Abstract
Salmonella enterica resistant to fluoroquinolones (FQs) and extended-spectrum cephalosporins (ESCs) has been deemed a high-priority pathogen by the WHO. Salmonella enterica serovar Saintpaul (S. Saintpaul) co-resistant to ESCs and FQs and harboring corresponding resistance genes (blaCTX–M–55 and qnrS1) have been previously reported. However, they have not been reported in China. Moreover, the genetic context and transferability of ESCs and FQs resistance genes in S. Saintpaul remain obscure. This study is the first study to characterize a multidrug-resistant (MDR) S. Saintpaul isolate (16Sal016) harboring plasmid-mediated blaCTX–M–55 and qnrS1 genes recovered from weever fish in China. The whole genome short- and long-read sequencing results identified the presence of 15 acquired antibiotic resistance genes encoding resistance to nine classes of antibiotics, as well as abundant mobile genetic elements residing on a 259,529 bp IncHI2 plasmid. The blaCTX–M–55 and qnrS1 genes were located in a 12,865 bp region, IS26-orf-orf-ISKpn19-qnrS1-IS3-Tn3-orf-blaCTX–M–55-ISEc9-orf-IS26. Similar structures have been identified in various bacterial species, indicating a high transferability of blaCTX–M–55 and qnrS1 genes within this gene cluster. The plasmid was found to be transferable to Escherichia coli (E. coli) J53 by conjugation and resulted in the acquisition of multiple resistances by the transconjugants. Genome sequence comparisons by core genome multilocus sequence typing (cgMLST) based on global 2,947 S. Saintpaul isolates indicated that strain 16Sal016 was epidemiologically linked with an isolate from the United Kingdom (UK). Our findings suggest that plasmids and IS26-mediated mobile genetic elements are carriers of blaCTX–M–55 and qnrS1 genes in S. Saintpaul, and highlight their potential transmission, which needs continuous investigations.
Keywords: S. Saintpaul, plasmid, bla CTX–M–55 , qnrS1, fish
Introduction
Salmonella is a leading cause of foodborne illness worldwide (Chen et al., 2020). Over the past two decades, multidrug-resistant (MDR) Salmonella strains have emerged in clinical settings and food products. These strains, especially those resistant to extended-spectrum cephalosporins (ESCs) and fluoroquinolones (FQs), are concerning because they encode resistance to critically important antibiotic classes commonly used to treat severe salmonellosis (Lu et al., 2019; Zhang et al., 2019).
Salmonella enterica serovar Saintpaul (S. Saintpaul) is an important gut pathogen, which has been reported to be associated with several outbreaks in many countries (Klontz et al., 2010; Hayford et al., 2015). It was among the top 20 most common serovars observed in the United States (CDC, 2021), and was reported as the second most common cause of outbreaks in Australia, 2001–2016 (Ford et al., 2018), as well as was noted to be among the top five most common serovars in Singapore in 2015 and 2016 (Aung et al., 2020). In China, S. Saintpaul has been reported in pets, seafood, poultry, and samples of human origin (Gong et al., 2014; Qi et al., 2019; Zhan et al., 2019; Wang et al., 2020; Wei et al., 2020). MDR resistance has been reported in S. Saintpaul isolates; however, co-resistance to ESCs and FQs is uncommon and has not been reported in China (Nadimpalli et al., 2019; Octavia et al., 2021). In China, various Salmonella serovars have been reported to acquire steadily increasing co-resistances to clinically important antibiotics (cefotaxime and ciprofloxacin) (Zhan et al., 2019). Consequently, it is important to pay concern to strains of S. Saintpaul acquiring those resistances as they may greatly limit current treatment options.
In this study, we characterized an ESCs- and FQs-resistant S. Saintpaul isolate recovered from a weever fish in Guangzhou, China, and analyzed the genetic context of corresponding resistance genes, their transferability, as well as the origin, in order to gain insight into their public health impact.
Materials and methods
Strains isolation and identification
During our routine surveillance of foodborne pathogens on various food products, Salmonella isolate, named GSJ/2016-Sal.-016 (hereafter 16Sal016), was recovered from a weever fish (Lateolabrax japonicus) in the Guangdong Province, China. The fish sample was collected from a retail market in Guangzhou, Southern China. The isolate was first identified by biochemical confirmation using API 20E test identification test strips (bioMérieux, France), and then 16S ribosomal RNA (rRNA) gene sequencing using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′ and 1,492R (5′-GGCTACCTTGTTACGACTT-3′). The serotype was determined by the slide agglutination test using Salmonella antisera (SSI Diagnostica, Denmark) according to the White–Kauffmann–Le Minor scheme (Issenhuth-Jeanjean et al., 2014).
Escherichia coli (E. coli) ATCC 25922 was used as the quality control strain for antimicrobial susceptibility testing and E. coli J53 (sodium azide resistant) was used as the recipient strain for conjugation. Strains were routinely grown for 12–24 h at 37°C in Luria–Bertani (LB) broth or agar (Guangdong Huankai Microbial Science and Technology Corporation Ltd., Guangzhou, China) supplemented with antibiotics when appropriate.
Antibiotic susceptibility testing
The isolate was tested for susceptibility to a panel of antimicrobial drugs by disk diffusion method (CLSI, 2017), including amikacin (30 μg), ampicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), ampicillin-sulbactam sodium (10/10 μg), azithromycin (15 μg), aztreonam (30 μg), cephalosporins IV [cefepime (30 μg)], cephalosporins III [cefotaxime (30 μg) and ceftazidime (30 μg)], cephalosporins II [cefoxitin (30 μg) and cefuroxime (30 μg)], cephalosporins I [cefazolin (30 μg)], chloramphenicol (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), ertapenem (10 μg), fosfomycin (200 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), netilmicin (30 μg), piperacillin (100 μg), streptomycin (10 μg), tigecycline (15 μg), tetracycline (30 μg), tobramycin (10 μg), and trimethoprim/sulfamethoxazole (23.75/1.25 μg) (Hangzhou Microbial Reagent Corporation Ltd., China). Minimal inhibitory concentrations (MICs) of the isolate to ciprofloxacin, nalidixic acid, polymyxin B/colistin, and cefotaxime (Sigma-Aldrich, St. Louis, MO, United States) were determined by broth microdilution method (CLSI, 2017). Production of extended-spectrum β-lactamase (ESBL) was confirmed by the disk diffusion clavulanate inhibition test using ceftazidime and cefotaxime (CLSI, 2017). Results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (The European Committee on Antimicrobial Susceptibility Testing [ECAST], 2017). The diameter of disks was presented as mean values from replications with SEs. The reference strain E. coli ATCC 25922 served as quality control.
An MDR isolate is defined as an isolate demonstrating resistance to three or more antibiotics belonging to different antibiotic classes (Magiorakos et al., 2012). All the measurements were performed in duplicates and each experiment was repeated three times.
Whole-genome sequencing and annotation
Genomic DNA was extracted using a commercial DNA extraction kit (Magen, Guangzhou, China) following the manufacturer’s recommendations. The whole genome of the isolate was sequenced by combining short-read and long-read technologies. The first set of reads was obtained on the Illumina Hiseq × 10 platform with a 150-bp paired-end reads approach (MajorBio Corporation, Shanghai, China). The second set of reads was obtained on a MinION Nanopore Sequencer (Oxford Nanopore, Oxford, United Kingdom), and a library was prepared with the 1 D ligation approach. The genome of the isolate was assembled de novo using both the short- and long-reads with SPAdes version 3.14.0 (Bankevich et al., 2012) and Unicycler hybrid assembler version 0.4.8 (Wick et al., 2017), and annotated by Prokka version 1.14.6 (Seemann, 2014).
Clonal analysis was assessed by multilocus sequence typing (MLST) version 2.0.1 The presence of acquired antibiotic resistance genes and mutations in the quinolone resistance-determining regions (QRDR) (gyrA, gyrB, parC, and parE) were assessed by ResFinder 4.1 (Zankari et al., 2012), and, subsequently, the genes were extracted from the genome sequences and further confirmed by BLAST nucleotide (BLASTn).2 PlasmidFinder version 2.1 and pMLST version 0.1.0 were used to identify plasmid replicon type and sequence type (ST) (Carattoli and Hasman, 2020). The complete plasmid sequence was BLASTn against the nr database with default parameters. Highly similar plasmids were selected for comparison. The map was generated by BRIG 0.95-dev.0004 (Alikhan et al., 2011).
Phylogenetic analysis of the genomic sequences
In order to assess the relatedness of 16Sal016 with other S. Saintpaul strains from different sources and countries, we retrieved all the 2,947 genome sequences of S. Saintpaul that have been released from EnteroBase databases (accessed on 8 March 2022) and performed core genome multilocus sequence typing (cgMLST) (cgMLST scheme available on EnteroBase).3 A minimum spanning tree was created from cgMLST allelic differences in EnteroBase using GrapeTree with the RapidNJ algorithm (Zhou et al., 2020).
Conjugation
The transferability of the plasmid was assessed by performing the conjugation experiment using sodium azide-resistant E. coil strain J53 as a recipient strain by solid mating on a filter (Whatman, Maidstone, United Kingdom) (Hammerum et al., 2016). Briefly, recipient and donor strains were cultured overnight and harvested, washed with saline, mixed together in a ratio of 1:1, and then spotted onto a 0.45-μm pore size filter (Millipore) on LB plates. They were also spotted individually on LB plates as controls. After overnight incubation at 37°C, mating spots were washed, resuspended in saline, and then serially diluted and plated on LB media containing 150 μg/ml sodium azide and 4, 8, or 16 μg/ml of cefotaxime to select transconjugants. Control spots were transferred to the same selective media to make sure that no growth was observed.
The conjugation frequency was calculated as the ratio of transconjugants over the number of recipients. The transfer of the plasmid was confirmed by PCR targeting blaCTX–M–55 gene with primer CTX-M-55-F (5′- AAGCACGTCAATGGGACGAT -3′) and CTX-M-55-R (5′- CCTTAGGTTGAGGCTGGGTG -3′), as well as the uidA household gene of E. coli with the primers U-F (5′-TGGAATTTCGCCGATTTTGC-3′) and U-R (5′-ATTGTTTGCCTCCCTGCTGC-3′) (Heijnen and Medema, 2016). Further, the plasmid DNA was extracted from a selected transconjugant by a commercial plasmid extraction kit (Magen, Guangzhou, China) following the manufacturer’s recommendations, and sequenced on the Illumina Hiseq platform (MajorBio Corporation, Shanghai, China).
Nucleotide sequence accession number
The Illumina sequence data were deposited in the Enterobase database under the barcode numbers SAL_KB2591AA. The assembly genome sequence of S. Saintpaul 16Sal016 was deposited in the National Center for Biotechnology Information (NCBI) database under the Biosample number SAMN12070262.
Results
Identification of Salmonella
The isolate 16Sal016 was confirmed as S. Saintpaul (4,12:e,h:1,2) by biochemical confirmation, 16S rRNA gene sequencing, and serotyping.
Antibiotic susceptibility
Antibiotic susceptibility testing showed that the isolate 16Sal016 was resistant to ampicillin, azithromycin, aztreonam, cefazolin, cefuroxime, cefotaxime, ceftazidime, chloramphenicol, doxycycline, gentamicin, nalidixic acid, streptomycin, tetracycline, and trimethoprim-sulfamethoxazole, ciprofloxacin (based on the EUCAST clinical breakpoint), intermediate resistant to ampicillin-sulbactam, and produced ESBL. The isolate exhibited MIC values of ciprofloxacin, nalidixic acid, and cefotaxime for 1, 128, and 128 mg/l, respectively.
General genomic features
The complete genome sequence of S. Saintpaul (16Sal016) contained a circular 4,785,442 bp chromosome with G + C content of 52.2%, and a 259,529 bp IncHI2 plasmid, denoted as pSal016. There were 4,736 predicted coding regions (CDs) in the whole-genome sequence. MLST analysis showed that 16Sal016 belonged to ST27.
A total of 15 acquired resistance genes were identified in 16Sal016 by ResFinder, which encodes resistance to nine different antimicrobial classes, including aminoglycoside, beta-lactam, FQs, macrolide-lincosamide-streptogramin B, phenicol, rifamycin, sulfonamide, tetracycline, and trimethoprim (Table 1). All the antimicrobial resistance genes were located on the plasmid.
TABLE 1.
The antibiotic resistance profile and drug resistance genes of S. Saintpaul 16Sal016, the selected transformant (16Sal016T), and the recipient (E. coli J53).
| MICa (mg/L) | Antibiotic resistance genes on plasmid | |||
|
|
||||
| Isolate | CTX | CIP | NAL | |
| 16Sal016 | 128 | 1 | 128 | blaLAP–2, blaTEM–1B, blaCTX–M–55, qnrS1, mph(A), lnu(F), tet(A), aph(6)-Id, aadA22, aac(3)-IId, aph(3′)-Ia, sul3, floR, arr-2, dfrA14 |
| 16Sal016T | 128 | 1 | 128 | blaLAP–2, blaTEM–1B, blaCTX–M–55, qnrS1, mph(A), lnu(F), tet(A), aph(6)-Id, aadA22, aac(3)-IId, aph(3′)-Ia, sul3, floR, arr-2, dfrA14 |
| E. coli J53 | <0.5 | 0.015 | 0.25 | |
aCTX, cefotaxime; CIP, ciprofloxacin; NAL, nalidixic acid.
Comparative analysis of plasmid pSal006
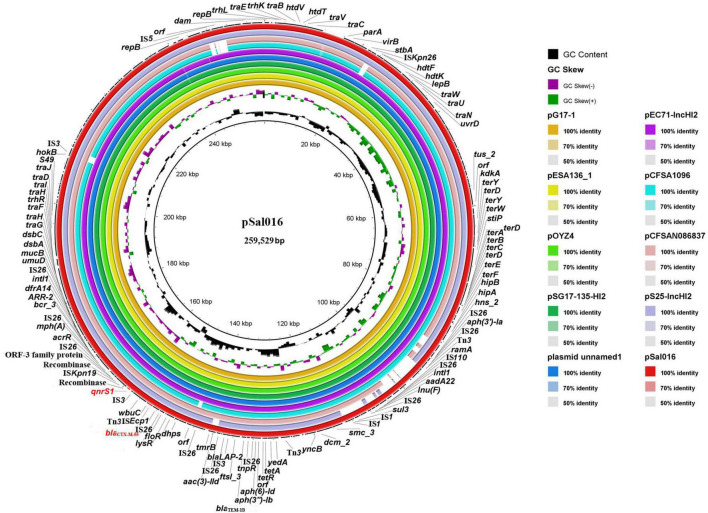
The pSal016 is a 259,529 bp plasmid, with 112 predicted coding regions (CDs) and an average GC content of 46.7%. Replicon typing of the plasmid by PlasmidFinder and pMLST showed that pSal016 was an IncHI2 plasmid and belongs to ST2. The plasmid includes the core region (traEBVC, traWUN, and traGHFIDJ), which served as the plasmid backbone and is involved in plasmid replication, horizontal transfer, stability, and maintenance functions (Figure 1; Call et al., 2010). Various antimicrobial resistance genes were identified on the plasmid, including aph(6)-Id, aadA22, aac(3)-IId, and aph(3′)-Ia for aminoglycoside, mph(A) for azithromycin, blaLAP–2, blaTEM–1B, and blaCTX–M–55 encoding beta-lactam resistance, qnrS1 for FQs resistance, lnu(F) for lincosamide, floR for phenicol, arr-2 for rifampicin, sul3 for sulfonamide, tet(A) for tetracycline, and dfrA14 for trimethoprim (Table 1). In addition, copies of transposases and recombinase/integrase family protein were observed on this plasmid (Figure 1).
FIGURE 1.
Sequence comparison of plasmid pSal016 identified in S. Saintpaul 16Sal016 with similar plasmids in BRIG. GC content and GC skew of pSal016 are depicted in the inner rings. The genes located on pSal016 are annotated at the outside black ring. The blaCTX–M–55 and qnrS1 genes are marked red.
BLAST nucleotide comparison of the entire plasmid sequence to microbial sequences in GenBank identified six highly similar plasmids with 99–100% sequence coverage and more than 99.9% nucleotide identity and several similar plasmids sharing similar backbones (Figure 1 and Table 2). These plasmids are all the IncHI2 types and belong to ST2, except pCFSA1096 and pCFSAN086837, which belong to ST3. Importantly, the highly similar plasmids were from different hosts than S. Saintpaul in different places and years, and most of them were from China (Table 2).
TABLE 2.
Characteristics of plasmids highly similar to pSal016.
| Plasmid name | Replication type | pMLST | Length (bp) | Host | Source | Country (region) | Year | Coverage (%) | Identity (%) | Accession No. |
| pSal016 | IncHI2 | 2 | 259,529 | S. Saintpaul | Fish | China (Guangzhou) | 2016 | – | – | SAMN12070262 |
| pG17-1 | IncHI2 | 2 | 264,084 | E. hormaechei | Duck | China (Henan) | 2021 | 100% | 99.98% | CP079936.1 |
| pESA136_1 | IncHI2 | 2 | 266,043 | E. albertii | Poultry | China (Shanxi) | 2018 | 100% | 99.99% | CP070297.2 |
| pOYZ4 | IncHI2 | 2 | 257,945 | Salmonella sp. | Duck | China (Guangzhou) | – | 100% | 99.96% | MN539018.1 |
| pSG17-135-HI2 | IncHI2 | 2 | 263,947 | S. Agona | Australian silver gull chick | Australia | 2017 | 100% | 99.98% | CP048776.1 |
| unnamed1 | IncHI2 | 2 | 263,947 | Salmonella sp. | Chicken | China (Jiangxi) | 2021 | 100% | 99.98% | CP084217.1 |
| pEC71-IncHI2 | IncHI2 | 2 | 269,592 | E. coli | Human urine | China (Guangzhou) | 2020 | 99% | 99.98% | CP085623.1 |
| pCFSA1096 | IncHI2 | 3 | 297,348 | S. enterica | Food | China (Hubei) | 2015 | 97% | 100% | CP033347.2 |
| pCFSAN086837 | IncHI2 | 3 | 255,725 | S. Newport | Chicken | Viet Nam | 2017 | 94% | 99.96% | CP039438.1 |
| pS25-IncHI2 | IncHI2 | 2 | 237,710 | S. Saintpaul | Human stool | China (Guangzhou) | 2014 | 93% | 100% | CP085697.1 |
| pPJM1 | IncHI2 | 2 | 263,947 | Salmonella sp. | Chicken | China (Guangzhou) | 2017 | 90% | 99.98% | MN539017.1 |
Genetic context of extended-spectrum cephalosporins and fluoroquinolones resistance genes
The blaCTX–M–55 and qnrS1 genes were located on a 12,865 bp IS26-mediated composite transposon unit IS26-orf-orf-ISKpn19-qnrS1-IS3-Tn3-orf-blaCTX–M–55-ISEc9-orf-IS26. The identical genetic structure was also found on plasmid pEC71-IncHI2 (GenBank accession number CP085623.1) from E. coli isolated from a human urine sample in China. Similar structures lacking one of the flanked IS26 were identified in many other plasmids from different bacterial species, including E. coli, Escherichia albertii, Enterobacter hormaechei, Salmonella spp., S. Agona, S. enterica, S. Newport, and S. Saintpaul. The core structure qnrS1-orf-IS3-Tn3-blaCTX–M–55 was also observed in many different bacterial species, which can be mediated by IS26 or ISKpn19 (Figure 2). In addition, for comparison, we assembled three S. Saintpaul isolates (F36, F3, and F47) reported containing blaCTX–M–55 and qnrS1 genes (Nadimpalli et al., 2019; Figure 2).
FIGURE 2.
Genetic environment of blaCTX–M–55 and qnrS1 genes from S. Saintpaul isolate 16Sal016 and different bacterial species. The arrows indicate open reading frames. Light gray shading denotes homology regions. Strains from China are in blue font.
Horizontal transfer of the plasmid
Polymerase chain reaction and sequencing results confirmed the successful transfer of the plasmid pSal016 from S. Saintpaul 16Sal016 to a plasmid-free recipient E. coli J53. Antimicrobial susceptibility testing revealed the acquisition of the plasmid by E. coli caused at least 256-fold increase for cefotaxime, 512-fold increase for nalidixic acid, and about 66-fold increase for ciprofloxacin (Table 1). The conjugation rate was 5.4 × 10–6 ± 0.6 transconjugant per recipient cell.
Phylogenetic analysis
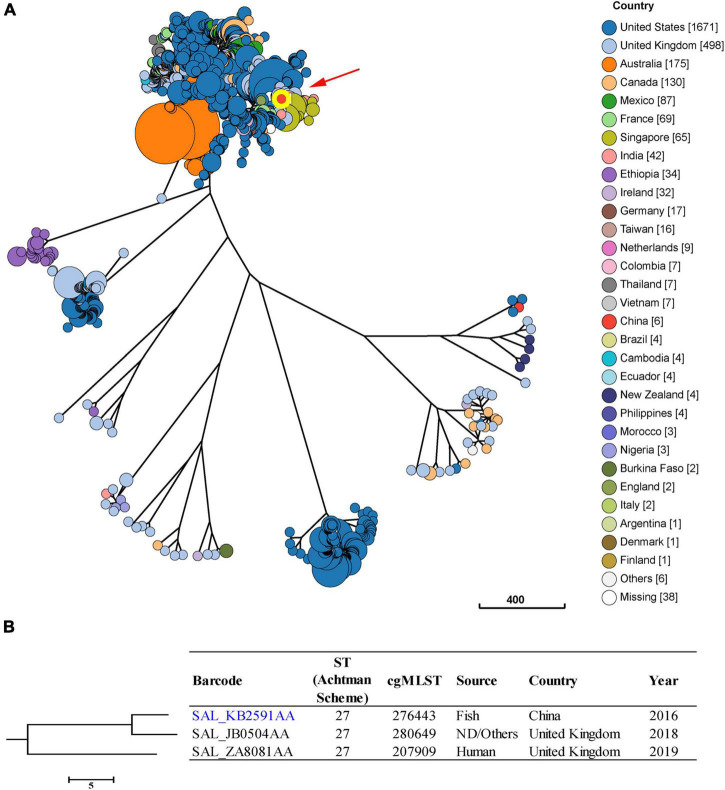
Isolates of S. Saintpaul were reported to be putatively polyphyletic and characterized by multilineages (Yin et al., 2020). In this study, we found seven major clusters among the 2,947 S. Saintpaul isolates from different countries (Figure 3A). These isolates belong to 47 classical MLST types, with the most frequent being ST50 (43.4%), ST27 (29.6%), and ST95 (11.3%) (Supplementary Figure 1).
FIGURE 3.
Phylogenetic analysis of 2,947 S. Saintpaul isolates from different sources and countries. (A) A minimum-spanning tree based on cgMLST analysis. The position of S. Saintpaul isolate 16Sal016 is indicated by red arrow and highlighted circle. Each circle represents a cgMLST group and the size of the circle is proportional to the number of isolates in that group. (B) Detailed information of strains in the branch contained 16Sal016. The isolate 16Sal016 is marked blue.
Core genome multilocus sequence typing and phylogenetic analysis showed that the isolate 16Sal016 harbored a unique cgST profile (Figure 3B and Supplementary Table 1), and displayed the closest relationship to an isolate (Barcode: SAL_JB0504AA) from the United Kingdom in 2018 (Figure 3B). The cgMLST results differentiated the isolate 16Sal016 from the closest related isolate up to the HC5 level (a maximum of 5 cgMLST allelic variations). Isolates belonging to the same HC5 cluster (up to 5 allelic variations between strains) were considered highly probably epidemiologically linked (Bonifait et al., 2021). Therefore, it is inferred that isolate 16Sal016 was highly epidemiologically linked with the isolate from the United Kingdom.
Discussion
Extended-spectrum β-lactamase-producing and FQs-resistant strains of Salmonella have been reported throughout the world, constituting a great public health concern (Castellanos et al., 2018; Nadimpalli et al., 2019; Zhang et al., 2019; Chattaway et al., 2021; Herdman et al., 2021). Contaminated animal-derived food products are an important route of transmission of Salmonella from animals to humans (Zhu et al., 2017; Zhang et al., 2018; Wang et al., 2020). In this study, we characterized a Chinese foodborne MDR S. Saintpaul isolate, which was found to be resistant to a number of antibiotics, and we further analyzed the genetic context of the corresponding resistance genes, their transferability, and tracked its source by global phylogenetic analysis.
Multidrug-resistant S. Saintpaul has been widely reported in many countries (Haq et al., 2017; Ding et al., 2018; Nadimpalli et al., 2019). However, concurrent resistance to ESCs and FQs has only been reported in a S. Saintpaul isolate from foals in Pakistan (Haq et al., 2017) and three S. Saintpaul isolates from fish in Cambodia (Nadimpalli et al., 2019). In this study, the isolated S. Saintpaul strain was found to be co-resistant to ESCs and FQs, as well as multiple antimicrobials, including the most prevalent ACSSuT MDR profile (defined as resistance to ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline in S. typhimurium) (Mølbak et al., 1999). To the best of our knowledge, this is the first time a foodborne S. Saintpaul isolates in China co-resistant to ESCs and FQs has been reported.
The ESCs and FQs resistance in 16Sal016, encoded by blaCTX–M–55 and qnrS1 genes, respectively, were located on an IncHI2 plasmid. Several IncHI2 plasmids carried blaCTX–M–55 gene were found to be closely related to pSal016 and most of them belong to pMLST type 2, suggesting that they are highly similar. It is worth noticing that these plasmids were from different bacterial species and most of them were from China, indicating that the circulating of IncHI2-type plasmid contributes to the increasing prevalence of blaCTX–M–55 and qnrS1 genes in China. Importantly, a similar plasmid pS25-IncHI2 was observed to reside in a clinical S. Saintpaul isolate collected in the same region, but different year in China. However, phylogenetic analysis of global S. Saintpaul isolates based on cgMLST did not reveal any epidemiological links between them, indicating a likely transmission of this plasmid among different S. Saintpaul isolates. As evaluated in the present study, pSal016 was transferable to E. coli; the conjugation ability of pSal016 may likely contribute to the acquisition of MDR among other bacterial species, which need continuous investigations.
Salmonella resistance to ESCs is reported to be associated with cross-resistance to FQs (Lu et al., 2019; Zhang et al., 2019). In our previous study, we identified an IS26-mediated composite transposon IS26-qnrS1-IS3-Tn3-orf-blaCTX–M–55-ISEcp1-IS26 that might contribute to the development of co-resistance to ESCs and FQs (Li et al., 2021). In the present study, we found a novel IS26-mediated composite transposon IS26-orf-orf-ISKpn19-qnrS1-IS3-Tn3-orf-blaCTX–M–55-ISEc9-orf-IS26, as a carrier for blaCTX–M–55 and qnrS1 genes in S. Saintpaul. An identical structure was also found in E. coli, suggesting that it is transferable. Similar structures were widely distributed among different bacterial species. Among them, a uniform unit (qnrS1-IS3-Tn3- orf-blaCTX–M–55) was observed as a core structure, which can be mediated by IS26 and ISKpn19 to transfer.
The global phylogenetic analysis showed that the strain 16Sal016 isolated in 2016 was highly epidemiologically linked with an isolate from the United Kingdom in 2018, suggesting that they might come from the same source. The identification of IS26-mediated composite transposon and transferable IncHI2 plasmid carrying blaCTX–M–55 and qnrS1 genes in Chinese S. Saintpaul represent potential clinical and food safety issues and need to be monitored, since they may transmit to humans through the food chain and may lead to reduced susceptibility of Salmonella to frontline drugs of choice for treating severe Salmonella infections, such as ESCs and FQs.
Conclusion
To summarize, this study reports for the first time a foodborne strain of S. Saintpaul in China co-resistant to ESCs and FQs and carried blaCTX–M–55 and qnrS1 genes. The blaCTX–M–55 and qnrS1 genes were located in a novel IS26-mediated composite transposon IS26-orf-orf-ISKpn19-qnrS1-IS3-Tn3-orf-blaCTX–M–55-ISEc9-orf-IS26 on a transferable IncHI2 plasmid. The transfer of the plasmid and the IS26-mediated composite transposon may contribute to the dissemination of blaCTX–M–55 and qnrS1 genes among different bacterial species and accelerate the development of isolates co-resistant to ESCs and FQs, and this warrants continuous investigations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LL performed the experiment and wrote the manuscript. RO revised the manuscript. JX contributed to reagents, materials, and analysis tools. HM conceptualized and designed the study. SP and LS provided the funding. All authors contributed to the article and approved the submitted version.
Footnotes
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers: 31901789 and 32001796), the Natural Science Foundation of Guangdong Province (grant numbers: 2022A1515011685 and 2020A1515010218), the Basic Research Project of Guangzhou (grant number: 202002030630020049), and Jiangsu Innovative and Enterpreneurial Talent Programme (JSSCBS20211458).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.899062/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Z. N., Beatson S. A. (2011). BLAST Ring Image Generator BRIG: Simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K. T., Khor W. C., Octavia S., Ye A., Leo J., Chan P. P., et al. (2020). Distribution of Salmonella serovars in humans, foods, farm animals and environment, companion and wildlife animals in Singapore. Int. J. Environ. Res. Publ. Health 17:5774. 10.3390/ijerph17165774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifait L., Thépault A., Baugé L., Rouxel S., Le Gall F., Chemaly M. (2021). Occurrence of Salmonella in the cattle production in France. Microorganisms 9:872. 10.3390/microorganisms9040872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call D. R., Singer R. S., Meng D., Broschat S. L., Orfe L. H., Anderson J. M., et al. (2010). blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54 590–596. 10.1128/AAC.00055-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Hasman H. (2020). PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in Whole-Genome sequencing WGS. Methods Mol. Biol. 2075 285–294. 10.1007/978-1-4939-9877-7_20 [DOI] [PubMed] [Google Scholar]
- Castellanos L. R., van der Graaf-van B. L., Donado-Godoy P., Leon M., Clavijo V., Arevalo A., et al. (2018). Genomic characterization of extended-spectrum cephalosporin-resistant Salmonella enterica in the colombian poultry chain. Front. Microbiol. 9:2431. 10.3389/fmicb.2018.02431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2021). National Antimicrobial Resistance Monitoring System NARMS Now: Human Data. Atlanta. Georgia: Department of Health and Human Services. Available online at: https://www.cdc.gov/narmsnow. [Accessed Nov 19, 2021]. [Google Scholar]
- Chattaway M. A., Gentle A., Nair S., Tingley L., Day M., Mohamed I., et al. (2021). Phylogenomics and antimicrobial resistance of Salmonella Typhi and Paratyphi A, B and C in England, 2016-2019. Microb. Genom. 78:000633. 10.1099/mgen.0.000633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Yang C., Dong N., Xie M., Ye L., Chan E., et al. (2020). Evolution of ciprofloxacin resistance-encoding genetic elements in Salmonella. Msystems 5:e1234–e1220. 10.1128/mSystems.01234-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI. [Google Scholar]
- Ding Y., Zwe Y. H., Chin S. F., Kohli G. S., Drautz-Moses D. I., Givskov M., et al. (2018). Characterization of a novel multidrug resistance plasmid pSGB23 isolated from Salmonella enterica subspecies enterica serovar Saintpaul. Gut Pathog. 10:20. 10.1186/s13099-018-0249-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L., Moffatt C. R. M., Fearnley E., Miller M., Gregory J., Sloan-Gardner T. S., et al. (2018). The epidemiology of Salmonella enterica outbreaks in Australia, 2001–2016. Front. Sustain. Food Syst. 2:86. 10.3389/fsufs.2018.00086 [DOI] [Google Scholar]
- Gong J., Zhang J., Xu M., Zhu C., Yu Y., Liu X., et al. (2014). Prevalence and fimbrial genotype distribution of poultry Salmonella isolates in China 2006 to 2012. Appl. Environ. Microbiol. 802 687–693. 10.1128/AEM.03223-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum A. M., Hansen F., Nielsen H. L., Jakobsen L., Stegger M., Andersen P. S., et al. (2016). Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 71 3117–3124. 10.1093/jac/dkw289 [DOI] [PubMed] [Google Scholar]
- Haq I., Durrani A. Z., Khan M. S., Mushtaq M. H., Ahmad I. (2017). Study of antimicrobial resistance and physiological biomarkers with special reference to Salmonellosis in diarrheic foals in Punjab. Pakistan. Acta. tropica. 176 144–149. 10.1016/j.actatropica.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Hayford A. E., Brown E. W., Zhao S., Mammel M. K., Gangiredla J., Abbott J. W., et al. (2015). Genetic and resistance phenotypic subtyping of Salmonella Saintpaul isolates from various food sources and humans: Phylogenetic concordance in combinatory analyses. Infect. Genet. Evol. 36 92–107. 10.1016/j.meegid.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Heijnen L., Medema G. (2016). Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J. Water Health 4 487–498. [PubMed] [Google Scholar]
- Herdman M. T., Karo B., Dave J., Katwa P., Freedman J., Do Nascimento V., et al. (2021). Increasingly limited options for the treatment of enteric fever in travellers returning to England, 2014-2019: A cross-sectional analytical study. J. Med. Microbiol. 708:001359. 10.1099/jmm.0.001359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issenhuth-Jeanjean S., Roggentin P., Mikoleit M., Guibourdenche M., de Pinna E., Nair S., et al. (2014). Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol 165 526–530. 10.1016/j.resmic.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Klontz K. C., Klontz J. C., Mody R. K., Hoekstra R. M. (2010). Analysis of tomato and jalapeño and serrano pepper imports into the United States from Mexico before and during a National Outbreak of Salmonella serotype Saintpaul infections in 2008. J. Food prot. 7311 1967–1974. 10.4315/0362-028x-73.11.1967 [DOI] [PubMed] [Google Scholar]
- Li L., Olsen R. H., Song A., Xiao J., Wang C., Meng H., et al. (2021). First report of a foodborne Salmonella enterica serovar Gloucester 4:i:l,w ST34 strain harboring blaCTX–M–55 and qnrS genes located in IS26-mediated composite transposon. Front. Microbiol. 12:646101. 10.3389/fmicb.2021.646101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Zeng M., Xu J., Zhou H., Gu B., Li Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. eBio Med. 42 133–144. 10.1016/j.ebiom.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mølbak K., Baggesen D. L., Aarestrup F. M., Ebbesen J. M., Engberg J., Frydendahl K., et al. (1999). An outbreak of multidrug resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 34119 1420–1425. 10.1056/NEJM199911043411902 [DOI] [PubMed] [Google Scholar]
- Nadimpalli M., Fabre L., Yith V., Sem N., Gouali M., Delarocque-Astagneau E., et al. (2019). CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemoth. 74 342–348. 10.1093/jac/dky451 [DOI] [PubMed] [Google Scholar]
- Octavia S., Chew K. L., Lin R., Teo J. (2021). Whole genome sequencing of Salmonella enterica serovar Saintpaul for elucidating the mechanisms of resistance to third generation cephalosporins. Pathology 53 768–772. 10.1016/j.pathol.2020.12.007 [DOI] [PubMed] [Google Scholar]
- Qi X., Li P., Xu X., Yuan Y., Bu S., Lin D. (2019). Epidemiological and molecular investigations on Salmonella responsible for gastrointestinal infections in the Southwest of Shanghai from 1998 to 2017. Front. Microbiol. 10:2025. 10.3389/fmicb.2019.02025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing [ECAST] (2017). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1. Available online at: http://www.eucast.org (accessed August 8, 2019). [Google Scholar]
- Wang W., Zhao L., Hu Y., Dottorini T., Fanning S., Xu J., et al. (2020). Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-Type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob. Agents Chemother. 647:e92–e20. 10.1128/AAC.00092-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Yang C., Shao W., Sun T., Wang J., Zhou Z., et al. (2020). Prevalence and drug resistance of Salmonella in dogs and cats in Xuzhou. China. J. Vet. Res. 642 263–268. 10.2478/jvetres-2020-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Liu J., Du B., Ruan H. H., Huo Y. X., Du Y., et al. (2020). Whole-genome based survey for polyphyletic serovars of Salmonella enterica subsp. enterica provides new insights into public health surveillance. Int. J. Mol. Sci. 21:5226. 10.3390/ijms21155226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Z., Xu X., Gu Z., Meng J., Wufuer X., Wang M., et al. (2019). Molecular epidemiology and antimicrobial resistance of invasive non-typhoidal Salmonella in China, 2007-2016. Infect. Drug Resist. 12 2885–2897. 10.2147/IDR.S210961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Z., Ding X. M., Lin X. L., Sun R. Y., Lu Y. W., Cai R. M., et al. (2019). The emergence of chromosomally located blaCTX-M-55 in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. 10.3389/fmicb.2019.01268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fu Y., Xiong Z., Ma Y., Wei Y., Qu X., et al. (2018). Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong. China. Front. Microbiol. 9:2104. 10.3389/fmicb.2018.02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Alikhan N. F., Mohamed K., Fan Y. Agama Study Group Achtman M. (2020). The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 30 138–152. 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lai H., Zou L., Yin S., Wang C., Han X., et al. (2017). Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 259 43–51. 10.1016/j.ijfoodmicro.2017.07.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.