Abstract
Oocyte maturation is pertinent to the success of in vitro maturation (IVM), which is used to overcome female infertility, and produced over 5000 live births worldwide. However, the quality of human IVM oocytes has not been investigated at single-cell proteome level. Here, we quantified 2094 proteins in human oocytes during in vitro and in vivo maturation (IVO) by single-cell proteomic analysis and identified 176 differential proteins between IVO and germinal vesicle oocytes and 45 between IVM and IVO oocytes including maternal effect proteins, with potential contribution to the clinically observed decreased fertilization, implantation, and birth rates using human IVM oocytes. IVM and IVO oocytes showed separate clusters in principal component analysis, with higher inter-cell variability among IVM oocytes, and have little correlation between mRNA and protein changes during maturation. The patients with the most aberrantly expressed proteins in IVM oocytes had the lowest level of estradiol per mature follicle on trigger day. Our data provide a rich resource to evaluate effect of IVM on oocyte quality and study mechanism of oocyte maturation.
Keywords: single-cell proteomics, oocyte maturation, human, heterogeneity, proteomics
Abbreviations: ART, assisted reproductive technology; FA, formic acid; FDR, false discovery rate; GO, gene ontology; GV, germinal vesicle; IVM, in vitro maturation; IVO, in vivo maturation; IVF, in vitro fertilization; KEGG, Kyoto Encyclopedia Genes and Genomes; LFQ, label-free quantification; MII, metaphase II; PCA, principal component analysis; P, progesterone; TFs, transcription factors
Graphical Abstract
Highlights
-
•
Single-cell proteomic profiling of human oocytes matured in vitro and in vivo.
-
•
Low correlation between protein and mRNA levels during human oocyte maturation.
-
•
In vitro matured (IVM) oocytes exhibit higher heterogeneity at the proteome level.
-
•
45 differentially expressed proteins between IVM and in vivo matured (IVO) oocytes.
In Brief
Here, we performed single-cell quantitative proteomic analysis of human germinal vesicle (GV), in vivo (IVO), and in vitro matured (IVM) oocytes and found low correlation between protein and mRNA levels. IVM oocytes showed higher heterogeneity in protein expression, which is related to the levels of estradiol per mature follicle on trigger day. This study provides a rich resource to characterize the mechanisms of oocyte maturation and to evaluate the quality heterogeneity of IVM oocytes at protein level.
Human oocyte maturation is a complex process that encompasses the development from the germinal vesicle (GV) stage through to the metaphase II (MII) stage, involving GV breakdown (transition from prophase I to MII) and extrusion of the first polar body (1). It involves several important structural and biochemical events in both the cytoplasm and nucleus that are necessary for achieving full maturity for fertilization and early embryo development (2). Oocyte in vitro maturation (IVM) is an assisted reproductive technology (ART) in clinics to treat female infertility and involves IVM of GV oocytes collected from antral follicles to MII stage (3). It is mainly used as an alternative treatment option for women with polycystic ovary syndrome to minimize the risk of ovarian hyperstimulation syndrome, for patients with resistant ovary syndrome to avoid repeated oocyte in vivo maturation (IVO) failure, and for fertility preservation in women diagnosed with cancers (4, 5). Although the success rates of IVM have significantly improved during the last 3 decades, resulting in over 5000 live births worldwide (6), structural and morphologic differences, including oocyte size at different developmental stages, have been observed when comparing IVO and IVM oocytes from polycystic ovary syndrome patients (7). And comparative studies have reported decreased fertilization rates, blastocyst formation rates, implantation rates, clinical pregnancy rates, and live birth rates following in vitro fertilization (IVF) of IVM oocytes versus standard IVF using IVO oocytes (8, 9, 10). Thus, the developmental potential of IVM oocytes may differ from that of IVO oocytes.
As each individual is developed from a single fertilized egg, and it is important to evaluate quality differences between oocytes matured in vitro and in vivo at single-cell level. Single-cell analysis of human oocytes was mainly performed at transcriptome level, because single-cell RNA-seq has become an established method in analyzing transcriptomic cell-to-cell variation (11). Single-cell transcriptomic studies of human IVM and IVO oocytes showed a cascade of competing and compensatory actions driven by genes encoding enzymes (12). Recent single-cell scTrioseq analysis further investigated potential epigenetic differences between human IVM and IVO oocytes (13). Considering the complex of transcriptional and posttranscriptional regulation during oocyte maturation, including early transcription, delayed translational activation, and regulated protein degradation, the abundance of proteins cannot be easily inferred from transcriptomic measurements (14, 15, 16, 17). Protein-level analysis can help better evaluate the quality of human IVM oocytes.
With the development of single-cell proteomics, the analysis of a limited number of samples has become more feasible, including approaches to profile the proteome of oocytes at the single-cell level. Virant-Klun et al. applied single-pot solid-phase–enhanced sample preparation technology and identified ∼450 proteins in a single human oocyte (18). Using Oil-Air-Droplet Chip–based single-cell proteomics analysis, Li et al. identified 355 proteins at the single mouse oocyte level (19). However, the heterogeneity of human oocytes, especially the comparison between IVO and IVM oocytes, has not been investigated at the proteome level. Here, we report the single-cell quantitative proteomic analysis of human oocytes matured in vitro and in vivo. With identification of 2382 proteins and quantification of 2094 proteins, we found complex protein regulation during human oocyte maturation and higher heterogeneity among human IVM oocytes.
Experimental Procedures
Experimental Design and Statistical Rationale
To investigate the quality differences between oocytes matured in vitro and in vivo, three types of human oocytes (GV, IVO oocytes: retrieval after controlled ovarian hyperstimulation regimens; IVM oocytes: cultured in vitro from GV) were used. Twelve biological replicates of each type were analyzed using our single-cell proteomic method (a total of 36 samples used).
The protein expression data were filtered according to the following criteria: i) only proteins with label-free quantification (LFQ) values in at least 50% of samples in any group were preserved for subsequent analysis; ii) samples that identified less than 70% of the total proteins were considered of low quality and discarded. Expression levels of each protein plus one were log2 transformed in the following analysis to avoid invalid values. To compensate for the missing quantitative value caused by low protein abundance, we imputed the data with quantile regression imputation of left-censored data algorithm (20). Linear mixed effects model was conducted using lmerTest package (21) to analyze the statistical significance of differentially expressed proteins among three groups, the formula was set as followed:
The response is denoted y, group is a fixed effect, and individual is a random effect which is nested within group. The types of oocytes were defined as group, that is GV, IVM and IVO, while individual refers to different donors. Only proteins with false discovery rate (FDR) q value (Benjamini-Hochberg) lower than 0.05 as well as fold change >1.5 were considered as differentially expressed among groups. Among the differentially expressed proteins, those with fold change >1.5 and Tukey’s posthoc p value <0.05 between GV and IVO or IVM and IVO oocytes were considered significant between groups. For transcriptomic data, the FDR q values between IVM and IVO groups were obtained from Zhao et. al.’s work (12); and for GV and IVO dataset, both the fold changes and FDR q values were calculated using deseq2 package (22). Only genes with fold change >1.5 and FDR q < 0.05 were considered significant between GV oocytes and oocytes matured in vivo (23) and between oocytes matured in vivo and in vitro (12).
Collection of GV, In vitro, and In vivo Matured Oocytes
This study included 36 human oocytes (12 GV oocytes, 12 IVM oocytes, and 12 IVO oocytes) from 13 donors. Informed consents were obtained from all donors in this study. This study has been approved by the Ethical Committee of Jiangsu Province Hospital (#2012-SR-128) and approved also by the Ethical Committee of Nanjing Medical University (# (2018)648). Studies in this work abide by the Declaration of Helsinki principles. All experiments and procedures involving animals were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University.
Retrieval and culture of human oocytes were performed as previously described (24). All donors underwent controlled ovarian hyperstimulation regimens, including GnRH agonist and GnRH antagonist, and B-ultrasound was used to monitor follicular growth and development. The donors were triggered with 250 μg r-hCG (Ovidrel, Merck Serono S.p.A.) or 0.2 mg GnRH agonist (Diphereline, Pharma Biotech) if there were two follicles’ diameter ≥18 mm or four follicles’ diameter ≥16 mm. The number of follicles ≥14 mm in diameter and the levels of E2 and progesterone (P) were assessed. The Estradiol/follicle ratio (E2/fol) was defined as estradiol level per mature follicle ≥14 mm in diameter. The oocytes were obtained by transvaginal puncture with an 18-gauge needle (Cook Medical) 36 to 38 h after the injection. The oocytes and surrounding cumulus oophorus were treated with 80 U/ml prewarmed hyaluronidase (SAGE, In-vitro Fertilization, Inc) to partially remove cumulus cells to facilitate the identification of GV-stage oocytes or IVO oocytes. To obtain IVM oocytes, GV oocytes with associated cumulus cells were cultured in oocyte maturation medium (IVM kit, SAGE) containing 0.075 IU/ml FSH and 0.075 IU/ml LH (Ferring Pharmaceuticals) in an incubator in 6% CO2, 5% O2 at 37 °C for 24 to 48 h by the presence of the first polar body in perivitelline space. All oocytes were frozen by vitrification using a blastocyst vitrification kit (COOK Medical) according to the manufacturer’s instructions in liquid nitrogen.
Before proteomic processing, vitrified oocytes (GV, IVO, and IVM) were warmed using a blastocyst warming kit (COOK Medical) and transferred to human tubal fluid (HTF, COOK) and cultured in the incubator of 6% CO2, 5% O2 at 37 °C for 1 h. The zona pellucida of each GV, IVO, and IVM oocyte was removed using Tyrode’s solution (Sigma), and oocytes were washed three times in PBS to remove culture media prior to proteomic analysis.
Proteomic Sample Preparation
To optimize the methods for single-oocyte proteomic analysis, single mouse MII oocytes were lysed either in a urea lysis buffer (8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, 1% (vol/vol) EDTA-free protease inhibitor, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate) followed by digestion with different amounts of trypsin (2 ng to 500 ng) according to a previously published method (25) or by RapiGest SF (Waters) buffer at different concentrations (0.01% to 0.1% RapiGest SF) followed by digestion with trypsin (100 ng to 500 ng). Protein and peptide yield from the different protocols were compared using LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific).
For single human oocyte proteomic analysis, each oocyte was lysed in 10 μl lysis buffer containing 0.02% RapiGest SF and stored on ice for 10 min. One microliter 50 mM DTT was added to a final concentration of 5 mM, followed by incubation at 60 °C for 40 min. Following reduction, 3 μl of 33 mM IAA solution in 167 mM ammonium bicarbonate was added to alkylate sulfhydryl groups by incubating for 40 min in the dark at room temperature. To avoid adhesion loss, a high concentration of trypsin (2 μl, 100 ng/μl) (26) was added and incubated at 37 °C for 5 h. The digestion was stopped by adding 0.5 μl of formic acid (FA), incubated at 37 °C for 30 min to cleave RapiGest SF. Finally, peptides were desalted by StageTips (ThermoFisher Scientific, SP301) and dried with a SpeedVac concentrator before MS analysis.
LC-MS/MS Analysis
The purified peptides were resuspended in 0.1% FA and loaded on an analytical column (75 μm × 25 cm, Acclaim PepMap RSLC C18 column, 2 μm, 100 Å; DIONEX) with direct injection mode based on EASY-nLC 1200 System (ThermoFisher Scientific). The HPLC solvent A was 0.1% FA, and solvent B was 80% acetonitrile, 0.1% FA. A 90-min linear gradient (3% to 8% buffer B for 4 min, 8% to 30% buffer B for 77 min, 30% to 100% buffer B for 5 min, 100% buffer B for 4 min) was applied.
For MS analyses, peptide analysis was performed on a Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (ThermoFisher Scientific) in the data-dependent acquisition mode. A full survey scan was obtained for the m/z range of 350 to 1500 at a resolution of 60,000. The automatic gain control target and maximum injection time were set at 3E6 and 20 ms. MS/MS spectra were acquired from the survey scan for the 40 most intense ions (as determined in real time by Xcalibur mass spectrometer software, version 4.3) with a resolution of 30,000. The automatic gain control target and maximum injection time were set at 5E4 and 100 ms. To minimize repeated sequencing, dynamic exclusion was set to a duration of 40 s.
Data Analysis
All raw files were processed using Maxquant (25, 27, 28) (version 1.2.2.5) for feature detection, database searching, and protein quantification. MS/MS spectra were searched against the UniProtKB human database (downloaded on July 18, 2018; 76,117 protein sequences) and commonly observed contaminants with N-terminal protein acetylation and methionine oxidation as variable modifications, and carbamidomethylation of cysteine residues as a fixed modification. Trypsin/P was selected as a protease. The peptide mass tolerances of the first search and main search (recalibrated) were <20 and 6 ppm, respectively. The match tolerance for MS/MS search was 20 ppm. The minimum peptide length was six amino acids, and the allowed missed cleavage for each peptide was 2. Both peptides and proteins were filtered with a maximum FDR of 0.01. To increase peptide/protein identification, the MBR feature with a match window of 0.7 min was used. LFQ was used to estimate protein abundance according to the previous procedure (25). Both unique and razor peptides were selected for protein quantification. All other parameters were the default settings of the Maxquant software.
Bioinformatics Analysis
Correlation analysis across samples was performed on corrPlot (29) package. The Principal Component Analysis (PCA) was performed using the stats package. The top two principal components of IVM group from PCA were used to build a two-dimensional space, the median of each dimension was defined as and , and group median = [, ], while IVMi = [, ], then the Euclidean distance Euci from IVMi to group median can be expressed as:
All heatmaps were derived using the ComplexHeatmap (30) package, and Z-score was used for standardization between samples. The volcano plots were by ggplot2 (31) package. ID conversion and annotation were conducted using clusterProfiler (32) and org.Hs.eg.db (33) packages. The annotation of transcriptional factors and cofactors were according to the “HumanTFs” website (34) and AnimalTFDB 3.0 (35), respectively, and epigenetic factor annotation was according to EpiFactors database (36). Gene ontology (GO), Kyoto Encyclopedia Genes and Genomes (KEGG), and Gene Set Enrichment Analysis analyses of differentially expressed proteins were also performed using clusterProfiler package (32). Reproductive phenotypes and embryo-related phenotypes were derived according to MGI database (37). The gene-phenotype network was conducted using igraph package (38). GRAVY scores were obtained against GRAVY calculator (http://www.gravy-calculator.de).
Results
Single-Cell Quantitative Proteomic Profiling of Human Oocyte Maturation Showed High Heterogeneity of IVM Oocytes
It is estimated the protein content in single human oocyte is about 100 ng, which is about 1000 folds compared to a typical human somatic cell (18). To optimize the method for single-cell proteomic analysis of oocytes, we first tested different conditions for protein lysis and trypsin digestion using mouse single-oocyte samples. We found that protein lysis in 0.02% RapiGest SF followed by protein digestion using 200 ng trypsin allowed for identification of the largest number of proteins and peptides (supplemental Fig. S1, A and B). Urea lysis buffer had miss-cleavages higher than 20% and were higher than those using RapiGest SF lysis buffer, and miss-cleavages using RapiGest SF were all less than 20% except that using 0.01% RapiGest SF (supplemental Fig. S1C). And the best sequence coverages were detected using 0.02% RapiGest SF lysis with 200 ng trypsin (supplemental Fig. S1D). For the peptide GRAVY to show hydrophobicity, we found the different concentrations of RapiGest SF and trypsin resulted in similar distribution of GRAVY scores (supplemental Fig. S1E). For the cellular component analysis, the terms enriched were similar between Urea and 0.01% or 0.02% RapiGest SF, while 0.05% and 0.1% RapiGest SF had less enriched terms such as “intracellular membrane-bounded organelle” or “cytoplasmic vesicle” (supplemental Fig. S1F). For the peak width analysis, RapiGest SF resulted in relatively wider peak widths than the urea lysis buffer, except that 0.10% RapiGest SF had significant wide peak widths (supplemental Fig. S1G). In view of all above analysis, 0.02% RapiGest SF lysis combined with 200 ng trypsin showed good performances, resulted in the largest number of protein and peptide identifications (supplemental Fig. S1, A and B), and was used for our subsequent human single-oocyte proteomic analysis.
To profile human oocyte maturation, we used a total of 36 oocytes collected from 13 donors undergoing assisted reproductive therapy. The sample included 12 GV oocytes from six donors, 12 IVM oocytes from eight donors, and 12 IVO oocytes from two donors (supplemental Table S1). For donors 5689, 5898, and 9432, both GV and IVM oocytes were collected. After removal of the zona pellucida and confirming normal morphology, each oocyte was subjected to trypsin digestion, LC-MS/MS analysis, and LFQ (Fig. 1, A and B). Totally, 2382 proteins and 20,528 peptides were identified (supplemental Table S2). Only proteins with expression in at least 50% of samples in at least one group were considered in the analysis, and oocyte samples with identification of greater than 70% of the total proteins were considered of high quality. After data filtering according to the standards mentioned in Experimental Procedures, 2094 proteins in 34 oocytes (GV: 11 oocytes, IVM: 12 oocytes, IVO: 11 oocytes) were quantified and subjected to the subsequent analysis (supplemental Table S2). The average number of proteins/peptides quantified per oocyte in GV, IVM, and IVO were 1947/12,589, 1900/12,063, and 1836/11,374, respectively (supplemental Fig. S2A). To assess the data quality, we compared our data with Virant-Klun et al.’s single-cell proteomics data (18), which identified 446 proteins in a single oocyte showed that 403 proteins (90.4%) were also identified in a single oocyte (IVO.4152.2) from our data (supplemental Fig. S2B), indicating consistent identification of human oocyte proteins.
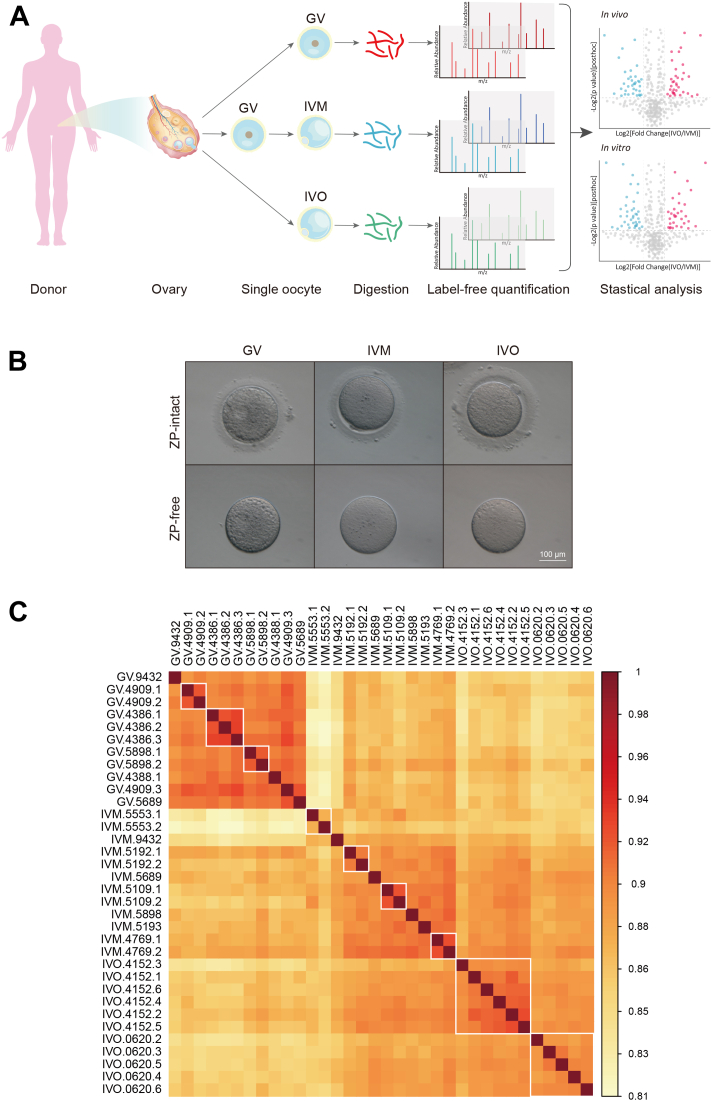
Fig. 1.
Single-cell quantitative proteomic profiling of human oocytes maturation.A, schematic illustration of single-cell quantitative proteomic profiling of human oocyte maturation in vivo and in vitro. B, representative images of GV, IVM, and IVO oocytes with (upper panel) and without (lower panel) zona pellucida (ZP). The scale bar represents 100 μm. C, pairwise correlation analysis among the 34 oocytes using log2 transformed LFQ values. White boxes identify oocytes from the same individual within each group. GV, germinal vesicle; IVM, in vitro maturation; IVO, in vivo maturation; LFQ, label-free quantification.
Reproducibility analysis of protein quantification among the oocyte samples indicated high reproducibility, with pairwise Pearson’s correlation coefficients ranging from 0.893 to 0.935, 0.844 to 0.924, and 0.872 to 0.916 among oocytes in GV, IVM, and IVO group, respectively (supplemental Fig. S2, C–E). The analysis showed relatively higher Pearson’s correlation coefficients within the GV samples, which appeared as a distinct group. This was expected as IVM and IVO oocytes are both MII oocytes and relatively close to each other. Additionally, for some of the donors, oocytes from the same individual tended to be more similar to each other with relatively higher Pearson’s correlation coefficients; for example, IVO oocytes from individual 4152 exhibited higher correlation coefficients to each other (Fig. 1C).
To better evaluate the similarity among oocytes, PCA was performed and outliers in each group were detected beyond the 95% confidence interval. Three clusters of oocytes were revealed, representing GV, IVM, and IVO oocyte clusters (Fig. 2A), indicating distinct proteome profiles among groups. Although GV oocytes were collected from six individuals, they were closely clustered together. IVO oocytes were also relatively closely clustered, indicating less heterogeneity among different in vivo oocytes in protein expression patterns. IVM oocytes were located between GV and IVO oocyte groups, but closer to the IVO oocytes, presumably, because IVM and IVO oocytes are both MII oocytes. The clustering of IVM and IVO oocytes into two clusters suggested different protein expression profiles between in vivo– and in vitro–matured oocytes. Furthermore, in contrast to the closely clustered patterns in GV and IVO groups, oocytes in the IVM group are less closely clustered, exhibiting more heterogeneity. IVM.5553.1 and IVM.5553.2 deviated most from other IVM oocytes and were detected as outliers beyond the 95% confidence interval. We found that oocytes from the same individual were largely similar to each other, for example, the oocytes from individual 4386 in the GV group, and those from individual 5553 in the IVM group, are closer to each other (Fig. 2A). Overall, our results show that the proteome profiles of oocytes were less heterogeneous for oocytes from the same donor than among different donors, and IVM oocytes exhibited overall higher heterogeneity between oocytes at the proteome level. It seems that oocytes cultured in vitro might have different and varied protein profiles, possibly related to the reported lower qualities of IVM oocytes.
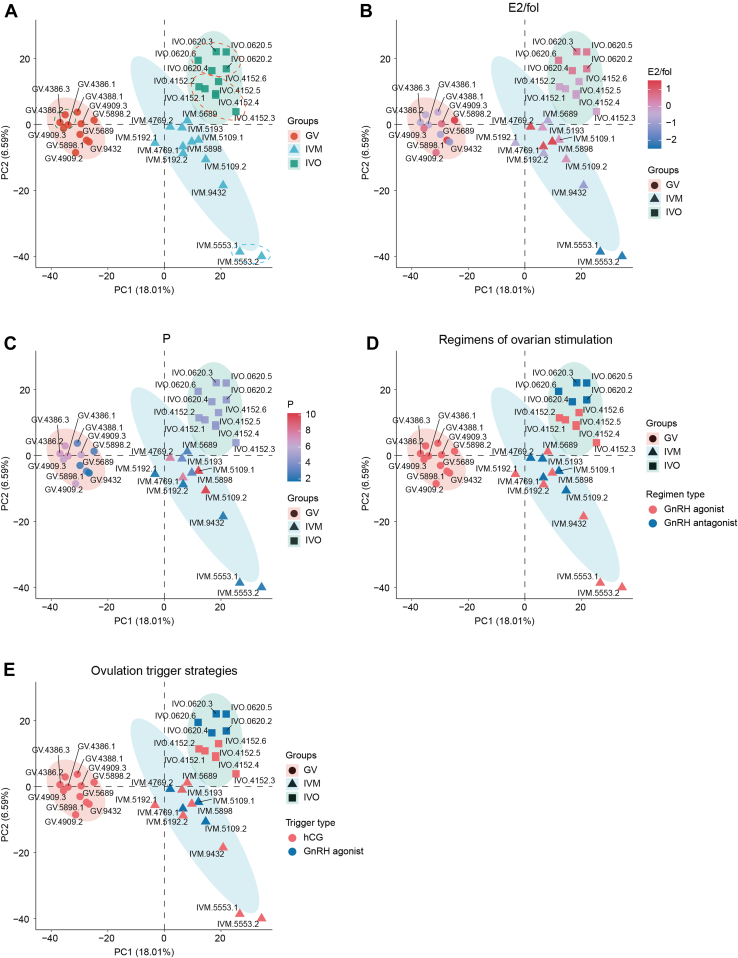
Fig. 2.
Principal component analysis and factors analysis of oocyte heterogeneity. PCA of the proteomic data from 34 human oocytes obtained from 13 individuals (A) with different colors representing the levels of E2/fol (B) and progesterone (C) on trigger day, different regimens of ovarian stimulation (D), and different ovulation trigger strategies (E). Ellipses show the 95% confidence interval in each group. Dotted circles denote oocytes obtained from the same individual. PCA, principal component analysis.
To explore the factors contributing to the heterogeneity of IVM oocytes, we analyzed the effects of the levels of estradiol per mature follicle (≥14 mm) (E2/fol), the levels of P on trigger day, the regimens of ovarian stimulation, and the ovulation trigger strategies (Fig. 2, B–E). Correlation analysis between the levels of E2/fol or P and the Euclidian distances from each oocyte to the group median showed that E2/fol was significantly correlated (p < 0.05, R = -0.625, supplemental Fig. S3A and supplemental Table S1). The oocytes from individual 5553 in the IVM group, which deviated most from other IVM oocytes, had the lowest levels of E2/fol (241.8 pmol/L for individual 5553) on trigger day (Fig. 2B and supplemental Table S1). No significant difference of the Euclidian distances from each oocyte to the group median was observed for different regimens of ovarian stimulation or ovulation trigger strategies (Figs 2, D and E, S3, C and D, supplemental Table S1). For example, although individual 5553 was subjected to GnRH agonist in ovarian stimulation, other individuals with their oocytes closely clustered, such as individuals 5193 and 4769, were also subjected to GnRH agonist treatment (Fig. 2D). A similar phenomenon can be observed in individuals subjected to different ovulation trigger strategies. It seems that the regimens of ovarian stimulation and the ovulation trigger strategies are not important factors contributing to the heterogeneity of oocytes from individual 5553.
Differential Protein Expression Analysis of Human Oocytes Reveals Inconsistent mRNA and Protein Changes
As GV, IVM, and IVO oocytes form three clusters in PCA, we quantified the expression levels of differentially expressed proteins between groups and identified 243 proteins that exhibited a fold change >1.5 and FDR q < 0.05 among the three groups (supplemental Table S2), including 176 differential proteins (supplemental Table S3) during maturation in vivo (IVO group compared to GV group) and 45 differential proteins (supplemental Table S4) in IVM versus IVO oocytes. Cluster and heatmap analysis of the 243 differentially expressed proteins showed five clusters (C1-C5) with different expression patterns (Fig. 3, A and B). Cluster 1 to 4 showed relatively decreased protein expression levels in IVO versus GV oocytes, Cluster 5 showed increased protein expression levels in IVO versus GV oocytes. Cluster 4 (59 proteins) and cluster 5 proteins (95 proteins) exhibited similar expression levels in IVM and IVO oocytes, suggesting similar regulation between IVM and IVO. Cluster 4 showed enrichment of proteins in “translational initiation”, and Cluster 5 in “mitotic nuclear division” as well as “negative regulation of chromosome organization”. Cluster 1 (64 proteins) had abnormally high levels in IVM oocytes versus IVO oocytes, suggesting possible abnormalities in protein degradation during IVM culture, and showed enrichment in “proteasomal ubiquitin-independent protein catabolic process”. Cluster 2 (11 proteins) and Cluster 3 proteins (14 proteins) exhibited lower expression levels in IVM oocytes when compared with GV oocytes and IVO oocytes. Notably, cluster 2 proteins had aberrantly low levels in three IVM oocytes of individuals 9432 and 5553 compared with other IVM and IVO oocytes, which may be related to the maturation quality and should be investigated in further functional studies.
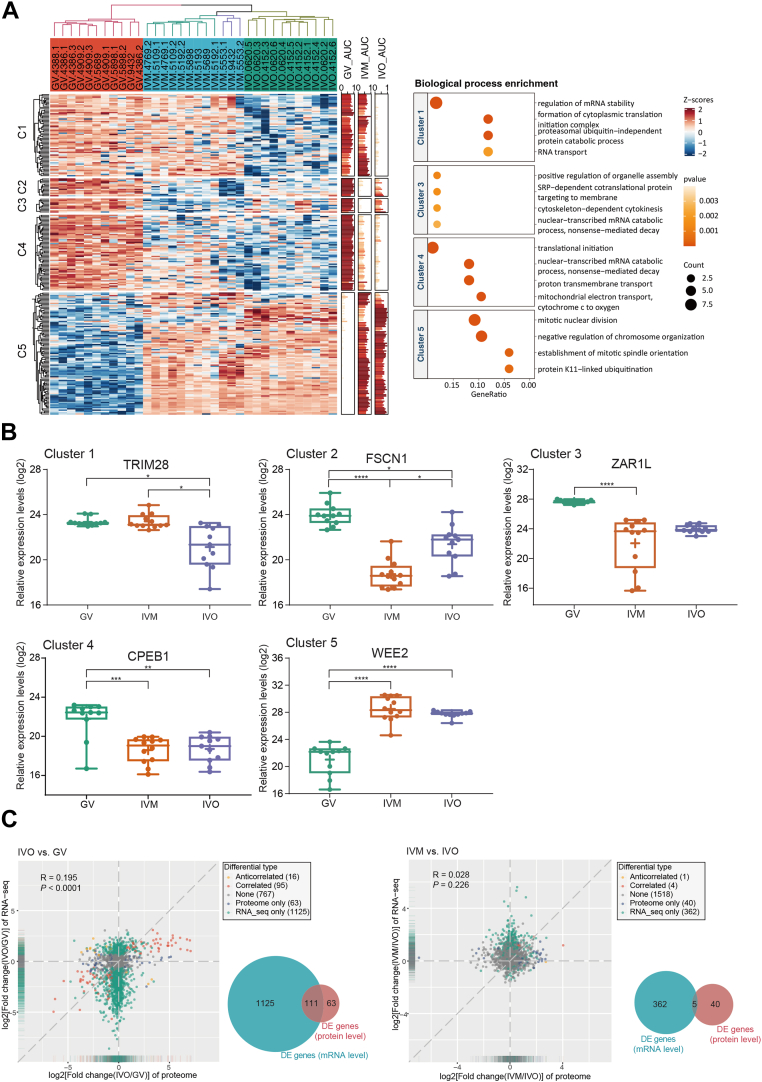
Fig. 3.
Differential protein expression analysis of human oocytes showed inconsistent mRNA and protein changes.A, heatmap (left) of 243 differentially expressed proteins among GV, IVM, IVO oocytes and enriched biological processes (right) in each cluster. Clusters of proteins (C1-C5) are indicated according to k-means. B, expression of representative proteins selected from each of the 5 clusters as interleaved boxes and whiskers with minimum and maximum values displayed. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001 (linear mixed effects model with Tukey’s posthoc test, fold change >1.5). C, scatter plot of the fold changes between the proteome and transcriptome during in vivo maturation (IVO, left) and during in vitro maturation (IVM, right) and Venn diagram of comparison of genes differentially expressed at the proteome (DE proteins) and transcriptome (DE genes) levels. Only genes identified in both transcriptome and proteome were considered. GV, germinal vesicle.
Posttranscriptional regulation during oocyte maturation is complex. The quantitative proteome profile data obtained in this study enabled a comparison of transcriptome and proteome changes in human oocytes. Therefore, we compared our data for proteins with quantified expression levels to the reported transcriptome profile data for in vivo (39) and in vitro oocyte maturation (12) in humans and found that the correlation coefficient of fold changes between proteins and RNAs was relatively low (R = 0.196, p < 0.0001). Among the 2066 genes identified in both proteome and transcriptome (IVO group compared with GV group) during IVO, 174 were differentially expressed at protein level and 1236 were differentially expressed at mRNA level (IVO group compared with GV group) during IVO, only 111 showed differential expression at both the mRNA and protein level (Fig. 3C). Of 73 genes that were upregulated at protein level in IVO oocytes versus GV oocytes, 26 (36%) did not exhibit upregulation of the corresponding mRNA and may therefore be subjected to translational activation during oocyte maturation. Of 101 gene that were downregulated at protein level in IVO oocytes versus GV oocytes, 53 (52%) did not show downregulation of the corresponding mRNA, suggesting that the levels of these proteins are regulated via protein stability. For example, AURKA (supplemental Fig. S4) was upregulated and TRIM28 (Fig. 3B) was downregulated at the protein level in IVO oocytes but neither showed a change at the mRNA level during maturation in vivo. The correlation coefficient of fold changes between proteins and RNAs following IVM and IVO was 0.028. Among the overlapped 1925 genes, 45 showed differential expression at protein level following IVM and IVO (IVM group versus IVO group), only 5 (11.1%) were also differentially expressed at the mRNA level (Fig. 3C). For example, FSCN1 and TRIM28 were differentially expressed only at the protein level in IVM oocytes (Fig. 3B). The mRNA and protein changes are inconsistent during human oocyte maturation.
Protein Regulation During IVO of Human Oocytes
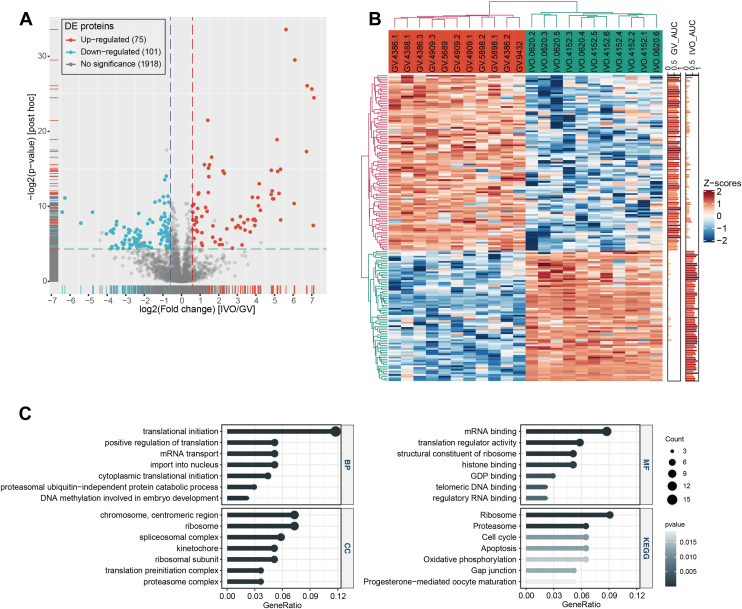
We used the single-cell protein expression data from GV and IVO oocytes as a model to investigate changes in proteins during oocyte maturation in physiological conditions and identified a total of 176 differential proteins (supplemental Table S3), including 75 upregulated proteins and 101 downregulated proteins in IVO oocytes (Fig. 4, A and B). Analysis of GO was performed (Fig. 4C and supplemental Table S5), and biological process annotation showed enrichment of “translational initiation (16 proteins)”, “mRNA transport (7 proteins)”, “import into nucleus (7 proteins)”, “proteasomal ubiquitin−independent protein catabolic process (4 proteins)”, and “DNA methylation involved in embryo development (3 proteins)”. Cellular component annotation showed enrichment of “chromosome, centromeric region (10 proteins)”, while molecular function annotation showed enrichment of “mRNA binding (12 proteins)”, “translation regulator activity (8 proteins)”, “structural constituent of ribosome (7 proteins)”, and “histone binding (7 proteins)”. The enriched terms indicate that these differential proteins may have important roles in epigenetic, transcriptional and protein translation, and degradation regulation in oocyte maturation or early embryo development.
Fig. 4.
Protein regulation during in vivo maturation of human oocytes.A, volcano plot comparing proteins from IVO versus GV oocytes. Differentially expressed proteins were defined by the following cutoff values: fold change >1.5, FDR q < 0.05, and posthoc p value <0.05. B, heatmap of differentially expressed proteins during in vivo maturation. Euclidean dis metric and complete linkage clustering algorithms were used. C, lollipop plot of enriched GO and KEGG pathway terms in differentially expressed proteins between GV and IVO oocytes. BP, biological process, CC, cellular components, MF, molecular function. GV, germinal vesicle; IVO, in vivo maturation; KEGG, Kyoto Encyclopedia Genes and Genomes.
To evaluate the pathways regulated during oocyte maturation, KEGG pathways were analyzed (Fig. 4C) and showed enrichment of differential proteins in the “Ribosome (7 proteins)” (supplemental Table S5); we observed that most identified ribosome subunits were downregulated, including RPL37A, RPS12, RPS3A, RPS13, RPS25, RPS27A, and RPS21. Meanwhile, an upregulation of the translation initiation inhibitor, TACC3 (supplemental Fig. S4), was upregulated, which is consistent with the transcriptional quiescence and active translation of specific transcripts in maturing oocytes (40).
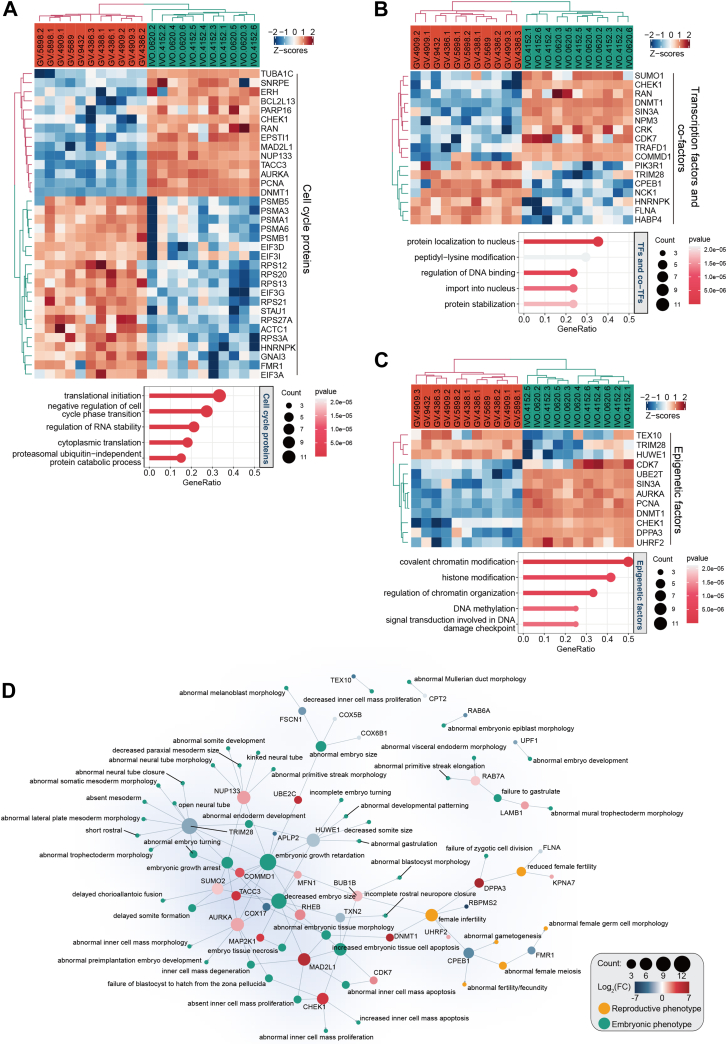
Among proteins exhibiting differential expression during oocyte maturation in vivo, we found 34 cell cycle proteins (supplemental Table S6), mainly involved in translation initiation, negative regulation of cell cycle phase transition, regulation of RNA stability and cytoplasmic translation, according to Cyclebase 3.0 database (Fig. 5A and supplemental Table S7) (41), suggesting that these proteins may play regulatory roles during the meiotic maturation of oocytes. Additionally, we identified three transcription factors (TFs) and 15 TF cofactors (supplemental Table S6) according to Lambert et al.’s and Hu et al.’s data (34, 35) (Fig. 5B). These proteins have roles in the regulation of DNA binding and protein stabilization according to GO enrichment analysis (supplemental Table S7) and including SUMO1 (supplemental Fig. S4), which is known to be essential for oocyte maturation and female fertility in mice (42). We found 12 differential epigenetic factors (supplemental Table S6) involved in epigenetic regulation mechanisms such as DNA methylation and histone modification according to the Epifactors database (Fig. 5C and supplemental Table S7) (36), including DPPA3/STELLA (supplemental Fig. S4) which is an important maternal factor protecting the maternal genome in mouse early embryo (43). These differential TFs/cofactors and epigenetic factors may regulate the chromatin state of oocytes and subsequent early embryonic development.
Fig. 5.
Functional annotations of differential human oocyte proteins during in vivo maturation.A-C, heatmap and GO annotations of cell cycle proteins (A), transcriptional factors and cofactors (B), as well as epigenetic factors (C). D, network of in vivo differential proteins and mouse reproductive and embryonic phenotypes. The color key ranging from blue to red identifies protein fold changes (IVO/GV). GV, germinal vesicle; IVO, in vivo maturation.
To analyze the in vivo functions of differentially expressed proteins in GV versus IVO oocytes, the proteins were annotated using the MGI database, identifying nine proteins with reproductive phenotypes and 29 proteins with embryonic phenotypes (Fig. 5D and supplemental Table S8). The reproductive phenotypes were mainly female infertility and reduced female fertility, and the embryonic phenotypes were mainly related to decreased embryo size, embryonic growth retardation, and embryonic growth arrest. Thus, differentially expressed proteins were not only important for oocyte maturation and functions but may function as maternal proteins that regulate early embryonic development.
Oocytes Matured In vitro and In vivo Exhibit Different Protein Expression Patterns
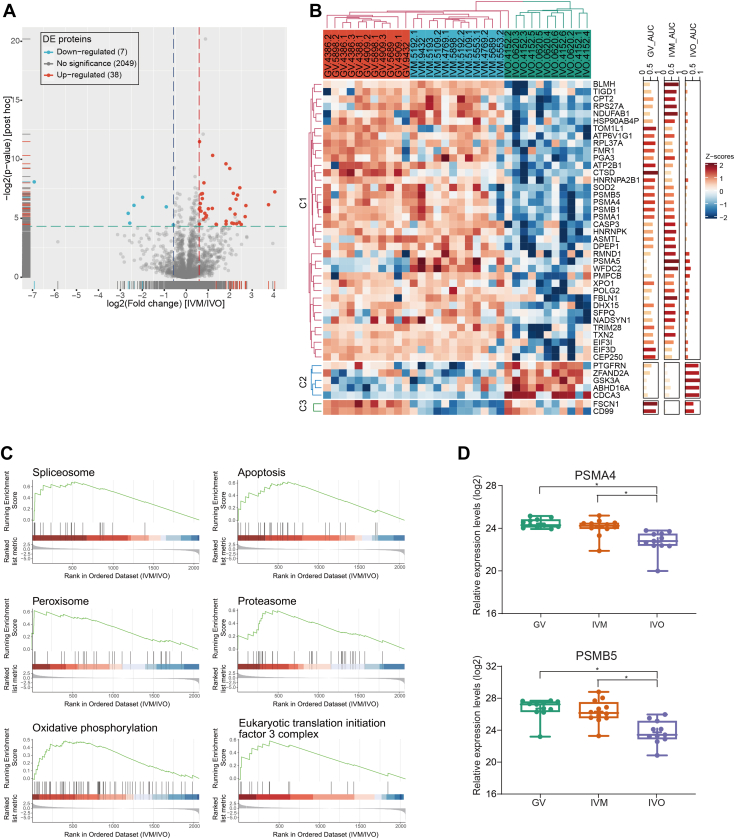
As the comparison of differential protein and mRNA expression of in vitro– and in vivo–matured oocytes showed little overlap (Fig. 3C), we evaluated the effects of IVM on the proteome level. Of 45 proteins with differential expression in IVM versus IVO oocytes (supplemental Table S4), seven were downregulated and 38 were upregulated in IVM oocytes (Fig. 6A). Heatmap analysis (Fig. 6B) showed that the expression pattern of most of these differential proteins in IVM oocytes (C1 and C2) resembled that of GV oocytes rather than that of IVO oocytes, for example, CDCA3 was upregulated in IVO oocytes but remained at a similarly low level in IVM oocytes to that in GV oocytes, while HNRNPK and CASP3 were downregulated in IVO oocytes but remained at a similarly high level in IVM oocytes to those in GV oocytes (supplemental Fig. S4).
Fig. 6.
Protein expression patterns during in vitro oocyte maturation.A, volcano plot of comparing protein expression between IVM and IVO oocytes. Differentially expressed proteins were defined by the following cutoff values: fold change >1.5, FDR q < 0.05, and posthoc p value <0.05. B, heatmap of differentially expressed proteins between IVM and IVO oocytes. C, GSEA enrichment plots of KEGG pathways between IVM and IVO oocytes. D, the expression values of PSMA4 and PSMB5 are presented as interleaved boxes and whiskers with minimum and maximum values displayed. ∗ p < 0.05, ∗∗ p < 0.01, (linear mixed effects model with Tukey’s posthoc test, fold change >1.5). IVM, in vitro maturation; IVO, in vivo maturation; KEGG, Kyoto Encyclopedia Genes and Genomes.
To further explore the functions of the proteins with abnormal expression levels in IVM oocytes, Gene Set Enrichment Analysis was applied. Among the enriched KEGG pathway gene sets (supplemental Table S9), proteins in the “spliceosome”, “proteasome”, “eukaryotic translation initiation factor 3 complex”, “peroxisome”, “oxidative phosphorylation”, and “apoptosis” pathways exhibited consistently higher levels in IVM versus IVO oocytes (Fig. 6C). For example, PSMA1, PSMA4, PSMA5, PSMB1, and PSMB5, which promote assembly of the 20S proteasome (44, 45, 46) involved in the proteolytic degradation of most intracellular proteins, exhibited higher levels in IVM oocytes (Figs 6D and S4).
Discussion
In clinics, even with ovarian stimulation, only around a dozen oocytes from a single individual can be retrieved in ART treatment (47). Single-cell global protein-level analysis of oocyte maturation in vivo and in vitro can contribute to a better understanding of oocyte development, providing a basis for the evaluation of oocyte quality and subsequent optimization of the IVM, a widely used ART technique. Due to the lack of amplification methods for global protein analysis (48) and limited human oocyte samples, only very few studies have explored the proteome of human oocytes at the single-cell level. Here, we have characterized the proteomes of GV, IVM, and IVO oocytes in a total of 36 human oocytes at the single-cell level, using optimized protein lysis and digestion conditions. Among the 34 oocytes (GV: 11 oocytes, IVM: 12 oocytes, IVO: 11 oocytes) passing quality control, we identified 2382 proteins and quantified 2094 proteins. Our single-oocyte proteomics data showed good reproducibility among different individual oocytes by the pairwise Pearson’s correlation coefficients analysis.
Single-cell proteomic analysis provides an approach to dissect the heterogeneity of protein expression within groups or even within individuals (49, 50), especially in rare cell samples such as human oocytes. PCA analysis showed relatively lower heterogeneity among GV and IVO oocytes than among IVM oocytes, suggesting homogenous oocyte states in vivo. However, IVM oocytes exhibited higher inter-cell variability, suggesting a higher variability in the quality of IVM oocytes than that of IVO oocytes, which is consistent with the clinical observations of lower cumulative biochemical pregnancy, clinical pregnancy, and live birth rates following ART using IVM oocytes (8). The higher heterogeneity of IVM oocytes is mainly caused by the deviation of two oocytes matured in vitro from individual 5553 according to PCA analysis and outlier detection. Additionally, three IVM oocytes from individuals 5553 and 9432 exhibited aberrantly low levels of the same subset of proteins (Cluster 2, Fig. 3A) that may be related to oocyte quality and clinical outcome. We explored the factors contributing to the variation of these oocytes and found significant correlation of the levels of E2/fol with the Euclidean distance from each oocyte to the group median, suggesting that the variation in E2/fol levels may contribute to the heterogeneity of in vitro–matured oocytes. Individual 5553 had the lowest level of E2/fol (241.8 pmol/L) on the hCG trigger day, and individual 9432 had the second lowest level of E2/fol (541.5 pmol/L). It is generally accepted that serum E2 is necessary for follicle/oocyte maturation (51). Suneeta et al. studied the level of E2/fol on trigger day and found that E2/fol level is positively correlated with qualities of oocytes and embryos (52). Hu et al. showed that implantation rate was significantly lower in the group with E2/fol <279.83 pg/ml on the hCG trigger day (51). Huang et al. provided evidence that low E2/fol on trigger day might reduce the oocyte retrieval rate and increase the risk of single and triple pronucleus (1PN and 3PN) formation and abortion (51). It is possible that the lowest levels of E2/fol on trigger day is a cause of the abnormalities of the two oocytes from individual 5553, which mainly showed aberrantly low expression of proteins in Cluster 2 (Fig. 3A). For ovarian stimulation regimens, compared to the GnRH agonist protocol, the GnRH antagonist protocol is more recommended given the comparable efficacy and higher safety (53), but no statistically significant differences were found in ovarian response or ongoing pregnancy rates between these two regimens (54). In our dataset, individual 5553 using GnRH agonists showed abnormal protein expression patterns, other samples using the same strategy showed no significant difference from those using GnRH antagonists, suggesting the heterogeneity among IVM oocytes might be unrelated to GnRH agonist. For oocyte ovulation triggering strategies, no difference was observed for pregnancy and implantation rates between the GnRH agonist triggering protocol and the hCG triggering protocol (55, 56, 57, 58). Although individual and 5553 was triggered by GnRH agonists, IVM oocytes from other individuals using GnRH agonists showed similar protein changes compared with those IVM oocytes using the hCG triggering protocol, indicating GnRH agonists might not be an important contributing factor to the proteome level perturbation observed in Cluster 2 (Fig. 3A). In addition, in our dataset, IVM oocytes from the same individual exhibited higher Pearson’s correlation coefficients, indicating higher inter-individual variability. Taken together, this single-oocyte proteomic quantification analysis enabled the evaluation of the quality of individual oocytes matured in vitro at the protein expression level and allowed for the evaluation of heterogeneity among different oocytes.
Our quantitative analysis of oocyte proteins identified a total of 176 proteins that were differentially expressed during maturation in vivo (IVO group compared to GV group). However, based on Yu et al.’s RNA-seq data, 1236 mRNAs are differentially expressed in IVO versus GV oocytes (39), and only 111 of them also had protein level changes. These apparent inconsistencies between changes at protein and mRNA levels imply complex posttranscriptional regulation. Consistently, GO analysis of differentially expressed proteins during human oocyte maturation in vivo identified enrichment of 12, 4, and 8 proteins in “mRNA binding”, “ssRNA binding”, and “cytoplasmic translation”, respectively. These differential proteins included the RNA-binding protein HNRNPF, which was upregulated during oocyte maturation. HNRNPF stabilizes mRNAs by binding to the 3′ UTR (59) and may therefore stabilize mRNAs for translation during the transcriptional suppression phase of oocyte maturation.
Our data indicate that oocyte maturation also involves differential expression of transcriptional and epigenetic factors, including three TFs, 15 transcription cofactors, and 12 epigenetic factors. These proteins may regulate the chromatin state of oocytes or regulate embryonic development. For example, SUMO1, DNMT1, and SIN3A (supplemental Fig. S4) were upregulated during oocyte maturation in vivo. As a member of the ubiquitin-like modifier family, SUMO1 plays crucial roles during meiotic oocyte maturation by regulating spindle organization, chromosome congression, and chromosome segregation (42). In addition to the role in oogenesis, SUMOylation is also required for the communication of the oocyte with the ovarian somatic cells (60). Deletion of the maternal factor DNMT1 in mouse oocytes causes lethality during the last third of gestation by affecting DNA methylation of imprinted genes (61). Inhibition of oocyte SIN3A, a scaffolding protein interacting with HDAC1/2, inhibits early embryonic development beyond the 2-cell stage (62). Furthermore, our results identified nine proteins with reproductive phenotypes and 29 proteins with embryonic phenotypes, also suggesting that the differential proteins may not only regulate oocyte maturation but also play important roles during early embryonic development. Maternal effect genes are oocyte-expressed genes important for embryo development (63). Among differential proteins with embryonic phenotypes, TRIM28, DNMT1, ZAR1, and DPPA3 were reported to be maternal effect proteins in mouse (64), while the differential protein WEE2 was found to be a maternal effect protein in human (65). Further studies of these factors are expected to help elucidate the mechanisms of human oocyte maturation and the roles of maternal proteins in early embryonic development.
IVM is a promising approach to conventional ART for those infertile women who cannot harvest in vivo–matured oocytes. Our proteome-level characterization of IVM and IVO oocytes showed that although IVM oocytes were more similar to IVO oocytes than to GV oocytes, IVM and IVO oocytes formed two distinct clusters and were not completely similar to each other. The IVM oocyte cluster was located between GV and IVO clusters, indicating that immature features were retained in IVM oocytes. This is supported by the phenomenon that many proteins retained expression patterns similar to GV oocytes according to heatmap analysis. As many mRNAs are transcribed at earlier stages of oocyte development and not actively translated until oocyte maturation (17), abnormally low levels of specific proteins in IVM oocytes retaining GV pattern may result from abnormalities in translation during oocyte maturation and might be related to the structural and morphologic differences between IVM and IVO oocytes (7). We also observed upregulation of translation initiation factor eIF3i, and ribosomal proteins RPL37A and RPS27A, which might be compensation for decreased translation activity in IVM oocytes.
Intriguingly, we observed that 40 of the 45 differential proteins (overlapped with mRNA) abnormally expressed in IVM oocytes showed no change at the corresponding mRNA level based on previously published data, suggesting complex translational regulation. Among the proteins aberrantly expressed in IVM oocytes, TRIM28 is a maternal effect protein in oocyte; its defective expression could lead to epigenetic changes of blastocyst, partial postimplantation embryonic loss, and no viable offspring (66). The abnormal expression of TRIM28 in IVM oocytes retaining GV expression pattern might also contribute to the reported lower live birth rates following IVF of IVM oocytes (8, 9, 10).
Some limitations may weaken the integrative analysis of RNA-seq and proteomics in this study. For example, the samples used in the published RNA level and our proteomic level analysis were not from the same individuals and may not pass the same quality control, and the sampling time point may impact on the RNA abundance during oocyte development. Future RNA and protein level analysis of oocytes from the same individuals or even the same cells could help better reveal the relationship between transcriptome and proteome during oocyte maturation. However, the maturing oocytes are transcriptionally quiescent, and their mRNAs contributing to growth, oocyte maturation, and downstream developmental stages are produced, stored in GV oocytes, and are relatively stable during maturation (40); the inconsistency between protein and mRNA expression levels during oocyte maturation in vivo and in vitro shown in our study indicates the importance of studies at protein levels to some extent.
Taken together, our single-cell proteomic profiling of human GV, IVM, and IVO oocytes has enabled a proteome-level evaluation of the quality of IVM oocytes. IVM oocytes had high inter-cell variation of protein expression patterns. Further functional studies of differential proteins between IVM and IVO oocytes may help identify approaches to improve the quality of IVM oocytes. The proteome profiles of human oocyte maturation will also be a rich resource to characterize the mechanisms of decreased quality of human IVM oocytes and identify the regulation of human oocyte maturation.
Data Availability
The proteomics data have been deposited in the ProteomeXchange Consortium via the proteomics identifications (PRIDE) database (Project accession: PXD023366).
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
We thank Yang Yu (Peking University Third Hospital, Beijing) for the statistical data of RNA-seq of human oocyte maturation in vivo and in vitro. This work was funded by National Key R&D Program of China (grant number 2021YFC2700200); Chinese National Natural Science Foundation Grants (grant numbers 32071133, 81971439, and 81771641); The Fok Ying Tung Education Foundation (grant number 161037), and the Open and Shared Research Projects of Large-scale Scientific Instruments of Jiangsu Province (grant number TC2021A010).
Author contributions
J. L., F. D., J. S., and X. G. conceptualization; L. C., L. M., and J. L. resources; L. C., L. M., and J. L. investigation; Y. G. and Y. Q. methodology; X. L., B. W., and H. Z. data curation; X. L., B. W., and H. Z. formal analysis; Y. G. and X. L. writing–original draft; F. D., J. S., and X. G. writing–review and editing.
Contributor Information
Feiyang Diao, Email: diaofeiyang@njmu.edu.cn.
Jiahao Sha, Email: shajh@njmu.edu.cn.
Xuejiang Guo, Email: guo_xuejiang@njmu.edu.cn.
Supplemental Data
References
- 1.Loutradis D., Kiapekou E., Zapanti E., Antsaklis A. Oocyte maturation in assisted reproductive techniques. Ann. New York Acad. Sci. 2006;1092:235–246. doi: 10.1196/annals.1365.020. [DOI] [PubMed] [Google Scholar]
- 2.Fulka J., Jr., First N.L., Moor R.M. Nuclear and cytoplasmic determinants involved in the regulation of mammalian oocyte maturation. Mol. Hum. Reprod. 1998;4:41–49. doi: 10.1093/molehr/4.1.41. [DOI] [PubMed] [Google Scholar]
- 3.De Vos M., Smitz J., Thompson J.G., Gilchrist R.B. The definition of IVM is clear-variations need defining. Hum. Reprod. 2016;31:2411–2415. doi: 10.1093/humrep/dew208. [DOI] [PubMed] [Google Scholar]
- 4.Chang E.M., Song H.S., Lee D.R., Lee W.S., Yoon T.K. In vitro maturation of human oocytes: its role in infertility treatment and new possibilities. Clin. Exp. Reprod. Med. 2014;41:41–46. doi: 10.5653/cerm.2014.41.2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chian R.C., Uzelac P.S., Nargund G. In vitro maturation of human immature oocytes for fertility preservation. Fertil. Steril. 2013;99:1173–1181. doi: 10.1016/j.fertnstert.2013.01.141. [DOI] [PubMed] [Google Scholar]
- 6.Sauerbrun-Cutler M.T., Vega M., Keltz M., McGovern P.G. In vitro maturation and its role in clinical assisted reproductive technology. Obstetrical Gynecol. Surv. 2015;70:45–57. doi: 10.1097/OGX.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 7.Walls M.L., Hart R., Keelan J.A., Ryan J.P. Structural and morphologic differences in human oocytes after in vitro maturation compared with standard in vitro fertilization. Fertil. Steril. 2016;106:1392–1398.e5. doi: 10.1016/j.fertnstert.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Walls M.L., Hunter T., Ryan J.P., Keelan J.A., Nathan E., Hart R.J. In vitro maturation as an alternative to standard in vitro fertilization for patients diagnosed with polycystic ovaries: a comparative analysis of fresh, frozen and cumulative cycle outcomes. Hum. Reprod. 2015;30:88–96. doi: 10.1093/humrep/deu248. [DOI] [PubMed] [Google Scholar]
- 9.Child T.J., Phillips S.J., Abdul-Jalil A.K., Gulekli B., Tan S.L. A comparison of in vitro maturation and in vitro fertilization for women with polycystic ovaries. Obstet. Gynecol. 2002;100:665–670. doi: 10.1016/s0029-7844(02)02193-2. [DOI] [PubMed] [Google Scholar]
- 10.Gremeau A.S., Andreadis N., Fatum M., Craig J., Turner K., McVeigh E., et al. In vitro maturation or in vitro fertilization for women with polycystic ovaries? A case-control study of 194 treatment cycles. Fertil. Steril. 2012;98:355–360. doi: 10.1016/j.fertnstert.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 11.Svensson V., Natarajan K.N., Ly L.H., Miragaia R.J., Labalette C., Macaulay I.C., et al. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods. 2017;14:381–387. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H., Li T., Zhao Y., Tan T., Liu C., Liu Y., et al. Single-cell transcriptomics of human oocytes: environment-driven metabolic competition and compensatory mechanisms during oocyte maturation. Antioxid. Redox Signal. 2019;30:542–559. doi: 10.1089/ars.2017.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye M., Yang Z.Y., Zhang Y., Xing Y.X., Xie Q.G., Zhou J.H., et al. Single-cell multiomic analysis of in vivo and in vitro matured human oocytes. Hum. Reprod. 2020;35:886–900. doi: 10.1093/humrep/deaa044. [DOI] [PubMed] [Google Scholar]
- 14.Piccioni F., Zappavigna V., Verrotti A.C. Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. C. R. Biol. 2005;328:863–881. doi: 10.1016/j.crvi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Smillie D.A., Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 16.Slavov N. Unpicking the proteome in single cells. Science. 2020;367:512–513. doi: 10.1126/science.aaz6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esencan E., Kallen A., Zhang M., Seli E. Translational activation of maternally derived mRNAs in oocytes and early embryos and the role of embryonic poly(A) binding protein (EPAB) Biol. Reprod. 2019;100:1147–1157. doi: 10.1093/biolre/ioz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virant-Klun I., Leicht S., Hughes C., Krijgsveld J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol. Cell Proteomics. 2016;15:2616–2627. doi: 10.1074/mcp.M115.056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z.Y., Huang M., Wang X.K., Zhu Y., Li J.S., Wong C.C.L., et al. Nanoliter-scale oil-air-droplet chip-based single cell proteomic analysis. Anal. Chem. 2018;90:5430–5438. doi: 10.1021/acs.analchem.8b00661. [DOI] [PubMed] [Google Scholar]
- 20.Wei R., Wang J., Su M., Jia E., Chen S., Chen T., et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 2018;8:663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pond R.S., McCool M.W., Bulla B.A. Multilevel modeling of interval-contingent data in neuropsychology Research using the lmerTest package in R. J. Pediatr. Neuropsychol. 2021;7:102–112. [Google Scholar]
- 22.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes J.M., Silva E., Chitwood J.L., Schoolcraft W.B., Krisher R.L., Ross P.J. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum. Reprod. 2017;32:2199–2208. doi: 10.1093/humrep/dex284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L., Cai L., Hu M., Wang J., Xie J., Xing Y., et al. Coenzyme Q10 supplementation of human oocyte in vitro maturation reduces postmeiotic aneuploidies. Fertil. Steril. 2020;114:331–337. doi: 10.1016/j.fertnstert.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Cheng Q., Li X., Hu F., Han L., Zhang H., et al. Loss of TIGAR induces oxidative stress and meiotic defects in oocytes from obese mice. Mol. Cell Proteomics. 2018;17:1354–1364. doi: 10.1074/mcp.RA118.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P., Gaffrey M.J., Zhu Y., Chrisler W.B., Fillmore T.L., Yi L., et al. Carrier-assisted single-tube processing approach for targeted proteomics analysis of low numbers of mammalian cells. Anal. Chem. 2019;91:1441–1451. doi: 10.1021/acs.analchem.8b04258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 28.Li L., Zhu S., Shu W., Guo Y., Guan Y., Zeng J., et al. Characterization of metabolic patterns in mouse oocytes during meiotic maturation. Mol. Cell. 2020;80:525–540.e9. doi: 10.1016/j.molcel.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesenti C., Navone S.E., Guarnaccia L., Terrasi A., Costanza J., Silipigni R., et al. The genetic landscape of human glioblastoma and matched primary cancer stem cells reveals intratumour similarity and intertumour heterogeneity. Stem Cell Int. 2019;2019 doi: 10.1155/2019/2617030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 31.Postma M., Goedhart J. PlotsOfData-A web app for visualizing data together with their summaries. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., Zheng Y., Li X., Hu X., Qi F., Luo J. Genome-wide mutation profiling and related risk signature for prognosis of papillary renal cell carcinoma. Ann. Transl. Med. 2019;7:427. doi: 10.21037/atm.2019.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., et al. The human transcription factors. Cell. 2018;175:598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 35.Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucl. Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medvedeva Y.A., Lennartsson A., Ehsani R., Kulakovskiy I.V., Vorontsov I.E., Panahandeh P., et al. EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database. 2015;2015 doi: 10.1093/database/bav067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bult C.J., Blake J.A., Smith C.L., Kadin J.A., Richardson J.E., Mouse Genome Database G. Mouse genome database (MGD) 2019. Nucl. Acids Res. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolaczyk E.D., Csárdi G. 2nd Ed. Springer Nature Switzerland AG; Switzerland AG: 2020. Statistical Analysis of Network Data with R. [Google Scholar]
- 39.Yu B., Doni Jayavelu N., Battle S.L., Mar J.C., Schimmel T., Cohen J., et al. Single-cell analysis of transcriptome and DNA methylome in human oocyte maturation. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes J.M., Ross P.J. Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip. Rev. RNA. 2016;7:71–89. doi: 10.1002/wrna.1316. [DOI] [PubMed] [Google Scholar]
- 41.Santos A., Wernersson R., Jensen L.J. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucl. Acids Res. 2015;43:D1140–1144. doi: 10.1093/nar/gku1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Y.F., Zhai R., Liu X.M., Khan H.A., Zhen Y.H., Huo L.J. SUMO-1 plays crucial roles for spindle organization, chromosome congression, and chromosome segregation during mouse oocyte meiotic maturation. Mol. Reprod. Dev. 2014;81:712–724. doi: 10.1002/mrd.22339. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 44.Groettrup M., Soza A., Eggers M., Kuehn L., Dick T.P., Schild H., et al. A role for the proteasome regulator PA28alpha in antigen presentation. Nature. 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 45.Yano M., Koumoto Y., Kanesaki Y., Wu X., Kido H. 20S proteasome prevents aggregation of heat-denatured proteins without PA700 regulatory subcomplex like a molecular chaperone. Biomacromolecules. 2004;5:1465–1469. doi: 10.1021/bm049957a. [DOI] [PubMed] [Google Scholar]
- 46.Rut W., Drag M. Human 20S proteasome activity towards fluorogenic peptides of various chain lengths. Biol. Chem. 2016;397:921–926. doi: 10.1515/hsz-2016-0176. [DOI] [PubMed] [Google Scholar]
- 47.Driscoll G.L., Tyler J.P., Hangan J.T., Fisher P.R., Birdsall M.A., Knight D.C. A prospective, randomized, controlled, double-blind, double-dummy comparison of recombinant and urinary HCG for inducing oocyte maturation and follicular luteinization in ovarian stimulation. Hum. Reprod. 2000;15:1305–1310. doi: 10.1093/humrep/15.6.1305. [DOI] [PubMed] [Google Scholar]
- 48.Couvillion S.P., Zhu Y., Nagy G., Adkins J.N., Ansong C., Renslow R.S., et al. New mass spectrometry technologies contributing towards comprehensive and high throughput omics analyses of single cells. Analyst. 2019;144:794–807. doi: 10.1039/c8an01574k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L., Vertes A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew. Chem. 2018;57:4466–4477. doi: 10.1002/anie.201709719. [DOI] [PubMed] [Google Scholar]
- 50.Budnik B., Levy E., Harmange G., Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:161. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu D., Dong X., Xiong M., Xiong T., Huang B., Zeng D., et al. Can the peak E2/follicle ratio be a quantitative indicator of pregnancy outcomes following assisted reproductive cycles? A retrospective study. Int. J. Clin. Exp. Med. 2015;8:10964–10970. [PMC free article] [PubMed] [Google Scholar]
- 52.Mittal S., Gupta P., Malhotra N., Singh N. Serum estradiol as a predictor of success of in vitro fertilization. J. Obstet. Gynaecol. India. 2014;64:124–129. doi: 10.1007/s13224-013-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Inany H.G., Youssef M.A., Aboulghar M., Broekmans F., Sterrenburg M., Smit J., et al. Gonadotrophin-releasing hormone antagonists for assisted reproductive technology. Cochrane Database Syst. Rev. 2011;4:CD001750. doi: 10.1002/14651858.CD001750.pub3. [DOI] [PubMed] [Google Scholar]
- 54.Martinez F., Racca A., Rodriguez I., Polyzos N.P. Ovarian stimulation for oocyte donation: a systematic review and meta-analysis. Human Reprod. Update. 2021;27:673–696. doi: 10.1093/humupd/dmab008. [DOI] [PubMed] [Google Scholar]
- 55.Acevedo B., Gomez-Palomares J.L., Ricciarelli E., Hernandez E.R. Triggering ovulation with gonadotropin-releasing hormone agonists does not compromise embryo implantation rates. Fertil. Steril. 2006;86:1682–1687. doi: 10.1016/j.fertnstert.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 56.Galindo A., Bodri D., Guillen J.J., Colodron M., Vernaeve V., Coll O. Triggering with HCG or GnRH agonist in GnRH antagonist treated oocyte donation cycles: a randomised clinical trial. Gynecol. Endocrinol. 2009;25:60–66. doi: 10.1080/09513590802404013. [DOI] [PubMed] [Google Scholar]
- 57.Melo M., Busso C.E., Bellver J., Alama P., Garrido N., Meseguer M., et al. GnRH agonist versus recombinant HCG in an oocyte donation programme: a randomized, prospective, controlled, assessor-blind study. Reprod. Biomed. Online. 2009;19:486–492. doi: 10.1016/j.rbmo.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Sismanoglu A., Tekin H.I., Erden H.F., Ciray N.H., Ulug U., Bahceci M. Ovulation triggering with GnRH agonist vs. hCG in the same egg donor population undergoing donor oocyte cycles with GnRH antagonist: a prospective randomized cross-over trial. J. Assist. Reprod. Genet. 2009;26:251–256. doi: 10.1007/s10815-009-9326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li F., Zhao H., Su M., Xie W., Fang Y., Du Y., et al. HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to its 3' UTR. EBioMedicine. 2019;45:208–219. doi: 10.1016/j.ebiom.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez A., Briley S.M., Patton B.K., Tripurani S.K., Rajapakshe K., Coarfa C., et al. Loss of the E2 SUMO-conjugating enzyme Ube2i in oocytes during ovarian folliculogenesis causes infertility in mice. Development. 2019;146 doi: 10.1242/dev.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howell C.Y., Bestor T.H., Ding F., Latham K.E., Mertineit C., Trasler J.M., et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 62.Jimenez R., Melo E.O., Davydenko O., Ma J., Mainigi M., Franke V., et al. Maternal SIN3A regulates reprogramming of gene expression during mouse preimplantation development. Biol. Reprod. 2015;93:89. doi: 10.1095/biolreprod.115.133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yatsenko S.A., Rajkovic A. Genetics of human female infertilitydagger. Biol. Reprod. 2019;101:549–566. doi: 10.1093/biolre/ioz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Condic M.L. The role of maternal-effect genes in mammalian development: are mammalian embryos really an exception? Stem Cell Rev. Rep. 2016;12:276–284. doi: 10.1007/s12015-016-9648-6. [DOI] [PubMed] [Google Scholar]
- 65.Yatsenko S.A., Rajkovic A. Genetics of human female infertility. Biol. Reprod. 2019;101:549–566. doi: 10.1093/biolre/ioz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B.B. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data have been deposited in the ProteomeXchange Consortium via the proteomics identifications (PRIDE) database (Project accession: PXD023366).