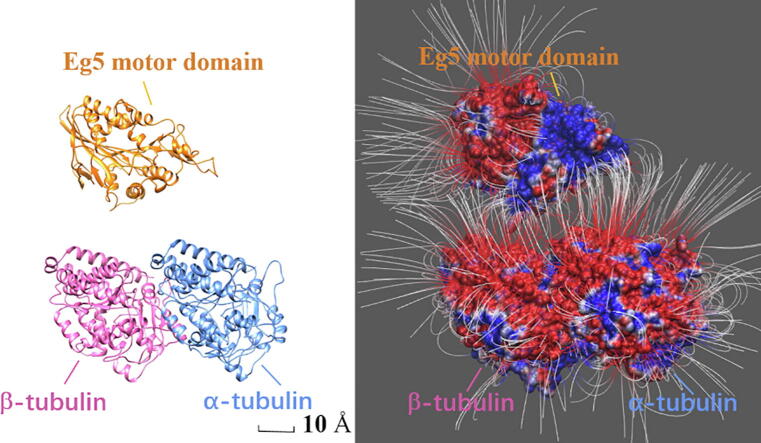
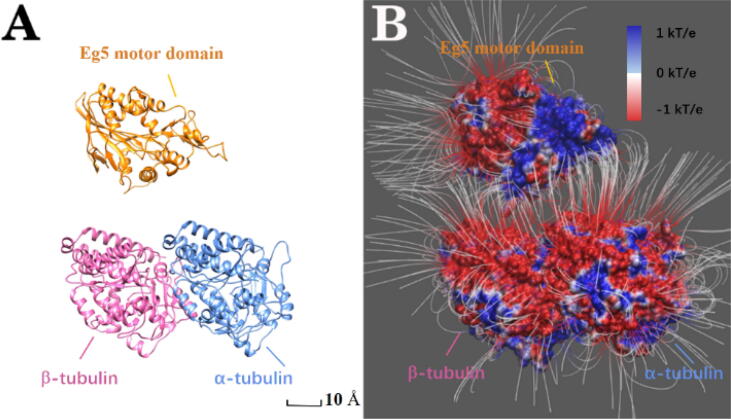
Graphical abstract
Keywords: Kinesin-5, Eg5, Tubulin, Microtubule, Electrostatic features, DelPhi, DelPhiForce, DelPhiPKa, Molecular dynamic simulation, Hydrogen bonds, Salt bridges
Abstract
Kinesins are microtubule-based motor proteins that play important roles ranging from intracellular transport to cell division. Human Kinesin-5 (Eg5) is essential for mitotic spindle assembly during cell division. By combining molecular dynamics (MD) simulations with other multi-scale computational approaches, we systematically studied the interaction between Eg5 and the microtubule. We find the electrostatic feature on the motor domains of Eg5 provides attractive interactions to the microtubule. Additionally, the folding and binding energy analysis reveals that the Eg5 motor domain performs its functions best when in a weak acidic environment. Molecular dynamics analyses of hydrogen bonds and salt bridges demonstrate that, on the binding interfaces of Eg5 and the tubulin heterodimer, salt bridges play the most significant role in holding the complex. The salt bridge residues on the binding interface of Eg5 are mostly positive, while salt bridge residues on the binding interface of tubulin heterodimer are mostly negative. Such salt bridge residue distribution is consistent with electrostatic potential calculations. In contrast, the interface between α and β-tubulins is dominated by hydrogen bonds rather than salt bridges. Compared to the Eg5/α-tubulin interface, the Eg5/β-tubulin interface has a greater number of salt bridges and higher occupancy for salt bridges. This asymmetric salt bridge distribution may play a significant role in Eg5′s directionality. The residues involved in hydrogen bonds and salt bridges are identified in this work and may be helpful for anticancer drug design.
1. Introduction
Kinesins are a superfamily of motor proteins that can be divided into 14 different subfamilies (Kinesin-1 through Kinesin-14) and an orphan group [1], [2]. Kinesins utilize chemical energy from ATP hydrolysis to interact with microtubules and generate movement inside cells [1], [3], [4], [5]. Kinesins play vital roles in various cellular processes ranging from intracellular transport to cell division [1], [3], [6], [7].
Kinesin-5 motors are responsible for the assembly and maintenance of spindle bipolarity during cell division [8], [9], [10], [11], [12]. Kinesin-5 motors are homo-tetramers with four identical heavy chains [13], [14], [15]. While human Kinesin-5 commonly exhibits plus-end-directed motility, Kinesin-5 from fungi can exhibit minus-end-directed motility [9], [16], [17], [18], [19], [20]. The tetramer structure with two pairs of motor domains at either end enables Kinesin-5 to interact with microtubules and generate forces which push and hold spindle poles apart [21], [22]. In addition, it has been confirmed that Kinesin-5 promotes microtubule polymerization [11], [23].
Cryo-electron microscopy (cryo-EM) is an important tool which can reveal the structural basis for kinesin motility on the microtubule. Many biochemical studies have been carried out to determine different kinesin-tubulin complex structures and explore the mechanisms of kinesin motility [22], [24], [25], [26], [27]. There are many known kinesin or kinesin-microtubule (MT) complex structures on the Protein Data Bank (PDB). Several experiments using computational approaches such as molecular dynamics (MD) simulations and Monte Carlo analysis have been used to study the binding mechanisms between kinesins and microtubules [28], [29], [30], [31]. Studies such as the one done by Scarabelli and Grant utilized MD simulations combined with bioinformatics-guided mutations to show the dynamic effects of nucleotide turnover and allosteric inhibition of the Kinesin-5 motor [32]. Grant et al. used atomistic Brownian dynamics simulations combined with experimental mutagenesis to elucidate a binding mechanism for kinesin-1 [28].
Even though many efforts have been undertaken to study molecular motors [33], [34], the mechanisms of kinesin motility on microtubules are still not completely characterized. In addition to in vivo and in vitro approaches, computational methods have been widely used to study the functions of biomolecules [35], [36], [37], [38], [39], [40], [41]. In this work, we used a multi-scale computational approach that combines DelPhi [42], [43], DelPhiForce [44], [45], DelPhiPKa [46], [47] and NAMD [48] to study the binding mechanism of Eg5 to the microtubule. Electrostatic features indicate that the charge distribution on the motor domain of Eg5 provides attractive interactions to the microtubule. Firstly, the electrostatic potential on the binding surface of the Eg5 motor domain shows a predominantly positive potential while the interface on the tubulin heterodimer shows a negative potential, suggesting the two proteins attract each other. Secondly, the electrostatic forces between the Eg5 motor domain and tubulin heterodimer are attractive at various distances. Moreover, the electric field lines between the two clearly show that Eg5 has a strong attractive binding force toward the tubulin heterodimer. Additionally, the pH effects on the binding energy for the Eg5 motor domain interacting with tubulin heterodimer and folding energy of Eg5 were analyzed using DelPhiPKa, which has been widely used to study pH dependence of folding and binding energies for biomolecules [49], [35], [50]. The dependence on pH reveals that the Eg5 motor domain performs its functions best when in a weak acidic environment (the optimal pH for the folding energy is 6 while the optimal pH for the binding energy is 4.5). Finally, the residues forming salt bridges and hydrogen bonds were identified with MD simulations. These residues are critical for the Eg5 motor domain to bind the microtubule. Knowing these specific residues may be helpful for anticancer drug design in future work.
2. Methods
2.1. Structure preparation
The motor domain of Eg5 in complex with the microtubule structure was obtained from the RCSB Protein Data Bank (PDB ID: 6TA4 [22]), and is based on the 6.10 Å resolution cryo-EM structure. In this structure, the motor domain of Eg5 was in the adenylyl imidodiphosphate (AMP-PNP) state and bound to tubulin proteins.
2.2. Electrostatic calculations
Two computational tools, DelPhi [42], [43] and DelPhiForce [44], [45], were used to calculate the electrostatic potential and force of the Eg5/microtubule complex. These two tools use the finite difference method to solve the Poisson-Boltzmann equation (PBE):
| (1) |
where ρ(r) is the permanent charge density, ε(r) is the dielectric permittivity, T is temperature, κ is the Debye-Huckel parameter and kB is the Boltzmann constant.
The electrostatic potential of the Eg5 motor domain and tubulin heterodimer was visualized by UCSF Chimera [51]. Positively and negatively charged regions are colored in blue and red, respectively. The color scale ranges from −1.0 to 1.0 kT/e (see Fig. 1). To see the directions and strength of electric fields, DelPhiForce was utilized to determine the electrostatic forces between the Eg5 motor domain and α,β-tubulin heterodimer. The Eg5 motor domain and the tubulin heterodimer were separated from 14 Å to 40 Å with the step size of 2 Å using StructureMan [52]. The electrostatic forces and electric field lines between the motor domain of Eg5 and the tubulin heterodimer were displayed using Visual Molecular Dynamics (VMD) [53].
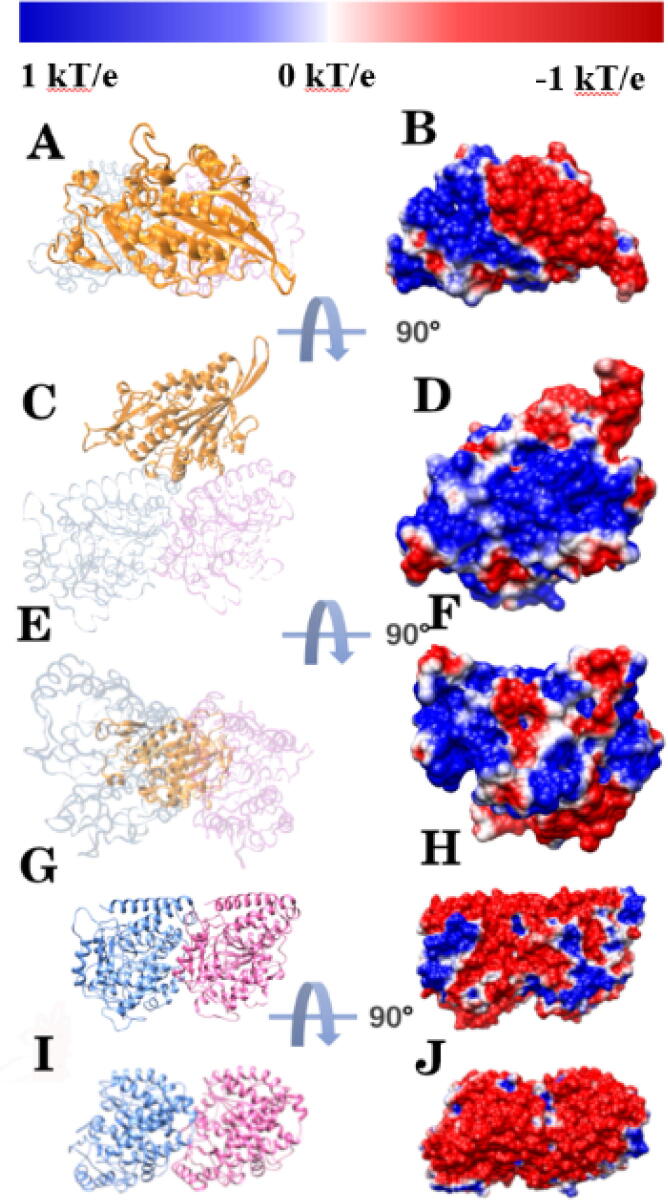
Fig. 1.
Structure and electrostatic potential on the surface of the Eg5 motor domain. Here, α tubulins, β tubulins, and the Eg5 motor domain are shown in blue, pink and orange, respectively. Positively and negatively charged regions are colored in blue and red, respectively. (A) Top view of the structure of Eg5. (B) Top view of the electrostatic potential on the surface of Eg5. (C) Front view of the structure of Eg5. (D) Front view of the electrostatic potential on the surface of Eg5. (E) Bottom view of the structure of Eg5. (F) Bottom view of the electrostatic potential on the surface of Eg5. (G) Front view of the structure of the tubulin heterodimer. (H) Front view of the electrostatic potential on the surface of the tubulin heterodimer. (I) Top view of the structure of the tubulin heterodimer. (J) Top view of the electrostatic potential on the surface of the tubulin heterodimer. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To simulate the Eg5 motor domain binding with a microtubule, we modeled a piece of microtubule structure based on PDB entry 6B0I [54]. Such a microtubule contains 75 tubulin dimers with a length of 400 Å. The Eg5 motor domain binding to the microtubule was modeled using 6TA4 [22]. The electrostatic binding forces between the Eg5 motor domain and the microtubule were calculated at a distance of 15 Å using DelPhiForce [44], [45].4 Folding free energy calculation
DelPhiPKa [46], [47] was used to calculate pKa values of the Eg5 motor domain and tubulin heterodimer. The pKa range was set from 0 to 14 with an interval of 0.5. First, we calculated the net charge of the unfolded state with the following equation:
| (2) |
where represents all titratable groups, y(i) is −1 for acidic groups, and y(i) is + 1 for basic groups.
Then, the pH dependence of the folding free energy was calculated using the following equation:
| (3) |
where represents the total net charge of the unfolded state (which is calculated in equation (2)), represents the total net charge of the folded state, R is the universal gas constant and T is the temperature.
2.3. Binding free energy calculation
Based on the pKa values calculated by DelPhiPKa [46], [47], the pH dependence of the binding free energy of the Eg5 motor domain and tubulin heterodimer was obtained by the following equation:
| (4) |
where represents binding free energy, , , and are the net charges of the complex, the Eg5 motor domain, and tubulin heterodimer, respectively, R is the universal gas constant, and T is the temperature.
2.4. MD simulations
MD simulations were performed on Stampede2 at the Texas Advanced Computing Center (https://www.tacc.utexas.edu). Three 60-ns simulations of the Eg5/microtubule complex were carried out. The minimization was set to 20,000 steps, temperature to 300 K and pressure to follow Langevin dynamics. The coordinates (x, y, z) of the full-system periodic electrostatics were fit to the grid size (115, 86, 116). In each simulation, residues with any atom within 15 Å from the binding interfaces were treated as interfacial residues. All the interfacial residues were set free while non-interfacial residues were constrained. Based on the Root Mean Square Deviation (RMSD) plots (see Supplementary Fig. 1), the last 20 ns (from 8000 frame to 12,000 frames) of the simulations were selected for analysis because the simulations in the last 20 ns were more stable than that in the first 40 ns. The simulations were visualized by VMD [53] (see Supplementary Movies 1–3).
To further explore the interactions between the Eg5 motor domain and the tubulin heterodimer, the hydrogen bonds and salt bridges between the two were analyzed. The threshold for hydrogen bonds was 3.2 Å while the threshold for salt bridges was 4 Å. The last 20 ns in each of the three simulations were selected and analyzed to identify the salt bridges and hydrogen bonds. In the analysis, these salt bridges and hydrogen bonds which were rarely formed (<10 % occupancy) in the whole simulations were ignored.
To expand the scope of our work, we introduced mutations on the loop 5 (L5) of Kinesin-5. Previous work has shown this element is critical to the functioning of the human Kinesin-5 motor domain [55]. Work done by Dr. Rosenfeld and his group investigated mutation sites on loop L5 of Eg5 [56]. To confirm the importance of the mutations on L5, four 10 ns simulations were performed. These involved wild-type, P131A, P121A, and P131A + P121A versions of Eg5. To focus on the flexibility of L5, a 10 ns simulation was performed on each structure. In each simulation, all the residues of Eg5 were constrained except for those in L5 (amino acids 117–134). The RMSFs were calculated based on the simulations.
3. Results and discussions
We investigated the electrostatic features of the Eg5 motor domain and tubulin heterodimer. Furthermore, the pH-dependent binding and folding energies of the Eg5-tubulin heterodimer complexes and their components were analyzed. Finally, the hydrogen bonds and salt bridges in each complex were investigated using MD simulations. We identified the residues involved in the hydrogen bonds and salt bridges between Eg5 and the αβ-tubulin complexes.
3.1. Electrostatic potential on surfaces
DelPhi [42], [43] was utilized to study the electrostatic features of the Eg5 motor domain and αβ -tubulin heterodimer. Fig. 1 shows the electrostatic potential of the Eg5 motor domain and the tubulin heterodimer. The positively and negatively charged regions are colored in blue and red, respectively. The color scale is from −1 kT/e to 1 kT/e. A bottom view of the motor domain (see Fig. 1E and 1F) shows the interfacial residues that contact the tubulin heterodimer, which is primarily positively charged. Fig. 1J shows the negative electrostatic potential of the tubulin heterodimer binding interface. Together, these figures imply the motor domain of Eg5 is attracted to the tubulin heterodimer because the two interfaces have opposite net charges. Electrostatic potential of the Eg5 motor domain and the αβ -tubulin heterodimer indicate that the electrostatic binding forces between them may enhance the stabilities of the complexes.
3.2. Electric field lines
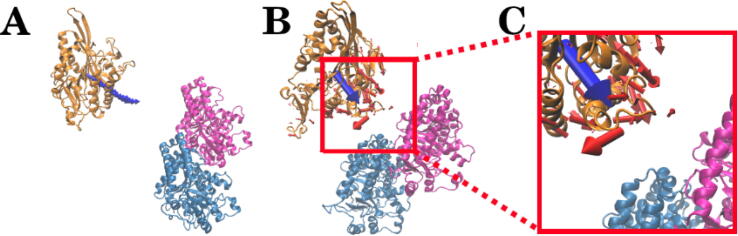
The original structure of Eg5 binding to the tubulin heterodimer was determined by electron microscopy [22]. To better visualize interactions between the components, we separated Eg5 from the tubulin heterodimer by 20 Å (Fig. 2A). The electric field lines between the Eg5 motor domain and tubulin heterodimer were calculated to investigate their electrostatic interactions (Fig. 2B). As shown in Fig. 2B, the interface of the Eg5 motor domain was separated from the tubulin heterodimer by 20 Å to visualize the electric field lines. Densities of field lines indicate the strengths of electrostatic interactions. The electric field line distributions clearly show that the Eg5 motor domain has strong, attractive binding forces to the tubulin heterodimer with dense field lines representing the strongest interactions. Such attractive binding forces are confirmed by a later section discussing electrostatic forces.
Fig. 2.
Structures of Eg5 and the tubulin heterodimer with the electric field lines. (A) Structures of Eg5 and tubulin at a 20 Å distance. The Eg5 motor domain is shown in orange, and α and β-tubulin are shown in blue and pink, respectively. (B) Electric field lines between the Eg5 motor domain and tubulin heterodimer. They are positioned the same as in figure (A). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Electrostatic forces
The electrostatic forces between the Eg5 motor domain and the tubulin heterodimer were quantified by DelPhiForce [44], [45] at distances ranging from 14 Å to 40 Å with a step size of 2 Å. Then the electrostatic forces between the Eg5 motor domain and the tubulin heterodimer were calculated at each position by implementing DelPhiForce [44], [45]. The blue arrows in Fig. 3 demonstrate the directions of net forces between the Eg5 motor domain and the tubulin heterodimer. Note that the blue arrows in Fig. 3A only represent the directions of the forces. The blue arrows are normalized to the same size for better visualization. Fig. 3A reveals that the net forces are attractive between the Eg5 motor domain and tubulin heterodimer. To analyze the system in more detail, the electrostatic forces on individual residues in the Eg5 motor domain at 20 Å are shown in Fig. 3B, where the blue arrow displays the net force between the Eg5 motor domain and tubulin heterodimer, and the red arrows represent forces on individual residues. The directions of the red arrows indicate the directions of the binding forces on residues. The sizes of the red arrows indicate the magnitudes of the forces on the residues.
Fig. 3.
Electrostatic forces on the Eg5 motor domain and tubulin heterodimer at distances from 14 Å to 40 Å. α-tubulin and β-tubulin are shown in blue and pink, respectively (A) Net direction of the electrostatic force on the Eg5 motor domain (orange) with tubulin heterodimer at distances from 14 Å to 40 Å with a step size of 2 Å. (B) Electrostatic forces between the Eg5 motor domain (orange) and tubulin heterodimer at 20 Å. (C) A close-up view of (B). The blue arrows show the net force, while the red arrows represent forces on individual residues of the Eg5 motor domain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
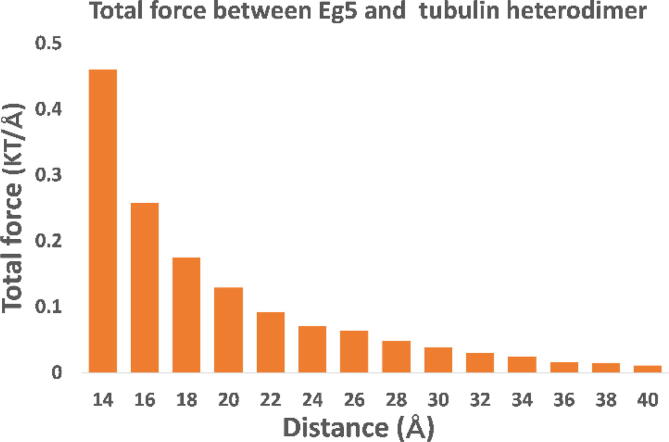
Besides the directions shown in Fig. 3A, the magnitudes of the net forces at different distances are presented in Fig. 4. As shown in the figure, the attractive force decreases when the distance between the motor domain and tubulin heterodimer is increased, which is expected due to Coulomb’s law.
Fig. 4.
The net electrostatic forces between the Eg5 motor domain and tubulin heterodimer at distances from 14 Å to 40 Å with a step size of 2 Å.
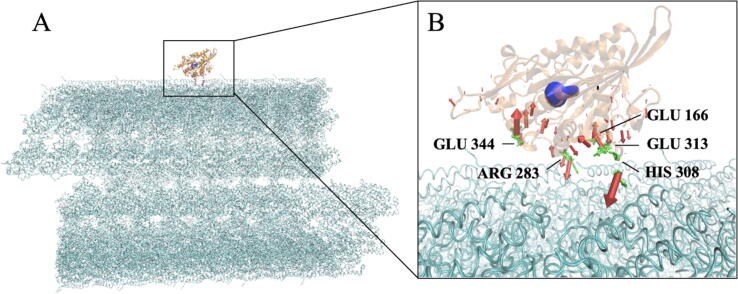
In addition to the three-protein system, we calculated the electrostatic forces between the Eg5 motor domain and the large-scale microtubule at 15 Å to study their interaction in a more physiologically relevant setup. The microtubule structure consisted of 75 α/β tubulin dimers. The total net force was shown in Fig. 5B with a blue arrow, which demonstrates the direction of net force. The arrow reveals the net force is mainly a sliding force between the Eg5 motor domain and the microtubule when the distance is 15 Å. This is reasonable because when the kinesin motor is detached from the microtubule, the trailing motor needs to be moved to be the heading motor. Such a sliding force helps the linker domain of kinesin to move the kinesin away from the binding pocket. The electrostatic forces on individual residues on the Eg5 motor domain are shown in Fig. 5B with red arrows. The sizes of the red arrows indicate the magnitudes of the forces on individual residues, while the direction represents the way of the forces on the residues on the Eg5 motor domain. The side chains of the five strongest individual forces are highlighted in green in Fig. 5B. Interestingly, three out of the five largest individual forces are GLU with negative charges. The positive HIS308 and ARG283 residues provide significant attractive forces for Eg5 to bind with tubulins. This is consistent with our electrostatic potential analysis. Because the interface of tubulin dimer is mainly negatively charged, as shown in Fig. 1.
Fig. 5.
The electrostatic forces between the Eg5 motor domain and microtubule at a 15 Å distance. (A) Electrostatic force of the Eg5 motor domain (orange) with microtubule at a 15 Å distance. (B) A close-up view of (A). The blue arrows show the net force, while the red arrows display forces on individual residues of the Eg5 motor domain. The side chains of the top five individual forces are highlighted in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Folding energies
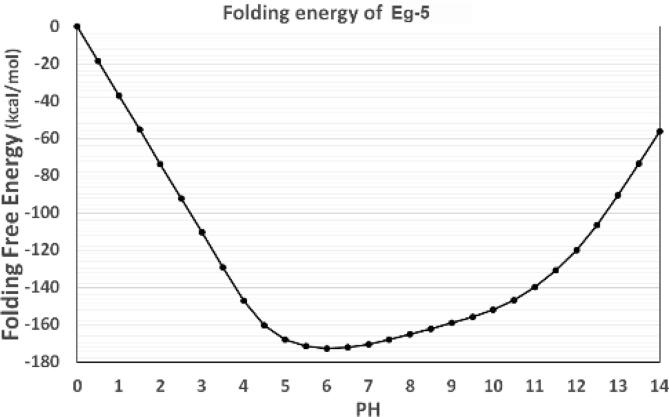
In the protein complex, the folding energies generally depend on pH. We calculated the net charges of the Eg5 motor domain with the DelPhiPKa web server [46], [47]. The pH range was set from 0.0 to 14.0 with an interval of 0.5. The method used in this work involves calculating relative folding energies to investigate the folding energy pH dependence. The absolute folding energies are not calculated in this work. Such relative folding energy at pH 0 is set as the reference, which is 0 kcal/mol. Fig. 6 shows the calculated folding energies of the Eg5 motor domain at different pH values. The curve of folding energy indicates that the optimum pH value for the motor domain is pH 6. Thus, the pH dependence of the folding free energy demonstrates that the Eg5 motor domain maintains its structure best when in a weakly acidic environment.
Fig. 6.
The pH-dependence of the folding energy of the Eg5 motor domain. The pH range was set from 0.0 to 14.0 with an interval of 0.5.
3.5. Binding energies
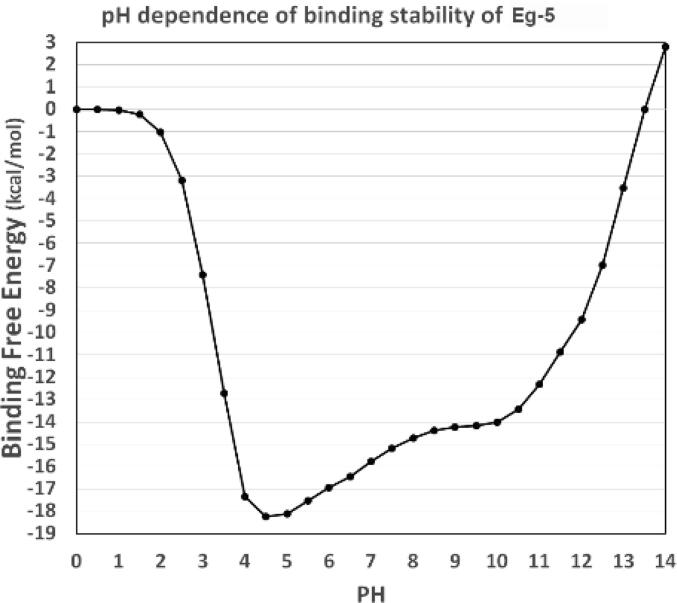
For different complexes, pKa shifts can be used to indicate the electrostatic contribution to the binding energies [57]. We calculated pKa using the DelPhiPKa web server [46], [47]. For the Eg5/tubulin complex, we ran one pKa calculation. For the motor domain and tubulin heterodimer, we performed two calculations. The pH ranges were set from 0.0 to 14.0 with an interval of 0.5. Like for the folding energy, we calculated the relative binding energies rather than absolute energies. The binding energy at pH 0 is set as the reference, which is 0 kcal/mol. The pH-optimum is defined as the pH at which the complex structure yields the most robust binding interactions. As shown in Fig. 7, the binding energy curve shows the strongest binding free energy at pH 4.5, which is the optimum pH for the binding of Eg5 and tubulin heterodimer. Combining the pH dependences of Eg5 folding energies and the Eg5: tubulin heterodimer binding energies reveals that the complex structure is most stable in the acidic environment.
Fig. 7.
The pH-dependence of the binding energy between the Eg5 motor domain and tubulin heterodimer. The pH range was set from 0.0 to 14.0 with an interval size of 0.5.
3.6. Hydrogen bonds
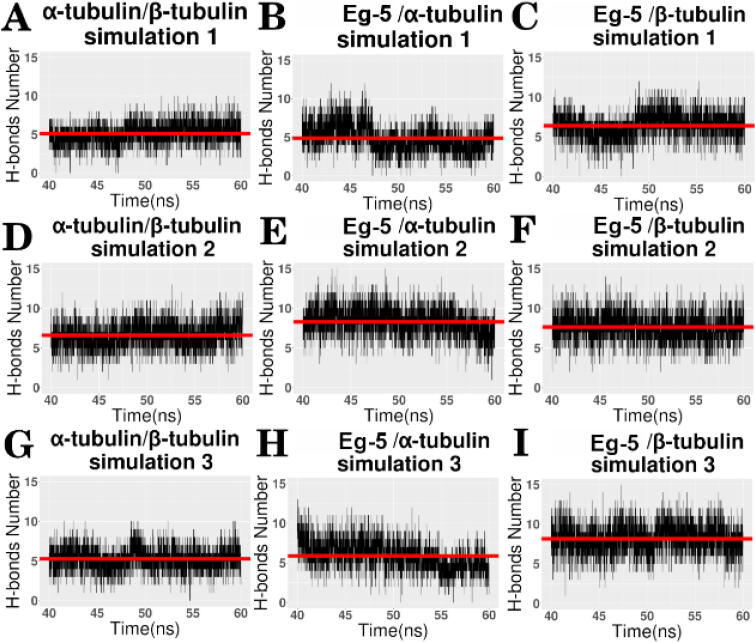
Hydrogen bonds among three complexes (α-tubulin/β-tubulin, Eg5/α-tubulin, Eg5/β tubulin) were analyzed based on the last 20 ns of MD simulations (see Fig. 8). The average number of hydrogen bonds are shown graphically with red lines in Fig. 8 and numerically in Table 1. By comparing average numbers of hydrogen bonds in the simulations of the three complexes (α: β tubulins, Eg5: α tubulin, Eg5: β tubulin), We found among the three complexes that the difference in the number of average hydrogen bonds between Eg5 and α tubulin was greatest. The average number ranges from 4.89 to 8.30. In the three simulations for the tubulin heterodimer, the difference of average hydrogen bonds number is only 1.5. This is likely because the α/β tubulin heterodimer is stable; but the binding interactions between Eg5 and α/β tubulin heterodimer are not permanent. The Eg5 need to be bound to and detached from the α/β tubulins periodically during the stepping process. Therefore, the hydrogen bonds between α and β tubulins are more stable. In the three complexes, the difference between the average number of hydrogen bonds between Eg5 and α tubulin is the largest, indicating that Eg5 and α tubulin may have the most flexible interfaces.
Fig. 8.
Hydrogen bonds at the interfaces of the Eg5 motor domain and tubulin heterodimer. The average number of hydrogen bonds is shown with red lines. (A) (D) (G) Number of hydrogen bonds between α tubulin and β tubulin. (B) (E) (H) Number of hydrogen bonds between the Eg5 motor domain and α tubulin. (C) (F) (I) Number of hydrogen bonds between the Eg5 motor domain and β tubulin. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Average numbers of hydrogen bonds.
| Simulation | α and β tubulin | Eg5 and α tubulin | Eg5 and β tubulin |
|---|---|---|---|
| 1 | 5.10 | 4.89 | 6.41 |
| 2 | 6.60 | 8.30 | 7.63 |
| 3 | 5.28 | 5.88 | 8.16 |
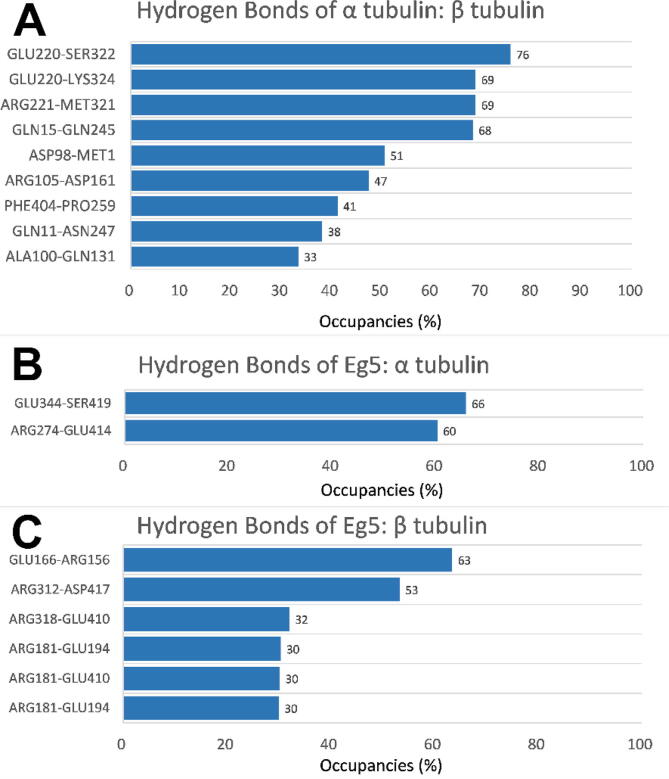
To analyze the average occupancies of hydrogen bonds formed in the three simulations for the Eg5/tubulin heterodimer complex, the three simulations were combined to calculate the occupancies for the hydrogen bonds. Occupancies of hydrogen bonds above 30 % are shown in Fig. 9. The hydrogen bonds with occupancies above 10 % are listed in the Supplementary Information (Table S1). We found that the total number of hydrogen bonds formed at the binding interface of Eg5/α-tubulin is similar to the number at the binding interface of Eg5/β-tubulin (see Supplementary Information Table S1). The total number of hydrogen bonds at the binding interface of α-tubulin/β-tubulin is the smallest—about half that of Eg5/α-tubulin or Eg5/β-tubulin. However, when the cutoff value was set at 30 %, as shown in Fig. 9, α-tubulin/β-tubulin was found to have more hydrogen bonds at the binding interface than the other two complexes. This observation shows the interaction between α and β-tubulins is more stable than the interaction between Eg5 and the tubulin heterodimer because high occupancy hydrogen bonds indicate strong binding interactions. High occupancy hydrogen bonds between Eg5 and α or β tubulin are crucial for exploring the binding mechanism of Eg5 with microtubules. Hydrogen bonds with high occupancy between the Eg5 motor domain and α tubulin include GLU344 (Eg5) – SER419 (α-tubulin) and ARG274 (Eg5) - GLU414 (α-tubulin). For Eg5/β-tubulin two such bonds are GLU166 (Eg5) - ARG156 (β-tubulin) and ARG312 (Eg5) - ASP417 (β-tubulin).
Fig. 9.
Occupancies of hydrogen bonds above 30% at interfaces among three complexes (α-tubulin/β-tubulin, Eg5/α-tubulin, Eg5/β tubulin). (A) The occupancies of hydrogen bonds between α-tubulin and β-tubulin (left residue from α tubulin). (B) The occupancies of hydrogen bonds between the interface of the Eg5 motor domain and α-tubulin (left residue from the Eg5 motor domain). (C) The occupancies of hydrogen bonds between the interface of the Eg5 motor domain and β tubulin (left residue from the Eg5 motor domain).
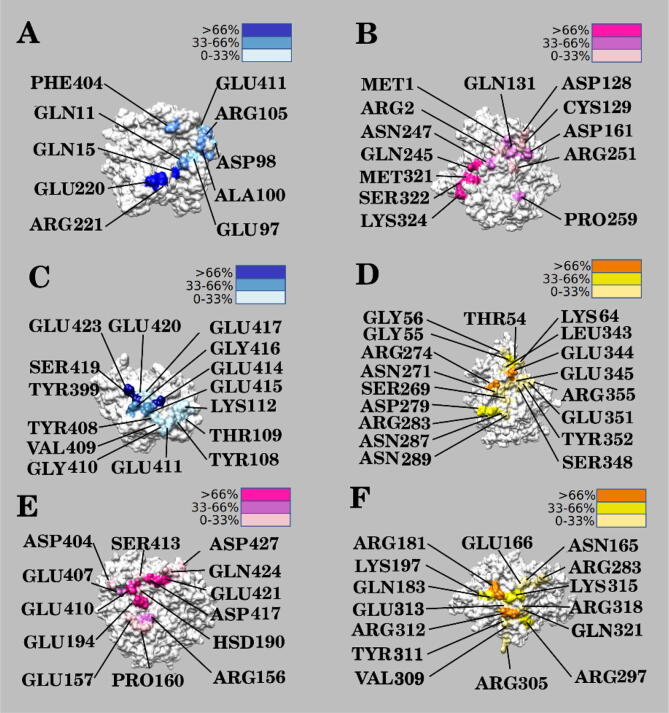
We calculated the occupancies of residues forming hydrogen bonds during the three simulations as well. Fig. 10 shows the distribution and occupancies of residues that form hydrogen bonds on the interfaces of the Eg5 motor domain, α-tubulin, and β-tubulin. Residues with high occupancies that likely contribute significantly to the binding interactions are marked in deep colors. As shown in the Fig. 10, all the high occupancy residues are on the binding interfaces.
Fig. 10.
The distribution of hydrogen bond residues colored by occupancy. (A) (B) The distribution of hydrogen bond residues between α-tubulin and β-tubulin. Figure (A) is α-tubulin while figure (B) is β-tubulin. (C) (D) The distribution of hydrogen bond residues between Eg5 and α-tubulin. Figure (C) is α-tubulin while figure (D) is Eg5. (E) (F) The distribution of hydrogen bond residues between Eg5 and β-tubulin. Figure (E) is β-tubulin while figure (F) is Eg5.
At the binding interface of α-tubulin/β-tubulin, the hydrogen bonds with occupancies over 66 % on α tubulin are GLU220, ARG221, and GLN15. Residues with occupancy greater than 66 % for β tubulin are SER322, LYS324, MET321 and GLN245. Between Eg5 and α-tubulin, the residues for α-tubulin are GLU414, SER419, and GLU423 while for Eg5 these are GLU344 and ARG274. Lastly, between Eg5 and β-tubulin the residues are GLU194, GLU410, GLU421 and ASP417 for β-tubulin while they are ARG181 and ARG312 for Eg5.
The residues which have crucial roles in forming hydrogen bonds and may significantly contribute to binding interactions are mostly distributed on the binding interfaces. Except for neutral residues, most of the hydrogen bond residues on the Eg5 interface are positively charged, while most hydrogen bond residues on the α-tubulin and β-tubulin interfaces are negatively charged. This is consistent with our previous analysis of the electrostatic potential on the interface of the Eg5 motor domain and tubulin heterodimer. The Eg5 interfacial residues are mainly positively charged, while the tubulin heterodimer binding interface shows many negatively charged residues. Thus, the Eg5 motor domain attracts the tubulin heterodimer due to opposite net charges.
3.7. Salt bridges
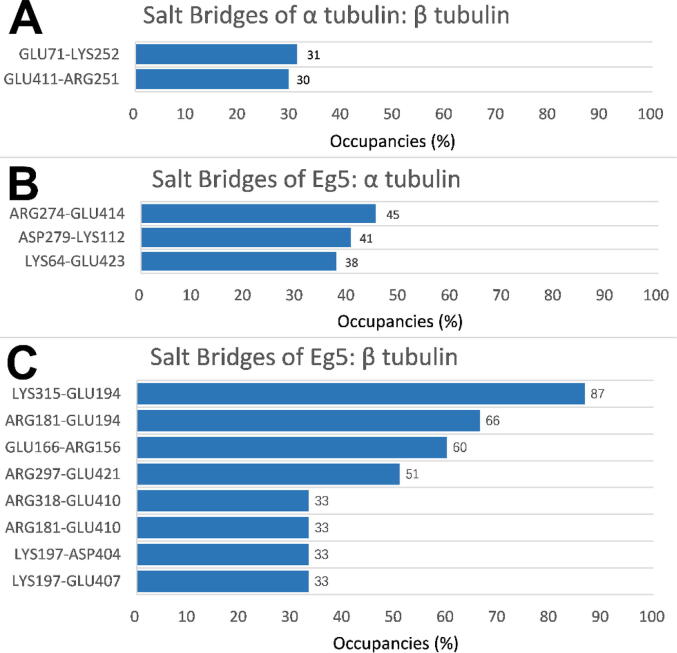
In order to analyze the average occupancies of salt bridges formed in the three simulations for the Eg5/heterodimer complex, the three simulations were combined to calculate the occupancies of the salt bridges. Occupancies of salt bridges above 30 % are shown in Fig. 11. The salt bridges with occupancies above 10 % are listed in the Supplementary Information (Table S2). Only two salt bridges were identified at the interface between α-tubulin and β-tubulin, while five salt bridges were identified at the interface between the Eg5 motor domain and α-tubulin. The binding interface of Eg5/β-tubulin was found to have the highest number of salt bridges with 11 pairs. Salt bridges with the maximum occupancy (86.68 %) were also found to be at the binding interface of Eg5/β-tubulin. In addition, the Eg5/β tubulin interface has four high occupancy salt bridges (occupancies greater than 50 %). In the other two complexes, the occupancies of all salt bridges are less than 50 %. These results indicate that the interaction of Eg5/β-tubulin is likely the strongest.
Fig. 11.
Occupancies of salt bridges above 30% at interfaces among three complexes (α-tubulin/β-tubulin, Eg5/α-tubulin, Eg5/β tubulin). (A) The occupancies of salt bridges between α-tubulin and β-tubulin (left residue from α-tubulin). (B) The occupancies of salt bridges between the Eg5 motor domain and α-tubulin (left residue from Eg5). (C) The occupancies of salt bridges between the interface of the Eg5 motor domain and β-tubulin (left residue from Eg5).
Very few salt bridges were found at the interface of α-tubulin/β-tubulin. Combined with the hydrogen bonds analysis, these facts demonstrate that the key interactions between α and β tubulins are the hydrogen bonds on the interface. On the Eg5/heterodimer interface, more salt bridges are observed. This indicates that the electrostatic interactions are the key factor for Eg5/microtubule interactions. The interactions of Eg5/α-tubulin and Eg5/β-tubulin are not symmetric. The salt bridge number for Eg5/β-tubulin is much higher than that of Eg5/α-tubulin. Furthermore, the occupancies of the salt bridges from Eg5/β tubulin are also higher than that of Eg5/α tubulin. Such asymmetric salt bridge distributions may contribute to the unidirectional motility of Eg5 moving along the microtubule. β-tubulin forms more salt bridges; therefore, it may provide stronger attractive forces to guide Eg5 towards the microtubule plus end.
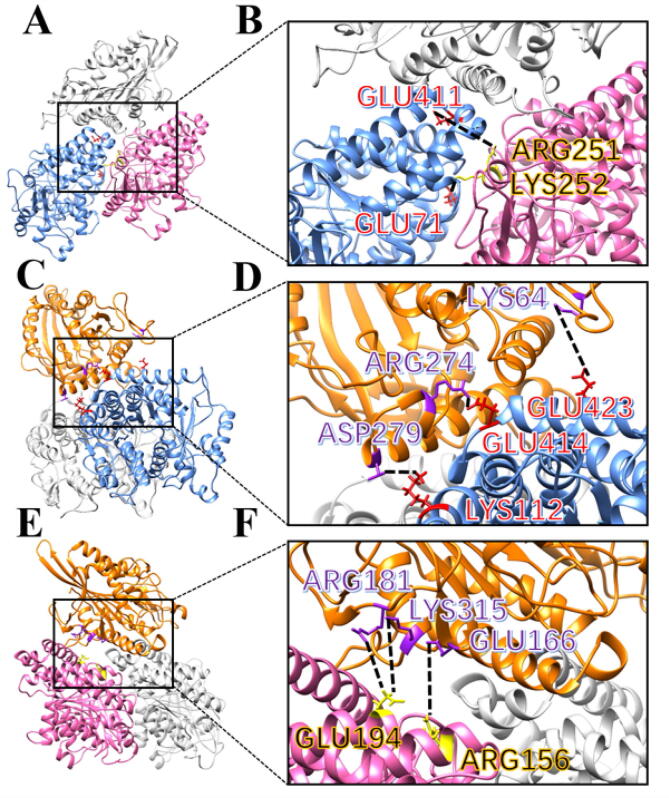
The top three salt bridges between each two of the monomers in the two-protein complexes are shown in Fig. 12. There are only two salt bridges between α-tubulin and β-tubulin, as shown in Fig. 12A and 12B. From the data, these high occupancy salt bridges are essential to study the binding mechanisms between Eg5 and the microtubule.
Fig. 12.
The top three salt bridges at interfaces among the Eg5 motor domain, α-tubulin, and β-tubulin (shown in orange, blue, and pink, respectively). (A) The salt bridges between α-tubulin and β-tubulin. (B) A close-up view of (A). (C) The top three salt bridges between Eg5 and α-tubulin. (D) A close-up view of (C). (E) The top three salt bridges between Eg5 and β-tubulin. (F) A close-up view of (E). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
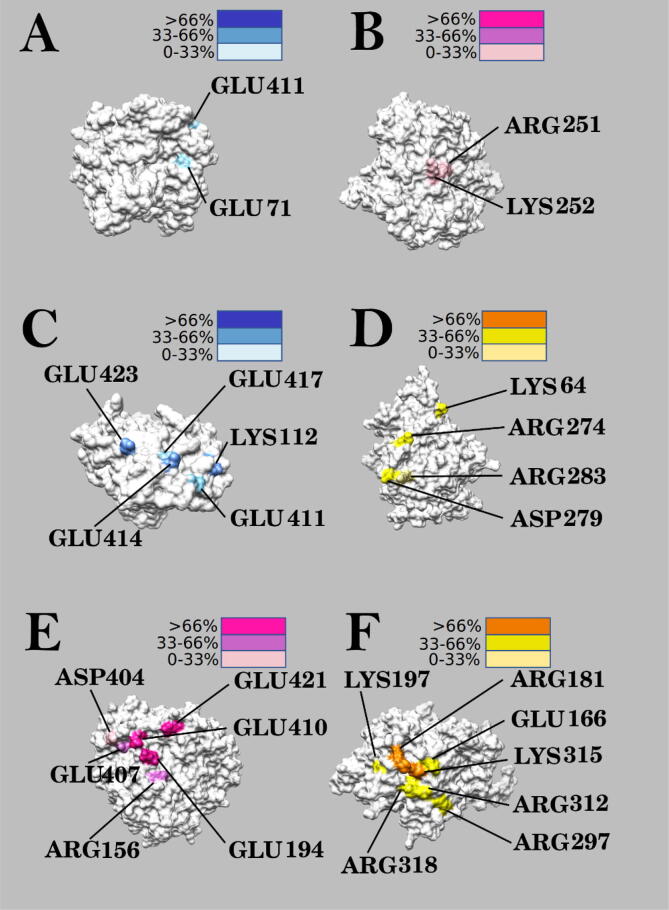
The distributions and occupancies of residues involved in salt bridges on the interfaces of the Eg5 motor domain, α-tubulin, and β-tubulin, are shown in Fig. 13. Darker colors show higher occupancies.
Fig. 13.
The distribution of residues involved in salt bridges are colored by occupancy (A) (B) The distribution of salt bridge residues between α-tubulin and β-tubulin. Figure (A) is α-tubulin while figure (B) is β-tubulin. (C) (D) The distribution of salt bridge residues between Eg5 and α-tubulin. Figure (C) is α-tubulin while figure (D) is Eg5. (E) (F) The distribution of salt bridge residues between Eg5 and β-tubulin. Figure (E) is β-tubulin while figure (F) is Eg5.
GLU411 (α-tubulin), GLU71 (α-tubulin), ARG251 (β-tubulin), and LYS252 (β-tubulin) are pivotal for forming salt bridges between α and β-tubulins. Residues that are involved in forming salt bridges at the interface of Eg5/α-tubulin include GLU414, GLU423, GLU417, GLU411 and LYS112 from α-tubulin, and ARG274, ASP279, LYS64 and ARG283 from Eg5. Except for LYS112, residues on the interface of α-tubulin, are all glutamates and negatively charged. On the Eg5 interface, most salt bridge residues positively charged—except ASP279. The residues GLU194, GLU410, GLU421, GLU407, ARG156 and ASP404 on β-tubulin, and ARG181, LYS315, LYS197, GLU166, ARG312, ARG297 and ARG318 on Eg5, play a vital role in forming salt bridges between the two proteins. Most of the residues on β-tubulin are negatively charged (except ARG156), while most of the residues on Eg5 are positively charged (except GLU166). This fact is in line with the results of the electrostatic potential analysis. Altogether, these salt bridges further explain how the Eg5 motor domain might be attracted to the tubulin heterodimer.
3.8. Explain previous experimental results
There are many experimental studies investigating the mutation sites on human Kinesin-5. We have conducted several computational analyses and compared the results with the experimental findings, which corroborate the mechanisms of previous studies. As discussed in the introduction, work done by Dr. Rosenfeld’s group investigated the mutation sites on loop L5 of Eg5 [56]. The loop 5 (L5) element is critical to the functioning of the human Kinesin-5 motor domain. To study the importance of mutations on L5, four 10 ns simulations were performed on Kinesin-5 variants (wild type, P131A, P121A, P131A + P121A). In each simulation, all the residues of Eg5 were constrained except for the L5 (residues 117 to 134). The RMSFs were calculated based on the simulations. The results show that the mutation P131A reduces the flexibility of L5 significantly (see Supplementary Fig. S2), which may weaken the function of L5. This is consistent with the experimental results [56].
Another paper from Dr. Kapoor’s group investigated two mutation sites located on β-tubulin which alter dynamic instability at the microtubule plus and minus-ends [58]. The two residues: ASP 417 and ARG 262, which were studied in this article, are all located on the interfacial surface in contact with the Eg5 motor domain. Our study found that residue ASP 417 forms hydrogen bonds with three residues: ARG312, TYR311, GLU313. The corresponding occupancies of the three hydrogen bonds are 53.34 %, 11.78 %, and 11.71 %, respectively. These data reveal that ASP417 plays a significant role of stabilizing the complex structure by forming hydrogen bonds (shown in Supplementary Fig. S3). ARG 262 forms a weak salt bridge with GLU313 with an occupancy of 2.23 %. In addition, several studies have probed mutated residues which confer resistance to Eg5-targeting drugs [59]. Dr. Dimitrios A. Skoufias and his group found that the expression of Eg5 single point mutants R119A, D130A, L132A, I136A, L214A, and E215A provided significant resistance to monastrol. The mutants involved in their study are drug-related amino acids, which are located at the ATP binding site. The mutation sites will be studied in our future work.
4. Conclusions
Electrostatic interactions between Eg5 and the microtubule were investigated using a comprehensive approach. The motor domain of Eg5 shows predominantly positive potential at the binding interface to attract the tubulin heterodimer which has negative potential on the binding interface. Electric field lines and electrostatic binding forces are provided, which demonstrate attractive forces between Eg5 and the tubulin heterodimer. For the folding energy of Eg5, the optimal pH value is found to be 6. The binding energy dependence on pH shows that the optimal pH value is 4.5. Together, these values mean that Eg5 is most stable and performs its function best when in an acidic environment.
Furthermore, residues forming salt bridges and hydrogen bonds are identified by MD simulations. In the three complexes (Eg5/α-tubulin, Eg5/β-tubulin, α-tubulin/β-tubulin), we find that Eg5/β-tubulin has the largest average number and highest occupancies of salt bridges, which indicates an asymmetric binding mechanism whereby Eg5 moves along the microtubule. Compared with hydrogen bonds, salt bridges play a more critical role in the Eg5/microtubule interaction. In contrast, the key forces between α and β-tubulins are the hydrogen bonds on the interface of the two proteins.
Between Eg5 and α-tubulin, most salt bridges involved GLU residues on α-tubulin. This contrasts with the salt-bridge-involved residues on Eg5 which were mostly positively charged. For Eg5 and β-tubulin, most of the residues on β-tubulin are negatively charged while most of the residues on Eg5 are positively charged. These findings are consistent with the electrostatic potential results, which show the residues on the interface of Eg5 are mainly positively charged while those on the heterodimer binding interface are predominantly negatively charged. These residues are critical for Eg5′s motility and binding to the microtubule. Furthermore, knowing how these residues function may help guide Eg5-focused anticancer drug design. We also conducted several calculations and compared the results with previous experimental studies, which revealed some mechanisms of Eg5 binding with microtubule. It also demonstrates that the combination of computational and experimental works is a promising direction to discover the fundamental mechanisms for biology problems.
CRediT authorship contribution statement
Wenhan Guo: Conceptualization, Data curation, Methodology, Software, Validation, Formal analysis, Supervision, Visualization, Writing – original draft, Writing – review & editing. Shengjie Sun: Validation, Formal analysis, Supervision, Writing – review & editing. Jason E. Sanchez: Writing – review & editing. Alan E. Lopez-Hernandez: . Tolulope A. Ale: Writing – review & editing. Jiawei Chen: Visualization. Tanjina Afrin: Writing – review & editing. Weihong Qiu: Writing – review & editing. Yixin Xie: Validation, Formal analysis, Supervision. Lin Li: Conceptualization, Data curation, Methodology, Software, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research is funded by Grant SC1GM132043 from National Institutes of Health (NIH) and Grant 5U54MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD), a component of the NIH.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.020.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hirokawa N., et al. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa N., Tanaka Y. Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp Cell Res. 2015;334(1):16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa N., Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 4.Palacios, I.M. and D.S. Johnston, Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. 2002. [DOI] [PubMed]
- 5.Lawrence C.J., et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167(1):19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vale R.D. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 7.Drummond D.R. Seminars in cell & developmental biology. Elsevier; 2011. Regulation of microtubule dynamics by kinesins. [DOI] [PubMed] [Google Scholar]
- 8.Ferenz N.P., Gable A., Wadsworth P. Seminars in cell & developmental biology. Elsevier; 2010. Mitotic functions of kinesin-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann B.J., Wadsworth P. Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 2019;29(1):66–79. doi: 10.1016/j.tcb.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Peters N.T., Kropf D.L. Kinesin-5 motors are required for organization of spindle microtubules in Silvetia compressa zygotes. BMC Plant Biol. 2006;6(1):1–10. doi: 10.1186/1471-2229-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Hancock W.O. Kinesin-5 is a microtubule polymerase. Nat Commun. 2015;6(1):1–10. doi: 10.1038/ncomms9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh S.K., et al. Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell Mol Life Sci. 2018;75(10):1757–1771. doi: 10.1007/s00018-018-2754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon D.M., Roof D.M. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem. 1999;274(40):28779–28786. doi: 10.1074/jbc.274.40.28779. [DOI] [PubMed] [Google Scholar]
- 14.Kashina A., et al. An essential bipolar mitotic motor. Nature. 1996;384(6606):225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholey J.E., et al. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife. 2014;3:e02217. doi: 10.7554/eLife.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roostalu, J., et al., Determinants of polar versus nematic organization in networks of dynamic microtubules and mitotic motors. Cell, 2018. 175(3): p. 796-808. e14. [DOI] [PMC free article] [PubMed]
- 17.Fridman V., et al. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013;126(18):4147–4159. doi: 10.1242/jcs.125153. [DOI] [PubMed] [Google Scholar]
- 18.Edamatsu M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem Biophys Res Commun. 2014;446(1):231–234. doi: 10.1016/j.bbrc.2014.02.106. [DOI] [PubMed] [Google Scholar]
- 19.Düselder A., et al. Deletion of the tail domain of the kinesin-5 cin8 affects its directionality. J Biol Chem. 2015;290(27):16841–16850. doi: 10.1074/jbc.M114.620799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britto, M., et al., Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proceedings of the National Academy of Sciences, 2016. 113(47): p. E7483-E7489. [DOI] [PMC free article] [PubMed]
- 21.Kapitein L.C., et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 22.Peña, A., et al., Structure of microtubule-trapped human kinesin-5 and its mechanism of inhibition revealed using cryoelectron microscopy. Structure, 2020. 28(4): p. 450-457. e5. [DOI] [PMC free article] [PubMed]
- 23.Chen, G.-Y., et al., Kinesin-5 promotes microtubule nucleation and assembly by stabilizing a lattice-competent conformation of tubulin. Current Biology, 2019. 29(14): p. 2259-2269. e4. [DOI] [PMC free article] [PubMed]
- 24.Gigant B., et al. Structure of a kinesin–tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20(8):1001. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- 25.Wang W., et al. Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat Commun. 2017;8(1):1–11. doi: 10.1038/s41467-017-00091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atherton J., et al. The mechanism of kinesin inhibition by kinesin-binding protein. Elife. 2020;9:e61481. doi: 10.7554/eLife.61481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asenjo A.B., et al. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 2013;3(3):759–768. doi: 10.1016/j.celrep.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 28.Grant B.J., et al. Electrostatically biased binding of kinesin to microtubules. PLoS Biol. 2011;9(11):e1001207. doi: 10.1371/journal.pbio.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M., Zheng W. Probing the structural and energetic basis of kinesin-microtubule binding using computational alanine-scanning mutagenesis. Biochemistry. 2011;50(40):8645–8655. doi: 10.1021/bi2008257. [DOI] [PubMed] [Google Scholar]
- 30.Li M., Zheng W. All-atom structural investigation of kinesin–microtubule complex constrained by high-quality cryo-electron-microscopy maps. Biochemistry. 2012;51(25):5022–5032. doi: 10.1021/bi300362a. [DOI] [PubMed] [Google Scholar]
- 31.Aprodu I., Soncini M., Redaelli A. Interaction forces and interface properties of KIF1A kinesin-αβ tubulin complex assessed by molecular dynamics. J Biomech. 2008;41(15):3196–3201. doi: 10.1016/j.jbiomech.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Scarabelli G., Grant B.J. Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule, and neck-linker binding sites. Biophys J. 2014;107(9):2204–2213. doi: 10.1016/j.bpj.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su X., et al. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43(5):751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimamoto Y., Forth S., Kapoor T.M. Measuring pushing and braking forces generated by ensembles of kinesin-5 crosslinking two microtubules. Dev Cell. 2015;34(6):669–681. doi: 10.1016/j.devcel.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W., et al. Electrostatic features for nucleocapsid proteins of SARS-CoV and SARS-CoV-2. Mathemat Biosci Eng MBE. 2021;18(3):2372. doi: 10.3934/mbe.2021120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallivan J.P., Dougherty D.A. A computational study of cation− π interactions vs salt bridges in aqueous media: implications for protein engineering. J Am Chem Soc. 2000;122(5):870–874. [Google Scholar]
- 37.Wang W., Kollman P.A. Computational study of protein specificity: the molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci. 2001;98(26):14937–14942. doi: 10.1073/pnas.251265598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xian Y., et al. The roles of electrostatic interactions in capsid assembly mechanisms of giant viruses. Int J Mol Sci. 2019;20(8) doi: 10.3390/ijms20081876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie, Y., et al., Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Frontiers in molecular biosciences, 2020. 7. [DOI] [PMC free article] [PubMed]
- 40.Sun S., et al. Computational study on the function of palmitoylation on the envelope protein in SARS-CoV-2. J Chem Theory Comput. 2021 doi: 10.1021/acs.jctc.1c00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y., et al. Computational study on DNA repair: the roles of electrostatic interactions Between uracil-DNA glycosylase (UDG) and DNA. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.718587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L., et al. DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012;5:9. doi: 10.1186/2046-1682-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., et al. On the dielectric “constant” of proteins: smooth dielectric function for macromolecular modeling and its implementation in DelPhi. J Chem Theory Comput. 2013;9(4):2126–2136. doi: 10.1021/ct400065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L., et al. DelPhiForce web server: electrostatic forces and energy calculations and visualization. Bioinformatics. 2017;33(22):3661–3663. doi: 10.1093/bioinformatics/btx495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Chakravorty A., Alexov E. DelPhiForce, a tool for electrostatic force calculations: Applications to macromolecular binding. J Comput Chem. 2017;38(9):584–593. doi: 10.1002/jcc.24715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L., Zhang M., Alexov E. DelPhiPKa web server: predicting pKa of proteins RNAs and DNAs. Bioinformatics. 2015;32(4):614–615. doi: 10.1093/bioinformatics/btv607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Li L., Alexov E. pKa predictions for proteins, RNAs, and DNAs with the Gaussian dielectric function using DelPhi pKa. Proteins Struct Funct Bioinf. 2015;83(12):2186–2197. doi: 10.1002/prot.24935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips J.C., et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L., et al. In silico investigation of pH-dependence of prolactin and human growth hormone binding to human prolactin receptor. Comm Comput Phys. 2013;13(1):207. doi: 10.4208/cicp.170911.131011s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie, Y., et al., The pH Effects on SARS-CoV and SARS-CoV-2 Spike Proteins in the Process of Binding to hACE2. Research square, 2021: p. rs. 3. rs-871118. [DOI] [PMC free article] [PubMed]
- 51.Pettersen E.F., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Xian Y., et al. Structure manipulation tool structureman: a structure manipulation tool to study large scale biomolecular interactions. Front Mol Biosci. 2020;7:476. doi: 10.3389/fmolb.2020.627087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.Benoit M.P., Asenjo A.B., Sosa H. Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-04044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waitzman J.S., et al. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101(11):2760–2769. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Behnke-Parks W.M., et al. Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J Biol Chem. 2011;286(7):5242–5253. doi: 10.1074/jbc.M110.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onufriev A.V., Alexov E. Protonation and pK changes in protein-ligand binding. Q Rev Biophys. 2013;46(2):181. doi: 10.1017/S0033583513000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ti S.-C., et al. Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus-and minus-ends. Dev Cell. 2016;37(1):72–84. doi: 10.1016/j.devcel.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tcherniuk S., et al. Mutations in the human kinesin Eg5 that confer resistance to monastrol and S-trityl-L-cysteine in tumor derived cell lines. Biochem Pharmacol. 2010;79(6):864–872. doi: 10.1016/j.bcp.2009.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.