Abstract
We present here data showing that the Avr proteins HrmA and AvrPto are secreted in culture via the native Hrp pathways from Pseudomonas syringae pathovars that produce these proteins. Moreover, their secretion is strongly affected by the temperature and pH of the culture medium. Both HrmA and AvrPto were secreted at their highest amounts when the temperature was between 18 and 22°C and when the culture medium was pH 6.0. In contrast, temperature did not affect the secretion of HrpZ. pH did affect HrpZ secretion, but not as strongly as it affected the secretion of HrmA. This finding suggests that there are at least two classes of proteins that travel the P. syringae pathway: putative secretion system accessory proteins, such as HrpZ, which are readily secreted in culture; and effector proteins, such as HrmA and AvrPto, which apparently are delivered inside plant cells and are detected in lower amounts in culture supernatants under the appropriate conditions. Because HrmA was shown to be a Hrp-secreted protein, we have changed the name of hrmA to hopPsyA to reflect that it encodes a Hrp outer protein from P. syringae pv. syringae. The functional P. syringae Hrp cluster encoded by cosmid pHIR11 conferred upon P. fluorescens but not Escherichia coli the ability to secrete HopPsyA in culture. The use of these optimized conditions should facilitate the identification of additional proteins traveling the Hrp pathway and the signals that regulate this protein traffic.
The hrp and hrc genes of plant-pathogenic bacteria belonging to the genera Erwinia, Pseudomonas, Ralstonia, and Xanthomonas encode a type III (Hrp) protein secretion system that is required for bacterial pathogenicity in host plants by compatible pathogens and elicitation of the hypersensitive response (HR) and other plant defenses in nonhost plants by incompatible pathogens (pathogens that can cause disease on different plants) (3). The HR is a programmed death of plant cells at the site of pathogen invasion and is associated with plant defense. Because this protein secretion system is required for pathogenicity, essential virulence proteins apparently travel this secretion pathway. Moreover, because nonhost plants often respond to pathogens with a functional Hrp system by inducing the HR, some of the proteins that travel this pathway can also act as elicitors of the defense response instead of contributing to disease.
The defense responses induced by an incompatible pathogen are at least partly due to the presence of avirulence (avr) genes in the pathogens that encode gene products that are recognized by the resistance (R) proteins present in the resistant plant (26). Many bacterial avr genes have been isolated from DNA libraries made from avirulent pathogens on the basis of their ability to convert a virulent pathogen to avirulence on a specific plant cultivar that contains the cognate R gene (13, 30). Within the last few years, it has been shown indirectly that many Avr proteins are delivered to the interior of the plant cell via the Hrp protein secretion system, and recognition of these proteins by plant R proteins occurs inside the plant cell (2, 42). There are examples where avr genes contribute significantly to virulence (13, 30). However, most mutants defective in specific avr genes show no detectable decrease in virulence, indicating either that they do not significantly contribute to disease or that their contribution is masked by genes that encode proteins that have similar functions. A current model predicts that Avr proteins are actually virulence proteins that collectively contribute to parasitism, but if the plant has coevolved the appropriate R gene product to recognize a specific virulence protein then that protein acts as an Avr protein (2, 26, 30).
The hrp and hrc genes of Pseudomonas syringae are clustered in a 25-kb region within the chromosome. A functional cluster of these genes from P. syringae pv. syringae 61 has been cloned onto cosmid pHIR11, and this cosmid enables nonpathogenic bacteria such as Escherichia coli and P. fluorescens to elicit an HR on tobacco (24). pHIR11 is capable of eliciting an HR in tobacco (and certain other plants) because it contains a functional set of hrp and hrc genes and at least one avr gene, hrmA (1). hrmA was not isolated as an avr gene in a screen for avirulence; rather, it was discovered because it flanks the hrp cluster carried on pHIR11 and is strictly required for a pHIR11-dependent HR on tobacco (21, 23). hrmA shares characteristics with classically isolated avr genes in that hrmA is sporadically present in different P. syringae pathovars, and an avr gene, avrPphE, resides in P. syringae pv. phaseolicola race 4 strain 1302A in the same location as hrmA in P. syringae pv. syringae (1, 31). Transient expression of hrmA in tobacco suspension cells is lethal to these cells in a manner consistent with HrmA traveling the Hrp pathway and having avirulence activity inside plant cells (4).
The Avr proteins AvrB and AvrPto can apparently be delivered into plant cells by the pHIR11 delivery system, as indicated by their dependence on a functional Hrp secretion system (16, 33). Moreover, the avirulence activity occurs inside plant cells when either transiently or transgenically expressed in plants containing the corresponding R genes (16, 40, 41). Furthermore, AvrPto has been shown to directly interact in the yeast two-hybrid system with its cognate R protein, Pto (40, 41).
Even though there is mounting indirect evidence that many Avr proteins are apparently delivered by the P. syringae Hrp secretion system to the interior of plant cells, none of these proteins have been shown to be secreted by P. syringae pathovars or by the heterologous P. syringae Hrp secretion system encoded by pHIR11. Similarly, AvrPphB was shown not to be secreted in culture by P. syringae pv. phaseolicola (35). However, both AvrB and AvrPto have been shown to be secreted in culture, using a Hrp secretion system from Erwinia chrysanthemi EC16 encoded by a cosmid in E. coli (18). The fact that an E. chrysanthemi Hrp system was capable of secreting these proteins in culture demonstrated that they are secreted via the Hrp secretion system and suggested that the E. chrysanthemi Hrp system was somehow more promiscuous in its secretion properties than the P. syringae Hrp system carried by pHIR11.
In this report, we demonstrate that HrmA and AvrPto can be detected in culture supernatants from different P. syringae pathovars, and pH and temperature were determined to be important factors for the Hrp-dependent secretion of these proteins. In addition, we detect secretion of HrmA from P. fluorescens(pHIR11) but not from E. coli(pHIR11), indicating that the heterologous Hrp system encoded by pHIR11 is sufficient to secrete Avr proteins in culture. Finally, we observed that P. syringae pv. glycinea also secretes heterologously expressed AvrPto while failing to secrete its native AvrB, demonstrating that Avr proteins differ in the ability to be secreted in culture. As described in Discussion, the name of HrmA has been changed to HopPsyA to reflect that this protein is a type III-secreted protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this work are listed in Table 1. E. coli strains were grown at 37°C in either LM or Terrific broth (39) unless otherwise noted. P. syringae pv. syringae 61, P. syringae pv. tomato DC3000, and P. syringae pv. glycinea race 0 were grown in King’s B broth at 30°C (28). For detection of the in vitro Hrp secretion of HrmA, AvrPto, AvrB, and HrpZ, P. syringae pathovars, P. fluorescens(pHIR11), and E. coli MC4100(pHIR11) were grown in hrp-derepressing fructose minimal medium at several different temperatures ranging from 22 to 28°C (25). Antibiotics were used at concentration of 100 (ampicillin), 20 (chloramphenicol), 10 (gentamicin), 50 (kanamycin), 100 (rifampin), 50 (spectinomycin), and 20 (tetracycline) μg/ml. Standard procedures (39) were used for DNA manipulations.
TABLE 1.
Strains and plasmids used
| Designation | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 Nalr | 19; Life Technologies (Grand Island, N.Y.) |
| MC4100 | F′ araD139 Δ(argF-lacZYA)U169 rpsL 150 relA1 flb-5301 ptsF25 deoC1 | 32 |
| Pseudomonas syringae | ||
| pv. syringae 61 | Wild type, Nalr | 24 |
| pv. syringae 61-2089 | hrcC::TnphoA | 23 |
| pv. tomato DC3000 | Wild type, Rifr | 12 |
| pv. glycinea race 0 | Wild type | N. T. Keen, University of California, Riverside |
| P. fluorescens 55 | Wild type, Nalr | 24 |
| Plasmids | ||
| pHIR11 | pLAFR3 derivative carrying P. syringae pv. syringae 61 hrp/hrc cluster, Tcr | 24 |
| pCPP2318 | pCPP30 derivative carrying blaM lacking signal peptide sequences, Tcr | 10 |
| pCPP2156 | pCPP19 derivative carrying E. chrysanthemi hrp/hrc cluster, Spr | 18 |
| pCPP2368 | pCPP2156::Tn5Cm that has HR− phenotype, Spr Cmr | 18 |
| pCPP3026 | pML123 carrying avrPto-Flag, Gmr | David Bauer, Cornell University |
| pCPP2089 | pHIR11 derivative containing TnphoA insert into hrcC, Tcr Kmr | 23 |
| pCPP2308 | pML122 carrying hrpL from P. syringae pv. syringae 61, Gmr | David Bauer |
To generate anti-HrmA and anti-AvrPto antibodies.
HrmA-Flag was purified from E. coli DH5α by affinity chromatography as described by Gopalan et al. (16). Fractions containing HrmA-Flag were pooled and concentrated in Centriprep-10 and Centricon-10 ultrafiltrations units (Amicon, Inc., Beverly, Mass.). N-terminally His6-tagged AvrPto was prepared from E. coli DH5α(pQE31::avrPto) essentially as described elsewhere (1). The samples were injected into different rabbits to generate anti-HrmA and anti-AvrPto polyclonal antibodies at the University of Illinois Immunological Resource Center. The crude antisera raised to both proteins were separately delipified and preabsorbed against E. coli DH5α extracts as described by Ham et al. (18).
Preparation of protein samples from cell-bound and supernatant fractions.
Pseudomonas spp. were grown overnight on King’s B plates at 30°C. Cells were washed and resuspended in hrp-derepressing fructose minimal medium to an initial optical density at 600 nm (OD600) of 0.15 and grown routinely at 22°C, unless otherwise noted, in a rotary shaking incubator at 220 rpm to an OD600 of 0.3 (25). Aliquots (80 ml) of the cultures were separated into cell-bound and supernatant fractions by centrifugation at 4°C. When protein samples were prepared from E. coli MC4100(pHIR11), bacterial cells were grown overnight on LM plates at 37°C, washed, resuspended in hrp-derepressing fructose minimal medium to an initial OD600 of 0.4, and cultured at 22°C (unless otherwise noted) in a rotary shaking incubator at 220 rpm to an OD600 of 0.500. When samples were prepared from P. fluorescens(pHIR11) cells, cultures were routinely inoculated in hrp-derepressing medium such that the initial OD600 was 0.3, and the cultures were harvested at an OD600 of 0.5. The cell-bound and supernatant fractions were separated by centrifugation at 4°C, and protein samples were prepared as described by Ham et al. (18). The total protein present in the cell-bound fractions was determined by the method of Bradford (8).
Protein analyses to detect in culture type III secretion.
Approximately 100 μg of protein from each cell-bound fraction was loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels. Based on the amount of total protein present in the cell-bound fraction, the amount of supernatant fractions that was loaded onto the gels was adjusted to reflect the total protein in each culture. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using standard procedures (39) and then transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore Co., Bedford, Mass.). HrmA, AvrPto, AvrB, HrpZ, and β-lactamase were detected with specific polyclonal antibodies raised to each protein, followed by goat anti-rabbit immunoglobulin G-alkaline phosphate conjugate (Sigma Chemical Co., St. Louis, Mo.). Membrane-bound secondary antibodies were visualized by chemiluminescence using a Western-Light chemiluminescence detection system (Tropix, Bedford, Mass.) and X-Omat X-ray film (Eastman Kodak, Rochester, N.Y.). AvrB and HrpZ were detected by using previously obtained polyclonal antibodies raised against AvrB-Flag and HrpZ, respectively (18, 20). Anti-β-lactamase polyclonal antibodies used in this study were purchased from 5 Prime→3 Prime Inc. (Boulder, Colo.).
RESULTS
P. syringae pv. syringae 61 secretes HrmA in culture via the Hrp (type III) protein secretion system.
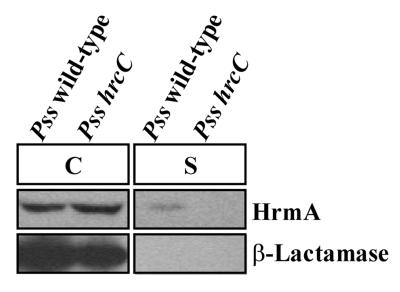
Because the P. syringae Avr proteins AvrB and AvrPto were found to be secreted by the type III secretion system encoded by the functional E. chrysanthemi hrp cluster carried on cosmid pCPP2156 expressed in E. coli (18), we sought to detect the secretion in culture of the Avr protein HrmA directly via the native Hrp system carried in P. syringae pv. syringae 61. P. syringae pv. syringae cultures grown in hrp-derepressing fructose minimal medium at 25°C were separated into cell-bound and supernatant fractions by centrifugation. Proteins present in the supernatant fractions were concentrated by trichloroacetic acid precipitation, and the cell-bound and supernatant samples were resolved by SDS-PAGE and analyzed by immunoblotting using anti-HrmA antibodies. A weak HrmA signal was detected in supernatant fractions from wild-type P. syringae pv. syringae 61 (Fig. 1). Importantly, HrmA was not detected in supernatant fractions from P. syringae pv. syringae 61-2089, which is defective in Hrp secretion, indicating that the HrmA signal in the supernatant was due specifically to type III protein secretion (Fig. 1). Since the level of HrmA secretion in culture was relatively low, we included a second control, to distinguish type III secretion from cell lysis. Both strains contained pCPP2318, which encodes the mature β-lactamase lacking its N-terminal signal peptide and provides a marker for cell lysis. The samples analyzed for HrmA secretion were also subjected to immunoblot analysis with anti-β-lactamase antibodies. β-Lactamase was detected only in the cell-bound fractions of these samples, clearly showing that cell lysis did not occur at a significant level (Fig. 1).
FIG. 1.
Distribution of HrmA and β-lactamase in cultures of P. syringae pv. syringae 61(pCPP2318) or hrp mutant P. syringae pv. syringae 61-2089(pCPP2318). Bacterial cultures were grown at 25°C in hrp-derepressing medium and separated into cell-bound (C) and supernatant (S) fractions. The cell-bound fractions were concentrated 13.4-fold and the supernatant fractions were concentrated 100-fold relative to the initial culture volumes. The samples were subjected to SDS-PAGE and immunoblot analysis, and HrmA and β-lactamase were detected with either anti-HrmA or anti-β-lactamase antibodies followed by secondary antibodies conjugated to alkaline phosphatase as described in Materials and Methods. Pss wild-type, P. syringae pv. syringae 61(pCPP2318); Pss hrcC, P. syringae pv. syringae 61-2089(pCPP2318). The image of the immunoblot was captured using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
Secretion of HrmA by P. syringae pv. syringae is strongly affected by the growth temperature and pH of the culture medium.
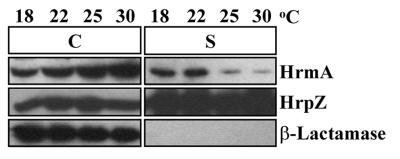
Secretion in culture of HrmA from wild-type P. syringae pv. syringae was detectable but weak. Therefore, we wanted to determine whether we could increase the amount of HrmA secreted in culture by subtly altering the growth conditions. Previously, it was shown that transcription of P. syringae hrp genes is sensitive to many different environmental factors, including the temperature and pH of the growth medium (25, 36, 47). To determine if temperature was an important factor in HrmA secretion, we grew P. syringae pv. syringae 61(pCPP2318) in hrp-derepressing medium at several different temperatures and assessed the distribution of HrmA between cell-bound and supernatant fractions. Each culture was grown to a final OD600 of 0.3 and separated by centrifugation into cell-bound and supernatant fractions. Total protein concentrations were determined by the method of Bradford (8) to be similar for all of the cell-bound fractions (data not shown). The cell-bound and supernatant samples were resolved by SDS-PAGE and subjected to immunoblot analysis with anti-HrmA antibodies or anti-β-lactamase antibodies, using the procedures described above. The temperature under which the culture was grown significantly affected the amount of HrmA detected in the supernatant. The Hrp secretion of HrmA from P. syringae pv. syringae was highest at 18 and 22°C (Fig. 2). At 25 and 30°C, the amounts of HrmA in the supernatant were substantially lower than the amounts detected at 18 and 22°C (Fig. 2). HrmA was found only in the cell-bound fractions from cultures of a P. syringae pv. syringae 61 mutant defective in type III secretion, confirming that the secreted HrmA was dependent on a functional Hrp secretion system (data not shown). The differences in the amount of HrmA in the supernatant could not be due to different amounts of cell lysis because the cytoplasmic marker β-lactamase remained entirely in the cell-bound fraction for each temperature (Fig. 2). Interestingly, the total amounts of HrmA produced at the different temperatures appeared to be approximately equal. Therefore, the enhancement of HrmA secretion was not due to greater levels of HrmA production.
FIG. 2.
The secretion in culture of HrmA (HopPsyA) is affected by temperature, whereas the secretion of HrpZ is not. P. syringae pv. syringae 61(pCPP2318) cultures were grown in hrp-derepressing medium at the temperatures indicated. Bacterial cultures were separated into cell-bound (C) and supernatant (S) fractions by centrifugation, and the supernatant fractions were adjusted to be 7.5 times more concentrated than the cell-bound fractions. After the samples were separated by SDS-PAGE and transferred to PVDF membranes, HrmA, HrpZ, and β-lactamase were detected by immunoblotting using anti-HrmA, anti-HrpZ, and anti-β-lactamase antibodies, respectively, followed by secondary antibodies conjugated to alkaline phosphatase. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
We also assessed whether the HrpZ harpin was found in the supernatant fraction of these samples to determine if temperature was an important factor in HrpZ secretion. HrpZ was the first protein determined to be secreted by the P. syringae Hrp system, is readily secreted in culture, and appears to be targeted to the plant cell wall (1, 9, 20, 22). Even at temperatures that resulted in less HrmA secreted, HrpZ was secreted in high amounts in culture, indicating that the temperature range used here did not affect the Hrp secretion of HrpZ (Fig. 2).
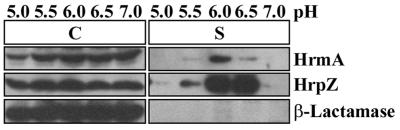
To determine the effect of pH of the culture medium on the secretion of HrmA, P. syringae pv. syringae 61 was grown in hrp-derepressing fructose minimal medium that was adjusted to pH 5.0, 5.5, 6.0, 6.5, and 7.0. P. syringae pv. syringae 61 cultures were grown in these conditions at the optimized temperature for HrmA secretion of 22°C to an OD600 of 0.3. Cell-bound and supernatant fractions were isolated by centrifugation and analyzed by SDS-PAGE and immunoblotting with anti-HrmA and anti-β-lactamase antibodies. P. syringae pv. syringae 61 secretion of HrmA was highest at pH 6.0. At pH 5.5 and 6.5, significantly less HrmA was secreted, and there was no detectable secretion at pH 5.0 and 7.0 (Fig. 3). HrmA was found only in the cell fractions of cultures from a P. syringae pv. syringae 61 mutant defective in type III secretion, indicating that the HrmA found in the supernatant fraction was due to a functional type III secretion system (data not shown). As observed with temperature, the increased secretion of HrmA via the type III protein secretion system at pH 6.0 was apparently not due to increased production of HrmA because all of the cell-bound fractions have approximately equal amounts of HrmA (Fig. 3). pH also affected the secretion in culture of HrpZ, but not to the same extent that different pH values affected the Hrp secretion of HrmA. For example, both HrmA and HrpZ were found in low amounts at pH 5.0 and 7.0. However, a greater percentage of the total HrpZ than of the total HrmA produced was found in the supernatant fractions from cultures grown at pH 5.5 and 6.5 (Fig. 3).
FIG. 3.
The pH of the growth medium affects the secretion in culture of HrmA (HopPsyA) and HrpZ via the Hrp secretion system. P. syringae pv. syringae 61(pCPP2318) cultures were grown at 22°C in hrp-derepressing media differing only in the pH of the medium. Bacterial cultures were separated into cell-bound (C) and supernatant (S) fractions by centrifugation, and proteins were separated by SDS-PAGE. Immunoblot analysis was carried out as described in Materials and Methods, using anti-HrmA, anti-HrpZ, or anti-β-lactamase antibodies followed by secondary antibodies conjugated to alkaline phosphatase. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
We also investigated whether secretion of HrmA could be enhanced by other conditions or factors, but we were unable to find additional conditions. For example, the hrpL gene encodes an alternate sigma factor required for transcription of many hrc and hrp genes (45, 46). We tested whether overexpression of hrpL would lead to increased production and secretion of HrmA by P. syringae pv. syringae 61. In experiments similar to those described above, we grew cultures of P. syringae pv. syringae and P. syringae pv. syringae with hrpL in trans carried on the construct pCPP2308 in hrp-derepressing media. Immunoblot analysis on fractions from these cultures showed the same levels of HrmA in both the cell-bound and supernatant fractions, indicating that hrpL in trans did not measurably affect HrmA secretion (data not shown).
Because HrmA was found conclusively to be a protein that traveled the Hrp pathway, the name of the hrmA gene was changed to hopPsyA to indicate that it encodes a protein that is secreted by the Hrp protein secretion system (see Discussion).
P. syringae pv. tomato DC3000 secretes AvrPto via the Hrp system in a manner that is affected by temperature and pH.
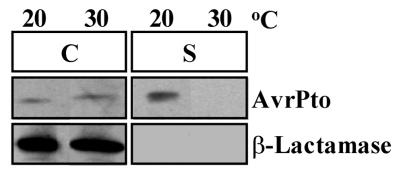
The ability to detect the secretion in culture of HopPsyA (HrmA) by P. syringae pv. syringae led us to investigate if we could detect similar secretion of other Avr proteins from their native P. syringae pathovars. We chose to determine if the Avr protein AvrPto was secreted from P. syringae pv. tomato because there is much indirect evidence that AvrPto is delivered into plant cells by the Hrp secretion system. We grew P. syringae pv. tomato DC3000(pCPP2318) in hrp-derepressing medium at temperatures of 20 and 30°C and isolated cell-bound and supernatant fractions from these cultures. Immunoblot analysis of these fractions with anti-AvrPto antibodies showed that AvrPto was secreted at 20°C but not at 30°C, while β-lactamase remained in the cell-bound fraction (Fig. 4). Therefore, the secretion of AvrPto, like that of HopPsyA, is enhanced at temperatures below 22°C. AvrPto was not found in the supernatant fractions of P. syringae pv. tomato mutants defective in type III secretion, indicating that differential secretion of AvrPto was type III dependent (data not shown). In contrast to HopPsyA, AvrPto was not found in high amounts at 30°C in either the cell-bound or supernatant fractions of P. syringae pv. tomato.
FIG. 4.
AvrPto is secreted in culture from P. syringae pv. tomato DC3000 via the Hrp secretion system at 20°C but not at 30°C. P. syringae pv. tomato DC3000(pCPP2318) cultures were grown in hrp-derepressing medium at 20 and 30°C. The supernatant (S) and cell-bound (C) fractions were isolated as before, separated by SDS-PAGE, and analyzed by immunoblotting with anti-AvrPto or anti-β-lactamase antibodies. The preparations of the protein samples resulted in supernatant fractions that were concentrated 7.5 times more than the cell-bound fractions. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
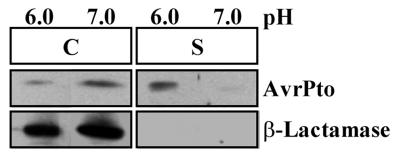
Since pH was an important factor for the type III secretion of HopPsyA, we tested whether pH had an effect on the type III secretion of AvrPto. We grew P. syringae pv. tomato DC3000(pCPP2318) in hrp-derepressing media that was adjusted to pH 6.0 or 7.0. Cell-bound and supernatant fractions were isolated as described above, and immunoblot analysis of these samples with anti-AvrPto antibodies showed that AvrPto was secreted at pH 6.0 but not pH 7.0 (Fig. 5). In both samples, β-lactamase remained in the cell-bound fraction, indicating that cell lysis did not occur at a significant level. Moreover, the secretion of AvrPto at pH 6.0 was dependent on type III secretion because AvrPto was not secreted from a P. syringae pv. tomato mutant defective in type III secretion (data not shown). Therefore, as observed for HopPsyA secretion from P. syringae pv. syringae, the type III secretion of AvrPto from P. syringae pv. tomato was enhanced at pH 6.0. AvrPto was produced at pH 7.0 in amounts approximately equal to those produced at pH 6.0. Therefore, the effect of pH on AvrPto secretion is not due to the regulation of avrPto at the transcriptional level.
FIG. 5.
AvrPto is secreted in culture from P. syringae pv. tomato DC3000 at pH 6.0 but not at pH 7.0. P. syringae pv. tomato DC3000(pCPP2318) cultures were grown at 20°C in hrp-derepressing medium adjusted to either pH 6.0 or pH 7.0. Isolated cell-bound (C) and supernatant (S) fractions were separated by SDS-PAGE and analyzed by immunoblotting using anti-AvrPto or anti-β-lactamase antibodies. Mature β-lactamase encoded by pCPP2318 was included as a control for cell lysis. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
Secretion of AvrB from P. syringae pv. glycinea cannot be detected in culture, even when heterologously expressed AvrPto is secreted.
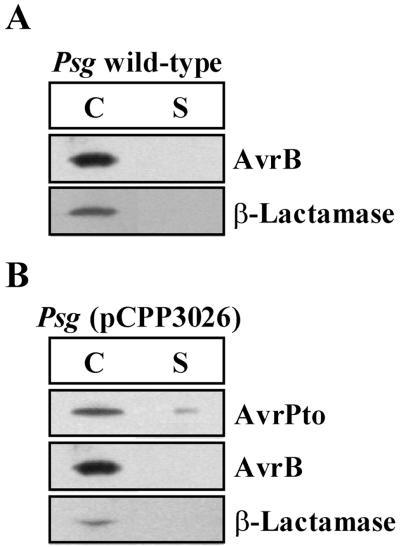
Another Avr protein, which based on indirect evidence appears to be translocated into plant cells, is the Avr protein AvrB. Recently, AvrB was found to be secreted in culture by the E. chrysanthemi Hrp system expressed in E. coli (18). However, AvrB has not been reported to be secreted from its native P. syringae pv. glycinea Hrp system or by P. fluorescens(pHIR11) (16). To determine if AvrB could be detected in P. syringae pv. glycinea culture supernatants, we grew P. syringae pv. glycinea race 0(pCPP2318) cultures in the optimal conditions described above and prepared the cell-bound and supernatant fractions in the same manner as used in the experiments with HopPsyA and AvrPto. The cell-bound and supernatant fractions were separated by SDS-PAGE and analyzed by immunoblotting with anti-AvrB antibodies. We were unable to detect any AvrB secreted in culture in the identical conditions that were sufficient to promote HopPsyA and AvrPto secretion (Fig. 6A). To determine if the failure to detect the type III secretion of AvrB in culture was due to a difference with the native Hrp system present in P. syringae pv. glycinea or if it was due directly to the secretion characteristics of the AvrB protein, we electroporated pCPP3026, which carries avrPto, into P. syringae pv. glycinea race 0. P. syringae pv. glycinea(pCPP3026, pCPP2318) cultures were grown as before and separated into cell-bound and supernatant fractions. Immunoblot analysis of these cultures detected AvtPto in the supernatant fractions of P. syringae pv. glycinea, while β-lactamase and AvrB remained in the cell-bound fraction (Fig. 6B). Therefore, the lack of AvrB in the supernatant fractions of P. syringae pv. glycinea cultures is not due to a difference in the Hrp secretion system but rather is apparently due to specific secretion properties of AvrB.
FIG. 6.
Native AvrB cannot be detected in culture supernatants from P. syringae pv. glycinea race 0, even though heterologously expressed AvrPto is secreted. (A) P. syringae pv. glycinea (Psg) race 0(pCPP2318) was grown at 20°C in hrp-derepressing medium and separated into cell-bound (C) and supernatant (S) fractions. The fractions were separated by SDS-PAGE, transferred to PVDF membranes, and analyzed by immunoblotting using either anti-AvrPto or anti-β-lactamase antibodies. (B) P. syringae pv. glycinea race 0(pCPP2318, pCPP3026) was grown at 20°C in hrp-derepressing medium and separated into cell-bound (C) and supernatant (S) fractions. pCPP3026 encodes AvrPto from P. syringae pv. tomato DC3000. After separation of proteins by SDS-PAGE, immunoblot analysis was carried out with anti-AvrB, anti-AvrPto, or anti-β-lactamase antibodies. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
P. fluorescens(pHIR11) secretes HopPsyA (HrmA) in culture in a Hrp-dependent manner, but E. coli(pHIR11) cannot secrete HrmA in detectable amounts.
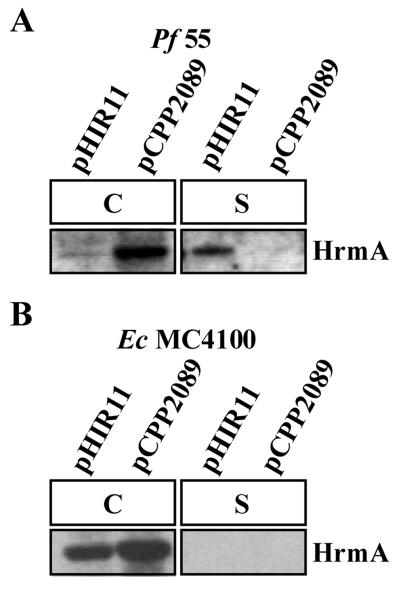
Previously it was reported that E. coli carrying cosmid pHIR11 was unable to secrete detectable amounts of AvrPto or HopPsyA in culture media (4, 18). Since we determined conditions and procedures that allowed for the detection of type III-secretion of HopPsyA in culture media, we checked if these same conditions would allow for the detection of HopPsyA secreted by bacteria carrying pHIR11. P. fluorescens 55(pHIR11) was grown in conditions similar to those used for the P. syringae secretion experiments described above. Immunoblot analysis with anti-HopPsyA antibodies revealed that HopPsyA was secreted in culture by P. fluorescens(pHIR11), while a pHIR11 derivative that carries a mutation in the hrcC gene and is defective in type III secretion, pCPP2089, did not secrete HopPsyA in culture (Fig. 7A). These samples were analyzed on Coomassie blue-stained gels after SDS-PAGE; this analysis did not detect many protein bands in the supernatant fraction, indicating that cell lysis did not occur at a significant level (data not shown). Interestingly, in contrast to the pattern of HopPsyA secretion from P. syringae, most of the HopPsyA is found in the supernatant fraction (Fig. 2 and 7). This pattern of HopPsyA secretion has been observed repeatedly. This may suggest that the native type III secretion system present in P. syringae has a mechanism to retain some of the HopPsyA in the cell-bound fraction, while the pHIR11 system may lack this mechanism and secrete most of the HopPsyA. Because there are reports of E. coli(pHIR11) being unable to secrete either HopPsyA or AvrPto in culture, we wanted to determine if HopPsyA could be secreted from E. coli(pHIR11) under the optimal conditions determined above (4, 18). Using the same conditions as employed in the P. fluorescens(pHIR11) secretion experiments, we tested whether E. coli MC4100(pHIR11) could secrete HopPsyA. Cell-bound and supernatant fractions were analyzed on immunoblots with anti-HopPsyA antibodies. HopPsyA was not detected in supernatant fractions, indicating that detectable levels of HopPsyA are not secreted in culture from the heterologous P. syringae pv. syringae type III secretion system expressed in E. coli (Fig. 7B). Thus, P. fluorescens carrying pHIR11 did secrete detectable amounts of HopPsyA in culture, while E. coli carrying pHIR11 did not.
FIG. 7.
HrmA (HopPsyA) is secreted in culture from P. fluorescens 55(pHIR11) via the Hrp pathway but not from E. coli MC4100(pHIR11). (A) P. fluorescens (Pf) 55 carrying either a functional hrp cluster from P. syringae pv. syringae (pHIR11) or a defective P. syringae pv. syringae hrp cluster (pCPP2089) was grown in hrp-derepressing medium at 22°C. Cell-bound (C) and supernatant (S) fractions were separated by SDS-PAGE and analyzed on immunoblots with anti-HrmA antibodies. (B) E. coli (Ec) MC4100 carrying either pHIR11 or pCPP2089 was grown at 25°C in hrp-derepressing medium and separated into cell-bound (C) and supernatant (S) fractions. After separation of the fractions by SDS-PAGE, the presence of HrmA from each fraction was determined by immunoblot analysis with anti-HrmA antibodies. The image of the immunoblot was captured by using the Bio-Rad Gel Doc 1000 UV fluorescent gel documentation system with the accompanying Multi-Analyst PC software. For figure construction, the image was manipulated by using Microsoft PowerPoint 97 and transferred to Adobe Photoshop 4.0 to meet the publisher’s specifications.
DISCUSSION
We report here conditions that allow the secretion in culture of the Avr proteins HopPsyA (HrmA) and AvrPto via their native Hrp (type III) protein secretion systems from the P. syringae pathovars that normally produce these proteins. We chose to study the secretion properties of these Avr proteins, along with the Avr protein AvrB, because they were among the first Avr proteins from P. syringae indirectly shown to be active inside plant cells. AvrPto and AvrB were previously shown to be secreted in culture, but via the E. chrysanthemi Hrp system encoded by a cosmid expressed in E. coli (18). The Erwinia amylovora DspA (DspE) protein had been shown previously to be secreted directly from the pathogen instead of a heterologous Hrp system (6, 15).
The P. syringae Hrp system appears to secrete at least two classes of proteins. One class consists of proteins that are either components of the extracellular secretion apparatus or may help in the deployment of the apparatus. Examples of these helper proteins are HrpA, which is the main protein component of a required pilus (37), and possibly HrpZ and HrpW (1, 9, 20), both of which probably are targeted to the plant cell wall. These proteins are readily secreted in culture via the Hrp pathway. The proteins of the other class are the actual effector proteins delivered by the Hrp system and consist of Avr proteins such as HopPsyA, AvrB, AvrPto, and probably other proteins that are not recognized by the R gene encoded antiparasite surveillance systems present in plants. Based on indirect evidence, many of these proteins are delivered directly into plant cells, probably upon contact. The secretion of the effector protein class appears to be more regulated than the secretion of the helper protein class. For example, HrpZ, HrpW, and HrpA can be detected easily in culture supernatant fractions separated on SDS-polyacrylamide gels stained with Coomassie blue (48). We were successful in finding conditions that allowed for the detection of HopPsyA and AvrPto in culture supernatants by using immunoblot analysis, but these proteins were not secreted at a level high enough to be observed on Coomassie blue-stained gels.
The fact that HrpZ is strongly secreted at temperatures that permit the secretion of only a low amount of HopPsyA and AvrPto suggests that these proteins are secreted differently by the Hrp secretion system. Temperature and pH may reflect actual cues for the secretion of these proteins that are sensed by the bacterium in the apoplast, or they may be conditions that inadvertently trigger the sensor proteins to release type III-secreted proteins. In the Yersinia type III system, virulence proteins (i.e., Yops [Yersinia outer proteins]) are secreted at 37°C in media lacking calcium. For many years, this so-called low-calcium response was thought to be a specific regulatory response and many genes that were implicated in this were designated lcr genes (5, 34). It now appears that many of these responses may be partly artifactual and that the low-calcium response may actually be due to artificially activating the sensors that would normally activate secretion upon contact with the eucaryotic cell (11, 38). It is premature to conclude that the effects of temperature and pH on the secretion of HopPsyA and AvrPto from P. syringae reflect a true regulatory response or if these conditions somehow artificially induce protein secretion. However, the temperatures these pathogens would encounter in nature are consistent with the low-temperature enhancement described in this report. Moreover, Agrobacterium has been shown to increase the production of a pilus required for its type IV secretion system in response to similar temperatures (14). Furthermore, the pH values that allowed for the highest amount of HrpZ, HopPsyA, and AvrPto secretion are within the estimated range of the pH of the apoplast of plant tissues (17).
E. coli carrying pHIR11 was unable to secrete detectable amounts of HopPsyA to the supernatant fraction, consistent with another published report (4). However, P. fluorescens(pHIR11) strongly secreted HopPsyA to the supernatant fraction. A possible explanation for this is that the Hrp secretion system from P. syringae is expressed better in another pseudomonad rather than an enterobacterium such as E. coli. This result may help in the interpretation of data from another recent paper. Ham et al. (18) found that AvrPto was secreted from E. coli carrying pCPP2156, which encodes the Hrp system from E. chrysanthemi, but not from E. coli(pHIR11). The data suggested that the E. chrysanthemi Hrp system may be inherently more promiscuous in its secretion properties than the P. syringae Hrp system encoded by pHIR11, possibly reflecting a difference in the pathogenic lifestyles of these contrasting pathogens.
A new designation for effector proteins that travel the Hrp pathway should now be helpful because the Avr nomenclature is not applicable to secreted proteins that are not recognized by an R gene product in a tester plant’s antiparasite surveillance system. HopPsyA was originally named HrmA because it was initially thought to be a regulator that modulated the HR (21). This name is no longer meaningful because HopPsyA does not appear to be a regulatory protein. We now know that HopPsyA is a protein that travels the Hrp pathway of P. syringae. In the prototypical type III system of Yersinia spp., proteins that travel the ysc-encoded type III apparatus are named Yops. We have proposed the adoption of an analogous nomenclatural system using a similar prefix: Hop (Hrp-dependent outer protein) (3). Thus, it is intended to be a prefix that could include proteins from other bacterial plant pathogens, which is consistent with the apparent mobility and functional interchangeability of avr-like genes among plant pathogens in different genera (7, 18, 27). To identify which pathogen a Hop is from, we have proposed adopting the system that Vivian and Mansfield proposed for avr genes, in which a suffix provides the first initial of the genus name and the first two initials of the pathovar name for the source bacterium (43). Using the above naming systems, we have renamed the hrmA gene as hopPsyA to identify it as a gene that encodes an Hrp-dependent outer protein that travels the Hrp pathway of P. syringae pv. syringae.
Based on the apparent abundance of avr genes that can be identified in single strains of plant pathogens on the basis of their interaction with R genes in differential cultivars of host or in nonhost plant species, we can expect that there are many Hop proteins that have yet to be identified (29, 30, 44). Thus, the identification of conditions that optimize the secretion of Hops will likely help in the identification of these cryptic Hops and will be important in determining how these proteins collectively interact with host plants to enable plant pathogenicity.
ACKNOWLEDGMENTS
We thank David Bauer for pCPP2308 and pCPP3026, Noel Keen for P. syringae pv. glycinea race 0, and Tanja Petnicki for reviewing the manuscript.
This work was supported by National Research Initiative Competitive Grants Program U.S. Department of Agriculture grant no. 98-35303-6464, and grants from the Applied Research Initiative of Nevada and the UNLV Office of Research to J.R.A. and by National Science Foundation grant MCB-9631530 and National Research Initiative Competitive Grants Program U.S. Department of Agriculture grant 97-35303-4488 to A.C.
REFERENCES
- 1.Alfano J R, Bauer D W, Milos T M, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally nonpolar hrpZ deletion mutants, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 2.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano J R, Kim H-S, Delaney T P, Collmer A. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol Plant-Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 5.Bergman T, Håkansson S, Forsberg A, Norlander L, Macellaro A, Backman T, Bolin I, Wolf-Watz H. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role for LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanove A J, Bauer D W, Beer S V. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J Bacteriol. 1998;180:2244–2247. doi: 10.1128/jb.180.8.2244-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogdanove A J, Kim J F, Wei Z, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 12.Cuppels D A. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1986;51:323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dangl J L. The enigmatic avirulence genes of phytopathogenic bacteria. Current Top Microbiol Immunol. 1994;192:99–118. doi: 10.1007/978-3-642-78624-2_5. [DOI] [PubMed] [Google Scholar]
- 14.Fullner K J, Lara J C, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 15.Gaudriault S, Malandrin L, Paulin J-P, Barny M-A. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol. 1997;26:1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x. [DOI] [PubMed] [Google Scholar]
- 16.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grignon C, Sentenac H. pH and ionic conditions in the apoplast. Annu Rev Plant Physiol Mol Biol. 1991;42:103–128. [Google Scholar]
- 18.Ham J H, Bauer D W, Fouts D E, Collmer A. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 20.He S Y, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 21.Heu S, Hutcheson S W. Nucleotide sequence and properties of the hrmA locus associated with the Pseudomonas syringae pv. syringae 61 hrp gene cluster. Mol Plant-Microbe Interact. 1993;6:553–564. doi: 10.1094/mpmi-6-553. [DOI] [PubMed] [Google Scholar]
- 22.Hoyos M E, Stanley C M, He S Y, Pike S, Pu X-A, Novacky A. The interaction of harpinPss with plant cell walls. Mol Plant-Microbe Interact. 1996;9:608–616. [Google Scholar]
- 23.Huang H-C, Hutcheson S W, Collmer A. Characterization of the hrp cluster from Pseudomonas syringae pv. syringae 61 and TnphoA tagging of genes encoding exported or membrane-spanning Hrp proteins. Mol Plant-Microbe Interact. 1991;4:469–476. [Google Scholar]
- 24.Huang H-C, Schuurink R, Denny T P, Atkinston M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 26.Keen N T. Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 27.Kim J F, Charkowski A O, Alfano J R, Collmer A C, Beer S V. Transposable elements and bacteriophage sequences flanking Pseudomonas syringae avirulence genes. Mol Plant-Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 28.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Med. 1954;22:301–307. [PubMed] [Google Scholar]
- 29.Kobayashi D Y, Tamaki S J, Keen N T. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc Natl Acad Sci USA. 1989;86:157–161. doi: 10.1073/pnas.86.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leach J E, White F F. Bacterial avirulence genes. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 31.Mansfield J, Jenner C, Hockenhull R, Bennett M A, Stewart R. Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol Plant-Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- 32.Oliver D B, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;78:1089–1099. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 33.Pirhonen M U, Lidell J C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 34.Plano G V, Barve S S, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puri N, Jenner C, Bennett M, Stewart R, Mansfield J, Lyons N, Taylor J. Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol Plant-Microbe Interact. 1997;10:247–256. doi: 10.1094/MPMI.1997.10.2.247. [DOI] [PubMed] [Google Scholar]
- 36.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosqvist R, Magnusson K-E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 41.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Physical interaction of AvrPto and the Pto kinase defines a recognition event involved in plant disease resistance. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 42.Van den Ackerveken G, Bonas U. Bacterial avirulence proteins as triggers of plant disease resistance. Trends Microbiol. 1997;5(10):394–398. doi: 10.1016/S0966-842X(97)01124-4. [DOI] [PubMed] [Google Scholar]
- 43.Vivian A, Mansfield J. A proposal for a uniform genetic nomenclature for avirulence genes in phytopathogenic pseudomonads. Mol Plant-Microbe Interact. 1993;6:9–10. [Google Scholar]
- 44.Whalen M C, Stall R E, Staskawicz B J. Characterization of a gene from a tomato pathogen determining hypersensitive resistance in non-host species and genetic analysis of this resistance in bean. Proc Natl Acad Sci USA. 1988;85:6743–6747. doi: 10.1073/pnas.85.18.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao Y, Hutcheson S W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y, Lu Y, Heu S, Hutcheson S W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan J, He S Y. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]