Abstract
The 2,3-dihydroxybiphenyl dioxygenase from Sphingomonas sp. strain BN6 (BphC1-BN6) differs from most other extradiol dioxygenases by its ability to oxidize 3-chlorocatechol to 3-chloro-2-hydroxymuconic semialdehyde by a distal cleavage mechanism. The turnover of different substrates and the effects of various inhibitors on BphC1-BN6 were compared with those of another 2,3-dihydroxybiphenyl dioxygenase from the same strain (BphC2-BN6) as well as with those of the archetypical catechol 2,3-dioxygenase (C23O-mt2) encoded by the TOL plasmid. Cell extracts containing C23O-mt2 or BphC2-BN6 converted the relevant substrates with an almost constant rate for at least 10 min, whereas BphC1-BN6 was inactivated significantly within the first minutes during the turnover of all substrates tested. Furthermore, BphC1-BN6 was much more sensitive than the other two enzymes to inactivation by the Fe(II) ion-chelating compound o-phenanthroline. The reason for inactivation of BphC1-BN6 appeared to be the loss of the weakly bound ferrous ion, which is the cofactor in the catalytic center. A mutant enzyme of BphC1-BN6 constructed by site-directed mutagenesis showed a higher stability to inactivation by o-phenanthroline and an increased catalytic efficiency for the conversion of 2,3-dihydroxybiphenyl and 3-methylcatechol but was still inactivated during substrate oxidation.
Extradiol dioxygenases play an important role in the degradation of aromatic compounds by bacteria. Enzymes such as catechol 2,3-dioxygenase, 2,3-dihydroxybiphenyl 1,2-dioxygenase (2,3-DHBPDO), and 1,2-dihydroxynaphthalene dioxygenase convert central intermediates in the degradation of toluene, xylene, biphenyl, and naphthalene. Many years ago it was generally accepted that these enzymes form a rather homogeneous group and that the catechol 2,3-dioxygenase encoded by the TOL plasmid (C23O-mt2) represents the archetypical extradiol dioxygenase. However, recent cloning and sequencing of various extradiol dioxygenases demonstrated that this superfamily of enzymes can be separated into different groups with very limited sequence homology (2, 3, 8, 12, 13, 26). Unfortunately, for most of these enzymes, only the deduced amino acid sequences are known and generally no or scant biochemical data are available to correlate the sequence data with the catalytical function of the enzymes.
We have recently cloned the genes for two extradiol dioxygenases from the naphthalenesulfonate-degrading bacterium Sphingomonas sp. strain BN6 which do not belong to the main group of catechol 2,3-dioxygenases or 2,3-DHBPDOs (14, 15). These enzymes, BphC1-BN6 and BphC2-BN6, were tentatively designated 2,3-DHBPDOs because they showed the highest catalytical efficiencies with 2,3-dihydroxybiphenyl (2,3-DHBP) from several tested aromatic ortho-diols. The natural function of these enzymes is still unknown. The gene encoding BphC2-BN6 is probably part of an operon which encodes a degradative pathway for aromatic compounds (reference 14 and unpublished results). In contrast, the gene encoding BphC1-BN6 does not appear to be part of an operon and lacks any Escherichia coli promoter- or ribosome-binding site-like sequences upstream of the open reading frame (15).
BphC1-BN6 differs significantly from most other extradiol dioxygenases by its small size and the ability to convert 3-chlorocatechol by a distal ring fission mechanism to 3-chloro-2-hydroxymuconic semialdehyde (15, 22). 3-Chlorocatechol has been repeatedly reported to act as a strong (suicide) inhibitor of extradiol dioxygenases (4, 18). Furthermore, the inactivation of extradiol dioxygenases by certain chlorinated catechols seems to be responsible for the inability of many natural communities to degrade chlorinated aromatics occurring singly or in the presence of methylated structural analogues (23, 27). Therefore, extradiol dioxygenases like BphC1-BN6 may find application in the productive degradation of chlorinated aromatics. In the present study, we compared the enzymatic characteristics of BphC1-BN6 and other extradiol dioxygenases and also attempted to gain some insight into the molecular basis of the unusual traits of BphC1-BN6 by site-directed mutagenesis.
MATERIALS AND METHODS
Bacterial strains.
The 2,3-DHBPDOs BphC1-BN6 and BphC2-BN6 were expressed in E. coli JM108(pGHS118) and E. coli JM109(pGHS201) (14, 15, 22). The source of C23O-mt2 was E. coli BL21(DE3)(pJF150). Plasmid pJF150 was constructed by cloning a 1.0-kb BamHI-HindIII fragment containing the xylE gene from pIJ4083 (7) into a derivative of pBluescript M13 (9).
Preparation of cell extracts.
Cell extracts were prepared and protein content was determined as described previously (19).
Enzyme assays.
One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min. The enzyme assays were performed spectrophotometrically and contained in a total volume of 1 ml in sodium phosphate-potassium phosphate (Na/K-phosphate) buffer (100 mM, pH 7.5), 0.2 mM catechol, or 0.1 mM 2,3-DHBP. Before initiating the experiments, BphC1-BN6 and the mutant enzyme were reactivated by the addition of Fe2+ ions (2 mM) and dithiothreitol (DTT) (5 mM). Generally, the reaction was started by the addition of 1 to 5 μl of enzyme solution and measured for 30 s. To examine the stability of the enzymes at low concentrations, cell extracts were first diluted (1:500, vol/vol) with Na/K-phosphate buffer (100 mM, pH 7.5) and the reaction was initiated at different time intervals by the addition of substrate. The wavelengths and reaction coefficients used have been described previously (15).
Analysis of enzyme kinetics.
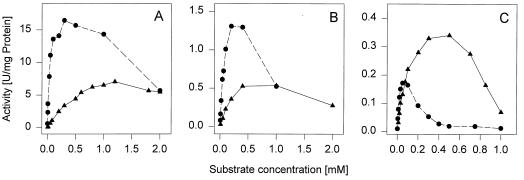
The reaction rates were determined by using the microcomputer regression analysis provided by a spectrophotometer (Kontron, Zurich, Switzerland), based on the first five absorbance measurements made at 6-s intervals. The kinetic data were calculated partly from the results shown in Fig. 4 according to the Michaelis-Menten equation, using Graph Pad Prism (GraphPad Software, San Diego, Calif.). For comparison of the substrate affinities of both enzymes, Vmax and Km values were estimated from those parts of the diagram shown in Fig. 4 which did not show a visible substrate inhibition effect.
FIG. 4.
Oxidation of various substrates by purified BphC1-BN6 and the Glu79His mutant. The reaction mixtures contained in 1 ml 100 mM Na/K-phosphate buffer (pH 7.5), the purified enzymes, and the indicated concentrations of substrates. The enzyme activities were calculated from the average reaction rates during the first 30 s after addition of the enzyme. The assays with BphC1-BN6 (▴) and 2,3-DHBP or 3-chlorocatechol contained 9 μg of protein, and the tests with 3-methylcatechol as the substrate contained 27 μg of protein. For the tests with the Glu79His mutant enzyme (●) with 3-methylcatechol and 3-chlorocatechol, 11 μg of protein was added; for those with 2,3-DHBP, 2 μg of protein was added. (A) 2,3-DHBP; (B) 3-methylcatechol; (C) 3-chlorocatechol.
The degree of enzyme inactivation during substrate turnover was quantified by calculating the apparent rate constant of enzyme inactivation, kinactγ, as described by Cerdan et al. (5, 6). The kinactγ values describe the deviation from linearity during the course of the enzymatic reaction, depending on the substrate concentration. The values were determined from the progress curve of the enzyme reaction for the first 30 s. Using kinactγ values for at least five different substrate concentrations between 50 and 1,000 μM, the rate constants of enzyme inactivation, kinact, were calculated.
Preincubation of cell extracts with inhibitors.
Cell extracts of E. coli JM109(pGHS201), E. coli JM108(pGHS118), and E. coli BL21(DE3)(pJF150) containing 10 to 62 mg of protein/ml were incubated with 3-chlorocatechol, Tiron (4,5-dihydroxy-1,3-benzenedisulfonate), or o-phenanthroline (1 mM each). The specific activities of BphC1-BN6 and BphC2-BN6 with 2,3-DHBP were 0.57 and 2.5 U/mg of protein, respectively. The specific activity of C23O-mt2 with catechol was 0.24 U/mg of protein. The cell extracts were diluted (1:5, vol/vol) with Na/K-phosphate buffer (100 mM, pH 7.5) and then incubated for 30 min with the appropriate inhibitor, and samples were removed at different time intervals. For enzyme assays with C23O-mt2, 2 μl (each) was removed and enzyme activity was determined with 0.2 mM catechol in 100 mM Na/K-phosphate buffer (pH 7.5) in a total volume of 1 ml at λ = 375 nm. When BphC1-BN6 and BphC2-BN6 were tested, 2 to 3 μl was removed from the incubation mixture and assayed in the same buffer system with 0.1 mM 2,3-DHBP at λ = 434 nm.
Enzyme purification.
BphC1-BN6 and the Glu79His mutant enzyme were purified by fast protein liquid chromatography essentially as described before (15). Because of the weaker expression of the enzymes from the plasmids used for this study, a gel filtration step and a Mono-Q step were added at the end of the purification protocol described previously (15). This resulted in final specific activities for BphC1-BN6 and the Glu79His mutant enzyme of 3.1 and 21.7 U/mg of protein, respectively.
Site-specific mutagenesis.
The mutants were produced from pGHS118 by using a QuikChange site-directed mutagenesis kit from Stratagene. The generated mutants were verified by sequencing of the mutated gene.
Chemicals and commercial enzymes.
The sources of all chemicals have been described previously (14, 15, 19).
RESULTS
Stability of the extradiol dioxygenases.
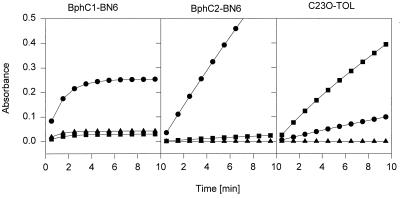
The stabilities of the 2,3-DHBPDOs BphC1-BN6 and BphC2-BN6 and of C23O-mt2 during the oxidation of 2,3-DHBP, 3-methylcatechol, or 3-chlorocatechol were compared. C23O-mt2 and BphC2-BN6 continuously oxidized 3-methylcatechol and 2,3-DHBP for at least 10 min with an almost linear rate; on the other hand, 3-chlorocatechol was not converted to a yellow meta-cleavage product. BphC1-BN6 oxidized all three compounds, although 3-methylcatechol and 3-chlorocatechol were converted with significant lower efficiencies than 2,3-DHBP. In contrast to the other two extradiol dioxygenases, BphC1-BN6 rapidly lost its activity during the oxidation of all three substrates (Fig. 1).
FIG. 1.
Oxidation of 2,3-DHBP (●), 3-methylcatechol (■), and 3-chlorocatechol (▴) by BphC1-BN6 (A), BphC2-BN6 (B), and C23O-mt2 (C). The cuvettes contained in 1 ml 0.1 mM substrate and 28 (A), 8 (B), and 120 (C) μg of protein.
The inactivated BphC1-BN6 could be reactivated by the addition of ferrous ions and DTT. This treatment resulted in a significant increase in the turbidity of the cuvettes by the formation of precipitates. Therefore, after the conversion of the substrate had terminated, the proteins were separated from the supernatant by ultrafiltration (cutoff molecular weight, 10,000). The concentrated protein solution (100 μl) was then reactivated by the addition of iron(II) ions (2 mM) and DTT (5 mM), and enzyme activity was assayed under the same conditions as before. This showed that the inactivation was reversible: about 60% of the initial activity was recovered after previous incubation with 3-methylcatechol or 3-chlorocatechol. In a control experiment, the cell extract was not incubated with substrate but otherwise treated in the same way. In this case, about 80% of the initial enzyme activity was recovered. It was thus concluded that the inactivation of BphC1-BN6 during substrate turnover was almost completely reversible.
The experiments described above suggested that the inactivation of BphC1-BN6 during turnover of the substrates was due to loss of the catalytically active ferrous ion. A further indication that the ferrous ion was only weakly bound to the enzyme was demonstrated by a dilution experiment in the absence of substrate. While the enzyme was nearly stable in the absence of substrate in cell extract with high protein content (>1 mg of protein/ml) for at least 10 min at room temperature, it lost about 30% of its activity within 1 min upon dilution of the cell extract with Na/K-phosphate buffer (100 mM, pH 7.5) to the same enzyme concentration as used for the spectrophotometric assays (<0.01 mg of protein/ml). In comparison, under the same conditions in the presence of 0.1 mM substrate, the enzyme lost about 55% of its activity after 1 min. Also, after inactivation by dilution, the enzyme could be reactivated by adding Fe2+ and DTT. This finding correlated well with the observation that the purified enzyme had to be consistently reactivated by the addition of Fe(II) ions during enzyme purification procedures (15). In contrast to BphC1-BN6, C23O-mt2 and BphC2-BN6 were not inactivated by dilution.
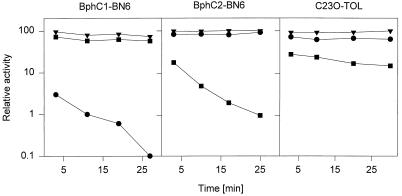
To confirm the lability of the binding of iron(II) ions to BphC1-BN6, we tested whether the enzyme was more susceptible than the other dioxygenases to known iron-chelating compounds. Incubation with o-phenanthroline resulted in rapid inactivation of BphC1-BN6 (<0.1% of the initial activity was found after 30 min of incubation), whereas this compound was almost ineffective with C23O-mt2 or BphC2-BN6 (Fig. 2). Furthermore, no effect on any of the enzymes was found with the Fe(III) ion-chelating compound Tiron.
FIG. 2.
Inactivation of BphC1-BN6, BphC2-BN6, and C23O-mt2 by various compounds. Residual enzyme activities after incubation with 1 mM o-phenanthroline (●), with 1 mM 3-chlorocatechol (■), and without further additions (▾) for various time periods are shown. Cell extracts with BphC1-BN6, BphC2-BN6, and C23O-mt2 contained 61.8, 10.0, and 13.5 mg, respectively, of protein per ml. Aliquots were taken at different time intervals, and enzyme activities were determined spectrophotometrically with catechol for C23O-mt2 or with 2,3-DHPB for BphC1-BN6 and BphC2-BN6.
Effect of 3-chlorocatechol on the three dioxygenases.
Whereas most (proximal-cleaving) extradiol dioxygenases are inactivated during turnover of 3-chlorocatechol (4, 18), BphC1-BN6 oxidizes this compound by a distal 1,6-cleavage mechanism (22). Therefore, we compared the effects of 3-chlorocatechol on BphC1-BN6, BphC2-BN6, and C23O-mt2. Incubating C23O-mt2 with 3-chlorocatechol resulted in a significant immediate reduction in the oxidation rate of catechol (Fig. 2). This effect was explained by a competitive inhibition of catechol oxidation by 3-chlorocatechol. The inhibition constant for 3-chlorocatechol was determined to be 50 nM, slightly higher than a previously reported value (30 nM [30]). The Km for catechol was determined to be 1.0 μM (literature value, 1.4 μM [30]).
In addition to the competitive inhibition, the C23O-mt2 was also inactivated by 3-chlorocatechol in a time-dependent reaction. The enzyme lost about half of its activity after 30 min of incubation with 3-chlorocatechol (Fig. 2).
The effect of 3-chlorocatechol on BphC2-BN6 was similar to the effect on C23O-mt2. It acted by a competitive inhibition (Ki = 0.22 mM; Km for 2,3-DHBP = 3 μM) and by a time-dependent inactivation mechanism. BphC2-BN6 was much more sensitive than C23O-mt2 to 3-chlorocatechol. After 30 min of incubation with 3-chlorocatechol, <1% of the initial activity was recovered.
After inactivation with 3-chlorocatechol, C23O-mt2 and BphC2-BN6 could be reactivated to less than 2% of their initial activity by the addition of Fe(II) ions (2 mM) and DTT (5 mM).
In contrast to the two other enzymes, BphC1-BN6 was not significantly inactivated by 3-chlorocatechol in this experiment (Fig. 2).
Site-specific mutagenesis of BphC1-BN6.
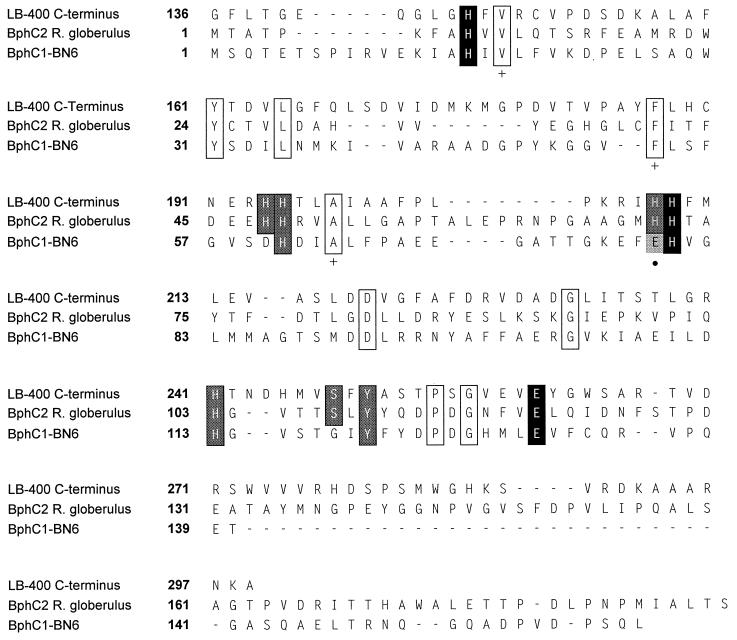
An alignment of BphC1-BN6 with two other extradiol dioxygenases revealed that the residues involved in binding of the iron ions are conserved in all three dioxygenases. In addition, the residues in the substrate binding site and those involved in the formation of hydrogen bonds with the iron-ligating amino acids are also conserved (Fig. 3).
FIG. 3.
Structure-based sequence alignment of the B. cepacia LB400 carboxy domain (LB-400 C-terminus) with the sequences of BphC2 from R. globerulus (BphC2 R. globerulus) (10) and BphC1-BN6. Identical residues are boxed. Fe ligands are in black boxes; conserved residues involved in hydrogen bonds with the ligands are in grey boxes. Other conserved residues in the substrate binding site are marked with a plus sign below the box; Glu79 of BphC1-BN6 is marked with a dot.
Based on the crystal structures of the DHBPDOs from Burkholderia (Pseudomonas) cepacia LB400 and Pseudomonas sp. strain KKS102, it was proposed that one of the His residues, conserved in the extradiol dioxygenase from LB400 and BphC2 from Rhodococcus globerulus but representing a Glu residue in BphC1-BN6, forms a bulge, thereby helping to place the iron-binding histidine residue near the Fe ion (10, 24) (Fig. 3).
The results presented above showed that BphC1-BN6 bound Fe2+ in a more labile fashion compared to other extradiol dioxygenases. Also, it had a limited efficiency for repeated catalytic cycles during turnover of its substrates. To determine whether the weak binding of the ferrous ion is related to the limited catalytic cycles and the ability to cleave 3-chlorocatechol distally, we constructed the Glu79His mutant by replacing the Glu residue at position 79 with His.
Enzyme activity and Fe2+-binding properties of the mutant enzyme Glu79His.
Cell extracts containing the mutant protein were compared with those containing the wild type for the ability to oxidize 2,3-DHBP, 3-methylcatechol, and 3-chlorocatechol (0.2 mM each). The Glu79His mutant enzyme showed a higher reaction rate for all three substrates tested. Both enzymes could be reactivated by the addition of Fe2+ ions and DTT. After reactivation, the mutant enzyme showed about fivefold-higher activities than the wild-type enzyme with 2,3-DHBP and 3-methylcatechol. In contrast, with 3-chlorocatechol the reactivated mutant enzyme was less active than the wild type. For both enzymes, no indications for the formation of 2-hydroxymuconate from 3-chlorocatechol were found by UV/visible light spectroscopy and high-pressure liquid chromatography (as described in reference 16). We therefore concluded that both enzymes converted 3-chlorocatechol only by a distal ring cleavage mechanism.
To analyze the strength of binding of the catalytically active ferrous ion to the active site of the enzyme, we diluted cell extracts of the wild-type and mutant enzymes to low protein concentrations (<0.01 mg of protein/ml) and determined the residual enzyme activities after 10 min. The mutant enzyme showed after this treatment more than 90% of the initial activity. In contrast, less than 10% of the initial activity could be recovered for BphC1-BN6. In addition, the effect of o-phenanthroline on the wild-type enzyme was compared with that on the Glu79His mutant. After a 30-min incubation with 1 mM o-phenanthroline, the wild-type enzyme lost almost all activity (<0.1% of the initial activity). In contrast, the mutant enzyme retained more than 70% of its initial activity. This was a strong indication that the Glu79His mutant enzyme binds the catalytically active ferrous ion with higher affinity.
Comparison of the catalytical properties of the purified Glu79His mutant enzyme with BphC1-BN6.
Since the mutation improved the binding of Fe(II) ions, we purified both proteins and compared them with respect to specificity, catalytical efficiency, and potential inhibitory effects. The basic catalytical constants were determined with 2,3-DHBP, catechol, and 3-methyl-, 3-chloro-, and 3,5-dichlorocatechol as substrates for both enzyme types. The mutant enzyme showed higher affinities for all substrates tested and a significant increase in the reaction rates with the majority of the substrates tested (Table 1); only 3-chlorocatechol and 3,5-dichlorocatechol were converted with a slightly lower reaction rate. The activities of both wild-type and mutant enzymes decreased with an increase in substrate concentration beyond a certain level (Fig. 4). The mutant enzyme was inhibited at lower substrate concentrations compared to the wild-type enzyme. This effect was most pronounced for the mutant enzyme with 3-chlorocatechol as the substrate.
TABLE 1.
Kinetic parameters of wild-type BphC1-BN6 and the Glu79His mutant
| Substrate | Kinetic parametera
|
|||
|---|---|---|---|---|
| Wild type
|
Glu79His mutant
|
|||
| Vmax (U mg−1) | Km (mM) | Vmax (U mg−1) | Km (mM) | |
| 2,3-DHBP | 11.6 ± 0.5 | 0.74 ± 0.06 | 17.5 ± 0.6 | 0.032 ± 0.004 |
| Catechol | 0.080 ± 0.004 | 1.13 ± 0.11 | 0.16 ± 0.01 | 0.67 ± 0.10 |
| 3-Methylcatechol | 0.68 ± 0.07 | 0.18 ± 0.05 | 1.95 ± 0.08 | 0.097 ± 0.008 |
| 3-Chlorocatechol | 0.42 ± 0.01 | 0.10 ± 0.01 | 0.27 ± 0.01 | 0.025 ± 0.002 |
| 3,5-Dichlorocatechol | 0.065 ± 0.008 | 0.030 ± 0.008 | 0.056 ± 0.007 | 0.007 ± 0.002 |
Calculated as described in Materials and Methods.
Inactivation during substrate conversion.
While determining the kinetic data shown in Fig. 4, we noticed that inactivation during substrate conversion (as shown in Fig. 1) was more pronounced at higher substrate concentrations. Therefore, we compared the effects of different substrate concentrations on the inactivation of the wild-type and mutant enzymes during substrate oxidation. We determined the apparent rate constants of enzyme inactivation at different substrate concentrations (kinactγ) for the Glu79His mutant and the wild-type enzyme as described by Cerdan et al. (5, 6). Furthermore, the concentration-independent kinact values were extrapolated from the kinactγ values (Table 2). A high rate constant of enzyme inactivation (kinact or kinactγ) corresponds to a rapid inactivation of the enzyme during substrate conversion. In all cases, the rate of enzyme inactivation (kinactγ) increased with increasing substrate concentration. For the wild-type and mutant enzymes, 3-chlorocatechol was the strongest and 2,3-DHBP was the weakest inactivator (Table 2). All tested substrates clearly had a more pronounced inactivating effect on the mutant enzyme than on the wild-type enzyme. In addition, this type of analysis confirmed that BphC1-BN6 and its mutant were much more susceptible to inactivation during substrate turnover than C23O-mt2 (Table 2).
TABLE 2.
Kinetic parameters for enzyme inactivation
| Substrate | Enzyme | Kinetic parameter (10−3 s−1)a
|
|||
|---|---|---|---|---|---|
|
kinactγ at substrate concn (mM) of:
|
kinact | ||||
| 0.1 | 0.3 | 0.5 | |||
| 3-Chlorocatechol | BphC1-BN6 | 23.2 | 42.9 | 67.7 | 190 |
| Glu79His | 62.5 | 148 | 240 | 437 | |
| 2,3-DHBP | BphC1-BN6 | 11.5 | 15.3 | 18.6 | 71 |
| Glu79His | 12.6 | 28.0 | 37.4 | 123 | |
| 3-Methylcatechol | BphC1-BN6 | 15.1 | 15.7 | 20.9 | 80 |
| Glu79His | 14.6 | 50.3 | 69.1 | 235 | |
| C23O-mt2 | 1.0 | 2.1b | ND | 3.1b | |
Calculated as described in Materials and Methods. ND, not determined.
From reference 6.
DISCUSSION
Our investigations showed several differences between BphC1-BN6, BphC2-BN6, and C23O-mt2: in contrast to the two other enzymes, BphC1-BN6 oxidized 3-chlorocatechol, while it only weakly bound the catalytically necessary iron(II) ions. Furthermore, the enzyme was rapidly inactivated during substrate turnover and demonstrated apparent substrate inhibition kinetics with increasing substrate concentrations. Surprisingly, the inactivation reactions were observed not only with 3-chlorocatechol but also with the other catechols tested. Our experiments with the wild-type and mutant enzymes enabled us to judge whether these factors were indispensably interrelated.
Our results suggested that the apparent substrate inhibition (as shown in Fig. 4) is mainly due to the time-dependent inactivation of the enzyme during substrate turnover. This could be deduced by comparing the results shown in Fig. 4 with the calculated kinactγ values shown in Table 2. In both cases, the most pronounced effect was observed with 3-chlorocatechol and the weakest effect was seen with 2,3-DHBP. This also explains why we were previously unable to calculate the apparent substrate inhibition of BphC1-BN6 by classical substrate inhibition kinetics (15). Furthermore, previous finding of substrate inhibition of other extradiol dioxygenases (1, 11) may have been caused by similar substrate- and time-dependent inactivation of the enzymes.
The inactivation of BphC1-BN6 during substrate turnover was largely reversible by the addition of ferrous iron. We therefore deduced from the results obtained with the wild-type enzyme that this inactivation was at least partially related to the weak binding of the catalytically active Fe2+ ions to the enzyme. This hypothesis was supported by the observation that merely diluting the enzyme to concentrations similar to those used during the spectrophotometric enzyme assays also caused a rapid loss of the activity. However, further experiments showed that this simple explanation was not sufficient to explain the overall kinetics of inactivation. This was shown by the increase in the rate of enzyme inactivation at higher substrate concentrations. An additional inactivation mechanism was also suggested by the observation that the mutant enzyme apparently showed a higher binding affinity for the iron while being more sensitive to inactivation during substrate turnover. Since the inactivated mutant enzyme could also be reactivated by the addition of Fe2+, this indicated that the substrate-dependent inactivation was also caused by the loss of the catalytically active ferrous ion. It should be mentioned at this point that the inactivation was not observed in the absence of oxygen (data not shown), which suggested that binding of the organic substrate and oxygen (25) or conversion of the substrate was necessary for the loss of the iron(II) cofactor.
Time-dependent inactivation phenomena similar to those shown here for BphC1-BN6 have been observed previously during the turnover of certain catechols (especially 4-ethylcatechol) by C23O-mt2 (5, 21) and are also known for other types of enzymes (17, 28). Although the inactivation of C23O-mt2 by 4-ethylcatechol has been described as suicide inactivation, it was also shown to be reversible by the addition of ascorbic acid plus FeSO4 (5). It is generally assumed that suicide inactivation involves an irreversible inactivation of the target enzyme (28, 29). The type of inactivation observed here for BphC1-BN6, and previously for the conversion of 4-ethylcatechol by C23O-mt2, is different from classical suicide inactivation mechanisms because it is reversible. Nevertheless, the inactivation mechanism of C23O-mt2 by 4-ethylcatechol and that of BphC1-BN6 by all substrates tested clearly resembled each other. The main difference was that BphC1-BN6 was much more susceptible than C23O-mt2 to this type of inactivation. The results of the present study and the study by Cerdan et al. (5) suggest that binding of the organic substrate (for C23O-mt2 observed mainly with 4-ethylcatechol) and oxygen results in a labilization of the catalytically necessary ferrous iron from the enzyme. This inactivation is not directly related to the strength of the binding of the ferrous iron to the enzyme in the absence of substrate. By random mutagenesis, we are currently trying to identify the amino acids responsible for the substrate-dependent enzyme inactivation.
There is growing interest in the extradiol cleavage of chlorinated catechols. Besides the distal cleavage of 3-chlorocatechol by BphC1-BN6 studied here, there are also recent reports demonstrating a proximal cleavage of 3-chlorocatechol to 2-hydroxymuconate. This reaction is catalyzed by a specific catechol 2,3-dioxygenase from Pseudomonas putida GJ31, which shows high relative activities with 3-chlorocatechol and was therefore termed chlorocatechol 2,3-dioxygenase (16, 20). These findings suggest that extradiol cleavage pathways may have a much higher potential for the degradation of chlorinated substrates than was previously expected.
REFERENCES
- 1.Adams R H, Huang C-M, Higson F K, Brenner V, Focht D D. Construction of a 3-chlorobiphenyl-utilizing recombinant from an intergeneric mating. Appl Environ Microbiol. 1992;58:647–652. doi: 10.1128/aem.58.2.647-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asturias J A, Eltis L D, Prucha M, Timmis K N. Analysis of three 2,3-dihydroxybiphenyl 1,2-dioxygenases found in R. globerulus P6. J Biol Chem. 1994;269:7807–7815. [PubMed] [Google Scholar]
- 3.Asturias J A, Timmis K N. Three different 2,3-dihydroxybiphenyl 1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J Bacteriol. 1993;175:4631–4640. doi: 10.1128/jb.175.15.4631-4640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from P. putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerdan P, Wasserfallen A, Rekik M, Timmis K N, Harayama S. Substrate specificity of catechol 2,3-dioxygenase encoded by TOL plasmid pWW0 of Pseudomonas putida and its relationship to cell growth. J Bacteriol. 1994;176:6074–6081. doi: 10.1128/jb.176.19.6074-6081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerdan P, Rekik M, Harayama S. Substrate specificity differences between two catechol 2,3-dioxygenases encoded by the TOL and NAH plasmids from Pseudomonas putida. Eur J Biochem. 1995;229:113–118. doi: 10.1111/j.1432-1033.1995.tb20445.x. [DOI] [PubMed] [Google Scholar]
- 7.Clayton T M, Bibb M J. Streptomyces promoter-probe plasmids that utilise the xylE gene of Pseudomonas putida. Nucleic Acids Res. 1990;18:1077. doi: 10.1093/nar/18.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer J. Entwicklung eines regulierbaren Expressionssystems zur effizienten Synthese rekombinanter Proteine in Streptomyces lividans. Ph.D. thesis. Stuttgart, Germany: Universität Stuttgart; 1996. [Google Scholar]
- 10.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 11.Happe B, Eltis L D, Poth H, Hedderich R, Timmis K N. Characterization of 2,2′,3-trihydroxybiphenyl dioxygenase, an extradiol dioxygenase from the dibenzofuran- and dibenzo-p-dioxin-degrading bacterium Sphingomonas sp. strain RW1. J Bacteriol. 1993;175:7313–7320. doi: 10.1128/jb.175.22.7313-7320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 13.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15328–15333. [PubMed] [Google Scholar]
- 14.Heiss G, Müller C, Altenbuchner J, Stolz A. Analysis of a new dimeric extradiol dioxygenase from a naphthalenesulfonate-degrading sphingomonad. Microbiology. 1997;143:1691–1699. doi: 10.1099/00221287-143-5-1691. [DOI] [PubMed] [Google Scholar]
- 15.Heiss G, Stolz A, Kuhm A E, Müller C, Klein J, Altenbuchner J, Knackmuss H-J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaschabek S R, Kasberg T, Müller D, Mars A E, Janssen D B, Reineke W. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J Bacteriol. 1998;180:296–302. doi: 10.1128/jb.180.2.296-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinemuchi H, Arai Y, Oreland L, Tiptom K F, Fowler C J. Time-dependent inhibition of monoamine oxidase by β-phenethylamine. Biochem Pharmacol. 1982;31:959–964. doi: 10.1016/0006-2952(82)90327-6. [DOI] [PubMed] [Google Scholar]
- 18.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhm A E, Stolz A, Ngai K-L, Knackmuss H-J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mars A E, Kasberg T, Kaschabek S R, van Agteren M H, Janssen D B, Reineke W. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos J L, Wasserfallen A, Rose K, Timmis K N. Redesigning metabolic routes: Manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987;235:593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- 22.Riegert U, Heiss G, Fischer P, Stolz A. Distal cleavage of 3-chlorocatechol to 3-chloro-2-hydroxymuconic semialdehyde by an extradiol dioxygenase. J Bacteriol. 1998;180:2849–2853. doi: 10.1128/jb.180.11.2849-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojo F, Pieper D H, Engesser K-H, Knackmuss H-J, Timmis K N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987;238:1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- 24.Senda T, Sugiyama K, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y. Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102. J Mol Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 25.Shu L, Chiou Y-M, Orville A M, Miller M A, Lipscomb J D, Que L., Jr X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 26.Spence E L, Kawamukai M, Sanvoisin J, Braven H, Bugg T D H. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J Bacteriol. 1996;178:5249–5256. doi: 10.1128/jb.178.17.5249-5256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taeger K, Knackmuss H-J, Schmidt E. Biodegradability of mixtures of chloro- and methylsubstituted aromatics: simultaneous degradation of 3-chlorobenzoate and 3-methylbenzoate. Appl Microbiol Biotechnol. 1988;28:603–608. [Google Scholar]
- 28.Tipton K F. Principles of enzyme assays and kinetic studies. In: Eisenthal R, Danson M J, editors. Enzyme assays. A practical approach. New York, N.Y: IRL Press; 1992. pp. 1–24. [Google Scholar]
- 29.Waley S G. Kinetics of suicide substrates. Biochem J. 1980;185:771–773. doi: 10.1042/bj1850771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasserfallen A, Rekik M, Harayama S. A Pseudomonas putida strain able to degrade m-toluate in the presence of 3-chlorocatechol. Bio/Technology. 1991;9:296–298. [Google Scholar]