-1 (serum) ↓ | Significantly lower levels of serum MMP-1 were found in PD patients, particularly in females | [270] | |
| PD | PD patients | GPR37 (Lewy bodies in midbrain) ↑ | GPR37 may be involved in the formation of Lewy bodies, mediating neurotoxicity in PD | [181] | |
| PD | PD patients |
GPR37 mRNA (substantia nigra) ↑; Ecto-GPR37 (CSF) ↑ |
Ecto-GPR37 in CSF is a potential biomarker for PD | [182] | |
| Epilepsy | TLE patients | Furin (temporal cortex) ↑ | There might be a correlation between furin expression and epilepsy | [25] | |
| Epilepsy |
KA-induced epileptic mice; PTZ-kindled epileptic mice |
Furin (cortex, hippocampus) ↑ | Furin may play a role in regulation of inhibitory synaptic transmission in epileptic mice | [25] | |
| Epilepsy | KA-induced epileptic mice | Furin mRNA (hippocampus) ↑ | Ngf mRNA (hippocampus) ↑; Bdnf mRNA (hippocampus) ↑ | Furin mRNA upregulation appears to be parallel to that of NGF and BDNF mRNAs following KA treatment | [12] |
| Epilepsy | TLE patients | BDNF/NGF/NT-3 mRNA (hippocampus) ↑ | There may be associations between increased neurotrophin mRNA levels in granule cells and damage to hippocampal neurons and synaptic plasticity in epilepsy | [276] | |
| Epilepsy | TLE patients | BDNF (temporal cortex) ↑ | The activity-dependent expression of BDNF in human subjects potentially contributes to the pathophysiology of human epilepsy | [277] | |
| Epilepsy | Pilocarpine-induced status epileptic mice | ProBDNF (hippocampus) ↑ | Rapid increases of proBDNF following epilepsy are due in part to reduced cleavage | [278] | |
| Epilepsy | Rats with limbic seizures induced by electrolytic lesion in DG |
Ngf mRNA (hippocampus) ↑; Ngf mRNA (cortex) ↑ |
The expression of NGF is affected by unusual physiological activity | [279] | |
| Epilepsy | KA-induced epileptic rats | Ngf mRNA (forebrain) ↑ | Seizure activity stimulates a transient increase of NGF expression by selective populations of forebrain neurons | [280] | |
| Epilepsy | Pilocarpine-induced status epileptic rats | ProNGF (hippocampus) ↑ | High levels of mRNA for both p75 receptors and proNGF are found in the epileptic model rats | [281] | |
| Epilepsy |
TLE patients; KA-induced epileptic mice |
Notch (hippocampus) ↑ | The effect of Notch signaling on seizures can be in part attributed to its regulation of excitatory synaptic activity in CA1 pyramidal neurons | [163] | |
| Epilepsy | Epilepsy patients |
MMP-2 (serum) ↓; MMP-3 (serum) ↓ |
Serum MMP-2 and MMP-3 are potential biomarkers for epilepsy | [283] | |
| Epilepsy | TLE patients; |
MMP2 mRNA (hippocampus) ↑; MMP3 mRNA (hippocampus) ↑; MMP14 mRNA (hippocampus) ↑ |
Increased MMP expression is a prominent hallmark of the human epileptogenic brain | [285] | |
| Epilepsy | Intractable epilepsy patients | MMP-9 (cortex) ↑ | Increased MMP-9 immunoreactivity was prominently upregulated at synapses in the cortex of intractable epilepsy patients | [286] | |
| Epilepsy | PTZ-induced kindled epileptic mice | MMP-9 (hippocampus) ↑ | MMP-9 is involved in the progression of epilepsy through cleavage of proBDNF to mBDNF in the hippocampus | [287] | |
| Cerebral ischemia | Global ischemic rats | Furin mRNA (hippocampus) ↑ | Furin may protect hippocampal neurons from ischemic damage | [296] | |
| Cerebral ischemia | Rats after MCAO | Furin mRNA (ischemic hemisphere) ↑ |
Mmp2 mRNA (ischemic hemisphere) ↑; Mmp14 mRNA (ischemic hemisphere) ↑ |
Furin activates MMP-14 and in turn enhances MMP-2 activation, contributing to the disruption of BBB in ischemia | [297] |
| Cerebral ischemia | Hypoxic-ischemic rats |
Furin mRNA (ipsilateral cortex) ↓; Furin mRNA (ipsilateral hippocampus) ↑ ↓ ↑ |
BDNF (ipsilateral cortex, hippocampus) ↓; Mmp9 mRNA (ipsilateral cortex) ↓ |
BDNF and its related enzymes such as furin play important roles in the pathogenesis of and recovery from hypoxic-ischemic brain damage | [298] |
| Cerebral ischemia | Rats after MCAO |
MMP-2 (ipsilateral cortex, striatum) ↑; MMP-9 (ipsilateral cortex, striatum) ↑ |
A specific spatial–temporal pattern of expression and activation of MMP-9 and MMP-2 may contribute to extracellular matrix degradation and BBB breakdown after transient focal cerebral ischemia | [299] | |
| Cerebral ischemia | Baboons after MCAO | MMP-2 (basal ganglia) ↑ | It is plausible that locally active MMP-2 contributes to early matrix degradation, loss of vascular integrity, neuron injury, and maturation of the ischemic lesion | [300] | |
| Cerebral ischemia | Mice after MCAO | MMP-9 (ischemic regions) ↑ | MMP-9 may play an active role in early vasogenic edema development after stroke | [302] | |
| Cerebral ischemia | Rats after MCAO | LRP1-ICD (ischemic areas) ↑ | Furin-mediated cleavage of LRP1 and changes in LRP1-ICD localization are involved in ischemic brain injury | [303] | |
| SCZ | SCZ patients | FURIN mRNA (prefrontal cortex) ↓ | Aberrant gene expression elucidates the functional impact of polygenic risk for SCZ | [304] | |
| SCZ | SCZ patients |
BDNF mRNA (cortex) ↓; BDNF (cortex) ↓ |
Cortical neurons may receive less trophic support in SCZ | [312] | |
| SCZ | SCZ patients | BDNF mRNA (cortex) ↓ | Decreased BDNF/TrkB signaling appears to underlie the dysfunction of inhibitory neurons in SCZ | [313] | |
| SCZ | SCZ patients |
BDNF (hippocampus) ↓; NT-3 (cortex) ↓ |
Alterations in expression of neurotrophic factors could be responsible for neural maldevelopment and disturbed neural plasticity in SCZ | [314] | |
| SCZ | SCZ patients | BDNF (serum) ↓ | BDNF may be involved in the pathophysiology of and cognitive impairment in SCZ | [316] [317] | |
| SCZ | Rats with ibotenic acid lesions in the hippocampus |
Bdnf mRNA (cortex) ↓; Bdnf mRNA (hippocampus) ↓ |
Alterations in BDNF render animals more susceptible to neurodegenerative insults | [318] | |
| SCZ | Dysbindin-1 mutant mice |
BDNF (cortex) ↓; BDNF (hippocampus) ↓ |
BDNF reduction leads to inhibitory synaptic deficits | [319] | |
| SCZ | SCZ patients |
NGF (serum) ↓; NT-3 (serum) ↓ |
SCZ is accompanied by an abnormal neurotrophin profile | [320] [321] [322] | |
| SCZ | SCZ patients | MMP-9 (serum) ↑ | Alterations in plasma MMP-9 are a biomarker for SCZ | [323] | |
| SCZ | SCZ patients | MMP-2 (CSF) ↑ | Increased CSF MMP-2 levels in SCZ may be associated with brain inflammation | [326] | |
| Depression | MDD patients | BDNF (serum) ↓ | Low BDNF levels may play a pivotal role in the pathophysiology of MDD | [327] [328] | |
| Depression | MDD patients |
BDNF (serum) ↓; mBDNF/proBDNF (serum) ↓ |
The changes in serum BDNF, TrkB, proBDNF and p75NTR may provide a diagnostic biomarker for MDD | [329] | |
| Depression | MDD patients |
MMP-9 (serum) ↑; MMP-2 (serum) ↓ |
MMP-2 and MMP-9 are involved in the pathophysiology of major depression | [323] | |
| Depression | Mood disorder patients | MMP-2 (serum) ↓ | A change in inflammatory homeostasis, as indicated by MMP-2 and MMP-9, could be related to mood disorders | [330] | |
| Depression | MDD patients |
MMP-2 (CSF) ↑; MMP-7 (CSF) ↑; MMP-10 (CSF) ↑ |
Increased MMP-2 levels in CSF are positively correlated with clinical symptomatic scores in MMD | [326] | |
| Depression | Rats after chronic unpredictable mild stress |
Lrp1 mRNA (hippocampus) ↑; LRP1 (hippocampus) ↑ |
LRP1 might impair the microtubule dynamics in depressive-like rats and is involved in the development of depression | [331] |
MCAO: middle cerebral artery occlusion
Furin in AD
AD overview
AD is a progressive neurodegenerative disease and the main cause of dementia in the elderly, affecting around 6% of the population over the age of 65 [223]. Currently, there is no effective prevention or treatment strategy for AD [20, 224, 225]. The major pathological hallmarks of AD are the accumulation of two aggregated proteins in the brain, Aβ and tau, leading to the formation of extracellular senile plaques and intracellular neurofibrillary tangles (NFTs), respectively [226, 227]. Aβ is produced from proteolytic cleavage of APP by β- and γ-secretases [139]. In contrast, APP cleaved by α-secretase produces sAPPα which shows neurotrophic and neuroprotective functions [226]. Both Aβ senile plaques and NFTs induce neuroinflammation and neuronal apoptosis, contributing to AD pathogenesis [226, 227]. Following Aβ and tau pathology, AD patients further exhibit synaptic damage and neuronal loss, particularly in the cortex and hippocampus, and show cognitive impairments as the disease progresses [228]. In addition to the Aβ cascade hypothesis, many other hypotheses have also been proposed to explain the pathologic process of AD, including the tau hypothesis [229], the blood–brain barrier (BBB) dysfunction hypothesis [230], the metal ion dysregulation hypothesis [231, 232], the inflammation hypothesis [233], the oxidative stress and mitochondrial cascade hypothesis [234, 235], and the insulin resistance hypothesis [236]. However, these hypotheses only explain certain aspects of the disease, and the mechanism of AD pathogenesis remains elusive.
Aberrant furin expression in AD
FURIN mRNA expression has been detected at a significantly lower level in the brains of AD patients and Tg2576 AD mouse model than in controls [13]. Notably, decreased mRNA expression of Furin is observed in cortices of both 4- and 24-month-old Tg2576 mice compared with their littermates, suggesting that furin reduction occurs in a relatively early age (prior to Aβ plaque formation) and may be involved in the pathogenesis of AD [13]. Moreover, this study also showed that injection of Furin-adenovirus into Tg2576 mouse brains markedly reduced Aβ production in the infected brain regions, which may be attributed to the enhancement of the α-secretase activity by furin cleavage of ADAM10 and tumor necrosis factor-α converting enzyme (TACE) [13]. Another study also showed decreased expression of furin and ADAM10 in the cortex of APP-C105 mouse model of AD compared to that of non-transgenic controls [23]. Moreover, treadmill exercise could elevate furin expression and suppress Aβ production in the APP-C105 mice [23]. While excess iron in AD brain induces disruption of furin activity, treadmill exercise alleviates cognitive decline and Aβ-induced neuronal cell death by promoting α-secretase-dependent processing of APP through low iron-induced enhancement of furin activity [23].
In addition to furin expression in the brain, the plasma furin also decreases significantly while serum Aβ increases in AD patients [237]. The decrease of plasma furin strongly correlates with the increase of plasma iron, thereby iron overload in plasma was proposed to be a possible contributor to the low level of furin, and the downstream reduction of α-secretase activity might account for the increase of Aβ [237]. Besides, studies have also reported that the bilateral injection of Aβ into the intracerebral ventricle of mice can induce furin expression compensatorily, which subsequently increases NGF via modulation of its maturation [238, 239].
Expression of substrates of furin in AD
Many proteins that are proteolytically processed by furin also show altered expression in AD. Numerous studies have indicated that the relative levels of BDNF mRNA and proteins are decreased in the hippocampus and cortex in AD patients [240–246]. Particularly, decreased mBDNF/proBDNF ratio has been found in the parietal cortex of subjects with mild cognitive impairment [246], suggesting that reduction of mBDNF occurs in early stages of AD and contributes to the impairment of synaptic plasticity and memory. In addition to AD patients, transgenic AD mouse models also show reduced mBDNF expression and decreased mBDNF/proBDNF levels in the hippocampus [247–249], indicating the involvement of altered cleavage of BDNF in AD pathology.
Similar to BDNF, NGF, Notch1, ADAM10, BACE1, MMPs, LRP1, BRI2 and sortilin also show altered expression or activity in AD. ProNGF increases markedly in the cortex and hippocampus of AD brains [94, 250, 251]. Notably, the increase of proNGF also appears in subjects with mild cognitive impairment [250]. These findings reflect that the decreased processing of proNGF to mNGF is involved in AD pathogenesis. Notch1 expression is increased in the hippocampus of AD patients, which may be linked to tau aggregation [252]. BACE1 expression has been found to be elevated in the cortex and CSF of AD patients as compared to the age-matched normal subjects [146], which is correlated with increased Aβ [253, 254]. MMP-1 levels are significantly elevated in AD patients in all cortical areas, which may contribute to the BBB dysfunction seen in AD [255]. MMP-2, MMP-9 and MMP-14 expression is up-regulated age-dependently in astrocytes and amyloid plaques in the hippocampus of 5× FAD mice [256]. Sortilin protein and the cytoplasmic domain of sortilin are found to be significantly increased in brains of AD patients, which contribute to the pathogenesis of AD by increasing cell death and impairing neuronal differentiation [199, 257]. LRP1 mRNA and protein are reported to be increased in neurons and GFAP-positive activated astrocytes associated with neuroinflammation in AD patients [258, 259]. Meanwhile, a decrease of LRP1 has also been reported in the midfrontal cortex of AD patients, playing a role in modulating Aβ deposition and AD susceptibility [260]. In addition, in the APP23 mouse model, LRP1 is increased in the cortex but decreased in the vascular endothelial cells, which may account for the imbalance between Aβ efflux and influx across the BBB [261]. The level of BRI2 containing the BRICHOS domain is increased in the hippocampus of early-stage AD patients, whereas the level of the BRI2-APP complex is decreased, accompanied by a decrease of furin, indicating that the aberrant processing of BRI2 may promote its deposition and affect its function in halting Aβ production and aggregation [212]
Potential role of furin in AD pathology
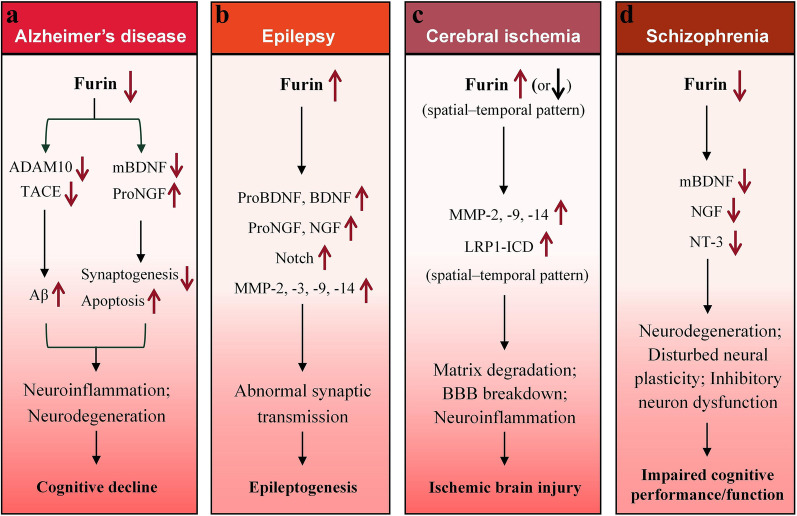
The above findings suggest an important role of furin in AD pathology. The downregulation of furin in AD patients or animal models likely leads to lower cleavage of ADAM10, TACE, proBDNF and proNGF. The decreased ADAM10 and TACE lead to reduced α-secretase activity, which in turn promotes Aβ generation and deposition; on the other hand, the low levels of mBDNF and high levels of proNGF cause neuronal death and synaptic damage (Fig. 3a). These alterations can account in part for the pathological symptoms of AD. In addition, the relationships between furin deregulation and changes in MMPs and LRP1 in AD pathology have yet to be investigated, and the causes of furin downregulation in AD need to be clarified.
Fig. 3.
Proposed working models of how aberrant furin expression participates in the pathogenesis of Alzheimer’s disease (a), epilepsy (b), cerebral ischemia (c) and schizophrenia (d)
Furin in PD
PD overview
PD is the second most common neurodegenerative disorder, pathologically characterized by abnormal deposition of α-synuclein aggregates in Lewy bodies and loss of nigrostriatal DA neurons [262, 263]. The striking clinical symptoms of PD are motor symptoms such as tremor, rigidity, bradykinesia and postural instability. Patients with severe motor symptoms often have difficulties moving their hands, or standing and walking due to the tremor and stiff muscles, which severely affects the quality of their lives [263]. PD patients also display non-motor signs and symptoms, such as olfactory loss, autonomic dysfunction and rapid eye movement sleep behavior disorder, which usually precede the motor symptoms but are often overlooked [264]. The mechanism of neurodegeneration in PD remains unclear, and currently there is no cure for PD.
Aberrant furin expression in PD
Currently, there is no report on the changes of furin expression in PD patients or murine models. However, in Parkinson’s-related Drosophila model, furin 1 has been found to be highly concentrated in TH-positive DA neurons [265], and furin 1 is translationally regulated by leucine-rich repeat kinase 2 (LRRK2) and involved in the impairment of synaptic plasticity and neurodegeneration [266]. In addition, in the paraquat-induced Drosophila model of PD, furin 1 expression is also enhanced by paraquat exposure in DA neurons [265]. These results highlight a potential role of furin in PD pathogenesis.
Expression of substrates of furin in PD
Aberrant BDNF expression has been found during the pathological processes of PD. The amount of 27-kDa BDNF is increased in the CSF samples of PD patients compared with normal controls [267], whereas serum BDNF levels are significantly lower in PD patients than in healthy controls, which are correlated with motor impairment and cognitive deficits in PD [268, 269]. MMP-2 levels are reduced in PD patients in the substantia nigra, but not in the cortex and the hippocampus [131]. MMP-3 levels are increased in a rat model of PD induced by injection of 6-hydroxydopamine into the substantia nigra [129], and MMP-3 may play a pivotal role in the progression of PD through digestion of α-synuclein in DA neurons and modulation of α-synuclein aggregation and Lewy body formation [129, 130]. Serum MMP-1 is significantly lower in PD patients than in controls, and the difference is more prominent in females [270]. Both mRNA and protein of GPR37 accumulate in Lewy bodies in the midbrain of PD patients [181, 182], and the increased Ecto-GPR37 in CSF is proposed as a potential biomarker for PD [182]. However, no studies currently exist regarding the specific relationship between furin expression and changes in these substrates in PD patients or animal models. Thus, deeper exploration of the underlying mechanisms remains essential in future studies, which may uncover new therapeutic targets for PD.
Potential role of furin in PD pathology
Although there is no report on the change of furin expression in vertebrate models of PD, the highly increased furin 1 expression in DA neurons of Parkinson’s-related Drosophila model indicates a potential role of furin in PD pathology. Furthermore, changes in the expression of some substrates of furin have been detected in PD models, such as increased GPR37 and MMP-3, highlighting the possible associations between furin and PD symptoms. Thus, the expression of furin in PD pathogenesis and associations with the change of its substrates in PD need to be clarified urgently.
Furin in epilepsy
Epilepsy overview
Epilepsy is a common chronic neurological disorder associated with abnormal synaptic transmission [271], inappropriate neuronal firing, and imbalance of excitation and inhibition of neuronal networks [272]. The etiology of epilepsy is mostly unclear, which possibly includes genetic risks, brain diseases, and systemic diseases. The abnormal neuronal firing is found to be closely related to mitochondrial dysfunction and abnormalities in neurotransmitters and ion channels [273, 274]. Due to the different starting sites and transmission modes of abnormal neuronal discharges, clinical manifestations of epilepsy are complex and diverse, including disorders in motor, sensory, and autonomic nervous systems and consciousness [275].
Aberrant furin expression in epilepsy
It has been reported that furin protein levels are increased in the temporal neocortex of patients with temporal lobe epilepsy (TLE) and in the cortex and hippocampus of kainic acid (KA)-induced and pentylenetetrazol (PTZ)-kindled epileptic mice [25]. Moreover, transgenic overexpression of furin in mice increases the susceptibility to epilepsy and increases the epileptic activity [25]. Furin has been identified to play a role in influencing the inhibitory synaptic transmission in epileptic mice [25]. In addition, an increase in Furin mRNA has been found in the hippocampus of KA-exposed mice [12], and the co-localizations of the increased Furin mRNA with Ngf and Bdnf mRNAs suggest a potentially important role of furin in the pathophysiology of epilepsy [12].
Expression of substrates of furin in epilepsy
Studies on animal models of epilepsy have proposed potential involvement of dysregulations of neurotrophins, such as BDNF, NGF and NT-3, in human epilepsy [276–282]. TLE patients with hippocampal sclerosis show increased mRNA levels of BDNF, NGF and NT-3 in granule cells of hippocampus, which are correlated with either hippocampal neuron loss or aberrant supragranular mossy fiber sprouting [276]. Patients with intractable TLE show a marked increase in protein levels of BDNF in the temporal neocortex [277]. Moreover, a rapid increase in the proBDNF level is found in principal neurons and astrocytes of all hippocampal subfields in pilocarpine-induced status epileptic mice, which is proposed to be associated with the reduced proBDNF cleavage machinery [278]. Similar to the changes in BDNF, Ngf mRNA increases in the hippocampus and neocortex of rats with limbic seizures [279]. The secreted proNGF is considered as a pathophysiological death-inducing ligand [280], while blocking proNGF can inhibit neuronal loss after seizures [281]. Notch signaling is activated in KA-induced epileptic mice and in human epileptogenic tissues, while activation of Notch signaling further promotes neuronal excitation of CA1 pyramidal neurons [163]. In addition, a large number of studies have shown that the expression levels of MMP-2, MMP-3, MMP-9 and MMP-14 in the brains of epilepsy patients or animal models are increased and dynamically regulated at different stages of epileptogenesis [283–289]. MMP inhibitors are considered as potential therapeutic drugs because of their anti-seizure and anti-epileptogenic effects [285, 290].
Potential role of furin in the pathology of epilepsy
The above findings suggest a crucial role of furin in the pathology of epilepsy. The upregulation of furin in epilepsy patients or animal models may promote the cleavage of proBDNF, proNGF, Notch receptor and MMPs. As a result, the inhibitory and excitatory synaptic transmissions are affected, leading to abnormal neuronal discharge, which contributes in part to the symptoms of epilepsy (Fig. 3b). However, the underlying mechanisms for furin upregulation and furin-mediated activities in epileptogenesis need to be determined.
Furin in cerebral ischemia
Overview of cerebral ischemia
Cerebral ischemia is a neurodegenerative disease caused by reduced blood supply to the brain tissue [291], and is currently a major cause of death and disability globally [292]. Cerebral ischemia causes reduced delivery of oxygen and glucose to the brain, and as a result, a loss of consciousness can occur [291]. The occurrence of metabolic disorders during ischemia or tissue hypoxia is relatively well established, but the subsequent reperfusion is the major events leading to cell and tissue dysfunctions [293]. Ischemia–reperfusion injury is the inexplicable aggravation of cellular dysfunction during the restoration of blood flow after a period of ischemia [294]. The reperfusion can lead to potentially very harmful effects, such as necrosis of irreversibly damaged cells, cell swelling, vascular and endothelial injury and mitochondrial dysfunction [295].
Aberrant furin expression in cerebral ischemia
It has been found that the Furin mRNA level in rat hippocampus at 24 h after transient global cerebral ischemia is two-fold of that in sham-operated controls, indicating a possible role furin may play [296]. In a focal ischemic rat model established by middle cerebral artery occlusion, increases in Furin mRNA and protein levels are found in the piriform cortex of the ischemic hemisphere 2 h after reperfusion compared with sham-operated animals, and it is predicted that the elevation of furin may contribute to the disruption of BBB during ischemia [297]. Another recent study found that the level of Furin mRNA in the ipsilateral cortex of hypoxic-ischemic rats had an insignificant increase at 6 h after ischemia, but then decreased significantly at 15 h and was sustained at a low level for 7 days [298], while Furin mRNA in the ipsilateral hippocampus was elevated at 6 h and 3 days but decreased at 15 and 24 h after injury compared with that of the control rats [298]. The change in furin expression is considered to account for the decrease of BDNF in the ipsilateral cortex and hippocampus of the rats [298]. An in vitro study also showed that the protein levels of furin and BDNF are upregulated in cultured rat astrocytes exposed to oxygen–glucose deprivation [64]. These findings indicate that furin may play important roles in the pathogenesis of cerebral ischemia and in the recovery from ischemia brain damage.
Expression of substrates of furin in cerebral ischemia
In addition to the changes in furin expression, the levels of Bdnf mRNA and protein in the ipsilateral cortex and hippocampus of hypoxic-ischemic rats are altered at different degrees at different time points after hypoxic-ischemic injury [298]. Many other studies have also reported changes of MMP levels, including levels of MMP-2, MMP-9 and MMP-14, in the model of focal ischemic rats [297, 299–302]. In particular, increased expression and activity of MMP-2 and MMP-9 are found in different models of focal cerebral ischemia, implying their potential roles in early matrix degradation, loss of vascular integrity, and neuronal injury in the ischemic lesion [300, 301]. In addition, a significant increase in the cleavage of LRP1 by furin has been found in rats after cerebral ischemia, which is predicted to aggravate neuroinflammation, and administration of a furin inhibitor inhibits the cleavage of LRP1 and decreases co-localization of ICD of LRP1 with furin in ischemic areas [303]. These findings imply that the furin-mediated cleavage of MMPs and LRP1 may be involved in the pathophysiology of ischemic brain injury.
Potential role of furin in the pathology of ischemia
The above observations imply the involvement of furin in the pathology of cerebral ischemia. Changes in furin expression may exist in varied temporal and spatial patterns after ischemic injury in the brain. The upregulation of furin in ischemic patients or animal models may promote the cleavage of MMPs, particularly MMP-2, MMP-9, and MMP-14. The activation of these MMPs leads to early matrix degradation and loss of vascular integrity, and finally contributes to BBB breakdown and neuronal injury in ischemic lesions (Fig. 3c). Moreover, the ICD of LRP1 is increased, which aggravates neuroinflammation. The relationship between changes of furin level and other molecules such as BDNF in ischemic brain injury needs to be elucidated in the future.
Furin in SCZ
SCZ overview
As one of the severe mental diseases, schizophrenia is characterized by cognitive distortions including impairments in concentration, thinking, speed of cognitive information processing, and verbal working memory [304]. These impairments in cognitive functions persist throughout the disease and determine the functional status of patients [305]. The etiology of schizophrenia is complex, commonly associated with genetic variants and changes in development-related factors and regulatory molecules [306].
Aberrant furin expression in SCZ
A study by Fromer et al. in 2016 using RNA sequencing data from the dorsolateral prefrontal cortex of post-mortem SCZ patients identified down-regulation of FURIN transcripts by risk allele [24]. They also found that depletion of furin in zebrafish model has the largest impact on head size, which can be attributed to the furin depletion-induced changes in neural cell proliferation and migration [24]. Furthermore, downregulation of furin expression specifically at the rs4702 G (in the 3' UTR of FURIN) allele by miR-338-3p reduces the production of mBDNF [307]. In addition, the association between pleiotropic effects of FURIN genetic loci and SCZ traits has been reported recently by several different studies [308–310]. A study using datasets from the Psychiatric Genomics Consortium related to SCZ, major depressive disorder (MDD) and bipolar disorder (BIP) patients identified rs8039305 in the FURIN gene as a novel pleiotropic locus across the three disorders [309]. Similarly, another study identified rs17514846, a variant within an intron of FURIN gene, as a common trait between SCZ and cardiometabolic disorder [310]. In addition, in C. elegans, the 3'UTR of kpc-1 (furin) promotes dendritic transport and local translation of mRNAs to regulate dendrite branching and self-avoidance [311]. These findings indicate the important role of furin in brain development and in the pathophysiology of SCZ.
Expression of substrates of furin in SCZ
The deregulation of BDNF expression has been extensively studied in SCZ patients and animal models [312–319]. Significant reductions of BDNF mRNA and protein have been observed in the dorsolateral prefrontal cortex of patients with SCZ compared to normal individuals [312]. The reduced BDNF/TrkB signaling in the prefrontal cortex appears to underlie the dysfunctions of inhibitory neurons in subjects with SCZ [313]. Studies have also shown significant reductions of BDNF in the hippocampus as well as NT-3 concentrations in the frontal and parietal cortical areas, in SCZ patients [314]. On the contrary, some studies have shown that the BDNF concentration is significantly increased in cortical areas of post-mortem SCZ patients [314, 315]. In addition, the plasma BDNF levels in schizophrenic patients are remarkably lower than those in the controls, which is predicted to be associated with the decreased hippocampal volume and cognitive impairments in first-episode and chronic SCZ [316, 317]. These findings suggest that the downregulation of neurotrophic factors could be responsible for neural maldevelopment and disturbed neural plasticity in the etiopathogenesis of schizophrenic psychoses. In schizophrenic animal models, reductions of Bdnf mRNA and protein levels have been observed in the cortex and the hippocampus [318, 319]. Decreased serum levels of NGF and NT-3 have been observed in SCZ as well [320–322]. In addition to the alterations of neurotrophins, plasma MMP-9 levels are also increased significantly in SCZ patients compared to controls [323], and MMP-9 gene polymorphisms in the brain are found to be associated with SCZ [324, 325]. Besides, increased MMP-2 levels in the CSF of SCZ patients are also reported [326].
Potential role of furin in SCZ pathology
The above findings uncover the involvement of furin in the pathology of SCZ. Furin expression in SCZ patients is downregulated, which in turn affects the maturation of neurotrophins, such as BDNF, NGF and NT3. The chronic low trophic support for neurons leads to neural maldevelopment, dysfunction of inhibitory neurons, disturbed neural plasticity and neurodegeneration, contributing to the impaired cognitive performance/function in SCZ (Fig. 3d). This hypothesis may in part explain the pathogenesis of SCZ. However, the relationships between furin deregulation and changes in MMPs and other furin substrates in SCZ pathology have yet to be investigated.
Furin in depression and anxiety
Currently, there is no report on the changes of furin expression in patients with depression and anxiety. However, the SNP rs8039305 in the FURIN gene has been indicated as a novel pleiotropic locus across the disorders of MDD, BIP and SCZ [309], indicating a potential role of furin in pathological mechanisms of the psychiatric disorders.
Aberrant expression of several substrates of furin has been reported in patients with depression. The serum BDNF level is significantly lower in MMD patients than in healthy controls [327–329]. The mBDNF/proBDNF ratio is also decreased [329], suggesting that the reduced BDNF maturation plays a pivotal role in the pathophysiology of MDD. Serum MMP-9 is found to be increased in MDD patients, while MMP-2 is decreased in MDD patients [323, 330], indicating the involvement of MMP-2 and MMP-9 in mood disorders. In addition, MMP-2 levels in the CSF are increased in MDD patients [326], and the state-dependent alterations of MMP-2 and activation of cascades involving MMP-2, MMP-7, and MMP-10 appear to play a role in the pathophysiology of MDD [326]. LRP1 has been reported to be up-regulated in the hippocampus of depressive-like rat model [331].
In anxiety-like disorders, aberrant BDNF expression has also been reported. In the social deprivation stress-triggered anxiety- and depressive-like mice, BDNF levels are reduced in the brain [332]. In serotonin transporter knockout rats with depressive- and anxiety-like behavior, a decrease in mBDNF in the prefrontal cortex has been reported as well [333]. The alterations of proBDNF and mBDNF expression have been indicated in many other diseases with anxiety- and depressive-like behavior [334–337], highlighting the association between aberrant BDNF expression and anxiety and depression disorders.
Furin-targeting strategies for neurological diseases
Currently, the use of furin-targeting strategies to diagnose or treat neurological disorders has not been reported in clinical studies. However, as described above, furin expression levels are altered in several neurodegenerative and neuropsychiatric diseases; for instance, serum furin level is decreased in AD mice. These highlight the great potential of furin to be a predictive diagnostic marker for neurological disorders in the future.
The potentials of furin-targeting strategies to treat neurological diseases have been suggested in several animal models (Table 2). In AD animal models, injection of Furin-adenovirus into the cortex of Tg2576 mice markedly increases the α-secretase activity of ADAM10 and TACE, which in turn reduces Aβ production [13]. Furin-transgenic mice with brain-specific overexpression of furin exhibit increased dendritic spine density and enhanced learning and memory, which are attributed to the increased mBDNF level caused by furin [26]. In aged APP-C105 mice, treadmill exercise attenuates AD-related symptoms, possibly by ameliorating iron dyshomeostasis and enhancing furin expression, thereby promoting α-secretase-directed processing of APP [23]. Gallic acid treatment in APP/PS1 mice has been shown to increase furin expression, which in turn promotes α-secretase activity and decreases Aβ production, partly reversing the learning and memory impairment in APP/PS1 mice [338]. In addition, cerebrolysin, a peptidergic mixture with neurotrophic-like properties, can improve the survival of neural stem cell grafts and alleviate Aβ deposition in the hippocampus of APP transgenic mice, and this protective effect also involves the activation of furin and increased BDNF expression [339]. On the other hand, knockdown of astrocytic Grin2a in rats reduces furin expression and in turn decreases the maturation and secretion of NGF, aggravating the Aβ-induced memory and cognitive deficits [238]. These findings suggest the potential of increasing furin expression as an effective approach for AD treatment, and open avenues for future targets and strategies for AD prevention and therapeutic interventions.
Table 2.
Treatment effects of modulation of furin expression on neurological diseases
| Disease | Model | Treatment | Targeted region | Furin expression | Effects | References |
|---|---|---|---|---|---|---|
| AD | Tg2576 mice | Furin adenovirus | Cortex | Cortex ↑ | Reduces Aβ production by increasing α-secretase activity of ADAM10 and TACE | [13] |
| Furin-Tg mice | Brain-specific transgenic overexpression of furin | Brain | Brain ↑ | Elevates production of mBDNF, enhances dendritic spine density and promotes learning and memory | [26] | |
| AD | APP-C105 mice | Treadmill exercise | Whole body | Cortex ↑ | Increases furin expression, promoted APP cleavage by α-secretase, and attenuates AD-related symptoms | [23] |
| AD | APP/PS1 mice | Gallic acid | Whole body | Brain ↑ | Increases furin expression, activates ADAM10, and reverses the loss of learning and memory | [338] |
| AD | APP transgenic mice | Cerebrolysin | Hippocampus | Hippocampus ↑ | Increases furin and BDNF expression, improves survival of neural stem cell grafts and alleviates Aβ deposition | [339] |
| PD | Paraquat-treated Drosophila | Transgenic knockdown of Fur1 | DA neurons | DA neurons ↓ | Protects DA neurons against the toxic effect of paraquat | [265] |
| PD | Drosophila with LRRK2 overexpression | Disruption one allele of Fur1 | Whole body | Whole body ↓ | Reduces the retrograde synaptic enhancement induced by postsynaptic overexpression of LRRK2 | [266] |
| PD | Drosophila with LRRK2 overexpression | Postsynaptic knockdown of Fur1 | Postsynaptic muscles | Neuromuscular junction ↓ | Reduces the retrograde synaptic enhancement induced by postsynaptic overexpression of LRRK2 | [266] |
| Epilepsy | KA-induced epileptic mice; PTZ-kindled epileptic mice | Lentivirus containing sh-Furin | Hippocampus | Hippocampus ↓ | Reduces the spontaneous rhythmic electrical activity of cerebral neurons and suppresses epileptic seizure activity and severity | [25] |
| Cerebral ischemia | Global ischemia rats | Monosialoganglioside; Flavanol epicatechin | Whole body | Hippocampus ↑ | Increases the levels of furin and NGF | [340] |
In paraquat-induced Drosophila model of PD, transgenic knockdown of Fur1 in DA neurons provides significant protection against the loss of DA neurons [265]. In Drosophila models with LRRK2 overexpression, disruption of one allele of Fur1 or postsynaptic knockdown of Fur1 using transgenic RNA interference approach can attenuate the LRRK2-induced retrograde synaptic enhancement [266]. These findings suggest potential involvement of furin in PD pathophysiology and treatment. However, great efforts are urgently needed to explore the role and pharmaceutical potential of furin in PD patients or murine models.
In both KA-induced and PTZ-kindled epileptic mouse models, lentivirus-mediated knockdown of furin in the hippocampus decreases the spontaneous rhythmic electrical activity of cerebral neurons, and suppresses epileptic seizure activity and severity [25]. This protective role is proposed to be associated with the regulation of synaptic transmission by altering the transcription level of postsynaptic gamma-amino butyric acid A receptor [25].
In a global ischemia/reperfusion rat model, monosialoganglioside or flavanol epicatechin treatment both can improve spatial memory retention and acquisition in experimental ischemic rats [340], and these neurotherapeutic effects are found to be related to the increases in furin and NGF levels [340]. In addition, application of furin inhibitor can protect primary cortical neurons from cell death induced by activated NMDA receptors [341], which is possibly attributed to the decrease of furin-mediated cleavage of LRP1 [303]. These findings suggest that manipulating furin expression is potentially a good strategy for the treatment of ischemic brain injury.
In addition, some furin activators and inhibitors have been identified with drug potentials. The small molecules phorbol esters dPPA (12-deoxyphorbol 13-phenylacetate 20-acetate) and dPA (12-deoxyphorbol 13-acetate) exhibit great effects in promoting furin expression via activation of the transcription factor CEBPβ in neuronal cells [34]. On the other hand, polyarginines, such as hexa-D-arginine, significantly inhibit furin activity in vivo [342, 343]. The therapeutic effects of these furin activators and inhibitors in prevention and treatment of neurological disorders need to be investigated further in the future.
Conclusions
A growing body of evidence has suggested the crucial role of furin in the pathophysiological conditions of neurodegenerative and neuropsychiatric diseases. Notably, reduced furin expression is closely associated with the pathogenesis of AD. Pharmaceutical targeting of furin expression has shown great promise for AD treatment. In addition to AD, alterations of furin expression also exist in patients or animal models of epilepsy, cerebral ischemia, or SCZ. Furthermore, changes in the expression of neurotrophins, such as BDNF and NGF, are common to these neurodegenerative and neuropsychiatric diseases, and many are related to the abnormal cleavage of proneurotrophins. In addition to neurotrophins, other substrates of furin such as MMPs and LRP1 also exhibit expression changes in these neurodegenerative and neuropsychiatric diseases. These lines of evidence highlight the important roles of furin and furin-mediated activities in the progression of these diseases, and render furin as a valuable therapeutic target. However, currently very little is known about the cellular and molecular mechanisms of furin regulation in these diseases. Future studies are needed to clarify the molecular mechanisms of furin deregulation and its involvement in the pathogenesis of these diseases, and to develop new diagnostic and treatment strategies.
Acknowledgements
Not applicable.
Abbreviations
- AD
Alzheimer’s disease
- SCZ
Schizophrenia
- PC
Proprotein convertase
- BDNF
Brain-derived neurotrophic factor
- NGF
Nerve growth factor
- MMPs
Multiple matrix metalloproteases
- PD
Parkinson’s disease
- HIF-1
Hypoxia-inducible factor-1
- TGFβ1
Transforming growth factor beta1
- ER
Endoplasmic reticulum
- DG
Dentate gyrus
- ADAMs
A disintegrin and metalloproteases
- BACE1
Beta-site APP cleaving enzyme 1
- LRP1
Low-density lipoprotein receptor-related protein 1
- GPR37
G-protein-coupled receptor
- mBDNF
Mature BDNF
- TrkB
Tropomyosin-related receptor kinase B
- PI3K
Phosphatidylinositol 3-kinase
- Akt
Protein kinase B
- PLCγ
Phospholipase C-gamma
- CaMK
Calcium-dependent protein kinase
- MAPK
Mitogen-activated protein kinase
- LTP
Long-term potentiation
- p75NTR
P75 neurotrophic receptor
- mNGF
Mature NGF
- TrkA
Tropomyosin-related receptor kinase A
- NT-3
Neurotrophin-3
- NT-4/5
Neurotrophin-4/5
- DA
Dopaminergic
- APP
Amyloid-β precursor protein
- sAPPα
αAPP
- Aβ
Amyloid-β
- CSF
Cerebrospinal fluid
- ICD
Intracellular domain
- NMDA
N-methyl-D-aspartate
- BRI2
Integral membrane protein 2
- NFTs
Neurofibrillary tangles
- BBB
Blood–brain barrier
- TACE
Tumor necrosis factor-α converting enzyme
- LRRK2
Leucine-rich repeat kinase 2
- TLE
Temporal lobe epilepsy
- KA
Kainic acid
- PTZ
Pentylenetetrazol
- MDD
Major depressive disorder
- BIP
Bipolar disorder
Author contributions
YZC and GG designed the work. YZ, XG and GG wrote a major portion of the manuscript and made all the figures. XB and SY wrote a small portion of the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 32170979 and 32070962), the Science and Technology Project of Hebei Education Department (grant number ZD2021327), and the Natural Science Foundation of Hebei Normal University (grant number L2021Z04).
Availability of data and materials
Available upon request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Yi Zhang and Xiaoqin Gao contributed equally to this work.
Contributor Information
Yan-Zhong Chang, Email: yzchang@hebtu.edu.cn.
Guofen Gao, Email: gaoguofen@hebtu.edu.cn.
References
- 1.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van de Ven WJM, Voorberg J, Fontijn R, Pannekoek H, van den Ouweland AMW, van Duijnhoven HLP, et al. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol Biol Rep. 1990;14(4):265–275. doi: 10.1007/BF00429896. [DOI] [PubMed] [Google Scholar]
- 3.Wise RJ, Barr PJ, Wong PA, Kiefer MC, Brake AJ, Kaufman RJ. Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc Natl Acad Sci U S A. 1990;87(23):9378–9382. doi: 10.1073/pnas.87.23.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun E, Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunol. 2019;8(8):e107-e. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res. 2020;50:102115. doi: 10.1016/j.scr.2020.102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discovery. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 9.He Z, Khatib AM, Creemers JWM. The proprotein convertase furin in cancer: more than an oncogene. Oncogene. 2022;41(9):1252–1262. doi: 10.1038/s41388-021-02175-9. [DOI] [PubMed] [Google Scholar]
- 10.Jaaks P, Bernasconi M. The proprotein convertase furin in tumour progression. Int J Cancer. 2017;141(4):654–663. doi: 10.1002/ijc.30714. [DOI] [PubMed] [Google Scholar]
- 11.Bresnahan PA, Leduc R, Thomas L, Thorner J, Gibson HL, Brake AJ, et al. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J Cell Biol. 1990;111(6 Pt 2):2851–2859. doi: 10.1083/jcb.111.6.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer A, Chrétien P, Massicotte G, Sargent C, Chrétien M, Marcinkiewicz M. Kainic acid increases the expression of the prohormone convertases furin and PC1 in the mouse hippocampus. Brain Res. 1996;732(1–2):121–132. doi: 10.1016/0006-8993(96)00502-1. [DOI] [PubMed] [Google Scholar]
- 13.Hwang EM, Kim SK, Sohn JH, Lee JY, Kim Y, Kim YS, et al. Furin is an endogenous regulator of α-secretase associated APP processing. Biochem Biophys Res Commun. 2006;349(2):654–659. doi: 10.1016/j.bbrc.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 14.Creemers JW, Ines Dominguez D, Plets E, Serneels L, Taylor NA, Multhaup G, et al. Processing of beta-secretase by furin and other members of the proprotein convertase family. J Biol Chem. 2001;276(6):4211–4217. doi: 10.1074/jbc.M006947200. [DOI] [PubMed] [Google Scholar]
- 15.Golubkov VS, Chernov AV, Strongin AY. Intradomain cleavage of inhibitory prodomain is essential to protumorigenic function of membrane type-1 matrix metalloproteinase (MT1-MMP) in vivo. J Biol Chem. 2011;286(39):34215–34223. doi: 10.1074/jbc.M111.264036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Pei D. Shedding of membrane type matrix metalloproteinase 5 by a furin-type convertase: a potential mechanism for down-regulation. J Biol Chem. 2001;276(38):35953–35960. doi: 10.1074/jbc.M103680200. [DOI] [PubMed] [Google Scholar]
- 17.Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, et al. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem. 2001;276(16):12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- 18.Thomas E, Joan M, Noel M, Thomas J, Joachim H. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem J. 1996;313(1):71–76. doi: 10.1042/bj3130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CS, Kavalali ET, Monteggia LM. BDNF signaling in context: From synaptic regulation to psychiatric disorders. Cell. 2022;185(1):62–76. doi: 10.1016/j.cell.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L, Zhang Y, Sterling K, Song W. Brain-derived neurotrophic factor in Alzheimer's disease and its pharmaceutical potential. Transl Neurodegener. 2022;11(1):4. doi: 10.1186/s40035-022-00279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camuso S, La Rosa P, Fiorenza MT, Canterini S. Pleiotropic effects of BDNF on the cerebellum and hippocampus: implications for neurodevelopmental disorders. Neurobiol Dis. 2022;163:105606. doi: 10.1016/j.nbd.2021.105606. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi A, Amooeian GV, Rashidi E. Dysfunction in brain-derived neurotrophic factor signaling pathway and susceptibility to schizophrenia, Parkinson’s and Alzheimer’s diseases. Curr Gene Ther. 2018;18(1):45–63. doi: 10.2174/1566523218666180302163029. [DOI] [PubMed] [Google Scholar]
- 23.Choi DH, Kwon KC, Hwang DJ, Koo JH, Um HS, Song HS, et al. Treadmill exercise alleviates brain iron dyshomeostasis accelerating neuronal amyloid-β production, neuronal cell death, and cognitive impairment in transgenic mice model of Alzheimer's disease. Mol Neurobiol. 2021;58(7):3208–3223. doi: 10.1007/s12035-021-02335-8. [DOI] [PubMed] [Google Scholar]
- 24.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, He M, Tian X, Guo Y, Liu F, Li Y, et al. Transgenic overexpression of furin increases epileptic susceptibility. Cell Death Dis. 2018;9(11):1058. doi: 10.1038/s41419-018-1076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B, Zhao L, Luo D, Xu D, Tan T, Dong Z, et al. Furin promotes dendritic morphogenesis and learning and memory in transgenic mice. Cell Mol Life Sci. 2018;75(13):2473–2488. doi: 10.1007/s00018-017-2742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roebroek AJM, Schalken JA, Bussemakers MJG, van Heerikhuizen H, Onnekink C, Debruyne FMJ, et al. Characterization of human c-fes/fps reveals a new transcription unit (fur) in the immediately upstream region of the proto-oncogene. Mol Biol Rep. 1986;11(2):117–125. doi: 10.1007/BF00364823. [DOI] [PubMed] [Google Scholar]
- 28.Fuller Robert S, Brake Anthony J, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246(4929):482–486. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- 29.Rockwell NC, Thorner JW. The kindest cuts of all: crystal structures of Kex2 and furin reveal secrets of precursor processing. Trends Biochem Sci. 2004;29(2):80–87. doi: 10.1016/j.tibs.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Ayoubi TA, Creemers JW, Roebroek AJ, Van de Ven WJ. Expression of the dibasic proprotein processing enzyme furin is directed by multiple promoters. J Biol Chem. 1994;269(12):9298–9303. doi: 10.1016/S0021-9258(17)37107-7. [DOI] [PubMed] [Google Scholar]
- 31.Lei RX, Shi H, Peng XM, Zhu YH, Cheng J, Chen GH. Influence of a single nucleotide polymorphism in the P1 promoter of the furin gene on transcription activity and hepatitis B virus infection. Hepatology. 2009;50(3):763–771. doi: 10.1002/hep.23062. [DOI] [PubMed] [Google Scholar]
- 32.Laprise M-H, Grondin F, Cayer P, McDonald PP, Dubois CM. Furin gene (fur) regulation in differentiating human megakaryoblastic Dami cells: involvement of the proximal GATA recognition motif in the P1 promoter and impact on the maturation of furin substrates. Blood. 2002;100(10):3578–3587. doi: 10.1182/blood.V100.10.3578. [DOI] [PubMed] [Google Scholar]
- 33.Dong X, Chiu H, Park YJ, Zou W, Zou Y, Özkan E, et al. Precise regulation of the guidance receptor DMA-1 by KPC-1/Furin instructs dendritic branching decisions. eLife. 2016 doi: 10.7554/eLife.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zha JS, Zhu BL, Liu L, Lai YJ, Long Y, Hu XT, et al. Phorbol esters dPPA/dPA promote furin expression involving transcription factor CEBPβ in neuronal cells. Oncotarget. 2017;8(36):60159–60172. doi: 10.18632/oncotarget.18569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111(2):924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z, Wang R, Yang X, Lu XY, Zhang Q, Wang YL, et al. The cAMP-responsive element binding protein (CREB) transcription factor regulates furin expression during human trophoblast syncytialization. Placenta. 2014;35(11):907–918. doi: 10.1016/j.placenta.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 37.McMahon S, Grondin F, McDonald PP, Richard DE, Dubois CM. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: impact on the bioactivation of proproteins. J Biol Chem. 2005;280(8):6561–6569. doi: 10.1074/jbc.M413248200. [DOI] [PubMed] [Google Scholar]
- 38.Blanchette F, Rudd P, Grondin F, Attisano L, Dubois CM. Involvement of Smads in TGFbeta1-induced furin (fur) transcription. J Cell Physiol. 2001;188(2):264–273. doi: 10.1002/jcp.1116. [DOI] [PubMed] [Google Scholar]
- 39.Ventura E, Weller M, Burghardt I. Cutting edge: ERK1 mediates the autocrine positive feedback loop of TGF-β and furin in glioma-initiating cells. J Immunol. 2017;198(12):4569–4574. doi: 10.4049/jimmunol.1601176. [DOI] [PubMed] [Google Scholar]
- 40.Bai L, Chang HM, Zhang L, Zhu YM, Leung PCK. BMP2 increases the production of BDNF through the upregulation of proBDNF and furin expression in human granulosa-lutein cells. FASEB J. 2020;34(12):16129–16143. doi: 10.1096/fj.202000940R. [DOI] [PubMed] [Google Scholar]
- 41.Osadchuk TV, Shybyryn OV, Kibirev VK. Chemical structure and properties of low-molecular furin inhibitors. Ukr Biochem J. 2016;88(6):5–25. doi: 10.15407/ubj88.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Gawlik K, Shiryaev SA, Zhu W, Motamedchaboki K, Desjardins R, Day R, et al. Autocatalytic activation of the furin zymogen requires removal of the emerging enzyme's N-terminus from the active site. PLoS One. 2009;4(4):e5031-e. doi: 10.1371/journal.pone.0005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehemtulla A, Dorner AJ, Kaufman RJ. Regulation of PACE propeptide-processing activity: requirement for a post-endoplasmic reticulum compartment and autoproteolytic activation. Proc Natl Acad Sci U S A. 1992;89(17):8235–8239. doi: 10.1073/pnas.89.17.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solovyeva NI, Gureeva TA, Timoshenko OS, Moskvitina TA, Kugaevskaya EV. Furin as proprotein convertase and its role in normal and pathological biological processes. Biochem Mosc Suppl B Biomed Chem. 2017;11(2):87–100. doi: 10.18097/PBMC20166206609. [DOI] [PubMed] [Google Scholar]
- 45.Leduc R, Molloy SS, Thorne BA, Thomas G. Activation of human furin precursor processing endoprotease occurs by an intramolecular autoproteolytic cleavage. J Biol Chem. 1992;267(20):14304–14308. doi: 10.1016/S0021-9258(19)49712-3. [DOI] [PubMed] [Google Scholar]
- 46.Denault J, Bissonnette L, Longpré J, Charest G, Lavigne P, Leduc R. Ectodomain shedding of furin: kinetics and role of the cysteine-rich region. FEBS Lett. 2002;527(1–3):309–314. doi: 10.1016/S0014-5793(02)03249-0. [DOI] [PubMed] [Google Scholar]
- 47.De Bie I, Savaria D, Roebroek AJ, Day R, Lazure C, Van de Ven WJ, et al. Processing specificity and biosynthesis of the Drosophila melanogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J Biol Chem. 1995;270(3):1020–1028. doi: 10.1074/jbc.270.3.1020. [DOI] [PubMed] [Google Scholar]
- 48.Gómez-Saladín E, Luebke AE, Wilson DL, Dickerson IM. Isolation of a cDNA encoding a Kex2-like endoprotease with homology to furin from the nematode Caenorhabditis elegans. DNA Cell Biol. 1997;16(5):663–669. doi: 10.1089/dna.1997.16.663. [DOI] [PubMed] [Google Scholar]
- 49.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 50.Li D, Liu X, Zhang L, He J, Chen X, Liu S, et al. COVID-19 disease and malignant cancers: the impact for the furin gene expression in susceptibility to SARS-CoV-2. Int J Biol Sci. 2021;17(14):3954–3967. doi: 10.7150/ijbs.63072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020 doi: 10.1126/science.aay5947. [DOI] [PubMed] [Google Scholar]
- 52.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 53.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021 doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dijk W, Ruppert PMM, Oost LJ, Kersten S. Angiopoietin-like 4 promotes the intracellular cleavage of lipoprotein lipase by PCSK3/furin in adipocytes. J Biol Chem. 2018;293(36):14134–14145. doi: 10.1074/jbc.RA118.002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hosaka M, Nagahama M, Kim WS, Watanabe T, Hatsuzawa K, Ikemizu J, et al. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266(19):12127–12130. doi: 10.1016/S0021-9258(18)98867-8. [DOI] [PubMed] [Google Scholar]
- 56.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267(23):16396–16402. doi: 10.1016/S0021-9258(18)42016-9. [DOI] [PubMed] [Google Scholar]
- 57.Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, et al. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125(24):4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 58.Liu ZW, Ma Q, Liu J, Li JW, Chen YD. The association between plasma furin and cardiovascular events after acute myocardial infarction. BMC Cardiovasc Disord. 2021;21(1):468. doi: 10.1186/s12872-021-02029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Xie Y, Qin D. Proteolytic cleavage of proBDNF to mBDNF in neuropsychiatric and neurodegenerative diseases. Brain Res Bull. 2021;166:172–184. doi: 10.1016/j.brainresbull.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 60.Farhat D, Ghayad SE, Icard P, Le Romancer M, Hussein N, Lincet H. Lipoic acid-induced oxidative stress abrogates IGF-1R maturation by inhibiting the CREB/furin axis in breast cancer cell lines. Oncogene. 2020;39(17):3604–3610. doi: 10.1038/s41388-020-1211-x. [DOI] [PubMed] [Google Scholar]
- 61.Ren K, Jiang T, Zheng XL, Zhao GJ. Proprotein convertase furin/PCSK3 and atherosclerosis: new insights and potential therapeutic targets. Atherosclerosis. 2017;262:163–170. doi: 10.1016/j.atherosclerosis.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez C, Rysä J, Almgren P, Nilsson J, Engström G, Orho-Melander M, et al. Plasma levels of the proprotein convertase furin and incidence of diabetes and mortality. J Intern Med. 2018;284(4):377–387. doi: 10.1111/joim.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Bai X, Zhang Y, Yao S, Cui Y, You LH, et al. Hippocampal iron accumulation impairs synapses and memory via suppressing furin expression and downregulating BDNF maturation. Mol Neurobiol. 2022 doi: 10.1007/s12035-022-02929-w. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Zhang J, Deng M. Furin mediates brain-derived neurotrophic factor upregulation in cultured rat astrocytes exposed to oxygen-glucose deprivation. J Neurosci Res. 2015;93(1):189–194. doi: 10.1002/jnr.23455. [DOI] [PubMed] [Google Scholar]
- 65.Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci. 2005;6(8):603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- 66.Yan Q, Rosenfeld RD, Matheson CR, Hawkins N, Lopez OT, Bennett L, et al. Expression of brain-derived neurotrophic factor protein in the adult rat central nervous system. Neuroscience. 1997;78(2):431–448. doi: 10.1016/S0306-4522(96)00613-6. [DOI] [PubMed] [Google Scholar]
- 67.Leal G, Bramham CR, Duarte CB. BDNF and hippocampal synaptic plasticity. Vitam Horm. 2017;104:153–195. doi: 10.1016/bs.vh.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Foltran RB, Diaz SL. BDNF isoforms: a round trip ticket between neurogenesis and serotonin? J Neurochem. 2016;138(2):204–221. doi: 10.1111/jnc.13658. [DOI] [PubMed] [Google Scholar]
- 69.Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22(3):123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizui T, Ishikawa Y, Kumanogoh H, Lume M, Matsumoto T, Hara T, et al. BDNF pro-peptide actions facilitate hippocampal LTD and are altered by the common BDNF polymorphism Val66Met. Proc Natl Acad Sci U S A. 2015;112(23):E3067–E3074. doi: 10.1073/pnas.1422336112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gray K, Ellis V. Activation of pro-BDNF by the pericellular serine protease plasmin. FEBS Lett. 2008;582(6):907–910. doi: 10.1016/j.febslet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 72.Niculescu D, Michaelsen-Preusse K, Güner Ü, van Dorland R, Wierenga CJ, Lohmann C. A BDNF-mediated push-pull plasticity mechanism for synaptic clustering. Cell Rep. 2018;24(8):2063–2074. doi: 10.1016/j.celrep.2018.07.073. [DOI] [PubMed] [Google Scholar]
- 73.Martins CC, Rosa SG, Recchi AMS, Nogueira CW, Zeni G. m-Trifluoromethyl-diphenyl diselenide (m-CF(3)-PhSe)(2) modulates the hippocampal neurotoxic adaptations and abolishes a depressive-like phenotype in a short-term morphine withdrawal in mice. Prog Neuropsychopharmacol Biol Psychiatr. 2020;98:109803. doi: 10.1016/j.pnpbp.2019.109803. [DOI] [PubMed] [Google Scholar]
- 74.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/S0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 75.Blum R, Konnerth A. Neurotrophin-mediated rapid signaling in the central nervous system: mechanisms and functions. Physiol (Bethesda) 2005;20:70–78. doi: 10.1152/physiol.00042.2004. [DOI] [PubMed] [Google Scholar]
- 76.Du Q, Zhu X, Si J. Angelica polysaccharide ameliorates memory impairment in Alzheimer's disease rat through activating BDNF/TrkB/CREB pathway. Exp Biol Med (Maywood) 2020;245(1):1–10. doi: 10.1177/1535370219894558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dechant G, Barde YA. The neurotrophin receptor p75(NTR): novel functions and implications for diseases of the nervous system. Nat Neurosci. 2002;5(11):1131–1136. doi: 10.1038/nn1102-1131. [DOI] [PubMed] [Google Scholar]
- 78.Zagrebelsky M, Korte M. Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology. 2014;76:628–38. doi: 10.1016/j.neuropharm.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 79.Choi SH, Bylykbashi E, Chatila ZK, Lee SW, Pulli B, Clemenson GD, et al. Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer's mouse model. Science. 2018 doi: 10.1126/science.aan8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie W, Meng X, Zhai Y, Ye T, Zhou P, Nan F, et al. Antidepressant-like effects of the Guanxin Danshen formula via mediation of the CaMK II-CREB-BDNF signalling pathway in chronic unpredictable mild stress-induced depressive rats. Ann Transl Med. 2019;7(20):564. doi: 10.21037/atm.2019.09.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Girotra P, Behl T, Sehgal A, Singh S, Bungau S. Investigation of the molecular role of brain-derived neurotrophic factor in Alzheimer's disease. J Mol Neurosci. 2022;72(2):173–186. doi: 10.1007/s12031-021-01824-8. [DOI] [PubMed] [Google Scholar]
- 82.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237(4819):1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 83.Fahnestock M, Yu G, Coughlin MD. ProNGF: a neurotrophic or an apoptotic molecule? Prog Brain Res. 2004;146:101–110. doi: 10.1016/S0079-6123(03)46007-X. [DOI] [PubMed] [Google Scholar]
- 84.Seidah NG, Benjannet S, Pareek S, Savaria D, Hamelin J, Goulet B, et al. Cellular processing of the nerve growth factor precursor by the mammalian pro-protein convertases. Biochem J. 1996;314:951–60. doi: 10.1042/bj3140951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan R, Yalinca H, Paoletti F, Gobbo F, Marchetti L, Kuzmanic A, et al. The structure of the pro-domain of mouse proNGF in contact with the NGF domain. Structure. 2019;27(1):78–89.e3. doi: 10.1016/j.str.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 86.Bruno MA, Cuello AC. Activity-dependent release of precursor nerve growth factor, conversion to mature nerve growth factor, and its degradation by a protease cascade. Proc Natl Acad Sci U S A. 2006;103(17):6735–6740. doi: 10.1073/pnas.0510645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fahnestock M, Yu G, Michalski B, Mathew S, Colquhoun A, Ross GM, et al. The nerve growth factor precursor proNGF exhibits neurotrophic activity but is less active than mature nerve growth factor. J Neurochem. 2004;89(3):581–592. doi: 10.1111/j.1471-4159.2004.02360.x. [DOI] [PubMed] [Google Scholar]
- 88.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294(5548):1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 89.Canu N, Amadoro G, Triaca V, Latina V, Sposato V, Corsetti V, et al. The intersection of NGF/TrkA signaling and amyloid precursor protein processing in Alzheimer's disease neuropathology. Int J Mol Sci. 2017;18(6):1319. doi: 10.3390/ijms18061319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10(3):381–391. doi: 10.1016/S0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 91.Beattie MS, Harrington AW, Lee R, Kim JY, Boyce SL, Longo FM, et al. ProNGF induces p75-mediated death of oligodendrocytes following spinal cord injury. Neuron. 2002;36(3):375–386. doi: 10.1016/S0896-6273(02)01005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harrington AW, Leiner B, Blechschmitt C, Arevalo JC, Lee R, Mörl K, et al. Secreted proNGF is a pathophysiological death-inducing ligand after adult CNS injury. Proc Natl Acad Sci U S A. 2004;101(16):6226–6230. doi: 10.1073/pnas.0305755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, et al. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427(6977):843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 94.Fahnestock M, Michalski B, Xu B, Coughlin MD. The precursor pro-nerve growth factor is the predominant form of nerve growth factor in brain and is increased in Alzheimer's disease. Mol Cell Neurosci. 2001;18(2):210–220. doi: 10.1006/mcne.2001.1016. [DOI] [PubMed] [Google Scholar]
- 95.Bruno MA, Leon WC, Fragoso G, Mushynski WE, Almazan G, Cuello AC. Amyloid beta-induced nerve growth factor dysmetabolism in Alzheimer disease. J Neuropathol Exp Neurol. 2009;68(8):857–869. doi: 10.1097/NEN.0b013e3181aed9e6. [DOI] [PubMed] [Google Scholar]
- 96.Tiveron C, Fasulo L, Capsoni S, Malerba F, Marinelli S, Paoletti F, et al. ProNGF\NGF imbalance triggers learning and memory deficits, neurodegeneration and spontaneous epileptic-like discharges in transgenic mice. Cell Death Differ. 2013;20(8):1017–1030. doi: 10.1038/cdd.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ip NY, Yancopoulos GD. Neurotrophic factors and their receptors. Ann Neurol. 1994;35(Suppl):S13–S16. doi: 10.1002/ana.410350706. [DOI] [PubMed] [Google Scholar]
- 98.Barbacid M. Neurotrophic factors and their receptors. Curr Opin Cell Biol. 1995;7(2):148–155. doi: 10.1016/0955-0674(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 99.Pathare-Ingawale P, Chavan-Gautam P. The balance between cell survival and death in the placenta: do neurotrophins have a role? Syst Biol Reprod Med. 2022;68(1):3–12. doi: 10.1080/19396368.2021.1980132. [DOI] [PubMed] [Google Scholar]
- 100.Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4(4):299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 101.Keefe KM, Sheikh IS, Smith GM. Targeting neurotrophins to specific populations of neurons: NGF, BDNF, and NT-3 and their relevance for treatment of spinal cord injury. Int J Mol Sci. 2017;18(3):548. doi: 10.3390/ijms18030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lai BQ, Bai YR, Han WT, Zhang B, Liu S, Sun JH, et al. Construction of a niche-specific spinal white matter-like tissue to promote directional axon regeneration and myelination for rat spinal cord injury repair. Bioact Mater. 2022;11:15–31. doi: 10.1016/j.bioactmat.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ozes B, Moss K, Myers M, Ridgley A, Chen L, Murrey D, Sahenk Z. AAV1.NT-3 gene therapy in a CMT2D model: phenotypic improvements in GarsP278KY/+mice. Brain Commun. 2021 doi: 10.1093/braincomms/fcab252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Müller ML, Peglau L, Moon LDF, Groß S, Schulze J, Ruhnau J, et al. Neurotrophin-3 attenuates human peripheral blood T cell and monocyte activation status and cytokine production post stroke. Exp Neurol. 2022;347:113901. doi: 10.1016/j.expneurol.2021.113901. [DOI] [PubMed] [Google Scholar]
- 105.Rodrigues-Amorim D, Iglesias-Martínez-Almeida M, Rivera-Baltanás T, Fernández-Palleiro P, Freiría-Martínez L, Rodríguez-Jamardo C, et al. The role of the second extracellular loop of norepinephrine transporter, neurotrophin-3 and tropomyosin receptor kinase C in T cells: a peripheral biomarker in the etiology of schizophrenia. Int J Mol Sci. 2021;22(16):8499. doi: 10.3390/ijms22168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Requena-Ocaña N, Araos P, Flores M, García-Marchena N, Silva-Peña D, Aranda J, et al. Evaluation of neurotrophic factors and education level as predictors of cognitive decline in alcohol use disorder. Sci Rep. 2021;11(1):15583. doi: 10.1038/s41598-021-95131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ribeiro D, Petrigna L, Pereira FC, Muscella A, Bianco A, Tavares P. The Impact of Physical Exercise on the Circulating Levels of BDNF and NT 4/5: A Review. Int J Mol Sci. 2021;22(16):8814. doi: 10.3390/ijms22168814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beroun A, Mitra S, Michaluk P, Pijet B, Stefaniuk M, Kaczmarek L. MMPs in learning and memory and neuropsychiatric disorders. Cell Mol Life Sci. 2019;76(16):3207–3228. doi: 10.1007/s00018-019-03180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ierusalimsky VN, Balaban PM. Type 1 metalloproteinase is selectively expressed in adult rat brain and can be rapidly up-regulated by kainate. Acta Histochem. 2013;115(8):816–826. doi: 10.1016/j.acthis.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 110.Rosenberg GA, Sullivan N, Esiri MM. White matter damage is associated with matrix metalloproteinases in vascular dementia. Stroke. 2001;32(5):1162–1168. doi: 10.1161/01.STR.32.5.1162. [DOI] [PubMed] [Google Scholar]
- 111.Wiera G, Nowak D, van Hove I, Dziegiel P, Moons L, Mozrzymas JW. Mechanisms of NMDA receptor- and voltage-gated L-type calcium channel-dependent hippocampal LTP critically rely on proteolysis that is mediated by distinct metalloproteinases. J Neurosci. 2017;37(5):1240–1256. doi: 10.1523/JNEUROSCI.2170-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vafadari B, Salamian A, Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J Neurochem. 2016;139(Suppl 2):91–114. doi: 10.1111/jnc.13415. [DOI] [PubMed] [Google Scholar]
- 113.Rosenberg GA. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol. 2009;8(2):205–216. doi: 10.1016/S1474-4422(09)70016-X. [DOI] [PubMed] [Google Scholar]
- 114.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, et al. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297(5584):1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 115.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stawowy P, Fleck E. Proprotein convertases furin and PC5: targeting atherosclerosis and restenosis at multiple levels. J Mol Med (Berl) 2005;83(11):865–875. doi: 10.1007/s00109-005-0723-8. [DOI] [PubMed] [Google Scholar]
- 117.Cao J, Rehemtulla A, Bahou W, Zucker S. Membrane type matrix metalloproteinase 1 activates pro-gelatinase a without furin cleavage of the N-terminal domain. J Biol Chem. 1996;271(47):30174–30180. doi: 10.1074/jbc.271.47.30174. [DOI] [PubMed] [Google Scholar]
- 118.Cao J, Rehemtulla A, Pavlaki M, Kozarekar P, Chiarelli C. Furin directly cleaves proMMP-2 in the trans-Golgi network resulting in a nonfunctioning proteinase. J Biol Chem. 2005;280(12):10974–10980. doi: 10.1074/jbc.M412370200. [DOI] [PubMed] [Google Scholar]
- 119.Mittal R, Patel AP, Debs LH, Nguyen D, Patel K, Grati M, et al. Intricate functions of matrix metalloproteinases in physiological and pathological conditions. J Cell Physiol. 2016;231(12):2599–2621. doi: 10.1002/jcp.25430. [DOI] [PubMed] [Google Scholar]
- 120.Valente MM, Allen M, Bortolotto V, Lim ST, Conant K, Grilli M. The MMP-1/PAR-1 axis enhances proliferation and neuronal differentiation of adult hippocampal neural progenitor cells. Neural Plast. 2015;2015:646595. doi: 10.1155/2015/646595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Allen M, Ghosh S, Ahern GP, Villapol S, Maguire-Zeiss KA, Conant K. Protease induced plasticity: matrix metalloproteinase-1 promotes neurostructural changes through activation of protease activated receptor 1. Sci Rep. 2016;6:35497. doi: 10.1038/srep35497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ogier C, Bernard A, Chollet AM, Diguardher TLE, Hanessian S, Charton G, et al. Matrix metalloproteinase-2 (MMP-2) regulates astrocyte motility in connection with the actin cytoskeleton and integrins. Glia. 2006;54(4):272–284. doi: 10.1002/glia.20349. [DOI] [PubMed] [Google Scholar]
- 123.Muir EM, Adcock KH, Morgenstern DA, Clayton R, von Stillfried N, Rhodes K, et al. Matrix metalloproteases and their inhibitors are produced by overlapping populations of activated astrocytes. Brain Res Mol Brain Res. 2002;100(1–2):103–117. doi: 10.1016/S0169-328X(02)00132-8. [DOI] [PubMed] [Google Scholar]
- 124.Woo MS, Park JS, Choi IY, Kim WK, Kim HS. Inhibition of MMP-3 or -9 suppresses lipopolysaccharide-induced expression of proinflammatory cytokines and iNOS in microglia. J Neurochem. 2008;106(2):770–780. doi: 10.1111/j.1471-4159.2008.05430.x. [DOI] [PubMed] [Google Scholar]
- 125.Bozdagi O, Nagy V, Kwei KT, Huntley GW. In vivo roles for matrix metalloproteinase-9 in mature hippocampal synaptic physiology and plasticity. J Neurophysiol. 2007;98(1):334–344. doi: 10.1152/jn.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nagy V, Bozdagi O, Matynia A, Balcerzyk M, Okulski P, Dzwonek J, et al. Matrix metalloproteinase-9 is required for hippocampal late-phase long-term potentiation and memory. J Neurosci. 2006;26(7):1923–1934. doi: 10.1523/JNEUROSCI.4359-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mroczko B, Groblewska M, Zboch M, Kulczyńska A, Koper OM, Szmitkowski M, et al. Concentrations of matrix metalloproteinases and their tissue inhibitors in the cerebrospinal fluid of patients with Alzheimer's disease. J Alzheimers Dis. 2014;40(2):351–357. doi: 10.3233/JAD-131634. [DOI] [PubMed] [Google Scholar]
- 128.Levin J, Giese A, Boetzel K, Israel L, Högen T, Nübling G, et al. Increased alpha-synuclein aggregation following limited cleavage by certain matrix metalloproteinases. Exp Neurol. 2009;215(1):201–208. doi: 10.1016/j.expneurol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 129.Sung JY, Park SM, Lee C-H, Um JW, Lee HJ, Kim J, et al. Proteolytic cleavage of extracellular secreted α-synuclein via matrix metalloproteinases. J Biol Chem. 2005;280(26):25216–25224. doi: 10.1074/jbc.M503341200. [DOI] [PubMed] [Google Scholar]
- 130.Choi DH, Kim YJ, Kim YG, Joh TH, Beal MF, Kim YS. Role of matrix metalloproteinase 3-mediated alpha-synuclein cleavage in dopaminergic cell death. J Biol Chem. 2011;286(16):14168–14177. doi: 10.1074/jbc.M111.222430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lorenzl S, Albers DS, Narr S, Chirichigno J, Beal MF. Expression of MMP-2, MMP-9, and MMP-1 and their endogenous counterregulators TIMP-1 and TIMP-2 in postmortem brain tissue of Parkinson's disease. Exp Neurol. 2002;178(1):13–20. doi: 10.1006/exnr.2002.8019. [DOI] [PubMed] [Google Scholar]
- 132.Mullooly M, McGowan PM, Crown J, Duffy MJ. The ADAMs family of proteases as targets for the treatment of cancer. Cancer Biol Ther. 2016;17(8):870–880. doi: 10.1080/15384047.2016.1177684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15(10):1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 134.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 135.Seipold L, Altmeppen H, Koudelka T, Tholey A, Kasparek P, Sedlacek R, et al. In vivo regulation of the a disintegrin and metalloproteinase 10 (ADAM10) by the tetraspanin 15. Cell Mol Life Sci. 2018;75(17):3251–3267. doi: 10.1007/s00018-018-2791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284(17):11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan XZ, Sun S, Tan CC, Yu JT, Tan L. The role of ADAM10 in Alzheimer's disease. J Alzheimers Dis. 2017;58(2):303–322. doi: 10.3233/JAD-170061. [DOI] [PubMed] [Google Scholar]
- 138.Manzine PR, Ettcheto M, Cano A, Busquets O, Marcello E, Pelucchi S, et al. ADAM10 in Alzheimer's disease: pharmacological modulation by natural compounds and its role as a peripheral marker. Biomed Pharmacother. 2019;113:108661. doi: 10.1016/j.biopha.2019.108661. [DOI] [PubMed] [Google Scholar]
- 139.Xu X. γ-Secretase catalyzes sequential cleavages of the AβPP transmembrane domain. J Alzheimers Dis. 2009;16:211–224. doi: 10.3233/JAD-2009-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hong L, Koelsch G, Lin X, Wu S, Terzyan S, Ghosh AK, et al. Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor. Science. 2000;290(5489):150–153. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- 141.Dislich B, Lichtenthaler SF. The membrane-bound aspartyl protease BACE1: molecular and functional properties in Alzheimer's disease and beyond. Front Physiol. 2012;3:8. doi: 10.3389/fphys.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Evin G, Barakat A, Masters CL. BACE: therapeutic target and potential biomarker for Alzheimer's disease. Int J Biochem Cell Biol. 2010;42(12):1923–1926. doi: 10.1016/j.biocel.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 143.Bennett BD, Denis P, Haniu M, Teplow DB, Kahn S, Louis JC, et al. A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer's beta -secretase. J Biol Chem. 2000;275(48):37712–37717. doi: 10.1074/jbc.M005339200. [DOI] [PubMed] [Google Scholar]
- 144.Neumann U, Ufer M, Jacobson LH, et al. TheBACE‐1 inhibitor CNP520 for prevention trials in Alzheimer's disease. EMBO Mol Med. 2018 doi: 10.15252/emmm.201809316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hampel H, Shen Y. Beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) as a biological candidate marker of Alzheimer's disease. Scand J Clin Lab Invest. 2009;69(1):8–12. doi: 10.1080/00365510701864610. [DOI] [PubMed] [Google Scholar]
- 146.Zhong Z, Ewers M, Teipel S, Bürger K, Wallin A, Blennow K, et al. Levels of beta-secretase (BACE1) in cerebrospinal fluid as a predictor of risk in mild cognitive impairment. Arch Gen Psychiatr. 2007;64(6):718–726. doi: 10.1001/archpsyc.64.6.718. [DOI] [PubMed] [Google Scholar]
- 147.Shen Y, Wang H, Sun Q, Yao H, Keegan AP, Mullan M, et al. Increased plasma beta-secretase 1 may predict conversion to Alzheimer's disease dementia in individuals with mild cognitive impairment. Biol Psychiatry. 2018;83(5):447–455. doi: 10.1016/j.biopsych.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weiss MM, Williamson T, Babu-Khan S, Bartberger MD, Brown J, Chen K, et al. Design and preparation of a potent series of hydroxyethylamine containing β-secretase inhibitors that demonstrate robust reduction of central β-amyloid. J Med Chem. 2012;55(21):9009–9024. doi: 10.1021/jm300119p. [DOI] [PubMed] [Google Scholar]
- 149.Lahiri DK, Maloney B, Long JM, Greig NH. Lessons from a BACE1 inhibitor trial: off-site but not off base. Alzheimers Dement. 2014;10(5 Suppl):S411–S419. doi: 10.1016/j.jalz.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yan R, Vassar R. Targeting the β secretase BACE1 for Alzheimer's disease therapy. Lancet Neurol. 2014;13(3):319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Moussa-Pacha NM, Abdin SM, Omar HA, Alniss H, Al-Tel TH. BACE1 inhibitors: current status and future directions in treating Alzheimer's disease. Med Res Rev. 2020;40(1):339–384. doi: 10.1002/med.21622. [DOI] [PubMed] [Google Scholar]
- 152.McDade E, Voytyuk I, Aisen P, Bateman RJ, Carrillo MC, De Strooper B, et al. The case for low-level BACE1 inhibition for the prevention of Alzheimer disease. Nat Rev Neurol. 2021;17(11):703–714. doi: 10.1038/s41582-021-00545-1. [DOI] [PubMed] [Google Scholar]
- 153.McKinzie DL, Winneroski LL, Green SJ, Hembre EJ, Erickson JA, Willis BA, et al. Discovery and early clinical development of LY3202626, a low-dose. CNS-penetrant BACE inhibitor J Med Chem. 2021;64(12):8076–8100. doi: 10.1021/acs.jmedchem.1c00489. [DOI] [PubMed] [Google Scholar]
- 154.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268(5208):225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 155.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95(14):8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lathia JD, Mattson MP, Cheng A. Notch: from neural development to neurological disorders. J Neurochem. 2008;107(6):1471–1481. doi: 10.1111/j.1471-4159.2008.05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 158.Alberi L, Hoey SE, Brai E, Scotti AL, Marathe S. Notch signaling in the brain: in good and bad times. Ageing Res Rev. 2013;12(3):801–814. doi: 10.1016/j.arr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 159.Wei Z, Chigurupati S, Arumugam TV, Jo DG, Li H, Chan SL. Notch activation enhances the microglia-mediated inflammatory response associated with focal cerebral ischemia. Stroke. 2011;42(9):2589–2594. doi: 10.1161/STROKEAHA.111.614834. [DOI] [PubMed] [Google Scholar]
- 160.Alberi L, Liu S, Wang Y, Badie R, Smith-Hicks C, Wu J, et al. Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron. 2011;69(3):437–44. doi: 10.1016/j.neuron.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Arumugam TV, Baik SH, Balaganapathy P, Sobey CG, Mattson MP, Jo DG. Notch signaling and neuronal death in stroke. Prog Neurobiol. 2018;165–167:103–116. doi: 10.1016/j.pneurobio.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sargin D, Botly LC, Higgs G, Marsolais A, Frankland PW, Egan SE, et al. Disrupting Jagged1-Notch signaling impairs spatial memory formation in adult mice. Neurobiol Learn Mem. 2013;103:39–49. doi: 10.1016/j.nlm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 163.Sha L, Wu X, Yao Y, Wen B, Feng J, Sha Z, et al. Notch signaling activation promotes seizure activity in temporal lobe epilepsy. Mol Neurobiol. 2014;49(2):633–644. doi: 10.1007/s12035-013-8545-0. [DOI] [PubMed] [Google Scholar]
- 164.Wolf BB, Lopes MB, VandenBerg SR, Gonias SL. Characterization and immunohistochemical localization of alpha 2-macroglobulin receptor (low-density lipoprotein receptor-related protein) in human brain. Am J Pathol. 1992;141(1):37–42. [PMC free article] [PubMed] [Google Scholar]
- 165.Herz J. The LDL receptor gene family: (un)expected signal transducers in the brain. Neuron. 2001;29(3):571–581. doi: 10.1016/S0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 166.Lin JP, Mironova YA, Shrager P, Giger RJ. LRP1 regulates peroxisome biogenesis and cholesterol homeostasis in oligodendrocytes and is required for proper CNS myelin development and repair. eLife. 2017 doi: 10.7554/eLife.30498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zlokovic BV, Deane R, Sagare AP, Bell RD, Winkler EA. Low-density lipoprotein receptor-related protein-1: a serial clearance homeostatic mechanism controlling Alzheimer's amyloid β-peptide elimination from the brain. J Neurochem. 2010;115(5):1077–1089. doi: 10.1111/j.1471-4159.2010.07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maier W, Bednorz M, Meister S, Roebroek A, Weggen S, Schmitt U, et al. LRP1 is critical for the surface distribution and internalization of the NR2B NMDA receptor subtype. Mol Neurodegener. 2013;8:25. doi: 10.1186/1750-1326-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nakajima C, Kulik A, Frotscher M, Herz J, Schäfer M, Bock HH, et al. Low density lipoprotein receptor-related protein 1 (LRP1) modulates N-methyl-D-aspartate (NMDA) receptor-dependent intracellular signaling and NMDA-induced regulation of postsynaptic protein complexes. J Biol Chem. 2013;288(30):21909–21923. doi: 10.1074/jbc.M112.444364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zurhove K, Nakajima C, Herz J, Bock HH, May P. γ-Secretase limits the inflammatory response through the processing of LRP1. Sci Signal. 2008 doi: 10.1126/scisignal.1164263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shinohara M, Tachibana M, Kanekiyo T, Bu G. Role of LRP1 in the pathogenesis of Alzheimer's disease: evidence from clinical and preclinical studies. J Lipid Res. 2017;58(7):1267–1281. doi: 10.1194/jlr.R075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.May P, Rohlmann A, Bock HH, Zurhove K, Marth JD, Schomburg ED, et al. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol Cell Biol. 2004;24(20):8872–8883. doi: 10.1128/MCB.24.20.8872-8883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gan M, Jiang P, McLean P, Kanekiyo T, Bu G. Low-density lipoprotein receptor-related protein 1 (LRP1) regulates the stability and function of GluA1 α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor in neurons. PLoS ONE. 2014;9(12):e113237. doi: 10.1371/journal.pone.0113237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Q, Trotter J, Zhang J, Peters MM, Cheng H, Bao J, et al. Neuronal LRP1 knockout in adult mice leads to impaired brain lipid metabolism and progressive, age-dependent synapse loss and neurodegeneration. J Neurosci. 2010;30(50):17068–17078. doi: 10.1523/JNEUROSCI.4067-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu CC, Hu J, Tsai CW, Yue M, Melrose HL, Kanekiyo T, et al. Neuronal LRP1 regulates glucose metabolism and insulin signaling in the brain. J Neurosci. 2015;35(14):5851–5859. doi: 10.1523/JNEUROSCI.5180-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Romeo R, Boden-El Mourabit D, Scheller A, Mark MD, Faissner A. Low-density lipoprotein receptor-related protein 1 (LRP1) as a novel regulator of early astroglial differentiation. Front Cell Neurosci. 2021;15:642521. doi: 10.3389/fncel.2021.642521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Safina D, Schlitt F, Romeo R, Pflanzner T, Pietrzik CU, Narayanaswami V, et al. Low-density lipoprotein receptor-related protein 1 is a novel modulator of radial glia stem cell proliferation, survival, and differentiation. Glia. 2016;64(8):1363–1380. doi: 10.1002/glia.23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Auderset L, Pitman KA, Cullen CL, Pepper RE, Taylor BV, Foa L, et al. Low-density lipoprotein receptor-related protein 1 (LRP1) is a negative regulator of oligodendrocyte progenitor cell differentiation in the adult mouse brain. Front Cell Dev Biol. 2020;8:564351. doi: 10.3389/fcell.2020.564351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mattila SO, Tuhkanen HE, Lackman JJ, Konzack A, Morató X, Argerich J, et al. GPR37 is processed in the N-terminal ectodomain by ADAM10 and furin. FASEB J. 2021;35(6):e21654. doi: 10.1096/fj.202002385RR. [DOI] [PubMed] [Google Scholar]
- 180.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105(7):891–902. doi: 10.1016/S0092-8674(01)00407-X. [DOI] [PubMed] [Google Scholar]
- 181.Murakami T, Shoji M, Imai Y, Inoue H, Kawarabayashi T, Matsubara E, et al. Pael-R is accumulated in Lewy bodies of Parkinson's disease. Ann Neurol. 2004;55(3):439–442. doi: 10.1002/ana.20064. [DOI] [PubMed] [Google Scholar]
- 182.Morató X, Garcia-Esparcia P, Argerich J, Llorens F, Zerr I, Paslawski W, et al. Ecto-GPR37: a potential biomarker for Parkinson's disease. Transl Neurodegener. 2021;10(1):8. doi: 10.1186/s40035-021-00232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Marazziti D, Mandillo S, Di Pietro C, Golini E, Matteoni R, Tocchini-Valentini GP. GPR37 associates with the dopamine transporter to modulate dopamine uptake and behavioral responses to dopaminergic drugs. Proc Natl Acad Sci U S A. 2007;104(23):9846–9851. doi: 10.1073/pnas.0703368104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lopes JP, Morató X, Souza C, Pinhal C, Machado NJ, Canas PM, et al. The role of parkinson's disease-associated receptor GPR37 in the hippocampus: functional interplay with the adenosinergic system. J Neurochem. 2015;134(1):135–146. doi: 10.1111/jnc.13109. [DOI] [PubMed] [Google Scholar]
- 185.Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat Commun. 2020;11(1):1220. doi: 10.1038/s41467-019-14198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yang HJ, Vainshtein A, Maik-Rachline G, Peles E. G protein-coupled receptor 37 is a negative regulator of oligodendrocyte differentiation and myelination. Nat Commun. 2016;7:10884. doi: 10.1038/ncomms10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dusonchet J, Bensadoun JC, Schneider BL, Aebischer P. Targeted overexpression of the parkin substrate Pael-R in the nigrostriatal system of adult rats to model Parkinson's disease. Neurobiol Dis. 2009;35(1):32–41. doi: 10.1016/j.nbd.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 188.Marazziti D, Golini E, Mandillo S, Magrelli A, Witke W, Matteoni R, et al. Altered dopamine signaling and MPTP resistance in mice lacking the Parkinson's disease-associated GPR37/parkin-associated endothelin-like receptor. Proc Natl Acad Sci U S A. 2004;101(27):10189–10194. doi: 10.1073/pnas.0403661101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang X, Mantas I, Fridjonsdottir E, Andrén PE, Chergui K, Svenningsson P. Deficits in motor performance, neurotransmitters and synaptic plasticity in elderly and experimental Parkinsonian mice lacking GPR37. Front Aging Neurosci. 2020;12:84. doi: 10.3389/fnagi.2020.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mandillo S, Golini E, Marazziti D, Di Pietro C, Matteoni R, Tocchini-Valentini GP. Mice lacking the Parkinson's related GPR37/PAEL receptor show non-motor behavioral phenotypes: age and gender effect. Genes Brain Behav. 2013;12(4):465–477. doi: 10.1111/gbb.12041. [DOI] [PubMed] [Google Scholar]
- 191.Veenit V, Zhang X, Ambrosini A, Sousa V, Svenningsson P. The effect of early life stress on emotional behaviors in GPR37KO mice. Int J Mol Sci. 2021;23(1):410. doi: 10.3390/ijms23010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Eggert S, Thomas C, Kins S, Hermey G. Trafficking in Alzheimer's disease: modulation of APP transport and processing by the transmembrane proteins LRP1, SorLA, SorCS1c, Sortilin, and Calsyntenin. Mol Neurobiol. 2018;55(7):5809–5829. doi: 10.1007/s12035-017-0806-x. [DOI] [PubMed] [Google Scholar]
- 193.Eggert S, Kins S, Endres K, Brigadski T. Brothers in arms: proBDNF/BDNF and sAPPα/Aβ-signaling and their common interplay with ADAM10, TrkB, p75NTR, sortilin, and sorLA in the progression of Alzheimer's disease. Biol Chem. 2022;403(1):43–71. doi: 10.1515/hsz-2021-0330. [DOI] [PubMed] [Google Scholar]
- 194.Yang M, Virassamy B, Vijayaraj SL, Lim Y, Saadipour K, Wang YJ, et al. The intracellular domain of sortilin interacts with amyloid precursor protein and regulates its lysosomal and lipid raft trafficking. PLoS ONE. 2013;8(5):e63049. doi: 10.1371/journal.pone.0063049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Takamura A, Sato Y, Watabe D, Okamoto Y, Nakata T, Kawarabayashi T, et al. Sortilin is required for toxic action of Aβ oligomers (AβOs): extracellular AβOs trigger apoptosis, and intraneuronal AβOs impair degradation pathways. Life Sci. 2012;91(23–24):1177–1186. doi: 10.1016/j.lfs.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 196.Jansen P, Giehl K, Nyengaard JR, Teng K, Lioubinski O, Sjoegaard SS, et al. Roles for the pro-neurotrophin receptor sortilin in neuronal development, aging and brain injury. Nat Neurosci. 2007;10(11):1449–1457. doi: 10.1038/nn2000. [DOI] [PubMed] [Google Scholar]
- 197.Lewin GR, Nykjaer A. Pro-neurotrophins, sortilin, and nociception. Eur J Neurosci. 2014;39(3):363–374. doi: 10.1111/ejn.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vaegter CB, Jansen P, Fjorback AW, Glerup S, Skeldal S, Kjolby M, et al. Sortilin associates with Trk receptors to enhance anterograde transport and neurotrophin signaling. Nat Neurosci. 2011;14(1):54–61. doi: 10.1038/nn.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Finan GM, Okada H, Kim TW. BACE1 retrograde trafficking is uniquely regulated by the cytoplasmic domain of sortilin. J Biol Chem. 2011;286(14):12602–12616. doi: 10.1074/jbc.M110.170217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mazella J, Borsotto M, Heurteaux C. The involvement of Sortilin/NTSR3 in depression as the progenitor of spadin and its role in the membrane expression of TREK-1. Front Pharmacol. 2018;9:1541. doi: 10.3389/fphar.2018.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pittois K, Deleersnijder W, Merregaert J. cDNA sequence analysis, chromosomal assignment and expression pattern of the gene coding for integral membrane protein 2B. Gene. 1998;217(1–2):141–149. doi: 10.1016/S0378-1119(98)00354-0. [DOI] [PubMed] [Google Scholar]
- 202.Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Thinakaran G, et al. Familial British dementia: expression and metabolism of BRI. Ann N Y Acad Sci. 2000;920:93–99. doi: 10.1111/j.1749-6632.2000.tb06909.x. [DOI] [PubMed] [Google Scholar]
- 203.Kim SH, Creemers JW, Chu S, Thinakaran G, Sisodia SS. Proteolytic processing of familial British dementia-associated BRI variants: evidence for enhanced intracellular accumulation of amyloidogenic peptides. J Biol Chem. 2002;277(3):1872–1877. doi: 10.1074/jbc.M108739200. [DOI] [PubMed] [Google Scholar]
- 204.Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Lynn DG, et al. Furin mediates enhanced production of fibrillogenic ABri peptides in familial British dementia. Nat Neurosci. 1999;2(11):984–988. doi: 10.1038/14783. [DOI] [PubMed] [Google Scholar]
- 205.Knight SD, Presto J, Linse S, Johansson J. The BRICHOS domain, amyloid fibril formation, and their relationship. Biochemistry. 2013;52(43):7523–7531. doi: 10.1021/bi400908x. [DOI] [PubMed] [Google Scholar]
- 206.Poska H, Haslbeck M, Kurudenkandy FR, Hermansson E, Chen G, Kostallas G, et al. Dementia-related Bri2 BRICHOS is a versatile molecular chaperone that efficiently inhibits Aβ42 toxicity in Drosophila. Biochem J. 2016;473(20):3683–3704. doi: 10.1042/BCJ20160277. [DOI] [PubMed] [Google Scholar]
- 207.Akiyama H, Kondo H, Arai T, Ikeda K, Kato M, Iseki E, et al. Expression of BRI, the normal precursor of the amyloid protein of familial British dementia, in human brain. Acta Neuropathol. 2004;107(1):53–58. doi: 10.1007/s00401-003-0783-1. [DOI] [PubMed] [Google Scholar]
- 208.Willander H, Presto J, Askarieh G, Biverstål H, Frohm B, Knight SD, et al. BRICHOS domains efficiently delay fibrillation of amyloid β-peptide. J Biol Chem. 2012;287(37):31608–31617. doi: 10.1074/jbc.M112.393157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Peng S, Fitzen M, Jörnvall H, Johansson J. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1–23) and amyloid beta-peptide (Abeta1-40): Implications for Bri2 effects on processing of amyloid precursor protein and Abeta aggregation. Biochem Biophys Res Commun. 2010;393(3):356–361. doi: 10.1016/j.bbrc.2009.12.122. [DOI] [PubMed] [Google Scholar]
- 210.Vidal R, Frangione B, Rostagno A, Mead S, Révész T, Plant G, et al. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399(6738):776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- 211.Matsuda S, Senda T. BRI2 as an anti-Alzheimer gene. Med Mol Morphol. 2019;52(1):1–7. doi: 10.1007/s00795-018-0191-1. [DOI] [PubMed] [Google Scholar]
- 212.Del Campo M, Hoozemans JJ, Dekkers LL, Rozemuller AJ, Korth C, Müller-Schiffmann A, et al. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer's disease. Neurobiol Aging. 2014;35(7):1596–1604. doi: 10.1016/j.neurobiolaging.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 213.Holthuis JC, Jansen EJ, Schoonderwoert VT, Burbach JP, Martens GJ. Biosynthesis of the vacuolar H+-ATPase accessory subunit Ac45 in Xenopus pituitary. Eur J Biochem. 1999;262(2):484–491. doi: 10.1046/j.1432-1327.1999.00396.x. [DOI] [PubMed] [Google Scholar]
- 214.Abbas YM, Wu D, Bueler SA, Robinson CV, Rubinstein JL. Structure of V-ATPase from the mammalian brain. Science. 2020;367(6483):1240–1246. doi: 10.1126/science.aaz2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang R, Long T, Hassan A, Wang J, Sun Y, Xie XS, et al. Cryo-EM structures of intact V-ATPase from bovine brain. Nat Commun. 2020;11(1):3921. doi: 10.1038/s41467-020-17762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Louagie E, Taylor NA, Flamez D, Roebroek AJ, Bright NA, Meulemans S, et al. Role of furin in granular acidification in the endocrine pancreas: identification of the V-ATPase subunit Ac45 as a candidate substrate. Proc Natl Acad Sci U S A. 2008;105(34):12319–12324. doi: 10.1073/pnas.0800340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brouwers B, Coppola I, Vints K, Dislich B, Jouvet N, Van Lommel L, et al. Loss of furin in β-cells induces an mTORC1-ATF4 anabolic pathway that leads to β-cell dysfunction. Diabetes. 2021;70(2):492–503. doi: 10.2337/db20-0474. [DOI] [PubMed] [Google Scholar]
- 218.Supek F, Supekova L, Mandiyan S, Pan YC, Nelson H, Nelson N. A novel accessory subunit for vacuolar H(+)-ATPase from chromaffin granules. J Biol Chem. 1994;269(39):24102–24106. doi: 10.1016/S0021-9258(19)51053-5. [DOI] [PubMed] [Google Scholar]
- 219.Hodi FS, Schmollinger JC, Soiffer RJ, Salgia R, Lynch T, Ritz J, et al. ATP6S1 elicits potent humoral responses associated with immune-mediated tumor destruction. Proc Natl Acad Sci U S A. 2002;99(10):6919–6924. doi: 10.1073/pnas.102025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Feng H, Cheng T, Pavlos NJ, Yip KH, Carrello A, Seeber R, et al. Cytoplasmic terminus of vacuolar type proton pump accessory subunit Ac45 is required for proper interaction with V(0) domain subunits and efficient osteoclastic bone resorption. J Biol Chem. 2008;283(19):13194–13204. doi: 10.1074/jbc.M709712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dulac A, Issa AR, Sun J, Matassi G, Jonas C, Chérif-Zahar B, et al. A novel neuron-specific regulator of the V-ATPase in Drosophila. eNeuro. 2021 [DOI] [PMC free article] [PubMed]
- 222.Jansen EJ, Timal S, Ryan M, Ashikov A, van Scherpenzeel M, Graham LA, et al. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun. 2016;7:11600. doi: 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vitória JJ, Trigo D. Revisiting APP secretases: an overview on the holistic effects of retinoic acid receptor stimulation in APP processing. Cel Mol Life Sci. 2022;79(2):1–5. doi: 10.1007/s00018-021-04090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jan AT, Azam M, Rahman S, Almigeiti AMS, Choi DH, Lee EJ, et al. Perspective insights into disease progression, diagnostics, and therapeutic approaches in Alzheimer's disease: a judicious update. Front Aging Neurosci. 2017;9:356. doi: 10.3389/fnagi.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cummings J, Lee G, Ritter A, Sabbagh M, Zhong K. Alzheimer's disease drug development pipeline: 2020. Alzheimers Dement (N Y) 2020;6(1):e12050. doi: 10.1002/trc2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314(5800):777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 227.Zhang Y, Song W. Islet amyloid polypeptide: another key molecule in Alzheimer's pathogenesis? Prog Neurobiol. 2017;153:100–120. doi: 10.1016/j.pneurobio.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 228.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 229.Yang Y, Wang JZ. Nature of Tau-associated neurodegeneration and the molecular mechanisms. J Alzheimers Dis. 2018;62(3):1305–1317. doi: 10.3233/JAD-170788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Erickson MA, Banks WA. Blood-brain barrier dysfunction as a cause and consequence of Alzheimer's disease. J Cereb Blood Flow Metab. 2013;33(10):1500–1513. doi: 10.1038/jcbfm.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.James SA, Churches QI, de Jonge MD, Birchall IE, Streltsov V, McColl G, et al. Iron, copper, and zinc concentration in Aβ plaques in the APP/PS1 mouse model of Alzheimer's disease correlates with metal levels in the surrounding neuropil. ACS Chem Neurosci. 2017;8(3):629–637. doi: 10.1021/acschemneuro.6b00362. [DOI] [PubMed] [Google Scholar]
- 232.Peters DG, Connor JR, Meadowcroft MD. The relationship between iron dyshomeostasis and amyloidogenesis in Alzheimer's disease: two sides of the same coin. Neurobiol Dis. 2015;81:49–65. doi: 10.1016/j.nbd.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Aisen PS, Davis KL. Inflammatory mechanisms in Alzheimer's disease: implications for therapy. Am J Psychiatry. 1994;151(8):1105–1113. doi: 10.1176/ajp.151.8.1105. [DOI] [PubMed] [Google Scholar]
- 234.Swerdlow RH, Khan SM. A "mitochondrial cascade hypothesis" for sporadic Alzheimer's disease. Med Hypoth. 2004;63(1):8–20. doi: 10.1016/j.mehy.2003.12.045. [DOI] [PubMed] [Google Scholar]
- 235.Liu Z, Zhou T, Ziegler AC, Dimitrion P, Zuo L. Oxidative stress in neurodegenerative diseases: from molecular mechanisms to clinical applications. Oxid Med Cell Longev. 2017;2017:2525967. doi: 10.1155/2017/2525967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Neth BJ, Craft S. Insulin resistance and Alzheimer's disease: bioenergetic linkages. Front Aging Neurosci. 2017;9:345. doi: 10.3389/fnagi.2017.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Guan C, Dang R, Cui Y, Liu L, Chen X, Wang X, et al. Characterization of plasma metal profiles in Alzheimer's disease using multivariate statistical analysis. PLoS ONE. 2017;12(7):e0178271. doi: 10.1371/journal.pone.0178271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Du Z, Song Y, Chen X, Zhang W, Zhang G, Li H, et al. Knockdown of astrocytic Grin2a aggravates β-amyloid-induced memory and cognitive deficits through regulating nerve growth factor. Aging Cell. 2021;20(8):e13437. doi: 10.1111/acel.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Song Y, Du Z, Chen X, Zhang W, Zhang G, Li H, et al. Astrocytic N-methyl-D-aspartate receptors protect the hippocampal neurons against amyloid-β142-induced synaptotoxicity by regulating nerve growth factor. J Alzheimers Dis. 2022;85(1):167–178. doi: 10.3233/JAD-210730. [DOI] [PubMed] [Google Scholar]
- 240.Phillips HS, Hains JM, Armanini M, Laramee GR, Johnson SA, Winslow JW. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer's disease. Neuron. 1991;7(5):695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- 241.Connor B, Young D, Yan Q, Faull RL, Synek B, Dragunow M. Brain-derived neurotrophic factor is reduced in Alzheimer's disease. Brain Res Mol Brain Res. 1997;49(1–2):71–81. doi: 10.1016/S0169-328X(97)00125-3. [DOI] [PubMed] [Google Scholar]
- 242.Holsinger RM, Schnarr J, Henry P, Castelo VT, Fahnestock M. Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: decreased levels in Alzheimer's disease. Brain Res Mol Brain Res. 2000;76(2):347–354. doi: 10.1016/S0169-328X(00)00023-1. [DOI] [PubMed] [Google Scholar]
- 243.Garzon D, Yu G, Fahnestock M. A new brain-derived neurotrophic factor transcript and decrease in brain-derived neurotrophic factor transcripts 1, 2 and 3 in Alzheimer's disease parietal cortex. J Neurochem. 2002;82(5):1058–1064. doi: 10.1046/j.1471-4159.2002.01030.x. [DOI] [PubMed] [Google Scholar]
- 244.Ferrer I, Marín C, Rey MJ, Ribalta T, Goutan E, Blanco R, et al. BDNF and full-length and truncated TrkB expression in Alzheimer disease. Implications in therapeutic strategies. J Neuropathol Exp Neurol. 1999;58(7):729–39. doi: 10.1097/00005072-199907000-00007. [DOI] [PubMed] [Google Scholar]
- 245.Michalski B, Fahnestock M. Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer's disease. Brain Res Mol Brain Res. 2003;111(1–2):148–154. doi: 10.1016/S0169-328X(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 246.Peng S, Wuu J, Mufson EJ, Fahnestock M. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer's disease. J Neurochem. 2005;93(6):1412–1421. doi: 10.1111/j.1471-4159.2005.03135.x. [DOI] [PubMed] [Google Scholar]
- 247.Gerenu G, Martisova E, Ferrero H, Carracedo M, Rantamäki T, Ramirez MJ, et al. Modulation of BDNF cleavage by plasminogen-activator inhibitor-1 contributes to Alzheimer's neuropathology and cognitive deficits. Biochim Biophys Acta Mol Basis Dis. 2017;1863(4):991–1001. doi: 10.1016/j.bbadis.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 248.Kimura R, Devi L, Ohno M. Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer's disease transgenic mice. J Neurochem. 2010;113(1):248–261. doi: 10.1111/j.1471-4159.2010.06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kaminari A, Giannakas N, Tzinia A, Tsilibary EC. Overexpression of matrix metalloproteinase-9 (MMP-9) rescues insulin-mediated impairment in the 5XFAD model of Alzheimer's disease. Sci Rep. 2017;7(1):683. doi: 10.1038/s41598-017-00794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Peng S, Wuu J, Mufson EJ, Fahnestock M. Increased proNGF levels in subjects with mild cognitive impairment and mild Alzheimer disease. J Neuropathol Exp Neurol. 2004;63(6):641–649. doi: 10.1093/jnen/63.6.641. [DOI] [PubMed] [Google Scholar]
- 251.Mufson EJ, He B, Nadeem M, Perez SE, Counts SE, Leurgans S, et al. Hippocampal proNGF signaling pathways and β-amyloid levels in mild cognitive impairment and Alzheimer disease. J Neuropathol Exp Neurol. 2012;71(11):1018–1029. doi: 10.1097/NEN.0b013e318272caab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nagarsheth MH, Viehman A, Lippa SM, Lippa CF. Notch-1 immunoexpression is increased in Alzheimer's and Pick's disease. J Neurol Sci. 2006;244(1–2):111–116. doi: 10.1016/j.jns.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 253.Holsinger RMD, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor β-secretase in Alzheimer's disease. Ann Neurol. 2002;51(6):783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 254.Li R, Lindholm K, Yang L-B, Yue X, Citron M, Yan R, et al. Amyloid β peptide load is correlated with increased β-secretase activity in sporadic Alzheimer's disease patients. Proc Natl Acad Sci U S A. 2004;101(10):3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Leake A, Morris CM, Whateley J. Brain matrix metalloproteinase 1 levels are elevated in Alzheimer's disease. Neurosci Lett. 2000;291(3):201–203. doi: 10.1016/S0304-3940(00)01418-X. [DOI] [PubMed] [Google Scholar]
- 256.Py NA, Bonnet AE, Bernard A, Marchalant Y, Charrat E, Checler F, et al. Differential spatio-temporal regulation of MMPs in the 5xFAD mouse model of Alzheimer's disease: evidence for a pro-amyloidogenic role of MT1-MMP. Front Aging Neurosci. 2014;6:247. doi: 10.3389/fnagi.2014.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fleitas C, Piñol-Ripoll G, Marfull P, Rocandio D, Ferrer I, Rampon C, et al. proBDNF is modified by advanced glycation end products in Alzheimer's disease and causes neuronal apoptosis by inducing p75 neurotrophin receptor processing. Mol Brain. 2018;11(1):68. doi: 10.1186/s13041-018-0411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Arélin K, Kinoshita A, Whelan CM, Irizarry MC, Rebeck GW, Strickland DK, et al. LRP and senile plaques in Alzheimer's disease: colocalization with apolipoprotein E and with activated astrocytes. Brain Res Mol Brain Res. 2002;104(1):38–46. doi: 10.1016/S0169-328X(02)00203-6. [DOI] [PubMed] [Google Scholar]
- 259.Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, et al. Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res. 2007;1161:116–123. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 260.Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, et al. Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106(9):1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shang J, Yamashita T, Tian F, Li X, Liu X, Shi X, et al. Chronic cerebral hypoperfusion alters amyloid-β transport related proteins in the cortical blood vessels of Alzheimer's disease model mouse. Brain Res. 2019;1723:146379. doi: 10.1016/j.brainres.2019.146379. [DOI] [PubMed] [Google Scholar]
- 262.Tofaris GK. Initiation and progression of α-synuclein pathology in Parkinson's disease. Cell Mol Life Sci. 2022;79(4):210. doi: 10.1007/s00018-022-04240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3(12):932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 264.Zhang J. Mining imaging and clinical data with machine learning approaches for the diagnosis and early detection of Parkinson's disease. NPJ Parkinson's disease. 2022;8(1):13. doi: 10.1038/s41531-021-00266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Maksoud E, Liao EH, Haghighi AP. A neuron-glial trans-signaling cascade mediates LRRK2-induced neurodegeneration. Cell Rep. 2019;26(7):1774–86.e4. doi: 10.1016/j.celrep.2019.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Penney J, Tsurudome K, Liao EH, Kauwe G, Gray L, Yanagiya A, et al. LRRK2 regulates retrograde synaptic compensation at the Drosophila neuromuscular junction. Nat Commun. 2016;7:12188. doi: 10.1038/ncomms12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Salehi Z, Mashayekhi F. Brain-derived neurotrophic factor concentrations in the cerebrospinal fluid of patients with Parkinson's disease. J Clin Neurosci. 2009;16(1):90–93. doi: 10.1016/j.jocn.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 268.Scalzo P, Kümmer A, Bretas TL, Cardoso F, Teixeira AL. Serum levels of brain-derived neurotrophic factor correlate with motor impairment in Parkinson's disease. J Neurol. 2010;257(4):540–545. doi: 10.1007/s00415-009-5357-2. [DOI] [PubMed] [Google Scholar]
- 269.Wang Y, Liu H, Zhang BS, Soares JC, Zhang XY. Low BDNF is associated with cognitive impairments in patients with Parkinson's disease. Parkinsonism Relat Disord. 2016;29:66–71. doi: 10.1016/j.parkreldis.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 270.Gupta V, Singh MK, Garg RK, Pant KK, Khattri S. Evaluation of peripheral matrix metalloproteinase-1 in Parkinson's disease: a case-control study. Int J Neurosci. 2014;124(2):88–92. doi: 10.3109/00207454.2013.824438. [DOI] [PubMed] [Google Scholar]
- 271.Thijs RD, Ryvlin P, Surges R. Autonomic manifestations of epilepsy: emerging pathways to sudden death? Nat Rev Neurol. 2021;17(12):774–788. doi: 10.1038/s41582-021-00574-w. [DOI] [PubMed] [Google Scholar]
- 272.Casillas-Espinosa PM, Powell KL, O'Brien TJ. Regulators of synaptic transmission: roles in the pathogenesis and treatment of epilepsy. Epilepsia. 2012;53(Suppl 9):41–58. doi: 10.1111/epi.12034. [DOI] [PubMed] [Google Scholar]
- 273.Manford M. Recent advances in epilepsy. J Neurol. 2017;264(8):1811–1824. doi: 10.1007/s00415-017-8394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zsurka G, Kunz WS. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol. 2015;14(9):956–966. doi: 10.1016/S1474-4422(15)00148-9. [DOI] [PubMed] [Google Scholar]
- 275.Conboy K, Henshall DC, Brennan GP. Epigenetic principles underlying epileptogenesis and epilepsy syndromes. Neurobiol Dis. 2021;148:105179. doi: 10.1016/j.nbd.2020.105179. [DOI] [PubMed] [Google Scholar]
- 276.Mathern GW, Babb TL, Micevych PE, Blanco CE, Pretorius JK. Granule cell mRNA levels for BDNF, NGF, and NT-3 correlate with neuron losses or supragranular mossy fiber sprouting in the chronically damaged and epileptic human hippocampus. Mol Chem Neuropathol. 1997;30(1–2):53–76. doi: 10.1007/BF02815150. [DOI] [PubMed] [Google Scholar]
- 277.Takahashi M, Hayashi S, Kakita A, Wakabayashi K, Fukuda M, Kameyama S, et al. Patients with temporal lobe epilepsy show an increase in brain-derived neurotrophic factor protein and its correlation with neuropeptide Y. Brain Res. 1999;818(2):579–582. doi: 10.1016/S0006-8993(98)01355-9. [DOI] [PubMed] [Google Scholar]
- 278.Thomas AX, Cruz Del Angel Y, Gonzalez MI, Carrel AJ, Carlsen J, Lam PM, et al. Rapid increases in proBDNF after pilocarpine-induced status epilepticus in mice are associated with reduced proBDNF cleavage machinery. eNeuro. 2016;3(1):452. doi: 10.1523/ENEURO.0020-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Gall CM, Isackson PJ. Limbic seizures increase neuronal production of messenger RNA for nerve growth factor. Science. 1989;245(4919):758–761. doi: 10.1126/science.2549634. [DOI] [PubMed] [Google Scholar]
- 280.Gall C, Murray K, Isackson PJ. Kainic acid-induced seizures stimulate increased expression of nerve growth factor mRNA in rat hippocampus. Brain Res Mol Brain Res. 1991;9(1–2):113–123. doi: 10.1016/0169-328X(91)90136-L. [DOI] [PubMed] [Google Scholar]
- 281.Mehrabi S, Janahamdi M, Joghataie MT, Barati M, Marzban M, Hadjighassem M, et al. Blockade of p75 neurotrophin receptor reverses irritability and anxiety-related behaviors in a rat model of status epilepticus. Iran Biomed J. 2018;22(4):264–274. doi: 10.29252/ibj.22.4.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Heinrich C, Lähteinen S, Suzuki F, Anne-Marie L, Huber S, Häussler U, et al. Increase in BDNF-mediated TrkB signaling promotes epileptogenesis in a mouse model of mesial temporal lobe epilepsy. Neurobiol Dis. 2011;42(1):35–47. doi: 10.1016/j.nbd.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 283.Wang R, Zeng GQ, Tong RZ, Zhou D, Hong Z. Serum matrix metalloproteinase-2: A potential biomarker for diagnosis of epilepsy. Epilepsy Res. 2016;122:114–119. doi: 10.1016/j.eplepsyres.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 284.Wang R, Zeng GQ, Liu X, Tong RZ, Zhou D, Hong Z. Evaluation of serum matrix metalloproteinase-3 as a biomarker for diagnosis of epilepsy. J Neurol Sci. 2016;367:291–297. doi: 10.1016/j.jns.2016.06.031. [DOI] [PubMed] [Google Scholar]
- 285.Broekaart DW, Bertran A, Jia S, Korotkov A, Senkov O, Bongaarts A, et al. The matrix metalloproteinase inhibitor IPR-179 has antiseizure and antiepileptogenic effects. J Clin Invest. 2021 doi: 10.1172/JCI138332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Konopka A, Grajkowska W, Ziemiańska K, Roszkowski M, Daszkiewicz P, Rysz A, et al. Matrix metalloproteinase-9 (MMP-9) in human intractable epilepsy caused by focal cortical dysplasia. Epilepsy Res. 2013;104(1–2):45–58. doi: 10.1016/j.eplepsyres.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 287.Mizoguchi H, Nakade J, Tachibana M, Ibi D, Someya E, Koike H, et al. Matrix metalloproteinase-9 contributes to kindled seizure development in pentylenetetrazole-treated mice by converting pro-BDNF to mature BDNF in the hippocampus. J Neurosci. 2011;31(36):12963–12971. doi: 10.1523/JNEUROSCI.3118-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kim GW, Kim HJ, Cho KJ, Kim HW, Cho YJ, Lee BI. The role of MMP-9 in integrin-mediated hippocampal cell death after pilocarpine-induced status epilepticus. Neurobiol Dis. 2009;36(1):169–180. doi: 10.1016/j.nbd.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 289.Gorter JA, Van Vliet EA, Rauwerda H, Breit T, Stad R, van Schaik L, et al. Dynamic changes of proteases and protease inhibitors revealed by microarray analysis in CA3 and entorhinal cortex during epileptogenesis in the rat. Epilepsia. 2007;48(Suppl 5):53–64. doi: 10.1111/j.1528-1167.2007.01290.x. [DOI] [PubMed] [Google Scholar]
- 290.Korotkov A, Broekaart DWM, van Scheppingen J, Anink JJ, Baayen JC, Idema S, et al. Increased expression of matrix metalloproteinase 3 can be attenuated by inhibition of microRNA-155 in cultured human astrocytes. J Neuroinflammation. 2018;15(1):211. doi: 10.1186/s12974-018-1245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371(9624):1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 292.Campbell BCV, Khatri P. Stroke. Lancet. 2020;396(10244):129–142. doi: 10.1016/S0140-6736(20)31179-X. [DOI] [PubMed] [Google Scholar]
- 293.Soares ROS, Losada DM, Jordani MC, Évora P, Castro ESO. Ischemia/reperfusion injury revisited: an overview of the latest pharmacological strategies. Int J Mol Sci. 2019;20(20):5034. doi: 10.3390/ijms20205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Carden DL, Granger DN. Pathophysiology of ischaemia-reperfusion injury. J Pathol. 2000;190(3):255–266. doi: 10.1002/(SICI)1096-9896(200002)190:3<255::AID-PATH526>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 295.Evora PR, Pearson PJ, Seccombe JF, Schaff HV. Ischemia-reperfusion lesion Physiopathologic aspects and the importance of the endothelial function. Arq Bras Cardiol. 1996;66(4):239–45. [PubMed] [Google Scholar]
- 296.Yokota N, Uchijima M, Nishizawa S, Namba H, Koide Y. Identification of differentially expressed genes in rat hippocampus after transient global cerebral ischemia using subtractive cDNA cloning based on polymerase chain reaction. Stroke. 2001;32(1):168–174. doi: 10.1161/01.STR.32.1.168. [DOI] [PubMed] [Google Scholar]
- 297.Yang Y, Estrada EY, Thompson JF, Liu W, Rosenberg GA. Matrix metalloproteinase-mediated disruption of tight junction proteins in cerebral vessels is reversed by synthetic matrix metalloproteinase inhibitor in focal ischemia in rat. J Cereb Blood Flow Metab. 2007;27(4):697–709. doi: 10.1038/sj.jcbfm.9600375. [DOI] [PubMed] [Google Scholar]
- 298.Xiong LL, Chen J, Du RL, Liu J, Chen YJ, Hawwas MA, et al. Brain-derived neurotrophic factor and its related enzymes and receptors play important roles after hypoxic-ischemic brain damage. Neural Regen Res. 2021;16(8):1453–1459. doi: 10.4103/1673-5374.303032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Planas AM, Solé S, Justicia C. Expression and activation of matrix metalloproteinase-2 and -9 in rat brain after transient focal cerebral ischemia. Neurobiol Dis. 2001;8(5):834–846. doi: 10.1006/nbdi.2001.0435. [DOI] [PubMed] [Google Scholar]
- 300.Chang DI, Hosomi N, Lucero J, Heo JH, Abumiya T, Mazar AP, et al. Activation systems for latent matrix metalloproteinase-2 are upregulated immediately after focal cerebral ischemia. J Cereb Blood Flow Metab. 2003;23(12):1408–1419. doi: 10.1097/01.WCB.0000091765.61714.30. [DOI] [PubMed] [Google Scholar]
- 301.Heo JH, Lucero J, Abumiya T, Koziol JA, Copeland BR, del Zoppo GJ. Matrix metalloproteinases increase very early during experimental focal cerebral ischemia. J Cereb Blood Flow Metab. 1999;19(6):624–633. doi: 10.1097/00004647-199906000-00005. [DOI] [PubMed] [Google Scholar]
- 302.Gasche Y, Fujimura M, Morita-Fujimura Y, Copin JC, Kawase M, Massengale J, et al. Early appearance of activated matrix metalloproteinase-9 after focal cerebral ischemia in mice: a possible role in blood-brain barrier dysfunction. J Cereb Blood Flow Metab. 1999;19(9):1020–1028. doi: 10.1097/00004647-199909000-00010. [DOI] [PubMed] [Google Scholar]
- 303.Yamada M, Hayashi H, Suzuki K, Sato S, Inoue D, Iwatani Y, et al. Furin-mediated cleavage of LRP1 and increase in ICD of LRP1 after cerebral ischemia and after exposure of cultured neurons to NMDA. Sci Rep. 2019;9(1):11782. doi: 10.1038/s41598-019-48279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Morozova A, Zorkina Y, Abramova O, Pavlova O, Pavlov K, Soloveva K, et al. Neurobiological highlights of cognitive impairment in psychiatric disorders. Int J Mol Sci. 2022;23(3):1217. doi: 10.3390/ijms23031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Xu MY, Wong AHC. GABAergic inhibitory neurons as therapeutic targets for cognitive impairment in schizophrenia. Acta Pharmacol Sin. 2018;39(5):733–753. doi: 10.1038/aps.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349(6255):1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Hou Y, Liang W, Zhang J, Li Q, Ou H, Wang Z, et al. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338-3p reduces BDNF production. Schizophr Res. 2018;199:176–180. doi: 10.1016/j.schres.2018.02.040. [DOI] [PubMed] [Google Scholar]
- 308.Korologou-Linden R, Leyden GM, Relton CL, Richmond RC, Richardson TG. Multi-omics analyses of cognitive traits and psychiatric disorders highlights brain-dependent mechanisms. Hum Mol Genet. 2021 doi: 10.1093/hmg/ddab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Wang H, Yi Z, Shi T. Novel loci and potential mechanisms of major depressive disorder, bipolar disorder, and schizophrenia. Sci China Life Sci. 2022;65(1):167–183. doi: 10.1007/s11427-020-1934-x. [DOI] [PubMed] [Google Scholar]
- 310.Perry BI, Bowker N, Burgess S, Wareham NJ, Upthegrove R, Jones PB, et al. Evidence for shared genetic aetiology between schizophrenia, cardiometabolic, and inflammation-related traits: genetic correlation and colocalization analyses. Schizophr Bull Open. 2022;3(1):sgac001. doi: 10.1093/schizbullopen/sgac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Zou Y, Shih M, Chiu H, Ferreira T, Suzuki N, Zou W, et al. The kpc-1 (furin) 3’ UTR promotes dendritic transport and local translation of mRNAs to regulate dendrite branching and self-avoidance of a nociceptive neuron. bioRxiv. 2021 doi: 10.1101/2021.08.03.453128. [DOI] [Google Scholar]
- 312.Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8(6):592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 313.Hashimoto T, Bergen SE, Nguyen QL, Xu B, Monteggia LM, Pierri JN, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25(2):372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Durany N, Michel T, Zöchling R, Boissl KW, Cruz-Sánchez FF, Riederer P, et al. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophr Res. 2001;52(1–2):79–86. doi: 10.1016/S0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- 315.Takahashi M, Shirakawa O, Toyooka K, Kitamura N, Hashimoto T, Maeda K, et al. Abnormal expression of brain-derived neurotrophic factor and its receptor in the corticolimbic system of schizophrenic patients. Mol Psychiatry. 2000;5(3):293–300. doi: 10.1038/sj.mp.4000718. [DOI] [PubMed] [Google Scholar]
- 316.Yang Y, Liu Y, Wang G, Hei G, Wang X, Li R, et al. Brain-derived neurotrophic factor is associated with cognitive impairments in first-episode and chronic schizophrenia. Psychiatry Res. 2019;273:528–536. doi: 10.1016/j.psychres.2019.01.051. [DOI] [PubMed] [Google Scholar]
- 317.Rizos EN, Papathanasiou M, Michalopoulou PG, Mazioti A, Douzenis A, Kastania A, et al. Association of serum BDNF levels with hippocampal volumes in first psychotic episode drug-naive schizophrenic patients. Schizophr Res. 2011;129(2–3):201–204. doi: 10.1016/j.schres.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 318.Ashe PC, Chlan-Fourney J, Juorio AV, Li XM. Brain-derived neurotrophic factor (BDNF) mRNA in rats with neonatal ibotenic acid lesions of the ventral hippocampus. Brain Res. 2002;956(1):126–135. doi: 10.1016/S0006-8993(02)03176-1. [DOI] [PubMed] [Google Scholar]
- 319.Yuan Q, Yang F, Xiao Y, Tan S, Husain N, Ren M, et al. Regulation of brain-derived neurotrophic factor exocytosis and gamma-aminobutyric acidergic interneuron synapse by the schizophrenia susceptibility gene dysbindin-1. Biol Psychiatry. 2016;80(4):312–322. doi: 10.1016/j.biopsych.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 320.Dai N, Jie H, Duan Y, Xiong P, Xu X, Chen P, et al. Different serum protein factor levels in first-episode drug-naive patients with schizophrenia characterized by positive and negative symptoms. Psychiatr Clin Neurosci. 2020;74(9):472–479. doi: 10.1111/pcn.13078. [DOI] [PubMed] [Google Scholar]
- 321.Qin XY, Wu HT, Cao C, Loh YP, Cheng Y. A meta-analysis of peripheral blood nerve growth factor levels in patients with schizophrenia. Mol Psychiatry. 2017;22(9):1306–1312. doi: 10.1038/mp.2016.235. [DOI] [PubMed] [Google Scholar]
- 322.Vargas HE, Gama CS, Andreazza AC, Medeiros D, Stertz L, Fries G, et al. Decreased serum neurotrophin 3 in chronically medicated schizophrenic males. Neurosci Lett. 2008;440(3):197–201. doi: 10.1016/j.neulet.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 323.Domenici E, Willé DR, Tozzi F, Prokopenko I, Miller S, McKeown A, et al. Plasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections. PLoS ONE. 2010;5(2):e9166. doi: 10.1371/journal.pone.0009166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lepeta K, Purzycka KJ, Pachulska-Wieczorek K, Mitjans M, Begemann M, Vafadari B, et al. A normal genetic variation modulates synaptic MMP-9 protein levels and the severity of schizophrenia symptoms. EMBO Mol Med. 2017;9(8):1100–1116. doi: 10.15252/emmm.201707723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Vafadari B, Mitra S, Stefaniuk M, Kaczmarek L. Psychosocial stress induces schizophrenia-like behavior in mice with reduced MMP-9 activity. Front Behav Neurosci. 2019;13:195. doi: 10.3389/fnbeh.2019.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Omori W, Hattori K, Kajitani N, Tsuchioka MO, Boku S, Kunugi H, et al. Increased matrix metalloproteinases in cerebrospinal fluids of patients with major depressive disorder and schizophrenia. Int J Neuropsychopharmacol. 2020;23(11):713–720. doi: 10.1093/ijnp/pyaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Shimizu E, Hashimoto K, Okamura N, Koike K, Komatsu N, Kumakiri C, et al. Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants. Biol Psychiatry. 2003;54(1):70–75. doi: 10.1016/S0006-3223(03)00181-1. [DOI] [PubMed] [Google Scholar]
- 328.Caldieraro MA, McKee M, Leistner-Segal S, Vares EA, Kubaski F, Spanemberg L, et al. Val66Met polymorphism association with serum BDNF and inflammatory biomarkers in major depression. World J Biol Psychiatry. 2018;19(5):402–409. doi: 10.1080/15622975.2017.1347713. [DOI] [PubMed] [Google Scholar]
- 329.Jiang H, Chen S, Li C, Lu N, Yue Y, Yin Y, et al. The serum protein levels of the tPA-BDNF pathway are implicated in depression and antidepressant treatment. Transl Psychiatry. 2017;7(4):e1079. doi: 10.1038/tp.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Shibasaki C, Takebayashi M, Itagaki K, Abe H, Kajitani N, Okada-Tsuchioka M, et al. Altered serum levels of matrix metalloproteinase-2, -9 in response to electroconvulsive therapy for mood disorders. Int J Neuropsychopharmacol. 2016;19(9):pyw019. doi: 10.1093/ijnp/pyw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wang H, Xiao L, Wang H, Wang G. Involvement of chronic unpredictable mild stress-induced hippocampal LRP1 up-regulation in microtubule instability and depressive-like behavior in a depressive-like adult male rat model. Physiol Behav. 2020;215:112749. doi: 10.1016/j.physbeh.2019.112749. [DOI] [PubMed] [Google Scholar]
- 332.Berry A, Bellisario V, Capoccia S, Tirassa P, Calza A, Alleva E, et al. Social deprivation stress is a triggering factor for the emergence of anxiety- and depression-like behaviours and leads to reduced brain BDNF levels in C57BL/6J mice. Psychoneuroendocrinology. 2012;37(6):762–772. doi: 10.1016/j.psyneuen.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 333.Sbrini G, Brivio P, Bosch K, Homberg JR, Calabrese F. Enrichment environment positively influences depression- and anxiety-like behavior in serotonin transporter knockout rats through the modulation of neuroplasticity, spine, and GABAergic markers. Genes. 2020;11(11):1248. doi: 10.3390/genes11111248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10(9):1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 335.Zhang Y, Bi YX, Chen J, Li S, Wu XX, Zhang L, et al. Association of Peroxiredoxin 1 and brain-derived neurotrophic factor serum levels with depression and anxiety symptoms in patients with irritable bowel syndrome. Gen Hosp Psychiatry. 2021;68:59–64. doi: 10.1016/j.genhosppsych.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 336.Malheiros RT, Delgado HO, Felber DT, Kraus SI, Dos Santos ARS, Manfredini V, et al. Mood disorders are associated with the reduction of brain derived neurotrophic factor in the hypocampus in rats submitted to the hipercaloric diet. Metab Brain Dis. 2021;36(1):145–151. doi: 10.1007/s11011-020-00625-z. [DOI] [PubMed] [Google Scholar]
- 337.Aksoy H, Ergun T, Akkiprik M, Peker Eyuboglu İ, Seckin Gencosmanoglu D, Cöbek Ünalan GP, et al. The impact of antipsoriatic treatment on serum pro-BDNF, BDNF levels, depression, anxiety scores, and quality of life. Dermatol Ther. 2021;34(2):e14872. doi: 10.1111/dth.14872. [DOI] [PubMed] [Google Scholar]
- 338.Mori T, Koyama N, Yokoo T, Segawa T, Maeda M, Sawmiller D, et al. Gallic acid is a dual α/β-secretase modulator that reverses cognitive impairment and remediates pathology in Alzheimer mice. J Biol Chem. 2020;295(48):16251–16266. doi: 10.1074/jbc.RA119.012330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rockenstein E, Desplats P, Ubhi K, Mante M, Florio J, Adame A, et al. Neuro-peptide treatment with Cerebrolysin improves the survival of neural stem cell grafts in an APP transgenic model of Alzheimer disease. Stem Cell Res. 2015;15(1):54–67. doi: 10.1016/j.scr.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 340.Choucry AM, Al-Shorbagy MY, Attia AS, El-Abhar HS. Pharmacological manipulation of Trk, p75NTR, and NGF balance restores memory deficit in global ischemia/reperfusion model in rats. J Mol Neurosci. 2019;68(1):78–90. doi: 10.1007/s12031-019-01284-1. [DOI] [PubMed] [Google Scholar]
- 341.Yamada M, Hayashi H, Yuuki M, Matsushima N, Yuan B, Takagi N. Furin inhibitor protects against neuronal cell death induced by activated NMDA receptors. Sci Rep. 2018;8(1):5212. doi: 10.1038/s41598-018-23567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cameron A, Appel J, Houghten RA, Lindberg I. Polyarginines are potent furin inhibitors. J Biol Chem. 2000;275(47):36741–36749. doi: 10.1074/jbc.M003848200. [DOI] [PubMed] [Google Scholar]
- 343.Sarac MS, Cameron A, Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect Immun. 2002;70(12):7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available upon request.