Key Words: cell migration, fate determination, Müller glia, Müller glia-derived progenitor, Notch signaling, photoreceptor, retina regeneration, Sox11, transcription factor, zebrafish
Abstract
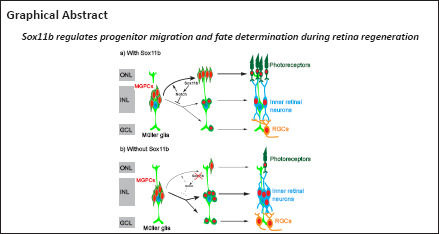
The transcription factor Sox11 plays important roles in retinal neurogenesis during vertebrate eye development. However, its function in retina regeneration remains elusive. Here we report that Sox11b, a zebrafish Sox11 homolog, regulates the migration and fate determination of Müller glia-derived progenitors (MGPCs) in an adult zebrafish model of mechanical retinal injury. Following a stab injury, the expression of Sox11b was induced in proliferating MGPCs in the retina. Sox11b knockdown did not affect MGPC formation at 4 days post-injury, although the nuclear morphology and subsequent radial migration of MGPCs were altered. At 7 days post-injury, Sox11b knockdown resulted in an increased proportion of MGPCs in the inner retina and a decreased proportion of MGPCs in the outer nuclear layer, compared with controls. Furthermore, Sox11b knockdown led to reduced photoreceptor regeneration, while it increased the numbers of newborn amacrines and retinal ganglion cells. Finally, quantitative polymerase chain reaction analysis revealed that Sox11b regulated the expression of Notch signaling components in the retina, and Notch inhibition partially recapitulated the Sox11b knockdown phenotype, indicating that Notch signaling functions downstream of Sox11b. Our findings imply that Sox11b plays key roles in MGPC migration and fate determination during retina regeneration in zebrafish, which may have critical implications for future explorations of retinal repair in mammals.
Introduction
Blinding eye illnesses (e.g., retinitis pigmentosa, glaucoma, and macular degeneration) often result in the death of retinal neurons, leading to severe visual loss. Similar to other components of the central nervous system, the mammalian retina exhibits little regenerative capacity after traumatic injury or degenerative diseases (Wilken and Reh, 2016; Roska and Sahel, 2018). In contrast, teleost fish (e.g., zebrafish) are able to regenerate an injured retina and even regain vision (Goldman, 2014). Key to this regeneration response is Müller glia (MG), the major glial cell type in the retina (Lenkowski and Raymond, 2014; Zhao et al., 2021; Campbell et al., 2022; Wohl, 2022). After injury, zebrafish MG undergo a reprogramming process and proliferate into multipotent progenitors (MG-derived progenitor cells, MGPCs) (Fausett and Goldman, 2006; Nagashima et al., 2013). These MGPCs then migrate to different retinal layers and differentiate into newborn retinal neurons, which participate in retinal repair (Gorsuch and Hyde, 2014). Although the mammalian retina cannot spontaneously regenerate, mammalian MG reportedly have neurogenic potential and can be engineered to serve as a source of functional retinal neurons (Jorstad et al., 2017; Yao et al., 2018). Thus, a thorough understanding of the principles behind effective retina regeneration in fish may provide critical information for future explorations of retinal repair in mammals.
Sox11 is a member of the SoxC subfamily of transcription factors, which are named for their shared Sry-related high mobility group (HMG)-box motif. Sox11 often functions in concert with other two SoxC group members, Sox4 and Sox12; it has been implicated as a critical regulator of cell proliferation, differentiation, and survival during development (Reiprich and Wegner, 2015; Chang and Hertz, 2017; Kavyanifar et al., 2018; Tsang et al., 2020). Furthermore, Sox11 plays important roles in regulating retinal neurogenesis during vertebrate eye development (Usui et al., 2013; Pillai-Kastoori et al., 2014; Chang et al., 2017; Kuwajima et al., 2017). Sox11 also promotes axonal regeneration in both the peripheral and central nervous systems (Jankowski et al., 2009; Jing et al., 2012; Wang et al., 2015; Norsworthy et al., 2017). During retina regeneration in zebrafish, Sox11b (a homolog of the mammalian Sox11) is expressed in MGPCs (Mu et al., 2017). However, the function of Sox11 in retina regeneration remains elusive.
Although the signaling pathways that regulate MG reprogramming and proliferation have been described in previous reports, considerably less is known regarding the mechanisms that govern migration and fate determination in MGPCs. Regardless of which retinal cell types are damaged, MG proliferate into multipotent progenitors that migrate to all retinal layers and replenish layer-specific cell types (Powell et al., 2016). Notably, MG proliferation and neuron regeneration are biased toward specific layers where neurons have been ablated (Powell et al., 2016; Ng Chi Kei et al., 2017; Lahne et al., 2020), implying that unknown feedback signals from surviving neurons influence fate determination in MGPCs. This study was performed to investigate the roles of Sox11b in MG proliferation, MGPC migration and fate determination in a mechanical retinal injury model in adult zebrafish.
Methods
Animals and eye injury
Wild-type (AB) zebrafish used in the study were obtained from the China Zebrafish Resource Center (http://www.zfish.cn). All zebrafish experiments were approved by the Nantong University Experimental Animal Center (approval No. 20170320-017, approval date March 20, 2017). Zebrafish were maintained at 28°C on a 14-/10-hour light/dark cycle in a zebrafish breeding system (Haisheng Biotech, Shanghai, China). Adult (4–6 months old) zebrafish of both sexes with body weights of 0.4 to 0.5 g were randomly assigned to groups of three zebrafish each.
Retinal stab injuries were performed as previously described (Zhang et al., 2020). Briefly, zebrafish were anesthetized in 0.02% Tricane (Sigma-Aldrich, St. Louis, MO, USA); a sterile 30-G needle was used to puncture the back of the right eye through the sclera, once in each quadrant. The needle was inserted to the length of the bevel to ensure that the same amount of retinal damage occurred in all quadrants. The intact left eye served as a negative control.
Lineage tracing of MGPCs
For lineage tracing, 20 μL of 20 mM 5-bromo-2’-deoxyuridine (BrdU) were intraperitoneally injected into the zebrafish to label MGPCs at 4 days post-injury (dpi). Zebrafish were then sacrificed at 7, 21, or 30 dpi; BrdU immunostaining was performed to identify the progeny cells of MGPCs at the injury site (Figure 1).
Figure 1.
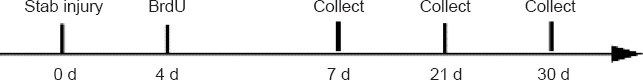
Schematic of the experimental design.
BrdU: 5-Bromo-2’-deoxyuridine.
Tissue preparation and immunofluorescence
Tissue preparation and retinal cryosectioning were performed in accordance with an established protocol (Zhang et al., 2020). Briefly, zebrafish were euthanized and the eyes were dissected. For each eye, the lens was removed and the eye cup was fixed in 4% paraformaldehyde at 4°C overnight. Fixed eyes were washed with PBS, incubated with sucrose (10% and 20%, 2 hours each), and embedded in SAKURA Tissue-Tek O.C.T. compound (4583, Sakura Finetek, Tokyo, Japan). Tissue samples were cryosectioned at a thickness of 12 μm, then collected on slides and dried at 50°C for 2 hours.
For immunofluorescence staining, slides were washed three times with phosphate-buffered saline (PBS), then incubated with the blocking solution (3% sheep serum, 0.1% Triton X-100, 1× PBS) at room temperature for 30 minutes. Slides were then incubated with primary antibodies in the blocking solution (described below) at 4°C overnight. On the next day, slides were washed with PBS and incubated with secondary antibodies in the blocking solution (described below) at room temperature for 2 hours. For BrdU and proliferating cell nuclear antigen (PCNA) staining, an antigen retrieval protocol (Zhang et al., 2016) was performed prior to standard immunostaining procedures. Briefly, slides were incubated with 2 M HCl at 37°C for 20 minutes; the HCl was neutralized with by incubation with 0.1 M Na2B4O7 (two neutralization incubations, each for 5 minutes at room temperature).
The following primary antibodies were used: rat anti-BrdU (1:500, Abcam, Cambridge, MA, USA, Cat# ab6326, RRID: AB_305426); rabbit anti-PCNA (1:500, GeneTex, Irvine, CA, USA, Cat# GTX14496, RRID:AB_11161916); mouse anti-Zpr1 (1:500, Abcam, Cat# ab174435); and mouse anti-Hu-antigen C/D (HuC/D; 1:500, Thermo Fisher Scientific, Waltham, MA, USA, Cat# A-21271, RRID: AB_221448). The following secondary antibodies were used: goat anti-rabbit IgG(H+L), Alexa Fluor 568 (1:500, Thermo Fisher Scientific, Cat# A-11036, RRID: AB_10563566); goat anti-rat IgG(H+L), Alexa Fluor 568 (1:500, Thermo Fisher Scientific, Cat# A-11077, RRID: AB_2534121); and goat anti-mouse IgG(H+L), Alexa Fluor 488 (1:500, Thermo Fisher Scientific, Cat# A-11029, RRID: AB_2534088).
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analysis was performed on fresh retinal cryosections, in accordance with the manufacturer’s instructions (PerkinElmer, Waltham, MA, USA). Briefly, slides were first incubated with 0.1% H2O2 at room temperature for 30 minutes to block endogenous peroxidase activity. Slides were washed with PBS, then incubated with 10 μg/mL proteinase K solution in Tris-ethylenediaminetetraacetic acid buffer for 5–10 minutes at 37°C. Subsequently, slides were washed with PBS and dehydrated in a graded ethanol series at room temperature. Digoxigenin-labeled sense or antisense RNA probes were added to the hybridization buffer at a final concentration of 1 μg/mL. Two hundred microliters of the hybridization solution were applied to each slide, then covered with parafilm to prevent evaporation; the covered slides were incubated in a sealed moisture chamber at 60°C overnight in the dark. On the next day, slides were washed with a graded saline-sodium citrate series and incubated with a sheep anti-digoxigenin-peroxidase antibody (1:500, Roche, Mannheim, Germany, Cat# 11207733910, RRID: AB_514500) at 4°C overnight. Slides were then washed in Tris-saline buffer (three times for 10 minutes each) and incubated with 100 μL of the tyramide working solution for 5–10 minutes at room temperature to enable signal visualization. Sense and antisense digoxigenin-labeled RNA probes against the zebrafish Sox11b transcript were transcribed using the digoxigenin-RNA labeling kit (Roche Applied Science, Penzberg, Upper Bavaria, Germany). No fluorescence signal above background was detected when using the sense Sox11b probe.
Morpholino and drug treatments
One microliter of 0.25 mM lissamine-tagged control or Sox11b morpholino (MO; Gene Tools, Philomath, OR, USA) was intravitreally injected with a Hamilton syringe (Hamilton Company Inc., Reno, NV, USA) to establish the experimental model. The method of MO electroporation was previously described (Fausett et al., 2008). The following two Sox11b translation-blocking MOs were used in the study: sox11b MO1 (5’-CAT GTT CAA ACA CAC TTT TCC CTC T-3’) (Veldman et al., 2007; Pillai-Kastoori et al., 2014) and sox11b MO2 (5’-TCC GTC TGC TGC ACC ATG TTC-3’).
For Notch inhibition, 1 μL of 250 μM of RO4929097 (MedChemExpress, Monmouth Junction, NJ, USA) was intravitreally injected through the front of the eye from 2 to 4 dpi, twice daily. One microliter of PBS was injected as a negative control.
Quantitative polymerase chain reaction
Zebrafish retinas were collected at the indicated time points and total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific). RNA was then reverse-transcribed and quantitative polymerase chain reaction (qPCR) was performed as previously described (Mu et al., 2017). qPCR was performed in triplicate using the Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China, Q712-02) with the following parameters: stage 1, 95°C for 30 seconds; stage 2, 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Relative mRNA levels were determined using the ΔΔCt method (Rao et al., 2013), then normalized to the mRNA levels of rp113. The primers used in this study are listed in Table 1.
Table 1.
Primers used for quantitative polymerase chain reaction analyses
| Gene | Forward primer (5'–3') | Reverse primer (5'–3') |
|---|---|---|
| dla | ACC TCT TAC CAG CAG TGC G | TGG GAT TGG TGA AAG AGC CA |
| dlb | CTC AAG CAT TCT GAG GAC GTG | CAT GAT GCT CTT TGG GCG AT |
| dlc | AGA AAT ACT GAG CGC GGA CT | TGC GTT CCA GGT TTC GAT TA |
| dld | TTC CCA ATC GTC TCC CTT TT | GTT TGC GTT GCC TGT CAC TC |
| notch1 | notch1 GGA CAG AGA GAA CGC CTA CA | TCG CAG ACA CAC TCA TAG CC |
| notch2 | GTG ACG CTT TAA CCT GAC GG | GTC CCG TTG CTT GAG ATG AC |
| notch3 | notch3 CCG TGC GAA AAT GGA GGA AT | GCT GCA TTT AGA CAA GGC GA |
| her4.1 | CTC TCA GAA AAG CAG TGC GG | TTT GCG TGT GTT TCA GTG GT |
| sox11a | TCC CAA ACT GAA AGC GAA CAA GAC | GCG CGG AGG AGG CGA AGT |
| sox11b | AGC AGC CGC AGG GAA GAA GT | CAC AGC ACA GTC AAT GTT TGG ACC |
dla: Delta-like protein A; dlb: delta-like protein B; dlc: delta-like protein C; dld: deltalike protein D; her4.1: hairy-related 4, tandem duplicate 1; notch1: neurogenic locus notch homolog protein 1; notch2: neurogenic locus notch homolog protein 2; notch3: neurogenic locus notch homolog protein 3; sox11ax: transcription - Sox11a; sox11b: transcription - Sox11b.
Microscopy
Fluorescence images of frozen retinal tissue were acquired using a Zeiss Imager M2 upright microscope (Carl Zeiss AG, Oberkochen, Germany) equipped with an Axiocam 506 monochrome camera. Images were captured with a 10× or 20× objective; ImageJ software (National Institutes of Health, Bethesda, MD, USA) (Schneider et al., 2012) was used to count cell number and measure the length/width ratio of MGPC nuclei. At the higher resolution, individual MGPC nuclei were identified on the basis of their morphology and staining pattern. Cell counts were performed using a series of sections that covered the injury region; the number of cells per injury was calculated as the sum of all cells in proximity to the injury site. One injury region was typically reconstructed in 25–30 sections; an injury in each of the four quadrants was counted separately.
Statistical analysis
No sample size calculations were performed; however, our sample sizes were similar to the numbers reported in a previous publication (Wan et al., 2012). All experiments in this study were performed in triplicate and repeated at least twice. Evaluators were blinded to their assignments to limit potential rater bias. For single comparisons, Student’s t-test (two-tailed, unpaired) was used. For multiple comparisons, a one-way analysis of variance was performed, followed by Tukey’s post hoc test. The chi-squared test with Bonferroni correction was used to compare the distributions of MGPCs among retinal layers. Error bars represent standard error (SEM) and statistical significance was defined as P < 0.05.
Results
Sox11b-expressing MGPCs preferentially migrate to the outer nuclear layer after stab injury
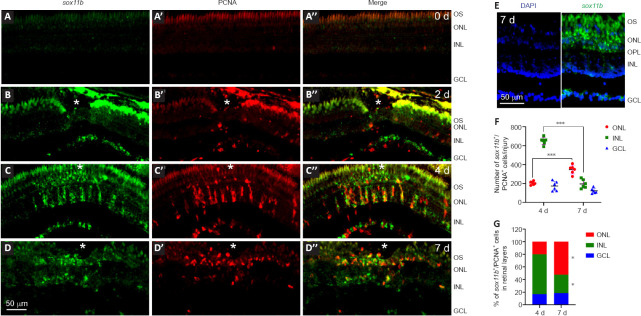
We have previously shown that after stab injury, sox11b is induced in the injured zebrafish retina and co-localizes with inner nuclear layer (INL) BrdU+ cells at 4 dpi (Mu et al., 2017). To better understand the expression pattern of sox11b, FISH using a sox11b antisense probe followed by PCNA immunofluorescence was performed in intact and injured zebrafish retinas at 2, 4 and 7 dpi. FISH showed that the sox11b mRNA signal was almost undetectable in intact retinas (Figure 2A–A’’). At 2 dpi, sox11b was induced in the ganglion cell layer (GCL) but was nearly undetectable in the INL (Figure 2B–B’’). Considering that MG entered the cell cycle at 2 days after stab injury (Fausett and Goldman, 2006; Zhang et al., 2020), our findings suggest that sox11b was induced in retinal ganglion cells (RGCs) rather than proliferating MG at 2 dpi. At 4 dpi, robust sox11b signals that co-localized with PCNA+ cell clusters were observed in the INL (Figure 2C–C’’). Because MGPCs reportedly constitute almost all proliferating cells in the INL at 4 dpi (Fausett and Goldman, 2006), we inferred that sox11b was predominantly expressed in the progenitor cells. At 7 dpi, there were fewer sox11b+/PCNA+ cells in the INL (compared with 4 dpi); instead, sox11b+/PCNA+ cells were preferentially located in the outer nuclear layer (ONL) (Figure 2D–D’’, and E–G). Thus, many of the sox11b-expressing MGPCs presumably migrated to the ONL at later time points, where they contribute to photoreceptor regeneration.
Figure 2.
Expression of sox11b in intact and injured zebrafish retinas.
Fluorescence in situ hybridization (FISH) and PCNA immunostaining analysis of sox11b mRNA expression in intact and injured zebrafish retinas. (A–A″) sox11b signal was nearly undetectable in intact retina (0 days). (B–B″) At 2 dpi, sox11b induction (green, fluorescein) was evident in the GCL near the injury site. PCNA+ cells (red, Alexa Fluor 568) in the INL exhibited marginal expression of sox11b. Fluorescence above the ONL constitutes autofluorescence from outer segments. (C–C″) At 4 dpi, strong sox11b signals were co-localized with PCNA in the INL. (D–D″) At 7 dpi, the sox11b signal decreased in PCNA+ cells in the INL, whereas it remained visible in PCNA+ cells in the ONL. White asterisks indicate sites of stab injury. (E) FISH combined with DAPI staining at 7 dpi showed that sox11b-expressing cells were preferentially located in the ONL. (F, G) Quantification of the number and distribution of sox11b+/PCNA+ cells across retinal layers. Data are expressed as mean ± SEM. Values were analyzed by one-way analysis of variance followed by Tukey’s post hoc test in F; they were analyzed by the chi-squared test with Bonferroni correction in G. *P < 0.05 vs. 4 dpi. ***P < 0.001. DAPI: 4’,6-Diamidino-2-phenylindole dihydrochloride; dpi: days post injury; GCL: ganglion cell layer; INL: inner nuclear layer; ONL: outer nuclear layer; OPL: outer plexiform layer; OS: outer segments; PCNA: proliferating cell nuclear antigen.
Sox11b knockdown does not affect MGPC formation but alters nuclear morphology in MGPCs
To investigate the role of Sox11b in MGPC formation, we electroporated a validated translation-blocking MO against Sox11b (sox11b MO1) (Veldman et al., 2007; Pillai-Kastoori et al., 2014) into the retina at the time of injury. Because most BrdU+ cells in the INL are MGPCs at 4 dpi (Fausett and Goldman, 2006), we quantified the number of these cells in the retina. BrdU immunofluorescence showed that Sox11b knockdown did not significantly affect the number of BrdU+ cells in the INL at the injury site compared to the control MO group (Figure 3A and B). The electroporation treatment did not significantly increase the numbers of INL proliferating cells, compared with the PBS control that lacked electroporation (Figure 3B), suggesting that electroporation itself did not cause extensive retinal damage. Notably, the electroporation of a second Sox11b translation-blocking MO (sox11b MO2) into the injured retina produced similar results (Figure 3A and B), confirming the above findings. Additionally, Sox11b knockdown did not alter the distribution of BrdU+ cells across the ONL and INL (Figure 3C). Although Sox11b knockdown did not affect MGPC formation, the nuclear morphology of MGPCs was altered in the INL (Figure 3A and D). Specifically, control MGPCs in the INL displayed an elongated, spindle-shaped nuclear morphology in which the long axis was largely perpendicular to the retinal layers (Figure 3D). In contrast, the nuclei of Sox11b-depleted MGPCs exhibited a more rounded shape (Figure 3D). Quantification of MGPC morphology revealed that Sox11b knockdown significantly decreased the length/width ratio for affected nuclei, compared with the ratio for control nuclei (Figure 3E). Because spindle-shaped MGPCs have been shown to migrate along the radial MG fibers to different retinal layers (Nagashima et al., 2013), the altered nuclear morphology observed in Sox11b-depleted MGPCs suggests that they exhibit defective radial migration.
Figure 3.
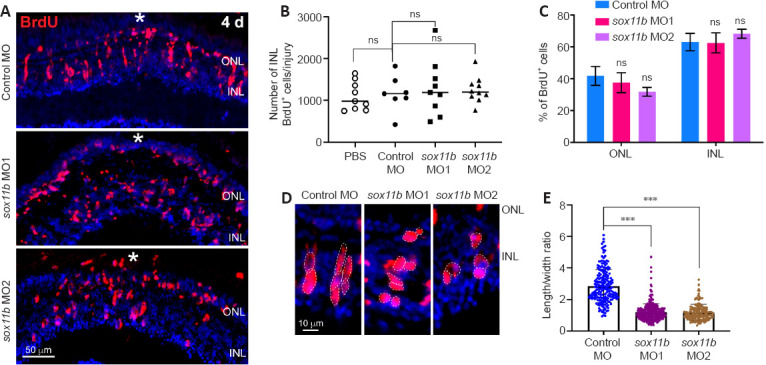
Sox11b knockdown does not affect MGPC cell number but causes changes in nuclear morphology at 4 dpi.
(A) BrdU immunofluorescence showed that MGPC formation was similar in control MO- and sox11b MO-treated retina at 4 dpi. White asterisks indicate sites of stab injury. (B) Quantification of BrdU+ cells at the injury site. (C) Proportions of BrdU+ cells located in the ONL or INL. (D) Higher-magnification images show differences in MGPC nuclear morphology, as indicated by white dotted lines, between control and Sox11b-depleted retinas at 4 dpi. (E) Quantification of the length/width ratio of MGPCs in the INL. Data are expressed as mean ± SEM (n = 3). Values were analyzed by one-way analysis of variance followed by Tukey’s post hoc test. ***P < 0.001. BrdU: 5-Bromo-2’-deoxyuridine; dpi: days post injury; GCL: ganglion cell layer; INL: inner nuclear layer; MGPC: Müller glia-derived progenitor cells; MO: morpholino; ns: not significant compared with control; ONL: outer nuclear layer.
Sox11b loss-of-function alters MGPC distribution during later stages of regeneration
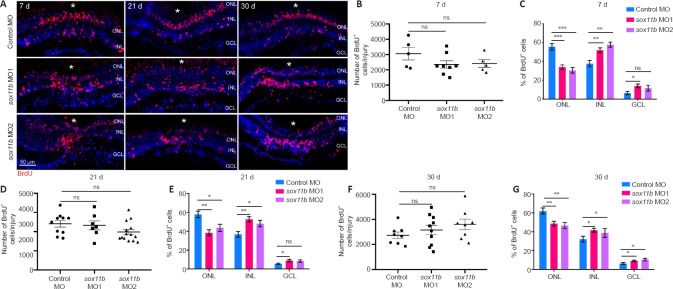
Because Sox11b exhibits late expression in MGPCs and has a nonessential role in MGPC formation, it may be involved in later stages of retinal regeneration (e.g., MGPC migration and fate determination). To explore this possibility, MGPCs were labeled with a pulse of BrdU at 4 dpi; the distributions of their progeny cells were examined at 7, 21, and 30 dpi. Consistent with the above findings at 4 dpi, Sox11b knockdown did not significantly affect the number of BrdU+ cells at 7 dpi (Figure 4A left panel, and 4B). However, assessment of BrdU+ cell locations revealed that Sox11b loss-of-function altered their distribution across retinal layers (chi-squared test, P < 0.05). In control retinas, BrdU-labeled nuclei were preferentially found in the ONL, rather than the INL and GCL, at 7 dpi (Figure 4A and C). In contrast, a greater proportion of the nuclei of Sox11b-depleted MGPCs was found in the INL, rather than the ONL (Figure 4A and C). Compared with control retinas, sox11b knockdown significantly reduced the proportion of MGPC nuclei in the ONL but increasing that proportion in the INL (Figure 4C). At 21 and 30 dpi, similar phenotypes were observed in retinas that had been treated with sox11b MOs (Figure 4A middle and right panels, and 4D–G). These results suggest that Sox11b regulates MGPC migration; Sox11b loss-of-function could lead to altered distributions of MGPCs at later time points.
Figure 4.
Sox11b loss-of-function alters MGPC distribution across retinal layers after 7 dpi.
(A) BrdU lineage tracing revealed greater proportions of Sox11b-depleted MGPCs in the inner retina at 7, 21, and 30 dpi. MGPCs were labeled with a pulse of BrdU at 4 dpi. White asterisks indicate sites of stab injury. (B, D, F) Quantification of the total number of BrdU+ cells at the injury site at indicated time points. (C, E, G) Quantification of the proportions of BrdU+ cells in the ONL, INL, or GCL at indicated time points. Data are expressed as mean ± SEM (n = 3). Values were analyzed by one-way analysis of variance followed by Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001. BrdU: 5-Bromo-2’-deoxyuridine; DAPI: 4’,6-diamidino-2-phenylindole dihydrochloride; dpi: days post injury; GCL: ganglion cell layer; INL: inner nuclear layer; MO: morpholino; ns: not significant compared with control MO; ONL: outer nuclear layer.
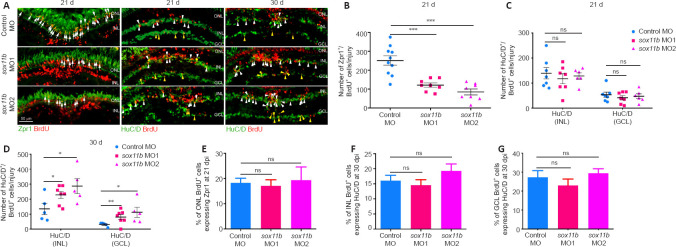
Sox11b biases MGPCs toward photoreceptor fates
To investigate the role of Sox11b in MGPC fate determination, MGPCs were labeled with a pulse of BrdU at 4 dpi; their subsequent differentiation was examined by immunostaining with BrdU and cell type markers (Zpr1, photoreceptors; HuC/D in INL, amacrine cells; HuC/D in GCL, RGCs) at 21 and 30 dpi. At 21 dpi, Sox11b knockdown significantly reduced the number of BrdU+/Zpr1+ cells in the ONL (Figure 5A and B) but had no effect on the numbers of BrdU+/ HuC/D+ cells in the INL or GCL (Figure 5A and C); these results suggested that Sox11b promotes the differentiation of MGPCs into photoreceptors. Because Sox11b did not appear to affect amacrine cell or RGC regeneration at 21 dpi, while the proportion of MGPCs in the INL was greater in Sox11b-depleted retinas (Figure 4), we examined the differentiation of MGPCs at 30 dpi. Indeed, immunostaining revealed that Sox11b knockdown led to increased numbers of BrdU+/ HuC/D+ cells in both the INL and GCL (Figure 5A, and D). These findings suggest that the influence of Sox11b knockdown on amacrine cell or RGC regeneration occurred between 21 and 30 dpi, which is later than the influence of Sox11b knockdown on photoreceptors.
Figure 5.
Sox11b biases MGPCs toward a photoreceptor fate.
(A) BrdU lineage tracing revealed that Sox11b knockdown biased MGPCs toward amacrine and RGC fates. Immunofluorescence showed that BrdU+ MGPCs co-localized with cell differentiation markers Zpr1 (photoreceptor, white arrows) or HuC/D (amacrine cells, white arrowheads, and RGCs, yellow arrowheads) in control MO- or sox11b MO-treated retinas at 21 and 30 dpi. Zpr1 and HuC/D, green color (Alexa Fluor 488); BrdU, red color (Alexa Fluor 568). (B–D) Quantification of the number of BrdU+ cells expressing specific cell type markers. (E–G) Calculation of the proportion of BrdU+ cells in each layer expressing specific cell type markers. Data are expressed as mean ± SEM (n = 3). Values were analyzed by one-way analysis of variance followed by Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001. BrdU: 5-Bromo-2’-deoxyuridine; dpi: days post injury; HuC/D: Hu-antigen C/D; GCL: ganglion cell layer; INL: inner nuclear layer; MO: morpholino; ns: not significant compared with control MO; ONL: outer nuclear layer; RGCs: retinal ganglion cells; Zpr1: ZPR1 zinc finger.
The similar effects of Sox11b silencing on MGPC distribution and neuronal regeneration across various retinal layers suggest that MGPC fates may be influenced by their location. Such influence would presumably cause the proportion of MGPCs that differentiate into each type of neuron within a single retinal layer to be comparable between controls and Sox11b morphants. Indeed, quantification revealed that the ratios of regenerated photoreceptors to ONL BrdU+ cells, amacrine cells to INL BrdU+ cells, and RGCs to GCL BrdU+ cells were all comparable between controls and Sox11b morphants (Figure 5E–G). Therefore, Sox11b may indirectly affect MGPC cell fate by regulating the radial migration and distribution of MGPCs.
Sox11b regulates injury-dependent induction of Notch signaling component genes
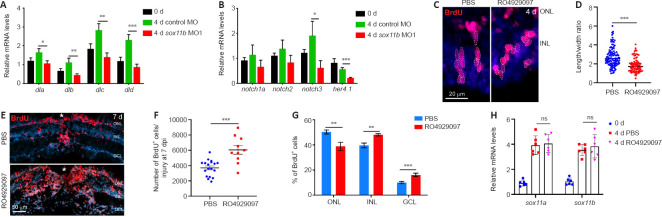
Notch signaling is an important regulator of neural progenitor proliferation and differentiation (Pinto-Teixeira and Desplan, 2014; Moore and Alexandre, 2020), as well as cortical neuron radial migration (Yang et al., 2017; Zhou et al., 2020b). Notch signaling components were reportedly induced in MGPCs, thereby regulating their differentiation, in injured zebrafish retinas (Wan et al., 2012). Because Sox11b loss-of-function resulted in defects in MGPC migration and fate determination, we examined its influence on the expression of Notch signaling components in the retina. At 4 dpi, qPCR revealed that Sox11b knockdown significantly reduced the expression levels of several Notch ligands, including dla, dlb, dlc, and dld; the Notch receptor notch3; and the Notch downstream target her4.1 (Wan et al., 2012) (Figure 6A and B). Thus, we inferred that Sox11b regulates Notch signaling in injured retinas. To investigate whether Notch inhibition might mimic the retinal phenotype of Sox11b morphants, zebrafish were intravitreally injected with PBS or the Notch inhibitor RO4929097 between 2 and 4 dpi; MGPC nuclear morphology was then examined via BrdU immunostaining. Indeed, Notch suppression altered MGPC nuclear morphology and decreased the nuclear length/width ratio in affected cells (Figure 6C and D), recapitulating the phenotype observed in Sox11b morphants. To examine the effect of Notch inhibition on subsequent MGPC distribution, MGPCs were labeled with a pulse of BrdU at 4 dpi; zebrafish were then treated with PBS or RO4929097 from 4 to 7 dpi. We found that Notch inhibition during this period led to an increased MGPC population (Figure 6E and F), consistent with the involvement of Notch in MGPC proliferation (Wan et al., 2012). Notably, Notch suppression resulted in greater proportions of MGPCs in the INL and GCL, compared with control retinas, while a smaller proportion of MGPCs was present in the ONL (Figure 6E and G); these results were similar to the findings in Sox11b-depleted retinas. qPCR analysis indicated that the retinal phenotype after Notch inhibition was not the product of altered Sox11 expression (Figure 6H). Taken together, these findings suggest that Sox11b regulates the expression patterns of Notch signaling components; it presumably controls MGPC migration and fate determination through this pathway.
Figure 6.
Sox11b regulates the expression of Notch signaling components in injured retinas.
(A, B) qPCR revealed the relative expression levels (normalized to 0 days) in the retina at 4 dpi: Notch ligands (A); Notch receptors and the reporter gene her4.1 (B). (C) BrdU immunofluorescence showed that RO4929097-treated MGPCs exhibited a more rounded morphology. Outlines of MGPC nuclei are indicated by white dotted lines. (D) Quantification of MGPC nuclear length/width ratio in the INL. (E) BrdU lineage tracing indicated that Notch inhibition altered the distribution and increased the number of MGPCs, compared with control retinas at 7 dpi. White asterisks indicate sites of stab injury. (F) Quantification of MGPC cell number at the injury site. (G) Quantification of the proportion of BrdU+ cells in each layer. (H) qPCR showed the relative mRNA expression levels (normalized to 0 days) of sox11a and sox11b in PBS- or RO4929097-treated retinas at 4 dpi. Data are expressed as mean ± SEM (n = 3). Values were analyzed by one-way analysis of variance followed by Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001. BrdU: 5-Bromo-2’-deoxyuridine; dpi: days post injury; GCL: ganglion cell layer; INL: inner nuclear layer; MO; morpholino; MGPC: Müller glia-derived progenitor cells; ns: not significant; ONL: outer nuclear layer; PBS: phosphate-buffered saline.
Discussion
In this study, we investigated the role of the transcription factor Sox11b in retina regeneration in zebrafish. FISH analysis showed that sox11b was expressed in proliferating MGPCs from 4 to 7 dpi. Functional studies revealed that Sox11b was not required for MGPC formation; instead, it regulated MGPC migration and differentiation, presumably through the Notch signaling pathway. Thus, Sox11b plays an important role in determining progenitor fate during retina regeneration.
Several previous studies have established that Sox11 is required for optic nerve regeneration (Mu et al., 2017; Norsworthy et al., 2017). In this study, we observed substantial sox11b expression in the GCL around the injury site at 2 and 4 dpi, indicating that the stab injury also caused damage to some RGC axons and produced a local regenerative response in the GCL. While sox11b was induced in the GCL at 2 dpi, its signal in the INL was nearly undetectable at this time point when MG entered the cell cycle (Fausett and Goldman, 2006); thus, Sox11b may not be essential for MG proliferation. Consistent with this hypothesis, we found that Sox11b knockdown did not affect MGPC formation at 4 dpi. This observation is inconsistent with previous findings that Sox11b was required for MG proliferation (Mu et al., 2017). The discrepancy is presumably related to the different amounts of Sox11b MOs used in the knockdown experiments; a considerably lower dose was used in the present study than in the prior study (0.25 mM vs. 1 mM). Similar to small interfering RNAs and short hairpin RNAs, MOs exhibit off-target effects in a dose-dependent manner (Stainier et al., 2017). Thus, the high concentration of MO (1 mM) used in the prior study was more likely to induce non-specific phenotypes. Additionally, we showed that the electroporation of a second Sox11b MO (sox11b MO2) into the injured retina produced similar results, confirming that Sox11b is not required for MG proliferation or MGPC formation.
Although Sox11b loss-of-function did not disrupt MGPC formation, detailed morphological analysis revealed the alteration of nuclear morphology in sox11b MO-treated retinas at 4 dpi. Further lineage tracing of MGPCs showed that Sox11b knockdown perturbed their migration into the ONL, such that greater proportions of these cells remained in the INL and GCL after 7 dpi. SoxC proteins play critical roles in the migration of precursor, epidermal, and tumor cells by regulating their cytoskeletal structure and epithelial-mesenchymal transition (Parvani and Schiemann, 2013; Kavyanifar et al., 2018; Miao et al., 2019). During cerebral development in mice, Sox11 inhibition caused precocious branching of neurites and suppressed the radial migration of cortical neurons (Hoshiba et al., 2016). The phenotype observed in the mouse cerebral cortex is similar to our findings that Sox11b knockdown reduced the length/width ratio of MGPC nuclei, altered their spindle-shape morphology to a more rounded form, and disrupted their radial migration pattern. However, because there is a lack of tools to specifically label the membrane structure in MGPCs, the effect of Sox11b knockdown on their overall morphology remains unknown; further investigations are needed. Future studies should also explore the effects of Sox11b knockdown on cytoskeletal structure and epithelial-mesenchymal markers in MGPCs.
At the molecular level, we found that Sox11b regulates the expression patterns of several Notch ligands, as well as notch3 and her4.1, in the zebrafish retina. A previous study showed that the Notch ligands dla, dlb, dlc and dld, as well as notch1 and her4, were expressed in proliferating MGPCs in stab-injured zebrafish retinas (Wan et al., 2012). A recent study demonstrated that Notch3 was expressed by quiescent MG, while the Notch ligand dlb was expressed in non-MG cells, in the zebrafish retina (Campbell et al., 2021). Because we have shown sox11b expression in proliferating MGPCs, we suspect that Sox11b promotes the expression of Notch ligands in MGPCs in a cell-autonomous manner; these Notch ligands bind to the Notch receptors on neighboring progenitors (Notch1) or MG (Notch3) to activate Notch signaling. Consistent with this hypothesis, the SoxC protein Sox4 was found to regulate the expression of Notch ligands, activate the Notch signaling pathway, and promote metastasis in prostate cancer cells (Moreno, 2010). Additionally, SoxC reportedly stimulated Notch signaling and influenced the cell fate of neighboring neural progenitors in sea urchin embryos (Mellott et al., 2017). Importantly, Notch inhibition produced a similar phenotype in terms of MGPC morphology and distribution in the retina, suggesting that Sox11b regulates MGPC migration via Notch signaling. Although Notch inhibition increased the number of BrdU+ MGPCs at 7 dpi, this effect was not observed in Sox11b-depleted retinas, implying that Notch signaling may not be the sole downstream effector of Sox11b; moreover, Notch signaling activity could be regulated by other molecules in these progenitor cells.
In the past several years, the transdifferentiation of MG into retinal neurons via genetic manipulation has been the focus of multiple studies (Jorstad et al., 2017; Yao et al., 2018; Hoang et al., 2020; Zhou et al., 2020a). Despite substantial progress, it remains challenging to achieve targeted differentiation of MG into specific types of retinal neurons in vivo. Our findings indicate that, in the zebrafish retina, Sox11b governs the migration of MGPCs and directs them toward a photoreceptor fate. Future research should investigate whether Sox11 can be used to direct mammalian MG differentiation; the present findings imply that Sox11 overexpression may drive MG toward photoreceptor fate, whereas Sox11 knockdown may bias them toward an inner retinal neuron fate. Additionally, the combination of Sox11 and other factors may result in more efficient MG reprogramming in the mammalian retina. Nevertheless, because this study only examined the role of Sox11b in zebrafish retina, we cannot exclude the possibility that Sox11 has a unique role in the mammalian retina.
Footnotes
Funding: This work was supported by the National Key Research and Development Project of China, Nos. 2017YFA0104100 (to JL), 2017YFA0701304 (to HX); and National Natural Science Foundation of China Nos. 81970820 (to HX), 31930068 (to JL).
Conflicts of interest: The authors declare no competing financial interests.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Chastain-Gross R, Yu J, Song LP; T-Editor: Jia Y
References
- 1.Campbell LJ, Hobgood JS, Jia M, Boyd P, Hipp RI, Hyde DR. Notch3 and DeltaB maintain Müller glia quiescence and act as negative regulators of regeneration in the light-damaged zebrafish retina. Glia. 2021;69:546–566. doi: 10.1002/glia.23912. [DOI] [PubMed] [Google Scholar]
- 2.Campbell LJ, Levendusky JL, Steines SA, Hyde DR. Retinal regeneration requires dynamic Notch signaling. Neural Regen Res. 2022;17:1199–1209. doi: 10.4103/1673-5374.327326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang KC, Hertz J. SoxC transcription factors in retinal development and regeneration. Neural Regen Res. 2017;12:1048–1051. doi: 10.4103/1673-5374.211178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang KC, Hertz J, Zhang X, Jin XL, Shaw P, Derosa BA, Li JY, Venugopalan P, Valenzuela DA, Patel RD, Russano KR, Alshamekh SA, Sun C, Tenerelli K, Li C, Velmeshev D, Cheng Y, Boyce TM, Dreyfuss A, Uddin MS, et al. Novel regulatory mechanisms for the SoxC transcriptional network required for visual pathway development. J Neurosci. 2017;37:4967–4981. doi: 10.1523/JNEUROSCI.3430-13.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman D. Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorsuch RA, Hyde DR. Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exp Eye Res. 2014;123:131–140. doi: 10.1016/j.exer.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, Saez C, Keuthan C, Palazzo I, Squires N, Campbell WA, Rajaii F, Parayil T, Trinh V, Kim DW, Wang G, et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 2020;370 doi: 10.1126/science.abb8598. eabb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshiba Y, Toda T, Ebisu H, Wakimoto M, Yanagi S, Kawasaki H. Sox11 balances dendritic morphogenesis with neuronal migration in the developing cerebral cortex. J Neurosci. 2016;36:5775–5784. doi: 10.1523/JNEUROSCI.3250-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jankowski MP, McIlwrath SL, Jing X, Cornuet PK, Salerno KM, Koerber HR, Albers KM. Sox11 transcription factor modulates peripheral nerve regeneration in adult mice. Brain Res. 2009;1256:43–54. doi: 10.1016/j.brainres.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing X, Wang T, Huang S, Glorioso JC, Albers KM. The transcription factor Sox11 promotes nerve regeneration through activation of the regeneration-associated gene Sprr1a. Exp Neurol. 2012;233:221–232. doi: 10.1016/j.expneurol.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature. 2017;548:103–107. doi: 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kavyanifar A, Turan S, Lie DC. SoxC transcription factors:multifunctional regulators of neurodevelopment. Cell Tissue Res. 2018;371:91–103. doi: 10.1007/s00441-017-2708-7. [DOI] [PubMed] [Google Scholar]
- 15.Kuwajima T, Soares CA, Sitko AA, Lefebvre V, Mason C. SoxC transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron. 2017;93:1110–1125.e5. doi: 10.1016/j.neuron.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahne M, Brecker M, Jones SE, Hyde DR. The regenerating adult zebrafish retina recapitulates developmental fate specification programs. Front Cell Dev Biol. 2020;8:617923. doi: 10.3389/fcell.2020.617923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenkowski JR, Raymond PA. Muller glia:Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellott DO, Thisdelle J, Burke RD. Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Development. 2017;144:3602–3611. doi: 10.1242/dev.151720. [DOI] [PubMed] [Google Scholar]
- 19.Miao Q, Hill MC, Chen F, Mo Q, Ku AT, Ramos C, Sock E, Lefebvre V, Nguyen H. SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat Commun. 2019;10:4042. doi: 10.1038/s41467-019-11880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore R, Alexandre P. Delta-Notch signaling:the long and the short of a neuron's influence on progenitor fates. J Dev Biol. 2020;8:8. doi: 10.3390/jdb8020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno CS. The Sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression:crosstalk with the Wnt, Notch, and PI3K pathways. Am J Pathol. 2010;176:518–527. doi: 10.2353/ajpath.2010.090657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu Z, Zhang S, He C, Hou H, Liu D, Hu N, Xu H. Expression of SoxC transcription factors during zebrafish retinal and optic nerve regeneration. Neurosci Bull. 2017;33:53–61. doi: 10.1007/s12264-016-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng Chi Kei J, Currie PD, Jusuf PR. Fate bias during neural regeneration adjusts dynamically without recapitulating developmental fate progression. Neural Dev. 2017;12:12. doi: 10.1186/s13064-017-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norsworthy MW, Bei F, Kawaguchi R, Wang Q, Tran NM, Li Y, Brommer B, Zhang Y, Wang C, Sanes JR, Coppola G, He Z. Sox11 expression promotes regeneration of some retinal ganglion cell types but kills others. Neuron. 2017;94:1112–1120.e4. doi: 10.1016/j.neuron.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvani JG, Schiemann WP. Sox4, EMT programs, and the metastatic progression of breast cancers:mastering the masters of EMT. Breast Cancer Res. 2013;15:R72. doi: 10.1186/bcr3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai-Kastoori L, Wen W, Wilson SG, Strachan E, Lo-Castro A, Fichera M, Musumeci SA, Lehmann OJ, Morris AC. Sox11 is required to maintain proper levels of Hedgehog signaling during vertebrate ocular morphogenesis. PLoS Genet. 2014;10:e1004491. doi: 10.1371/journal.pgen.1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto-Teixeira F, Desplan C. Notch activity in neural progenitors coordinates cytokinesis and asymmetric differentiation. Sci Signal. 2014;7:e26. doi: 10.1126/scisignal.2005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell C, Cornblath E, Elsaeidi F, Wan J, Goldman D. Zebrafish Müller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons. Sci Rep. 2016;6:24851. doi: 10.1038/srep24851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 31.Reiprich S, Wegner M. From CNS stem cells to neurons and glia:Sox for everyone. Cell Tissue Res. 2015;359:111–124. doi: 10.1007/s00441-014-1909-6. [DOI] [PubMed] [Google Scholar]
- 32.Roska B, Sahel JA. Restoring vision. Nature. 2018;557:359–367. doi: 10.1038/s41586-018-0076-4. [DOI] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ:25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, Ingham PW, Schulte-Merker S, Yelon D, Weinstein BM, Mullins MC, Wilson SW, Ramakrishnan L, Amacher SL, Neuhauss SCF, Meng A, Mochizuki N, Panula P, Moens CB. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017;13:e1007000. doi: 10.1371/journal.pgen.1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang SM, Oliemuller E, Howard BA. Regulatory roles for SOX11 in development, stem cells and cancer. Semin Cancer Biol. 2020;67:3–11. doi: 10.1016/j.semcancer.2020.06.015. [DOI] [PubMed] [Google Scholar]
- 36.Usui A, Mochizuki Y, Iida A, Miyauchi E, Satoh S, Sock E, Nakauchi H, Aburatani H, Murakami A, Wegner M, Watanabe S. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development. 2013;140:740–750. doi: 10.1242/dev.090274. [DOI] [PubMed] [Google Scholar]
- 37.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Reynolds A, Kirry A, Nienhaus C, Blackmore MG. Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J Neurosci. 2015;35:3139–3145. doi: 10.1523/JNEUROSCI.2832-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilken MS, Reh TA. Retinal regeneration in birds and mice. Curr Opin Genet Dev. 2016;40:57–64. doi: 10.1016/j.gde.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Wohl SG. View on microRNAs as a potential tool to fight blindness:focus on Müller glia and gliosis. Neural Regen Res. 2022;17:1501–1502. doi: 10.4103/1673-5374.330610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Li PF, Chen RC, Wang J, Wang S, Shen Y, Wu X, Fang B, Cheng X, Xiong ZQ. ADAM10-initiated release of notch intracellular domain regulates microtubule stability and radial migration of cortical neurons. Cereb Cortex. 2017;27:919–932. doi: 10.1093/cercor/bhx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, Demb JB, Chen B. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature. 2018;560:484–488. doi: 10.1038/s41586-018-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S, Mu Z, He C, Zhou M, Liu D, Zhao XF, Goldman D, Xu H. Antiviral drug ganciclovir is a potent inhibitor of the proliferation of müller glia-derived progenitors during zebrafish retinal regeneration. Invest Ophthalmol Vis Sci. 2016;57:1991–2000. doi: 10.1167/iovs.15-18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Hou H, Yu S, Zhou C, Zhang X, Li N, Zhang S, Song K, Lu Y, Liu D, Lu H, Xu H. Inflammation-induced mammalian target of rapamycin signaling is essential for retina regeneration. Glia. 2020;68:111–127. doi: 10.1002/glia.23707. [DOI] [PubMed] [Google Scholar]
- 46.Zhao N, Yu HD, Feng Z, Ding JY, Liu XZ. Salidroside inhibits apoptosis of retinal Müller cells induced by high glucose in rats. Zhongguo Zuzhi Gongcheng Yanjiu. 2021;25:1664–1669. [Google Scholar]
- 47.Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z, Xiao Q, Wang B, Wu W, Sun Y, Zhou Y, Tang C, Liu F, Wang L, Feng C, Liu M, Li S, Zhang Y, Xu H, Yao H, et al. Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020a;181:590–603.e16. doi: 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 48.Zhou ZW, Kirtay M, Schneble N, Yakoub G, Ding M, Rüdiger T, Siniuk K, Lu R, Jiang YN, Li TL, Kaether C, Barzilai A, Wang ZQ. NBS1 interacts with Notch signaling in neuronal homeostasis. Nucleic Acids Res. 2020b;48:10924–10939. doi: 10.1093/nar/gkaa716. [DOI] [PMC free article] [PubMed] [Google Scholar]