Abstract
In the mammalian retina, a metabolic ecosystem exists in which photoreceptors acquire glucose from the choriocapillaris with the help of the retinal pigment epithelium (RPE). While the photoreceptor cells are primarily glycolytic, exhibiting Warburg-like metabolism, the RPE is reliant on mitochondrial respiration. However, the ways in which mitochondrial metabolism affect RPE cellular functions are not clear. We first used the human RPE cell line, ARPE-19, to examine mitochondrial metabolism in the context of cellular differentiation. We show that nicotinamide induced rapid differentiation of ARPE-19 cells, which was reversed by removal of supplemental nicotinamide. During the nicotinamide-induced differentiation, we observed using quantitative PCR, Western blotting, electron microscopy, and metabolic respiration and tracing assays that (1) mitochondrial gene and protein expression increased, (2) mitochondria became larger with more tightly folded cristae, and (3) mitochondrial metabolism was enhanced. In addition, we show that primary cultures of human fetal RPE cells responded similarly in the presence of nicotinamide. Furthermore, disruption of mitochondrial oxidation of pyruvate attenuated the nicotinamide-induced differentiation of the RPE cells. Together, our results demonstrate a remarkable effect of nicotinamide on RPE metabolism. We also identify mitochondrial respiration as a key contributor to the differentiated state of the RPE and thus to many of the RPE functions that are essential for retinal health and photoreception.
Keywords: nicotinamide, mitochondria, RPE, differentiation, retina
Abbreviations: COX4, cytochrome c oxidase 4; CRB2, crumbs cell polarity complex component 2; DMEM, Dulbecco's modified Eagle's medium; ECAR, extracellular acidification rate; FCCP, carbonilcyanide p-triflouromethoxyphenylhydrazone; hfRPE, human fetal RPE; MEM-Nic, nicotinamide-containing minimum essential medium; MPC, mitochondrial pyruvate carrier; OCR, oxygen consumption rate; PGC1α, peroxisome proliferator–activated receptor-gamma coactivator-1 alpha; RPE, retinal pigment epithelium; RT, room temperature; TCA, tricarboxylic acid; ZO-1, zonula occludens-1
In the early 20th century, the German physiologist Otto Warburg investigated the metabolism of rapidly dividing cancer cells, which he showed utilized aerobic glycolysis to fuel their anabolic activities, including DNA replication and lipid synthesis (1, 2). Interestingly, Warburg (3, 4) also observed this type of metabolism in the postmitotic cells of the retina. Since then, there have been several studies demonstrating that the light-sensitive photoreceptor cells of the retina are among the most energy-demanding cells in the body (5, 6). As in cancer cells, this is likely because of their large anabolic activity, including the continual synthesis of the phototransductive disk membranes of the outer segment (7, 8), as well as maintenance of the dark current (9). To fuel this type of metabolism, the photoreceptor cells are heavily dependent on an adjacent monolayer of cells called the retinal pigment epithelium (RPE).
The apical surface of the RPE faces the outer segment of the photoreceptors, whereas its basal surface is juxtaposed to the nutrient-rich blood supply of the choriocapillaris. The polarity of this epithelium is essential to the many functions that it performs to maintain the health of the photoreceptors, including (1) vectorial transport of water, ions, nutrients, and waste products, (2) regeneration of the visual chromophore, (3) growth factor secretion, and (4) phagocytosis of distal photoreceptor outer segment disk membranes (10, 11).
In the retinal metabolic ecosystem, glucose supplied by the choriocapillaris enters the RPE, which then spares this fuel molecule, and exports it to the photoreceptor cells (12). In return, the photoreceptor cells transport lactate, the major byproduct of incomplete glucose oxidation, to the RPE, where it may serve as a fuel source. Lactate in the RPE can be converted to pyruvate, via lactate dehydrogenase, which is then able to enter the mitochondrial matrix and completely oxidized to power ATP production via oxidative phosphorylation. Interestingly, this complementary metabolic relationship between photoreceptor and RPE cells is similar to that between neurons and glial cells, which rely on lactate and glucose as fuel sources, respectively (13, 14).
In contrast to photoreceptor cells, the RPE has been shown to be heavily dependent on oxidative phosphorylation and mitochondrial metabolism. This is presumably because RPE cells suppress glucose consumption to allow a sufficient amount of it to pass through to the photoreceptors. Instead, the RPE utilizes lactate (12), succinate (15), glutamine (16), proline (17), and fatty acids derived from ingested photoreceptor outer segment disk membranes (18, 19) as fuel sources for mitochondrial respiration. Studies have shown that RPE cells that switch to a glycolytic-based metabolism become less supportive of their neighboring photoreceptor cells, eventually causing progressive rod and cone degeneration (20). In addition, disruption of mitochondrial oxidative phosphorylation or biogenesis has been shown to induce dedifferentiation in mouse RPE in vivo (21). These studies suggest a link between the mitochondrial metabolism of the RPE and its state of differentiation as a polarized epithelium.
In the present study, we used the human immortalized RPE cell line, ARPE-19, and primary cultures of human fetal RPE (hfRPE) cells to examine mitochondrial metabolism in the context of RPE cell differentiation. We took advantage of the default state of ARPE-19 cells, which exist in an undifferentiated fusiform form when cultured under standard conditions. We have previously identified the vitamin B3 nicotinamide and nicotinamide riboside as rapid inducers of ARPE-19 differentiation (22). Moreover, nicotinamide is often used in media for the differentiation of pluripotent stem cells into RPE (23, 24). Our current study explores the effect of nicotinamide on the metabolism of RPE mitochondria. Importantly, our results show that mitochondrial metabolism is a driver of the differentiation of human RPE cells into a polarized epithelium.
Results
Morphology of ARPE-19 cells in the presence of nicotinamide
When the human immortalized cell line, ARPE-19, was first derived in 1996, it exhibited many characteristics of native RPE, including pigmentation, cobblestone appearance, and expression of RPE-specific markers (25). Following extensive passaging of the cell line over time, many of these characteristics have been lost; today, ARPE-19 cells cultured in standard media have a flat morphology that resembles fibroblastic cells. We have previously reported a culturing method and medium that rapidly differentiates fibroblastic-like ARPE-19 cells into cultures with RPE-like characteristics, including cobblestone morphology, expression of RPE-specific markers, and polarized secretion of proteins (22). Moreover, we identified the vitamin B3, nicotinamide, as an essential component of our culture medium that supported this rapid differentiation of ARPE-19 cells; in the absence of nicotinamide, cultures did not differentiate into a polarized epithelium even after 6 or more weeks in culture.
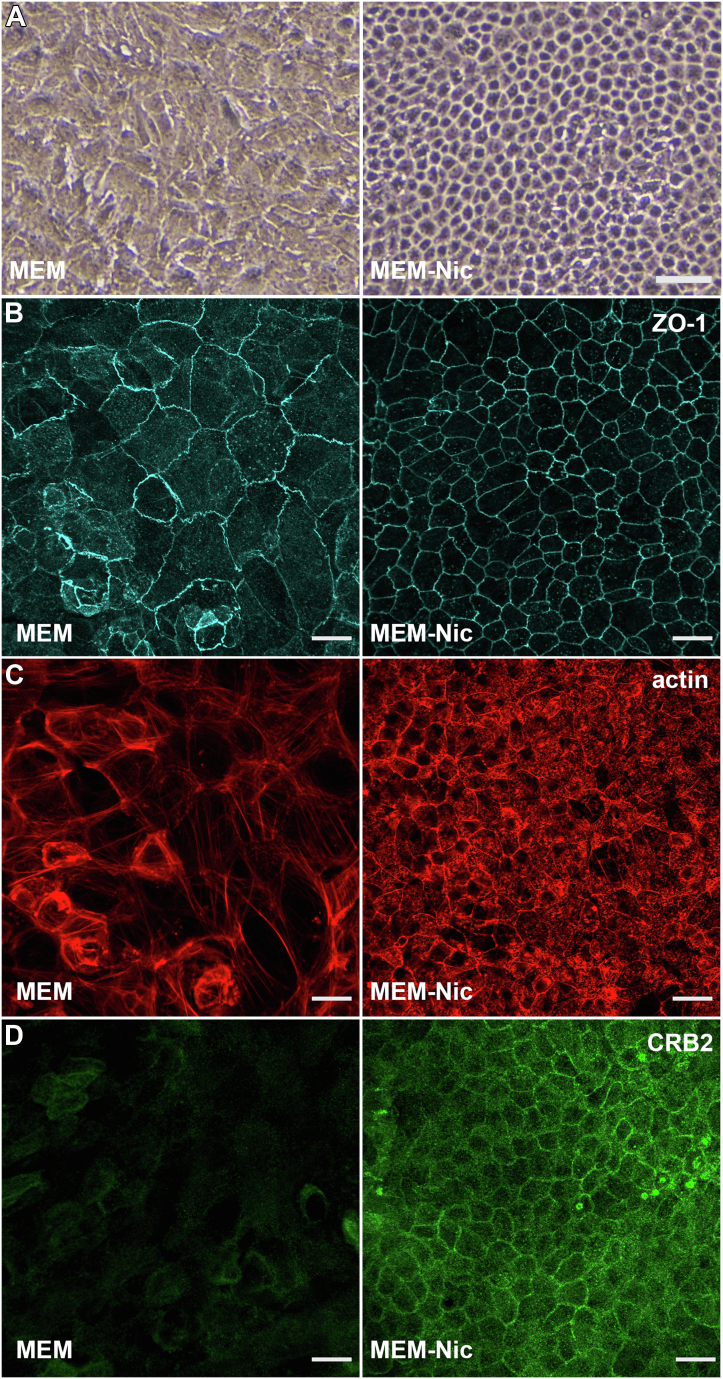
When cultured on plastic surfaces in the presence of nicotinamide, ARPE-19 cells acquired a cobblestone morphology that resembles that of a polarized epithelium (Fig. 1A). To test for a differentiated morphology, we cultured ARPE-19 cells on laminin-coated Transwell inserts in the presence or the absence of nicotinamide, and determined cytoskeletal organization, as well as polarity (Fig. 1, B–D). Immunolabeling of the junctional protein, zonula occludens-1 (ZO-1), revealed a compact and organized cobblestone morphology only in ARPE-19 cells cultured with nicotinamide (Fig. 1B). Because actin filaments have a contrasting arrangement in a polarized and unpolarized epithelium, we labeled ARPE-19 cells cultured in the presence or the absence of nicotinamide with phalloidin. We observed that the actin cytoskeleton in ARPE-19 cells, cultured without nicotinamide, consisted mainly of stress fibers, a characteristic of fibroblastic cells, whereas those cultured with nicotinamide possessed a cortical belt of actin filaments that is normally found in differentiated RPE cells (Fig. 1C). In a recent study, Paniagua et al. (26) demonstrated that crumbs cell polarity complex component 2 (CRB2) was essential for the development and the maintenance of a polarized epithelial state in RPE cells. We therefore tested the expression and localization of CRB2 in ARPE-19 cells cultured in the presence or the absence of nicotinamide. Cells not exposed to nicotinamide showed low expression of CRB2 and primarily cytoplasmic localization of the protein, whereas those cultured with nicotinamide showed robust expression and peripheral localization of CRB2 at the plasma membrane of the cells (Fig. 1D). These experiments demonstrate the role of nicotinamide in promoting the differentiation of ARPE-19 cells into a well-polarized epithelium.
Figure 1.
Cellular morphology of ARPE-19 cells cultured in the presence or the absence of nicotinamide.A, brightfield micrographs of ARPE-19 cells cultured for 2 weeks in MEM or MEM-Nic. B, immunofluorescence micrographs of ZO-1 labeling in ARPE-19 cells cultured on laminin-coated Transwell inserts for 3 weeks in the same media as for A. Cells cultured in MEM-Nic exhibited compact and cobblestone morphology. C, phalloidin labeling showed cortical arrangement of actin filaments in cells cultured in MEM-Nic. Cells cultured in the absence of nicotinamide (MEM) contained actin filaments arranged in stress fibers. D, immunofluorescence micrographs of CRB2 labeling in ARPE-19 cells cultured on laminin-coated Transwell inserts for 3 weeks in the same media as for A. Cells differentiated in MEM-Nic showed higher expression of CRB2 as well as junctional localization at the apical plasma membrane. The scale bars represent 50 μm (A) and 20 μm (B–D). All images are representative from at least three independent experiments. CRB2, crumbs cell polarity complex component 2; MEM-Nic, nicotinamide-containing minimum essential medium; ZO-1, zonula occludens-1.
We next asked whether the effects of nicotinamide on ARPE-19 cell differentiation are reversible. In one experiment (Fig. 2A), we differentiated ARPE-19 cells in nicotinamide-containing minimum essential medium (MEM-Nic) for 2 weeks, during which they formed a compact monolayer with a cobblestone appearance (Fig. 2B). The monolayer of cells was dissociated enzymatically with trypsin, and an equivalent number of cells was seeded in MEM or MEM-Nic. The cultures were maintained for an additional 2 weeks and then imaged. ARPE-19 cells that were seeded in MEM-Nic reformed a homogeneous monolayer with cobblestone morphology (Fig. 2C). In contrast, ARPE-19 cells that were no longer in the presence of nicotinamide (MEM) failed to reform the cobblestone appearance and instead adopted a fusiform morphology uncharacteristic of polarized epithelial cells (Fig. 2D).
Figure 2.
The effects of nicotinamide on ARPE-19 cell differentiation are reversible.A–D, schematic diagram (A) of an experiment in which ARPE-19 cells cultured in MEM-Nic for 2 weeks (B) were trypsinized, passaged, and then plated in MEM-Nic (C) or MEM (D) media for 2 weeks. Cells switched to media lacking nicotinamide (MEM; D) lost their cobblestone morphology. E–H, schematic diagram (E) of an experiment in which two separate cultures of ARPE-19 cells were grown in MEM-Nic for 1 week (F), and on day 7, one of the cultures was kept in MEM-Nic for an additional week (G), whereas the other was switched to MEM (H). Cells that exhibited cobblestone morphology after 1 week in MEM-Nic lost that appearance when nicotinamide was removed from the medium. The scale bars represent 300 μm (B–D and F–H). MEM-Nic, nicotinamide-containing minimum essential medium.
To further test the effects of nicotinamide on ARPE-19 cell differentiation, we adopted a slightly different experimental scheme (Fig. 2E). Two ARPE-19 cultures were seeded and maintained in MEM-Nic for 1 week. The cells began to differentiate and take on the cobblestone appearance as expected (Fig. 2F). On day 7, one of the cultures was kept in MEM-Nic, whereas the other culture was switched to an identical medium lacking nicotinamide (MEM). The ARPE-19 culture kept in the presence of nicotinamide continued to differentiate, and on day 14, it exhibited a compact morphology with cobblestone appearance (Fig. 2G). In contrast, the ARPE-19 culture that was no longer exposed to nicotinamide lost the cobblestone appearance it had on day 7 and appeared to have undergone dedifferentiation (Fig. 2H).
These experiments highlight the role of nicotinamide in promoting an ARPE-19 cell morphology resembling that of a differentiated and polarized epithelium. Moreover, they show that nicotinamide was necessary for not only establishing but also maintaining the epithelial morphology of ARPE-19 cells, as its removal from the culture medium resulted in dedifferentiation of the cells.
Expression of mitochondrial genes and proteins in the presence of nicotinamide
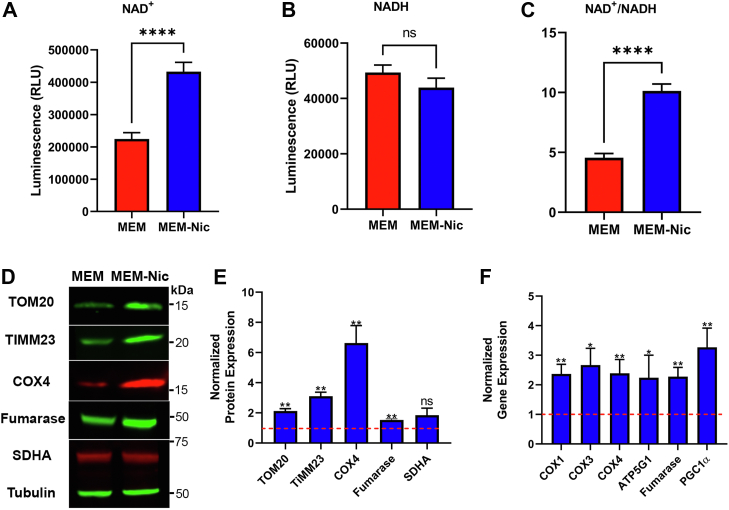
Nicotinamide is a water-soluble member of the vitamin B3 family. Among its prominent biological roles, nicotinamide has been shown to be a key player in the biosynthetic salvage pathway used by cells to generate NAD+ (27). NAD+ is one of the most abundant metabolites in the body and plays a vital role in numerous cellular processes, including mitochondrial metabolism, redox homeostasis, DNA repair, RNA processing, and epigenetic pathways (28). We therefore began by testing whether nicotinamide altered the redox status of the NAD+/NADH pool in ARPE-19 cells. Undifferentiated ARPE-19 cells were seeded on 96-well culture plates in MEM lacking nicotinamide. The following day, some of the cultures were switched to MEM-Nic medium containing 10 mM nicotinamide. After 24 h, the cells were then lysed to measure the pools of NAD+ and NADH separately using a luciferase-based assay. The results showed a two-fold increase in the levels of NAD+ when the ARPE-19 cells were exposed to nicotinamide in the culture medium (Fig. 3A). In contrast, there was no significant difference in the levels of the reduced form, NADH (Fig. 3B). Several studies have previously demonstrated that nicotinamide and its related form, nicotinamide riboside, increase the ratio of NAD+/NADH in different types of mammalian cells (29, 30, 31). In agreement with those studies, we have found that nicotinamide increased the pool of NAD+ and therefore the ratio of NAD+/NADH in ARPE-19 cells (Fig. 3C).
Figure 3.
Nicotinamide increases the intracellular NAD+pool as well as the expression of mitochondrial proteins and genes in ARPE-19 cells.A–C, quantification of the NAD+ (A) and NADH (B) pools in undifferentiated ARPE-19 cells exposed to 10 mM nicotinamide over a 24 h period using a luciferase-based assay; the quantification was used to calculate an NAD+/NADH ratio (C). D, immunolabeled Western blot revealing the expression of several mitochondrial proteins in ARPE-19 cells cultured for 2 weeks in MEM or MEM-Nic. Tubulin was used as a loading control. The positions of apparent molecular mass markers are indicated on the right. E, quantification of band intensity on the immunoblot showed significantly higher abundance of mitochondrial proteins in cells exposed to nicotinamide, including TOM20, TIMM23, COX4, and fumarase. The normalized expression of these proteins in cells cultured in MEM (control group) was set to 1, indicated by the red-dotted line on the graph. The mean of the SDHA expression level was elevated under MEM-Nic conditions, but statistical analysis of our data determined p = 0.14. Mitochondrial protein expression was normalized to tubulin expression. F, quantification of real-time PCR for mitochondrial-related genes showed significantly higher expression in ARPE-19 cells cultured for 2 weeks under MEM-Nic conditions. The normalized expression of these genes in cells cultured in MEM (control group) was set to 1, indicated by the red-dotted line on the graph. Both Western blot and real-time PCR data are collated from at least three independent experiments. Error bars in (A–C, E, and F) represent ±SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001 using Student’s t test. COX4, cytochrome c oxidase 4; MEM-Nic, nicotinamide-containing minimum essential medium; SDHA, succinate dehydrogenase complex flavoprotein subunit A; TIMM23, translocase of inner mitochondrial membrane 23; TOM20, translocase of outer membrane 20 kDa subunit.
Boosting NAD+ levels by supplementation with NAD+ precursors, including nicotinamide riboside, has been shown to increase mitochondrial gene expression in some other cell types (29, 32). We therefore tested the expression of mitochondrial proteins and genes in ARPE-19 cells differentiated in the presence of nicotinamide. Immunoblotting showed a significant increase in the expression of mitochondrial importer proteins, including translocase of outer membrane 20 kDa subunit (TOM20) and translocase of inner mitochondrial membrane 23 (TIMM23), as well as mitochondrial enzymes, including cytochrome c oxidase 4 (COX4) and the tricarboxylic acid (TCA) cycle enzyme, fumarase, after 2 weeks of differentiation in MEM-Nic (Fig. 3, D and E). To test our hypothesis using a different approach, we cultured ARPE-19 cells in the presence or the absence of nicotinamide for 2 weeks and harvested total RNA to test the expression of mitochondria-related genes. Consistent with the immunoblotting data, we found that nicotinamide significantly increased the expression of genes encoding mitochondrial enzymes, including COX1, COX3, COX4, the ATP synthase subunit (ATP5G1; ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 [subunit 9]), and fumarase (Fig. 3F). We also observed a significant increase in the mitochondria-related transcription factor peroxisome proliferator–activated receptor-gamma coactivator-1 alpha (PGC1α), which has previously been shown to promote an increase in expression of mitochondria-related genes (33, 34) (Fig. 3F). These results demonstrate a biologically significant effect of nicotinamide, enhancing the expression of mitochondrial proteins and genes in differentiated ARPE-19 cells.
Given the reversible effects of nicotinamide on ARPE-19 morphology we observed, we next tested whether the changes to mitochondrial protein expression were also reversible. We adopted an experimental scheme in which ARPE-19 cells seeded in MEM-Nic were no longer exposed to nicotinamide after 7 or 14 days of differentiation (Fig. 4A). Cells were cultured in MEM without nicotinamide for an additional 7 or 14 days, respectively, and harvested for total protein. Immunoblotting for COX4 (Fig. 4B) revealed that, during culture in medium without nicotinamide, expression of this mitochondrial enzyme was decreased by 54% in the 7-day protocol (Fig. 4C), and by 51% in the 14-day protocol (Fig. 4D). These results demonstrate that, as with the morphology phenotype, nicotinamide exhibits reversible effects on the expression of mitochondrial proteins.
Figure 4.
Removal of nicotinamide results in decreased expression of mitochondrial protein, COX4.A, schematic diagram of experiments in which ARPE-19 cells were seeded on plastic surfaces in MEM-Nic and cultured for 1 or 2 weeks. On day 7 or day 14, the media were switched to an identical medium lacking nicotinamide (MEM), and the cells were continued to be cultured for an additional 1 or 2 weeks, respectively, before being harvested for total protein. B, immunolabeled Western blot revealing expression of COX4 in ARPE-19 cells following removal of nicotinamide from the medium after 1 or 2 weeks of differentiation in MEM-Nic. The positions of apparent molecular mass markers are indicated on the right. C and D, quantification of COX4 band intensity on immunoblots showed a significant decrease in the levels of COX4 when nicotinamide was removed from the culture medium after 1 (C) or 2 (D) weeks of differentiation in MEM-Nic. COX4 expression was normalized to β-tubulin expression. Error bars in (C and D) represent ±SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, using Student’s t test. COX4, cytochrome c oxidase 4; MEM-Nic, nicotinamide-containing minimum essential medium.
Mitochondrial morphology
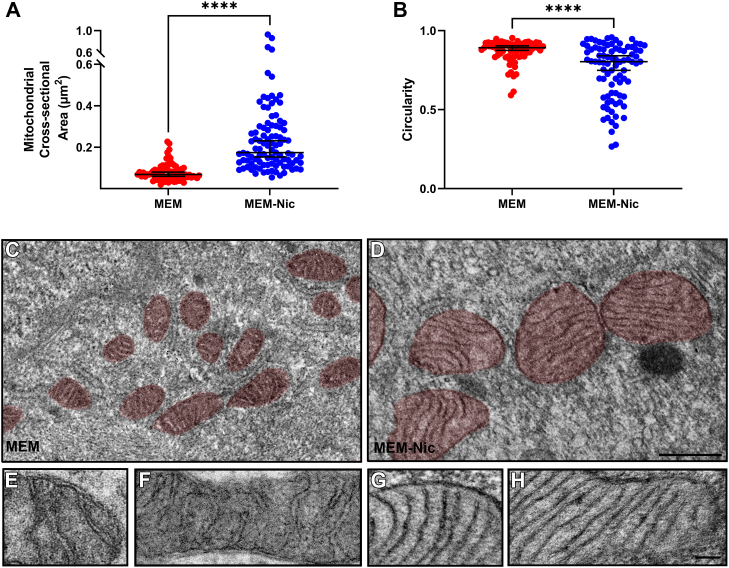
The morphology of mitochondria is an important characteristic that has been linked to specific mitochondrial metabolic profiles that reflect respiratory activity in different cell types (35, 36, 37). Large and filamentous mitochondria are frequently associated with higher rates of oxidative phosphorylation compared with smaller fragmented mitochondria (38, 39). The changes in expression of mitochondrial proteins in ARPE-19 cells differentiated with nicotinamide led us to investigate the morphology of the mitochondria in these cells. We used transmission EM to image ARPE-19 cells differentiated on Transwell inserts for 2 weeks in the presence or the absence of nicotinamide. ARPE-19 cells differentiated with nicotinamide had significantly larger mitochondria in comparison with cells that were not exposed to nicotinamide (Fig. 5, A, C and D). The mitochondria in these cells were also less circular in shape than those in cells that were not exposed to nicotinamide (Fig. 5B). In addition, higher magnification micrographs indicated that the mitochondrial cristae were more numerous and more tightly folded in cells differentiated with nicotinamide (Fig. 5, E–H). These findings revealed that in addition to an altered morphology of the ARPE-19 cells themselves, supplementation with nicotinamide affected the morphological characteristics of their mitochondria.
Figure 5.
Nicotinamide alters the size and morphology of mitochondria in ARPE-19 cells.A, quantification of mitochondrial cross-sectional area revealed that the mitochondria in cells treated with nicotinamide were larger in size compared with those in cells not exposed to nicotinamide. B, quantification of mitochondrial circularity revealed that the mitochondria in ARPE-19 cells cultured with nicotinamide were less circular in shape than those in ARPE-19 cells not exposed to nicotinamide. A and B, each dot represents a measurement made from an individual mitochondrion, and all the measurements were made from a total of 17 EM micrographs acquired from three separate experiments. Horizontal bar indicates median of the data with 95% confidence intervals (CIs). C and D, EM micrographs of mitochondria (highlighted in red) in ARPE-19 cells cultured on laminin-coated Transwell inserts for 2 weeks in the absence (C) or the presence (D) of nicotinamide. E–H, higher magnification EM micrographs of mitochondria in ARPE-19 cells cultured on laminin-coated Transwell inserts for 2 weeks in the absence (E and F) or the presence (G and H) of nicotinamide. The mitochondrial cristae in ARPE-19 cells exposed to nicotinamide were more numerous and more tightly folded than those in cells not exposed to nicotinamide. C and D are the same magnification, as are E–H. The scale bars represent 0.5 μm (C and D) and 0.1 μm (E–H). Confidence intervals: (A) MEM: 95% CI, 0.06 to 0.08. MEM-Nic: 95% CI, 0.15 to 0.23. B, MEM: 95% CI, 0.88 to 0.90. MEM-Nic: 95% CI, 0.75 to 0.84. ∗∗∗∗p < 0.0001, using the Mann–Whitney U test. MEM-Nic, nicotinamide-containing minimum essential medium.
Mitochondrial respiration
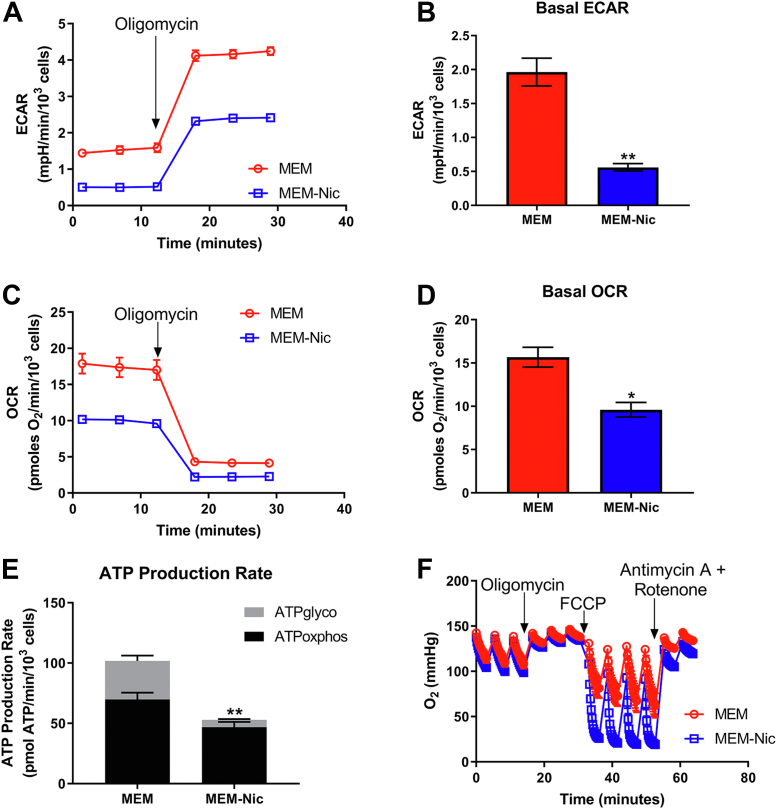
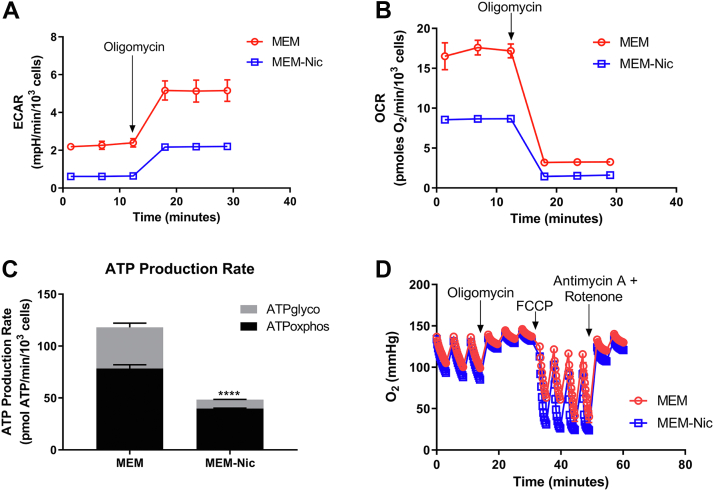
Given the changes in mitochondrial morphology and protein expression we observed in ARPE-19 cells in the presence of nicotinamide, we then tested the hypothesis that these cells were becoming more reliant on oxidative phosphorylation to meet their energy demand. We cultured ARPE-19 cells for 2 weeks in the presence or the absence of nicotinamide and used the Seahorse XF Analyzer to measure rates of both glycolysis and mitochondrial respiration. Measuring the basal extracellular acidification rate (ECAR) served as an indirect measurement of glycolysis and revealed that it was significantly lower in the cells cultured with nicotinamide (Fig. 6, A and B). This difference suggested that differentiated ARPE-19 cells were less dependent on glycolysis to meet their energy demand. Next, we measured the oxygen consumption rate (OCR) and found that it too was significantly lower in differentiated ARPE-19 cells (Fig. 6, C and D). While this finding may be counterintuitive, it likely reflects a low-energy demand upon differentiation as seen in several other terminally differentiated and postmitotic cells (40). Indeed, this is clear when comparing rates of cellular ATP production from glycolysis (ATPglyco) and oxidative phosphorylation (ATPoxphos), as the differentiated ARPE-19 cells have a lower ATP demand but a pronounced shift toward oxidative phosphorylation (41). In ARPE-19 cells exposed to nicotinamide, oxidative phosphorylation contributes to 89% of the total ATP pool but only 69% in ARPE-19 cells cultured without nicotinamide (Fig. 6E). A caveat to these experiments, however, is that the media lack fatty acids that are available in serum that may be an important substrate for RPE cells. Interestingly, the cells cultured in the absence of nicotinamide had a larger overall ATP production rate. In culturing the ARPE-19 line, we have consistently observed that the cells cultured with nicotinamide differentiated into a single monolayer of cells, becoming quiescent once they reached full confluency. In contrast, ARPE-19 cells cultured in the absence of nicotinamide had a higher propensity to divide, and grow on top of each other, even after reaching full confluency. Given the high-energy cost of cell division, this may explain why the undifferentiated cells exhibited a higher overall ATP production rate (42).
Figure 6.
Enhanced mitochondrial metabolism in ARPE-19 cells by nicotinamide. ARPE-19 cells cultured in MEM or MEM-Nic for 2 weeks and analyzed by a Seahorse XF Analyzer to measure cellular respiration. A, representative ECAR trace. B, basal ECAR. C, representative OCR trace. D, basal respiration. E, ATP production rate. F, representative trace of oxygen depletion in the well demonstrating that ARPE-19 cells cultured in MEM-Nic have a higher oxygen consumption, but that the OCR was underestimated because of rapid oxygen depletion in the well. All measurements were derived from three independent experiments, each with four replicates per condition. In all experiments, the following were used at the indicated concentration: oligomycin (2–2.5 μM), FCCP (0.45 μM for first injection and 0.90 μM for second injection), and antimycin A/rot (2 μM/2 μM). Error bars represent ±SEM. ∗p < 0.05, ∗∗p < 0.01 using Student’s t test (B and D) or two-way ANOVA followed by Sidak’s multiple comparisons test (E) to compare mean difference of ATPglyco and ATPoxphos production. ECAR, extracellular acidification rate; FCCP, carbonilcyanide p-triflouromethoxyphenylhydrazone; glyco, glycolysis; MEM-Nic, nicotinamide-containing minimum essential medium; OCR, oxygen consumption rate; oxphos, oxidative phosphorylation.
To demonstrate that the reduced rates of oxidative phosphorylation were due to a reduced energy demand, we measured respiration after addition of the ATP synthase inhibitor, oligomycin, and the uncoupler, carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP) (Fig. 6F). This estimates the maximal capacity for mitochondrial substrate oxidation by decoupling oxygen consumption from any rate limitations set by the ATP demand (40). Maximal respiration rates in the ARPE-19 cells cultured with nicotinamide were so high that oxygen was depleted in the XF24 microchamber well prior to the completion of the 3-min measurement time. The depletion of oxygen prior to the completion of the measurement cycle causes an artifactual and untrustworthy rate reported by the instrument software, making quantification impossible (43). However, the rapid depletion of oxygen clearly shows a substantially increased maximal respiratory capacity upon differentiation in the presence of nicotinamide. Collectively, these experiments support the hypothesis that ARPE-19 cells, in the presence of nicotinamide, differentiate into polarized epithelial cells that undergo a metabolic switch by which they become less reliant on glycolysis and more dependent on oxidative phosphorylation for their energy demands.
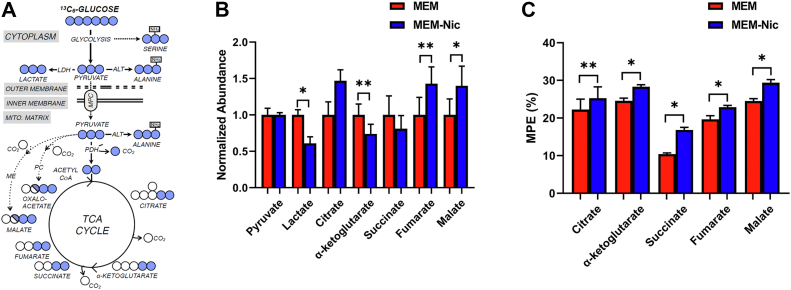
Stable isotope tracing
To further test our hypothesis that the ARPE-19 cells differentiated with nicotinamide were more reliant on oxidative phosphorylation, we provided them with uniformly labeled 13C-glucose and utilized gas GC/MS to trace incorporation of glucose into intermediary metabolism (Fig. 7A). In addition to decreased abundance of lactate (Fig. 7B), there was increased enrichment of glucose-derived carbon in TCA cycle intermediates in ARPE-19 cells cultured with nicotinamide (Fig. 7C), further supporting our bioenergetics data of a switch toward oxidative phosphorylation and away from glycolysis.
Figure 7.
Increased carbon shunting from glucose into TCA intermediates in the presence of nicotinamide.A, schematic diagram depicting metabolic tracing of labeled glucose. B, mass spectrometry quantification of the abundance of metabolites in ARPE-19 cells differentiated in MEM or MEM-Nic. C, mass spectrometry quantification of the mole percent enrichment (MPE) of TCA intermediates in ARPE-19 cells differentiated in MEM or MEM-Nic. Error bars represent ±SEM. ∗p < 0.05, ∗∗p < 0.01 using Student’s t test. MEM-Nic, nicotinamide-containing minimum essential medium; TCA, tricarboxylic acid.
Mitochondrial metabolism drives ARPE-19 cell differentiation
RPE cells in vivo have a very high-energy demand, and recent studies have shown that they are heavily dependent on their mitochondrial metabolism (12, 21, 44, 45). Given the increased mitochondrial oxidation of glucose we observed by glucose tracing in ARPE-19 differentiated with nicotinamide (Fig. 7), we asked whether the polarization of the cells, and concomitant changes in their morphology, were driving the alterations of mitochondrial metabolism, or whether enhanced mitochondrial metabolism was permitting the cells to differentiate into a polarized epithelium. To test this, we utilized a chemical inhibitor, UK5099, specific to the mitochondrial pyruvate carrier (MPC), a protein that transports pyruvate into the mitochondria where it undergoes oxidation to feed the TCA cycle (46) (Fig. 8A). To confirm the efficacy of the inhibitor, we first cultured ARPE-19 cells in the presence of nicotinamide, and two different concentrations of UK5099 (2.5 and 5.0 μM) (41), for 10 days before using the Seahorse XF Analyzer to test their mitochondrial respiration. Both concentrations of the UK5099 inhibitor were sufficient to significantly lower the OCR of the ARPE-19 cells and therefore impair their mitochondrial respiration (Fig. 8B). Next, we cultured ARPE-19 cells in the presence of nicotinamide and 2.5 μM or 5.0 μM of UK5099 on plastic surfaces and Transwell inserts to test if they would differentiate into a polarized epithelial monolayer. Gross morphological examination of the cells cultured on plastic surfaces revealed that perturbation of mitochondrial respiration by the UK5099 inhibitor impeded the ARPE-19 cells from acquiring a uniform cobblestone morphology even in the presence of nicotinamide (Fig. 8C). These observations were further confirmed when ARPE-19 cells cultured on Transwell inserts in the presence of UK5099 were immunolabeled for the junctional protein, ZO-1. As shown in Figure 8D, cells exposed to the inhibitor exhibited noncontinuous ZO-1 labeling at their borders and appeared to have lost their compact cobblestone morphology. These experiments support the hypothesis that enhanced mitochondrial respiration, mediated by the addition of nicotinamide, is a contributing mechanism that is allowing the ARPE-19 cells to differentiate into a polarized epithelium and acquire native RPE characteristics.
Figure 8.
The oxidative phosphorylation inhibitor, UK5099, attenuates the effects of nicotinamide on ARPE-19 differentiation.A, schematic diagram illustrating inhibition of MPC, the importer of pyruvate into the mitochondria, by UK5099. B, representative trace of oxygen depletion (normalized to cell number) in ARPE-19 cells cultured in MEM-Nic for 10 days with or without UK5099 (2.5 and 5.0 μM). The oxygen level remained stable in the presence of the UK5099, indicating that the inhibitor was effective in reducing mitochondrial respiration. C, brightfield micrographs of ARPE-19 cultured as in B showed that addition of UK5099 prevented the cells from differentiating and acquiring a compact and cobblestone morphology. D, immunofluorescence micrographs of ZO-1 labeling in ARPE-19 cells cultured as in B confirmed that the cells’ borders were wider and not as compact in the presence of the UK5099 inhibitor. The scale bars represent 100 μm (C) and 20 μm (D). MEM-Nic, nicotinamide-containing minimum essential medium; MPC, mitochondrial pyruvate carrier; ZO-1, zonula occludens-1.
Effects of nicotinamide on differentiation of primary human RPE
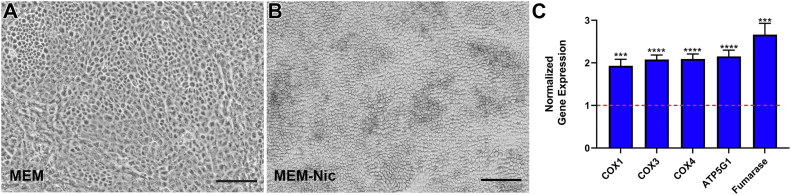
To test whether nicotinamide has a similar effect on a different source of RPE, we complemented our ARPE-19 studies with experiments on primary hfRPE cells from three different donors. The RPE cells from each donor were cultured separately in the presence or the absence of nicotinamide for 2 weeks. As with the ARPE-19 cells, hfRPE cultures matured more rapidly in the presence of nicotinamide, exhibiting a more uniform cobblestone morphology and a higher proportion of pigmented cells than those lacking nicotinamide (Fig. 9, A and B). Pigmentation is a notable characteristic of mature RPE cells in vivo and one that does not develop in ARPE-19 cells. Therefore, the hfRPE cells provided additional and novel evidence that nicotinamide promotes key characteristics of in vivo RPE.
Figure 9.
Rapid differentiation of primary human RPE and increased expression of mitochondrial genes in the presence of nicotinamide.A and B, representative brightfield micrographs of hfRPE cultured in the absence (A) or the presence (B) of 10 mM nicotinamide for 2 weeks. Cells exhibited a more uniform cobblestone appearance and pigmentation at this time point only in the presence of nicotinamide. C, quantification of real-time PCR for mitochondria-related genes showed significantly higher expression in hfRPE cells cultured for 4 weeks under MEM-Nic conditions. The normalized expression of these genes in cells cultured in MEM (control group) was set to 1, indicated by the red-dotted line on the graph. Real-time PCR data are collated from at least three independent experiments. The scale bars represent 100 μm (A and B). Error bars in (C) represent ±SEM. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 using Student’s t test. hfRPE, human fetal RPE; MEM-Nic, nicotinamide-containing minimum essential medium; RPE, retinal pigment epithelium.
Next, we tested whether nicotinamide had a comparable effect on the expression of mitochondria-related genes in hfRPE cells that we observed in the ARPE-19 cell line. We cultured hfRPE cells in the presence or the absence of nicotinamide for 4 weeks and harvested total RNA to compare the relative expression of mitochondrial genes. As with the ARPE-19 cell line, nicotinamide significantly increased the expression of mitochondrial genes, including those that are nuclear encoded and mitochondrial encoded (Fig. 9C).
Finally, we tested the effect of nicotinamide on mitochondrial function in hfRPE cells. Similar to the ARPE-19 cell line, hfRPE cells exhibited reduced ECAR in the presence of nicotinamide, thus indicating a decreased dependence on glycolysis (Fig. 10A). Furthermore, respirometry measurements revealed a similar phenotype to that observed in ARPE-19 cells. Basal OCR was decreased in the hfRPE cells cultured with nicotinamide, likely reflecting a low energy demand upon differentiation of the cells (Fig. 10B). Consistent with this hypothesis, we also observed a lower overall ATP demand in the hfRPE cells cultured with nicotinamide. Similar to the ARPE-19 cell line, the hfRPE cells exhibited a pronounced shift toward oxidative phosphorylation, which contributed to 82% of the ATP pool in cells cultured with nicotinamide, but only 67% in those cultured without nicotinamide (Fig. 10C). Finally, as was observed in the ARPE-19 cell line, respirometry measurements after the addition of oligomycin and FCCP revealed a rapid depletion of oxygen from the microchamber well when the hfRPE cells were cultured with nicotinamide, indicating a profound increase in the maximal respiratory capacity (Fig. 10D).
Figure 10.
Nicotinamide enhances mitochondrial metabolism in primary human RPE. Fetal human RPE cells cultured in MEM or MEM-Nic for 2 weeks (as for Fig. 9, A and B) and analyzed by a Seahorse XF Analyzer to measure cellular respiration. A, representative ECAR trace. B, representative OCR trace. C, ATP production rate. D, representative trace of oxygen depletion in the well demonstrating that hfRPE cells cultured in MEM-Nic have a higher oxygen consumption, but that the OCR was underestimated because of rapid oxygen depletion in the well. The following were used at the indicated concentration: oligomycin (2 μM), FCCP (0.45 μM for first injection and 0.90 μM for second injection), and antimycin A/rot (2 μM/2 μM). Error bars represent ±SEM. ∗∗∗∗p < 0.0001 using two-way ANOVA followed by Sidak’s multiple comparisons test (C) to compare mean difference of ATPglyco and ATPoxphos production. ECAR, extracellular acidification rate; FCCP, carbonilcyanide p-triflouromethoxyphenylhydrazone; glyco, glycolysis; hfRPE, human fetal RPE; MEM-Nic, nicotinamide-containing minimum essential medium; OCR, oxygen consumption rate; oxphos, oxidative phosphorylation; RPE, retinal pigment epithelium.
Collectively, these experiments demonstrate that similar to ARPE-19 cells, primary hfRPE cells mature more rapidly in the presence of nicotinamide and undergo a metabolic switch whereby they become less dependent on glycolysis and more dependent on oxidative phosphorylation.
Discussion
Beginning with the observation that nicotinamide promotes the rapid differentiation and polarization of ARPE-19 cells, we sought to determine the metabolic consequences of nicotinamide treatment and thus gain insight into the relationship between RPE differentiation and metabolism. Early changes in NAD+/NADH ratio, and then, 2 weeks later, in mitochondrial gene and protein expression and morphology, indicate mitochondrial respiration as a driver of differentiation, rather than vice versa. Similar results with primary hfRPE cells suggest that this relationship is a general RPE characteristic. Inhibition of the rapid differentiation by inhibiting the MPC provided further support that the metabolic changes induced by nicotinamide promoted differentiation of ARPE-19 cells. These findings indicate that the RPE is not only important for its role in the retinal metabolic ecosystem (47) but also that metabolism is key for establishing its own polarized organization, on which many essential RPE functions depend.
The ARPE-19 cell line has been notorious for generating findings that have little relevance to human RPE cell biology because the cells have often been studied in an undifferentiated state (11). Nonetheless, the ease of using this cell line has made it a popular model among both vision and nonvision science laboratories worldwide. As shown in Figure 1, nicotinamide promoted rapid induction of RPE differentiation in ARPE-19 cells. Nicotinamide's addition to the cell culture medium promoted a compact cobblestone morphology, epithelial-like arrangement of the actin cytoskeleton, and expression of polarity-specific proteins, including ZO-1 and CRB2. Therefore, the characteristics induced by nicotinamide make the ARPE-19 cell line a more physiologically relevant model without sacrificing the ease by which the cells can be cultured in vitro. Furthermore, we confirmed that nicotinamide has a similar effect on the differentiation of primary hfRPE, rapidly promoting a uniform cobblestone morphology as well as pigmentation. The use of well-differentiated RPE cells has been underscored by many studies. Specific to metabolism, studies have shown that, unlike native RPE, poorly differentiated ARPE-19 as well as human telomerase reverse transcriptase RPE-1 cells are incapable of utilizing the amino acid proline as a nutrient source (17, 48). Our current study demonstrates a clear link between RPE differentiation and mitochondrial-related metabolism.
Similar to its effects on some other mammalian cell types, nicotinamide increased the NAD+/NADH ratio as well as the expression of mitochondrial-specific genes and proteins in ARPE-19 and hfRPE cells (Figs. 3 and 9). Furthermore, nicotinamide addition altered the size and shape of the mitochondria, making them larger and less spherical. They also contained cristae that were more abundant and tightly folded (Fig. 5). We hypothesize that the increased expression of PGC1α in the differentiated ARPE-19 cells may promote the expression of mitochondrial proteins, which, because of the observed elaboration of the cristae, can be packed at a larger density in a single mitochondrion. Nonetheless, these effects on mitochondrial morphology and protein expression suggest an enhancement of the RPE mitochondrial metabolic profile. Similar increases in mitochondrial protein expression have been shown to be a part of metabolic switches that occur during the differentiation of several cell types, including neurons (49), mature osteoblasts (50), and myotubes (51). Moreover, the findings from metabolic assays on ARPE-19 and hfRPE cells, including respirometry and glucose tracing (Figs. 6, 7 and 10), further supported the hypothesis that these cells are undergoing a metabolic switch as they differentiate in the presence of nicotinamide. Previous studies have demonstrated that oxidative phosphorylation and mitochondrial-based metabolism are essential for RPE function (12, 21, 44). Here, we found that perturbation of mitochondrial metabolism by the MPC inhibitor, UK5099, impeded the differentiation of ARPE-19 cells, even in the presence of nicotinamide, indicating that the promotion of differentiation by nicotinamide occurs via mitochondrial function.
Interestingly, we have found the effects of nicotinamide to be reversible with respect to both ARPE-19 differentiation and mitochondrial protein expression. As shown in Figure 2, simply removing nicotinamide from the ARPE-19 culture medium caused the cells to dedifferentiate and revert to a fibroblast-like morphology. Furthermore, loss of nicotinamide from the cell culture medium significantly reduced the expression of the mitochondrial protein COX4 (Fig. 4). This would suggest that nicotinamide might serve as an autonomous reagent to manipulate mitochondrial protein expression in cultured RPE cells. Mitochondrial function has been shown to dramatically decline with normal aging (52) and especially under pathological conditions. Mitochondrial dysfunction has been implicated in age-related macular degeneration, the number one cause of blindness in people over the age of 50 (53, 54, 55, 56). In addition, RPE dedifferentiation has been implicated in proliferative vitreoretinopathy (57). Further studies are necessary to test whether nicotinamide, and other members of the vitamin B3 family, can be used to restore RPE mitochondrial function and its differentiated state under pathological conditions.
The differentiated state of the RPE is essential for many of its functions (10, 11). Here, we have studied mitochondrial metabolism in relation to RPE differentiation. Our results show that mitochondrial metabolism both drives and maintains the RPE differentiation state, thus demonstrating a key effect of mitochondrial function in supporting retinal function and viability.
Experimental procedures
Conditions for cell culture
ARPE-19 cells, obtained from American Type Culture Collection (passage 19), and hfRPE from three different donors were maintained as described previously (22). Briefly, the cells were cultured in MEM-Nic (MEM alpha with GlutaMAX [Fisher Scientific]), 1% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin, 1% N2 supplement (Sigma–Aldrich), taurine (83 mg/ml) (Sigma–Aldrich), hydrocortisone (20 μg/ml) (Sigma–Aldrich), triiodothyronine (13 ng/ml) (Sigma–Aldrich), and 10 mM nicotinamide (Sigma–Aldrich). The MEM was identical to the one above but was lacking nicotinamide. The media were changed every 2 or 3 days. In all experiments, ARPE-19 cells were used within five additional passages, whereas the hfRPE cells were used after no more than two passages. For experiments on Transwells, 6.5-mm inserts with polyester membranes (Costar; catalog no.: 3470) were coated (10 μg/cm2) with natural mouse laminin (Fisher Scientific). Transepithelial resistance of ARPE-19 cells on coated Transwell inserts was ∼40 Ω cm2 (22). For some experiments, ARPE-19 cells were cultured in the presence of the MPC inhibitor, UK5099 (Sigma), at final concentrations of 2.5 or 5.0 μM.
Immunocytochemistry
ARPE-19 cells cultured on laminin-coated Transwell inserts were washed 3× with Dulbecco’s PBS (Fisher Scientific) before being fixed with 4% formaldehyde for 10 min at room temperature (RT). Cell permeabilization was then achieved with 0.25% Triton X-100 for 15 min at RT. Nonspecific antibody binding was blocked by incubating the cells with 4% bovine serum albumin for 1 h at RT. Cells were then incubated with Dulbecco’s PBS plus 1% bovine serum albumin and one or a combination of primary antibodies overnight at 4 °C. The appropriate Alexa Fluor–conjugated secondary antibodies were then used for 1 h at RT in the dark. For actin labeling, the cells were incubated with Phalloidin–TRITC (Sigma–Aldrich). To prepare the samples for imaging, membranes of the Transwell inserts were excised and mounted on frosted microscope slides using Fluoro-Gel II mounting medium containing 4′,6-diamidino-2-phenylindole (Electron Microscopy Sciences). Imaging was performed on a FluoView 1000 Olympus confocal microscope with a 60× numerical aperture of 1.40 oil objective, using FluoView FV10-ASW 4.2 software (Olympus). Primary antibodies used included ZO-1 (1:500 dilution) (Invitrogen; catalog no.: 402200), and CRB2 (1:100 dilution) (Thermo Fisher; catalog no.: PA5-25628).
NAD+/NADH-Glo assay
Undifferentiated ARPE-19 cells were seeded on to 96-well culture plates in MEM. The following day, some of the wells were switched to nicotinamide-containing medium (MEM-Nic) and allowed to grow for 24 h. The cells were lysed with 0.2 N NaOH, and the lysates were transferred to a white-walled tissue culture plate. A double extraction was performed to measure the pools of NAD+ and NADH separately, using the NAD/NADH-Glo Assay (Promega), according to the manufacturer’s instructions. Luminescence was measured using a SpectraMax iD3 microplate reader (Molecular Devices).
Western blot
Total protein was collected from ARPE-19 cells cultured on plastic surfaces in radioimmunoprecipitation assay buffer, and 15 to 20 μg of protein were separated by SDS-PAGE. Semidry transfer was used to transfer the proteins on to polyvinylidene difluoride membranes. The membranes were then blocked with Odyssey blocking buffer (LI-COR) for 1 h at RT. Primary antibody incubation was performed overnight at 4 °C in Odyssey buffer with 0.1% Tween-20. For signal detection, the membranes were incubated with species-matched secondary IRDye 680RD and 800CW antibodies diluted in Odyssey blocking buffer with 0.1% Tween-20 and 0.01% SDS for 1 h at RT in the dark. Imaging was performed using the Odyssey CLx Imaging System (LI-COR), and blots were processed using Image Studio Lite, version 5.2 software (LI-COR). Primary antibodies used included translocase of outer membrane 20 (1:1000 dilution) (Cell Signaling; catalog no.: 42406), TIMM23 (1:1000 dilution) (Abcam; catalog no.: ab230253), COX4 (1:500 dilution) (Molecular Probe; catalog no.: A21348), fumarase (1:1000 dilution) (ABclonal Technology; catalog no.: A5688), succinate dehydrogenase A (1:10,000 dilution) (Abcam; catalog no.: ab14715), α-tubulin (1:2000 dilution) (Sigma; catalog no.: T9026), and β-tubulin (1:2000 dilution) (Cell Signaling; catalog no.: 2128).
Real-time PCR
Total RNA was harvested from ARPE-19 cells cultured on plastic surfaces for 2 weeks and hfRPE cells cultured for 4 weeks in MEM or MEM-Nic, using the PureLink RNA Mini Kit (Fisher Scientific). Reverse transcription was performed with the SuperScript IV First-Strand Synthesis System (Fisher Scientific) to generate complementary DNA from approximately 3 to 5 μg of total RNA. The complementary DNA was amplified using KAPA SYBR Green mastermix (KAPA Biosystems) on the Roche LightCycler 480 Instrument II. Thermal cycling conditions were performed as follows: one cycle at 95 °C for 5 min; 45 cycles at 95 °C for 10 s, 53 °C for 10 s, and 72 °C for 10 s; and one cycle at 72 °C for 10 min. The primer sequences include COX1, 5′-ACGTTGTAGCCCACTTCCAC-3′ and 5′-GGTTTGGTCTAGGGTGTAGCC-3′; COX3, 5′-AGTAAAACCCAGCCCATGACC-3′ and 5′-TTCTCGTGTTACATCGCGCC-3′; COX4, 5′-CAGGGTATTTAGCCTAGTTGGC-3′ and 5′-GCCGATCCATATAAGCTGGGA-3′; ATP5G1, 5′-TTCCAGACCAGTGTTGTCTCC-3′ and 5′-GACGGGTTCCTGGCATAGC-3′; Fumarase, 5′-GGAGGTGTGACAGAACGCAT-3′ and 5′-CATCTGCTGCCTTCATTATTGC-3′; PGC1α, 5′-GTGGTGCAGTGACCAATCAG-3′ and 5′-CTGTCAGCGCATCAAATGAG-3′; 18S rRNA, 5′-GGCCCTGTAATTGGAATGAGTC-3′ and 5′-CCAAGATCCAACTACGAGCTT-3′.
EM
ARPE-19 cells were fixed with 2% formaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 24 h at 4 °C. The samples were then rinsed with 0.1 M cacodylate buffer containing 1% tannic acid and postfixed with 1% OsO4 (w/v) diluted in ultrapure water for 1 h in the dark. After a wash with distilled water, the samples were dehydrated using a graded ethanol series, with the final step in propylene oxide followed by resin infiltration. Samples were then embedded in Araldite 502 resin (Electron Microscopy Sciences), and ultrathin sections (70 nm) were obtained with a PowerTome X ultramicrotome (RMC Boeckeler). Uranyl acetate and lead citrate were then used for enhanced contrast. Micrographs were acquired using a JEM-1400 Plus (JEOL) electron microscope equipped with an Orius SC1000A (Gatan) camera and Gatan Microscopy Suite Software. Minor contrast and brightness adjustments to whole images were performed with Adobe Photoshop CS6 software. The mitochondria identified in EM sections were outlined on the ImageJ software (National Institutes of Health), which was then used to measure the area of their profile in section, as well as the circularity of that profile (calculated by the formula: 4π × area/perimeter2).
Seahorse assay
Respirometry assays were performed on a Seahorse XFe24 Extracellular Flux Analyzer (Agilent Technologies). Cells were seeded at 1.66 × 105 cells/cm2 on to a laminin-coated Seahorse XF24 microplate and maintained in an incubator at 37 °C in 5% CO2. On the day of the assay, cells were washed twice with Seahorse assay medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 8 mM glucose, 2 mM glutamine, 2 mM sodium pyruvate, and 5 mM Hepes; pH 7.4) and brought to a final volume of 500 μl per well. The XF24 plate was incubated at 37 °C for 30 min in an incubator without CO2 before the start of the assay. During the assay, oligomycin was injected from port A to a final concentration of 2 to 2.5 μM, FCCP was injected from ports B and C to final concentrations of 0.45 and 0.90 μM, respectively, and antimycin A and rotenone were injected from port D to a final concentration of 2 μM.
Seahorse data were normalized to cell number per well by quantifying Hoechst-stained nuclei. After the assay, the cells were stained with Hoechst (1 μg/ml) and quantified with an Operetta High-Content Imaging System (PerkinElmer). Normalized rates are presented as pmol O2/min/103 cells. Data are presented as the average compiled data from three independent experiments. Statistical analysis was conducted by t test or two-way ANOVA, and statistical significance (p ≤ 0.05) is denoted with an asterisk (∗).
Data were analyzed by first subtracting nonmitochondrial respiration (lowest rate after injection of antimycin A and rotenone) from all OCRs. Basal OCR and ECAR were calculated as the average of the two measurements before injection of oligomycin. ATP production rates from glycolysis (glyco) and oxidative phosphorylation (oxphos) were calculated as described previously (41).
Stable isotope tracing assay
ARPE-19 cells were cultured on 6-well plates in MEM or MEM-Nic for 2 weeks. On the day of the experiment, the cells were washed with DMEM lacking glucose, glutamine, and phenol red (Gibco; catalog no.: A14430). The cells were then cultured for 24 h in DMEM supplemented with 10 mM uniformly labeled 13C6-glucose, 10% (v/v) fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 3 mg/l phenol red. Cells were prepared for GC/MS as previously described (58). Briefly, they were kept on ice and quickly washed with ice-cold 0.9% (w/v) NaCl. Cells were then scraped in ice-cold methanol and water containing 1 μg norvaline, an internal standard. Chloroform was then added to the samples, which were then vortexed for 1 min, and centrifuged at 10,000g for 5 min at 4 °C. The top layer was removed, and the samples were dried overnight using a refrigerated CentriVap vacuum concentrator (LabConco). For quantification purposes, metabolites (50 nmol to 23 pmol) were extracted alongside the cell samples to generate a standard curve.
The dried samples were reconstituted in 2% (w/v) methoxyamine in pyridine prior to a 45-min incubation at 37 °C. Subsequently, an equal volume of MTBSTFA and 1% TBDMSCI with 1% tert-butyldimethylchlorosilane was added to samples, followed by an additional 45-min incubation at 37 °C. Samples were analyzed using Agilent MassHunter software. For stable isotope tracing, the data were analyzed with FluxFix software (http://fluxfix.science) to correct the abundance of natural heavy isotope against a reference set of unlabeled metabolite standards (59).
Data availability
All data are contained within the article.
Conflict of interest
A. S. D. has previously served as a paid consultant for Agilent Technologies. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful for Barry Burgess for general laboratory support, and Jane Hu and Dean Bok for providing the human fetal RPE cells. Diagrams in Figures 2, 4, and 8 were generated using BioRender.com.
Author contributions
R. A. H. and D. S. W. conceptualization; R. A. H. and A. S. D. methodology; R. A. H., A. E. P., L. T., K. Y., K. K. O. K., L. S., and A. S. D. investigation; R. A. H. and D. S. W. writing–original draft; A. S. D. writing–review & editing; D. S. W. supervision; D. S. W. funding acquisition.
Funding and additional information
The study was supported by the National Institutes of Health grants R01EY027442, R21EY031109, and P30EY00331 (to D. S. W.) as well as National Institutes of Health grants R35GM138003 and P30DK063491 and support from the W.M. Keck Foundation (to A. S. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Kirill Martemyanov
References
- 1.Warburg O.P., Negelein E. On the metabolism of carcinoma cells. Biochem. Z. 1924;152:309–344. [Google Scholar]
- 2.Rajala R.V.S. Aerobic glycolysis in the retina: functional roles of pyruvate kinase isoforms. Front. Cell Dev. Biol. 2020;8:266. doi: 10.3389/fcell.2020.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 5.Niven J.E., Laughlin S.B. Energy limitation as a selective pressure on the evolution of sensory systems. J. Exp. Biol. 2008;211:1792–1804. doi: 10.1242/jeb.017574. [DOI] [PubMed] [Google Scholar]
- 6.Wong-Riley M. Energy metabolism of the visual system. Eye Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Punzo C., Xiong W., Cepko C.L. Loss of daylight vision in retinal degeneration: are oxidative stress and metabolic dysregulation to blame? J. Biol. Chem. 2012;287:1642–1648. doi: 10.1074/jbc.R111.304428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinchore Y., Begaj T., Wu D., Drokhlyansky E., Cepko C.L. Glycolytic reliance promotes anabolism in photoreceptors. Elife. 2017;6 doi: 10.7554/eLife.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okawa H., Sampath A.P., Laughlin S.B., Fain G.L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008;18:1917–1921. doi: 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 11.Lakkaraju A., Umapathy A., Tan L.X., Daniele L., Philp N.J., Boesze-Battaglia K., et al. The cell biology of the retinal pigment epithelium. Prog. Retin. Eye Res. 2020;78 doi: 10.1016/j.preteyeres.2020.100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordone M.P., Salman M.M., Titus H.E., Amini E., Andersen J.V., Chakraborti B., et al. The energetic brain – a review from students to students. J. Neurochem. 2019;151:139–165. doi: 10.1111/jnc.14829. [DOI] [PubMed] [Google Scholar]
- 14.Russo G.L., Sonsalla G., Natarajan P., Breunig C.T., Bulli G., Merl-Pham J., et al. CRISPR-mediated induction of neuron-enriched mitochondrial proteins boosts direct glia-to-neuron conversion. Cell Stem Cell. 2021;28:524–534.e7. doi: 10.1016/j.stem.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisbach C.M., Hass D.T., Robbings B.M., Rountree A.M., Sadilek M., Sweet I.R., et al. Succinate can shuttle reducing power from the hypoxic retina to the O2-rich pigment epithelium. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J., Yanagida A., Knight K., Engel A.L., Vo A.H., Jankowski C., et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14710–14715. doi: 10.1073/pnas.1604572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yam M., Engel A.L., Wang Y., Zhu S., Hauer A., Zhang R., et al. Proline mediates metabolic communication between retinal pigment epithelial cells and the retina. J. Biol. Chem. 2019;294:10278–10289. doi: 10.1074/jbc.RA119.007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes-Reveles J., Dhingra A., Alexander D., Bragin A., Philp N.J., Boesze-Battaglia K. Phagocytosis-dependent ketogenesis in retinal pigment epithelium. J. Biol. Chem. 2017;292:8038–8047. doi: 10.1074/jbc.M116.770784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adijanto J., Du J., Moffat C., Seifert E.L., Hurley J.B., Philp N.J. The retinal pigment epithelium utilizes fatty acids for ketogenesis. J. Biol. Chem. 2014;289:20570–20582. doi: 10.1074/jbc.M114.565457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurihara T., Westenskow P.D., Gantner M.L., Usui Y., Schultz A., Bravo S., et al. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife. 2016;5 doi: 10.7554/eLife.14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazim R.A., Volland S., Yen A., Burgess B.L., Williams D.S. Rapid differentiation of the human RPE cell line, ARPE-19, induced by nicotinamide. Exp. Eye Res. 2019;179:18–24. doi: 10.1016/j.exer.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idelson M., Alper R., Obolensky A., Ben-Shushan E., Hemo I., Yachimovich-Cohen N., et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Hazim R.A., Karumbayaram S., Jiang M., Dimashkie A., Lopes V.S., Li D., et al. Differentiation of RPE cells from integration-free iPS cells and their cell biological characterization. Stem Cell Res. Ther. 2017;8:1–17. doi: 10.1186/s13287-017-0652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn K.C., Aotaki-Keen A.E., Putkey F.R., Hjelmeland L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996;62:155–160. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 26.Paniagua A.E., Segurado A., Dolón J.F., Esteve-Rudd J., Velasco A., Williams D.S., et al. Key role for CRB2 in the maintenance of apicobasal polarity in retinal pigment epithelial cells. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.701853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braidy N., Berg J., Clement J., Khorshidi F., Poljak A., Jayasena T., et al. Role of nicotinamide adenine dinucleotide and related precursors as therapeutic targets for age-related degenerative diseases: rationale, biochemistry, pharmacokinetics, and outcomes. Antioxid. Redox Signal. 2019;30:251–294. doi: 10.1089/ars.2017.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie N., Zhang L., Gao W., Huang C., Huber P.E., Zhou X., et al. Vol. 5. 2020. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential; p. 227. (Signal Transduct. Target. Ther.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canto C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y., et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C.F., Chavez J.D., Garcia-Menendez L., Choi Y., Roe N.D., Chiao Y.A., et al. Normalization of NAD+ redox balance as a therapy for heart failure. Circulation. 2016;134:883–894. doi: 10.1161/CIRCULATIONAHA.116.022495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh C.-L., Hsieh S.-Y., Huang H.-M., Lu S.-L., Omori H., Zheng P.-X., et al. Nicotinamide increases intracellular NAD+ content to enhance autophagy-mediated group A streptococcal clearance in endothelial cells. Front. Microbiol. 2020;11:117. doi: 10.3389/fmicb.2020.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schondorf D.C., Ivanyuk D., Baden P., Sanchez-Martinez A., De Cicco S., Yu C., et al. The NAD+ precursor nicotinamide riboside rescues mitochondrial defects and neuronal loss in iPSC and fly models of Parkinson's disease. Cell Rep. 2018;23:2976–2988. doi: 10.1016/j.celrep.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 34.Lehman J.J., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benard G., Rossignol R. Ultrastructure of the mitochondrion and its bearing on function and bioenergetics. Antioxid. Redox Signal. 2008;10:1313–1342. doi: 10.1089/ars.2007.2000. [DOI] [PubMed] [Google Scholar]
- 36.Picard M., Shirihai O.S., Gentil B.J., Burelle Y. Mitochondrial morphology transitions and functions: implications for retrograde signaling? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;304:R393–R406. doi: 10.1152/ajpregu.00584.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia G.C., Bartol T.M., Phan S., Bushong E.A., Perkins G., Sejnowski T.J., et al. Mitochondrial morphology provides a mechanism for energy buffering at synapses. Sci. Rep. 2019;9:18306. doi: 10.1038/s41598-019-54159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Vermulst M., Wang Y.E., Chomyn A., Prolla T.A., McCaffery J.M., et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glancy B., Kim Y., Katti P., Willingham T.B. The functional impact of mitochondrial structure across subcellular scales. Front. Physiol. 2020;11:541040. doi: 10.3389/fphys.2020.541040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Divakaruni A.S., Paradyse A., Ferrick D.A., Murphy A.N., Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- 41.Desousa B.R., Kim K.K.O., Hsieh W.Y., Jones A.E., Swain P., Morrow D.H., et al. Calculating ATP production rates from oxidative phosphorylation and glycolysis during cell activation. bioRxiv. 2022 doi: 10.1101/2022.04.16.488523. [preprint] [DOI] [Google Scholar]
- 42.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Divakaruni A.S., Jastroch M. A practical guide for the analysis, standardization, and interpretation of oxygen consumption measurements. Nat. Metab. 2022 doi: 10.1038/s42255-022-00619-4. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keeling E., Chatelet D.S., Tan N.Y.T., Khan F., Richards R., Thisainathan T., et al. 3D-Reconstructed retinal pigment epithelial cells provide insights into the anatomy of the outer retina. Int. J. Mol. Sci. 2020;21:8408. doi: 10.3390/ijms21218408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinha T., Naash M.I., Al-Ubaidi M.R. The symbiotic relationship between the neural retina and retinal pigment epithelium is supported by utilizing differential metabolic pathways. iScience. 2020;23 doi: 10.1016/j.isci.2020.101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halestrap A.P. The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem. J. 1975;148:85–96. doi: 10.1042/bj1480085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurley J.B. Retina metabolism and metabolism in the pigmented epithelium: a busy intersection. Annu. Rev. Vis. Sci. 2021;7:665–692. doi: 10.1146/annurev-vision-100419-115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J., Zhu S., Lim R.R., Chao J.R. Proline metabolism and transport in retinal health and disease. Amino Acids. 2021;53:1789–1806. doi: 10.1007/s00726-021-02981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arrázola M.S., Andraini T., Szelechowski M., Mouledous L., Arnauné-Pelloquin L., Davezac N., et al. Mitochondria in developmental and adult neurogenesis. Neurotox. Res. 2018;36:257–267. doi: 10.1007/s12640-018-9942-y. [DOI] [PubMed] [Google Scholar]
- 50.Zheng C.-X., Sui B.-D., Qiu X.-Y., Hu C.-H., Jin Y. Mitochondrial regulation of stem cells in bone homeostasis. Trends Mol. Med. 2020;26:89–104. doi: 10.1016/j.molmed.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Fortini P., Iorio E., Dogliotti E., Isidoro C. Coordinated metabolic changes and modulation of autophagy during myogenesis. Front. Physiol. 2016;7:237. doi: 10.3389/fphys.2016.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomes A.P., Price N.L., Ling A.J., Moslehi J.J., Montgomery M.K., Rajman L., et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrington D.A., Ebeling M.C., Kapphahn R.J., Terluk M.R., Fisher C.R., Polanco J.R., et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017;13:255–265. doi: 10.1016/j.redox.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong J., Cai H., Noggle S., Paull D., Rizzolo L.J., Del Priore L.V., et al. Stem cell-derived retinal pigment epithelium from patients with age-related macular degeneration exhibit reduced metabolism and matrix interactions. Stem Cells Transl. Med. 2019;9:364–376. doi: 10.1002/sctm.19-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaarniranta K., Uusitalo H., Blasiak J., Felszeghy S., Kannan R., Kauppinen A., et al. Mechanisms of mitochondrial dysfunction and their impact on age-related macular degeneration. Prog. Retin. Eye Res. 2020;79 doi: 10.1016/j.preteyeres.2020.100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.La Cunza N., Li Xuan T., Thamban T., Germer C.J., Rathnasamy G., Toops K.A., et al. Mitochondria-dependent phase separation of disease-relevant proteins drives pathological features of age-related macular degeneration. JCI Insight. 2021;6 doi: 10.1172/jci.insight.142254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamiya S., Kaplan H.J. Role of epithelial–mesenchymal transition in proliferative vitreoretinopathy. Exp. Eye Res. 2016;142:26–31. doi: 10.1016/j.exer.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Cordes T., Metallo C.M. In: High-Throughput Metabolomics: Methods and Protocols. D'Alessandro A., editor. Springer New York; New York, NY: 2019. Quantifying intermediary metabolism and lipogenesis in cultured mammalian cells using stable isotope tracing and mass spectrometry; pp. 219–241. [DOI] [PubMed] [Google Scholar]
- 59.Trefely S., Ashwell P., Snyder N.W. FluxFix: automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinformatics. 2016;17:485. doi: 10.1186/s12859-016-1360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.