Abstract
cAMP is a ubiquitous second messenger with many functions in diverse organisms. Current cAMP sensors, including Föster resonance energy transfer (FRET)-based and single-wavelength-based sensors, allow for real time visualization of this small molecule in cultured cells and in some cases in vivo. Nonetheless the observation of cAMP in living animals is still difficult, typically requiring specialized microscopes and ex vivo tissue processing. Here we used ligand-dependent protein stabilization to create a new cAMP sensor. This sensor allows specific and sensitive detection of cAMP in living zebrafish embryos, which may enable new understanding of the functions of cAMP in living vertebrates.
Cyclic adenosine 3′–5′-monophosphate (cAMP) is an essential second messenger that amplifies environmental signals received by G-protein-coupled receptors (GPCRs).1 The importance of cAMP is underscored by the multitude of physiological processes that it regulates including heartbeat, learning, and memory,2−4 and it can initiate a wide range of cellular responses including proliferation, differentiation, and death.5 The kinetics and function of cAMP depend on many factors including the cell type and subcellular compartment where it accumulates and interacts with other signaling molecules,6,7 which highlights the importance of sensors to visualize cAMP in cultured cells and intact organisms.
Fluorescent sensors for cAMP based on Föster resonance energy transfer (FRET) respond rapidly to local changes in cAMP concentration.4,8 Although useful in cultured cells, FRET-based sensors are characterized by low signal-to-noise ratio, are prone to photobleaching, and are hampered by light scattering in intact tissues, limiting their use for in vivo imaging.1,8−10 Single wavelength cAMP sensors using circularly permuted fluorescent proteins, such as Pink Flamindo, R-FlincA, and cAMPr, can mitigate some of the disadvantages of FRET-based sensors.11−14 Single wavelength sensors have been used to track fast cAMP dynamics in dissected Drosophila brains, in C. elegans neurons, and in mouse astrocytes.12,15,16 However, there is often need for dissection and an ex vivo imaging, and it is not clear whether the current tools are useful for long-term imaging.
Ligand-dependent protein stabilization is a strategy that can be used to generate single-wavelength biosensors for small molecules.17 This approach is based on a protein that is stable only when bound to its cognate ligand.17−20 This sensor is metabolically unstable and degraded by the proteasome in its unbound state, but engagement by its cognate ligand prevents degradation and leads to a dose-dependent fluorescent signal when the sensor is fused to an appropriate partner such as GFP. Here we report the application of this approach to develop a genetically encoded single-wavelength cAMP sensor for in vivo imaging using the zebrafish model system (Figure 1; Figure S1).
Figure 1.
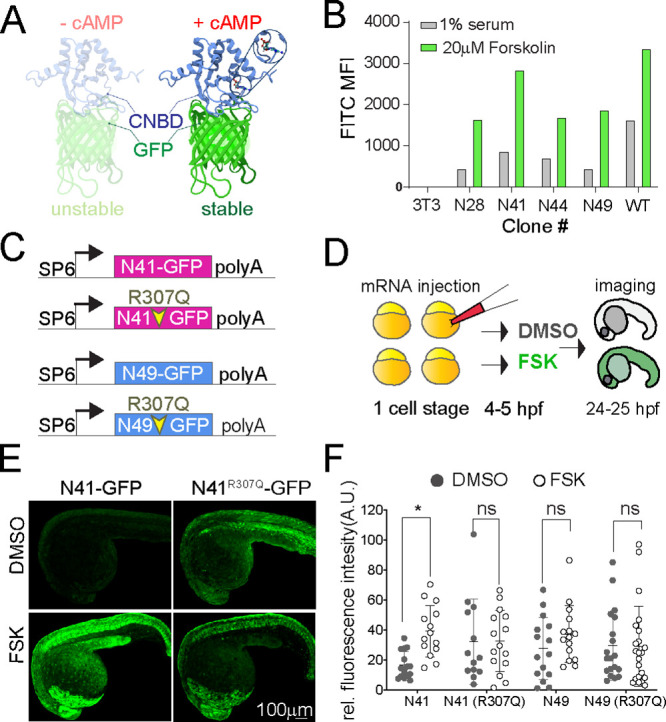
(A) Ribbon diagram of the cAMP sensor composed of CNBD-GFP protein that is unstable without cAMP but stabilized upon cAMP binding: solution structure of a bacterial cyclic nucleotide-activated K+ channel binding domain in cAMP-free form on the left (PBD 2KXL) and bound to cAMP on the right (PBD 2K0G). (B) NIH 3T3 cells stably expressing CNBD-GFP derived from error-prone PCR were treated with 1% serum or 20 μM forskolin for 17 h. (C) Synthetic mRNA encoding the sensor variants N41 and N49 CNBD and the corresponding cAMP-insensitive controls (R307Q). (D) mRNA was injected into zebrafish embryos at the one-cell stage. Embryos were treated at 4–5 hpf with FSK or DMSO for 20 h, then imaged at 24–25 hpf. (E) Representative images showing 24 hpf embryos injected with N41-GFP and N41R307Q-GFP mRNA at one-cell stage and treated with DMSO or 20 μM FSK starting at 4 hpf. (F) Each dot represents mean GFP intensity of one embryo. Error bars indicate SD: *p < 0.5 by one-way ANOVA (with Šídák’s multiple comparisons), n = 13–22 embryos each condition; ns = not significant. AU = arbitrary unit.
We sought to identify conditionally stable mutants of the cyclic nucleotide binding domain (CNBD) from the MlotiK1 bacterial channel, which binds cAMP with high specificity and sensitivity in mammalian cells21,22 (Figure 1A). We started with a codon-optimized CNBD sequence and generated a library of sequence variants using error-prone PCR. This library of CNBD domains was then fused to GFP to enable a cell-based screen for mutants stabilized by cAMP (Figure S1A,B). NIH3T3 cells were stably transduced with the library and subjected to serial rounds of FACS sorting after application or withdrawal of the adenylyl cyclase agonist forskolin (FSK). We selected clones with high GFP signal in the presence of FSK and very low GFP signal in the absence of FSK (Figure S1B). DNA sequencing of selected clones identified many variants encoding unique missense mutations in CNBD, and two variants in particular displayed 3-fold (N41) to 4-fold (N49) increases in GFP intensity upon forskolin treatment (Figure 1B; Figure S1C,D). The N49 variant contains a single mutation in the CNBD domain (Figure S1C,D; blue), whereas N41 contains four mutations spread across the CNBD protein structure (Figure S1C,D; pink).
To test the forskolin responsivity of these CNBD variants in vivo, we injected synthetic mRNA encoding the N41-GFP and N49-GFP sensors into zebrafish embryos, allowing ubiquitous and transient expression of the sensor proteins (Figure 1C,D). Four or five hours after injection of mRNA, we treated embryos with 20 μM FSK for 20 h and subsequently imaged the GFP signal (Figure 1E). Both variants displayed a visible fluorescent signal when expressed in zebrafish embryos, but only N41 increased in response to FSK treatment (Figure 1F). To test whether cAMP binding is required for the response of the N41 variant to forskolin, we compared FSK treatment of N41-GFP with another variant, N41R307Q-GFP, which contains a mutation in a conserved arginine that reduces the efficiency of cAMP binding (Figure 1C; Figure S1C, red circle).21−23 FSK treatment significantly increased GFP signal in embryos injected with N41-GFP, whereas embryos expressing the cAMP-insensitive N41R307Q-GFP control did not display significant changes in GFP fluorescence (Figure 1E,F). The R307Q mutation appears to stabilize the N41R307Q-GFP protein, because DMSO-treated N41R307Q-GFP embryos displayed greater GFP signal than DMSO-treated embryos expressing N41-GFP (Figure 1E,F). These analyses provided evidence that the N41-GFP sensor protein is selectively stabilized upon binding to cAMP in vivo, and we hereafter refer to this engineered sensor as DDcAMP (destabilized detector of cAMP).
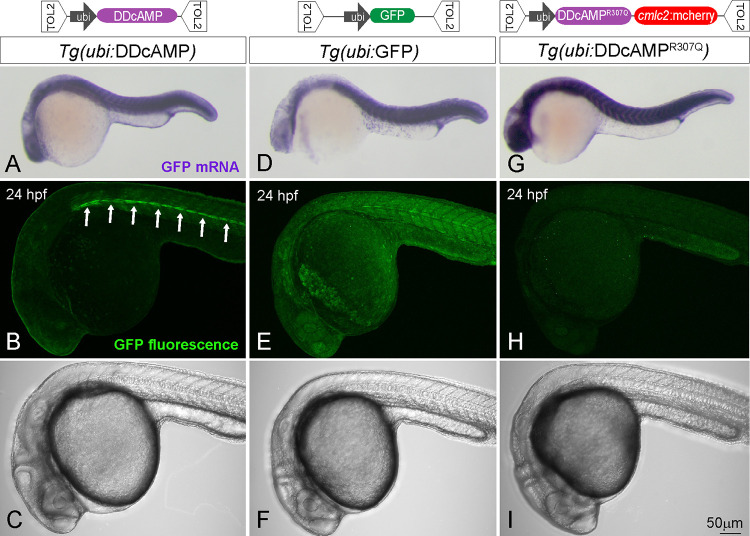
To further characterize this cAMP sensor, we generated stable transgenic fish lines expressing DDcAMP under control of the regulatory sequences from the ubiquitin gene (abbreviated as Tg(ubi:DDcAMP), Figure 2A–C), which drives widespread expression in the embryo.24 As controls, we generated transgenic fish lines expressing ubiquitin-GFP with no CNBD (Figure 2D–F) or the cAMP-insensitive variant DDcAMPR307Q (Figure 2G–I). In situ hybridization demonstrated that the transgenes expressed GFP mRNA at similar levels at 24 h postfertilization (hpf) (Figure 2A,D,G), whereas confocal imaging revealed differences in GFP signal. In contrast to the control transgenes (Figure 2E,F,H,I), embryos expressing the sensor displayed discrete GFP signal localized to the horizontal myoseptum of the somites (Figure 2B,C; white arrows). Quantification confirmed that a region containing muscle pioneers displayed much more GFP signal than a reference area not containing muscle pioneers in Tg(ubi:DDcAMP) transgenic embryos, but not in embryos expressing the cAMP-insensitive variant cAMP-DDcAMPR307Q (Figure S2C). In Tg(ubi:DDcAMP) embryos from different transgenic founders and in transgenic embryos expressing the N49 sensor variant Tg(ubi:N49-GFP), GFP expression was enriched in the muscle pioneers and in the slow superficial fibers (Figure S2Ba–c; yellow arrowheads), cells known to require cAMP signaling for proper specification and differentiation.25−27 Thus, the comparison of sensor and control transgenic animals provides evidence that DDcAMP is specifically stabilized in a subpopulation of muscle cells.
Figure 2.
(top) Diagram of the TOL2 plasmids containing N41-GFP, N41R307Q-GFP, and GFP DNA under control of ubi promoter. (bottom) In situ hybridization for GFP mRNA in Tg(ubi:DDcAMP) (A), Tg(ubi:GFP) (D) and Tg(ubi:DDcAMPR307Q) (G) embryos at 24 hpf. Confocal acquisition of GFP signal in Tg(ubi:DDcAMP) (B, C), Tg(ubi:GFP) (E, F), and Tg(ubi:DDcAMPR307Q) (H, I) embryos at 24 hpf. Horizontal myoseptum is indicated by white arrows in panel B.
To test the specificity and sensitivity of DDcAMP to cAMP, we treated Tg(ubi:DDcAMP) transgenic embryos with FSK for 20 h starting at 4–5 hpf and imaged fluorescent signal in anterior somites using confocal microscopy (Figure 3A,B). FSK significantly increased GFP expression in Tg(ubi:DDcAMP) but not in Tg(ubi:DDcAMPR307Q) embryos, and the overall GFP signal in Tg(ubi:DDcAMPR307Q) embryos was low (Figure 3B). Transgenic sensor animals Tg(ubi:DDcAMP) showed increased GFP intensity in the enveloping layer (EVL) of the tail (Figure 3C) when treated with increasing concentrations of the cell-permeant cAMP analog 8-cpt-cAMP for 24 h (Figure 3D). The sensor was not activated by treatment with the cGMP analog 8-cpt-cGMP, and treatment with 8-cpt-cAMP did not increase signal in Tg(ubi:DDcAMPR307Q) control animals (Figure 3D, Figure S3B). Taken together, these experiments provide evidence that DDcAMP is a specific and sensitive sensor for cAMP in the developing embryo.
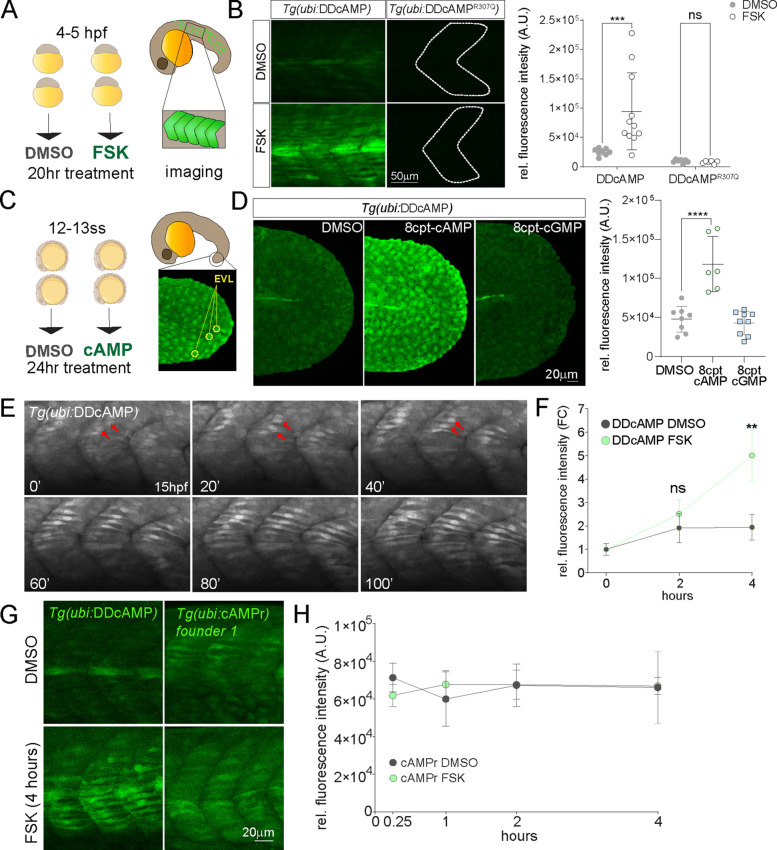
Figure 3.
(A) Diagram of DMSO and 20 μM FSK treatment on Tg(ubi:DDcAMP) and Tg(ubi:DDcAMPR307Q) embryos beginning at 4–5 hpf for 20 h. Anterior somites from treated embryos were imaged at 24 hpf, and GFP intensity was measured in somites 7–11. (B) Images of somites from DMSO and FSK-treated Tg(ubi:DDcAMP) and Tg(ubi:DDcAMPR307Q) embryos at 24 hpf. The graph represents the mean fluorescence intensity of five somites per embryo, and each point corresponds to one embryo. Error bars indicate SD: ***p < 0.001 by two-way ANOVA (Šídák’s multiple comparisons), n = 6–11 animals for each condition. ns = not significant. AU = arbitrary unit. (C) Diagram of DMSO and 8-cpt-cAMP treatment of Tg(ubi:DDcAMP) starting at 12–13 somite stage (12–13ss) and imaged after 24 h of incubation. The tip of the tail was imaged for GFP intensity and the EVLs were quantified. (D) Tg(ubi:DDcAMP) embryos from the same clutch were treated with DMSO, 100 μM 8-cpt-cAMP, or 100 μM 8-cpt-cGMP; signal from 20 EVL cells was averaged per animal. Each dot in the graph represents one animal. Error bars indicate SD; ****p < 0.0001 one-way ANOVA (with Bonferroni’s multiple comparisons), n = 6–9 animal per condition. AU = arbitrary unit. (E) Frames from a confocal time lapse image with Airyscan 2 processing of Tg(ubi:DDcAMP) embryo starting at 15 hpf (t = 0 min). One frame every 20 min is shown as representation of the time lapse. GFP-expressing muscle cells are indicated with red arrows. (F) Graph represents the fold change of mean fluorescence intensity measured in five somites per embryo at time 0 and after 2 and 4 h of treatment. Error bars indicate SEM; **p < 0.01 two-way ANOVA (with Šídák’s multiple comparisons), n = 6–10 animals for each condition. ns = not significant, FC = fold change. (G) Confocal images of somites from Tg(ubi:DDcAMP) and Tg(ubi:cAMPr) embryos after 4 h of FSK and DMSO treatment show GFP expression in muscle cells but different response to FSK treatment. (H) Graph represents the mean fluorescence intensity measured in five somites per embryo at the time points indicated. Error bars indicate SEM; there is no significance among the groups; n = 4–5 animals for each condition.
Time course and time lapse analyses of Tg(ubi:DDcAMP) indicate that GFP signal appeared in adaxial cells of developing somites at 15 hpf (Movie 1; Figure 3E, red arrow; Figure S3C, white arrows). The signal increased in the somites over time (Figure S3C), pointing to a potential time window of cAMP production during somitogenesis. Further kinetic analyses on Tg(ubi:DDcAMP) animals showed that the sensor provides a 3-fold increase in fluorescence intensity when treated with FSK for 4 h starting from 14 to 18 hpf (Figure 3F). To compare DDcAMP with the existing cAMP sensor cAMPr,12 we generated a stable transgenic fish line expressing cAMPr driven by the ubi promoter Tg(ubi:cAMPr). Transgenic cAMPr originating from one founder expressed GFP in slow muscle cells, similar to DDcAMP (Figure 3G), whereas cAMPr transgenic fish from another line expressed detectable GFP in skin but not muscle cells (Figure S3D). Signal from cAMPr increases after few minutes of FSK treatment in cell culture,12 so we examined cAMPr transgenic fish from both founders treated with FSK for times ranging from 15 min to 4 h (Figure 3H, Figure S3D). In contrast to Tg(ubi:DDcAMP), GFP intensity did not significantly increase in response to FSK in Tg(ubi:cAMPr) animals at any of these time points (Figure 3H).
Previous studies show that cAMP regulates muscle cell specification.25,26 In zebrafish, secretion of Sonic Hedgehog (Shh) from the notochord instructs adjacent adaxial cells to differentiate into muscle pioneers, slow muscle fibers, or medial fast fibers.28−31 The patterning of somitic cells depends in part on the level and timing of cAMP–protein kinase A (PKA) activity, which inhibits the response to Shh.25,26,32 Analysis of doubly transgenic embryos expressing both Tg(ubi:DDcAMP) and the Shh reporter Tg(gli:mCherry-NLS) revealed that N41-GFP signal appeared in Shh-responsive muscle pioneers (mp) and slow muscle cells (smc) (Figure S4A).33 This colocalization was not evident in the controls with Tg(ubi:DDcAMPR307Q) or Tg(ubi:GFP) (Figure S4B).
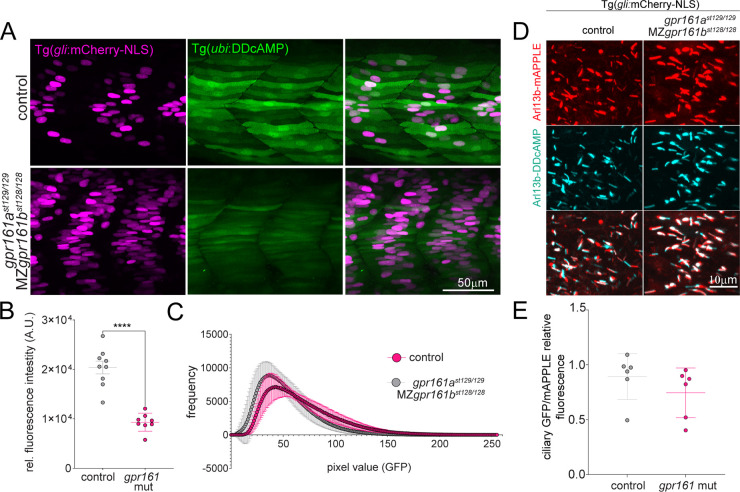
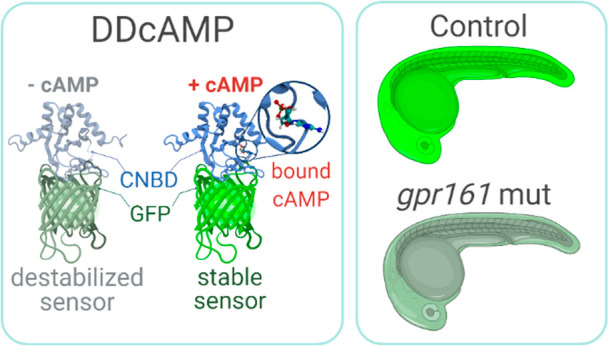
There is evidence that the ciliary GPCR Gpr161 negatively regulates Shh signaling by increasing cAMP concentration in the cilium,34−36 and gpr161b is expressed in developing adaxial cells of zebrafish embryos.37 To determine if the DDcAMP sensor detects cAMP generated by Gpr161 in developing muscle cells, we used CRISPR-Cas9 to generate presumed null mutations in the duplicated genes encoding the Gpr161a and Gpr161b proteins (gpr161a and gpr161b, Figure S4C). In accordance with previous work, the number of Shh responsive cells in the somites was increased in the double mutant for gpr161ast129/129 and maternal zygotic MZgpr161b128/128, hereafter referred to as “gpr161 mutants” (Figure 4A, magenta).36 In gpr161 mutants, the Tg(ubi:DDcAMP) GFP signal in somites was reduced, providing evidence that DDcAMP accurately reports on the reduced levels of endogenous cAMP produced by Gpr161 in developing muscle (Figure 4B,C). Imaging indicated that DDcAMP is detectable in the nucleus and cytoplasm, whereas Gpr161 is most active in cilia.27,35,38 In an effort to quantify cAMP levels in cilia, a cilium-localizing peptide was appended to the N-terminus of DDcAMP (Arl13b-DDcAMP). The cilia-localized Arl13b-DDcAMP displayed similar levels of GFP signal to the corresponding ciliary control (Arl13b-DDcAMPR307Q), FSK treatment did not significantly affect the GFP signal of either protein (Figure S4D,E), and ciliary DDcAMP signal was similar in wild-type and gpr161 mutants (Figure 4D,E). These results indicate that the sensor does not specifically detect Gpr161-dependent cAMP in the cilium, perhaps because ciliary localization impedes efficient degradation of the unliganded DDcAMP protein.
Figure 4.
(A) Confocal images of cAMP (green) and Shh reporter (magenta) signals in control fish and double mutants forgpr161a (st129 allele) and MZgpr161b (st128 allele). MZ = maternal zygotic. Control fish include gpr161ast129/+, gpr161b128/+ and gpr161ast129/129, gpr161b128/+. (B) Graphs show GFP fluorescence intensity measured in the somite area corresponding to the muscle pioneers in Tg(gli:mCherry-NLS); Tg(ubi:DDcAMP) (control) and in Tg(gli:mCherry-NLS); Tg(ubi:DDcAMP); gpr161ast129/129; MZgpr161b128/128 (gpr161 mut) embryos. Error bars indicate SD: ****p < 0,0001 by t-test. (C) Mean of the pixel values in each image acquired in panel A plotted as a function of their frequency. gpr161 double mutant fish display darker values compared to the controls. n = 4–5 animals per genotype. (D) Magnification of somites from 24 hpf Tg(gli:mCherry-NLS) control and Tg(gli:mCherry-NLS); gpr161ast129/129; MZgpr161b128/128 embryos injected at one-cell stage with Arl13b-DDcAMP (cyan) and Arl13b-mApple (red cilia). (E) Ratio of ciliary GFP/ciliary mAPPLE in over 20 cilia per animal in Tg(gli:mCherry-NLS) (control) and Tg(gli:mCherry-NLS); gpr161ast129/129; MZgpr161b128/128 (gpr161 mut) embryos.
Ligand-dependent protein stabilization is a general strategy that has been used to develop a portfolio of reagents that allow users to tunably regulate expression levels of a wide variety of proteins using cell permeable small molecules.18,39,40 Feng et al. demonstrated that the same strategy could be extended to develop protein-based sensors for secondary metabolites such as progesterone in yeast and digoxin in plants.17 Given that existing cAMP sensors are not suitable for imaging in living zebrafish, we used ligand-dependent protein stabilization to generate a genetically encoded single wavelength sensor that allows long-term imaging of endogenous cAMP in zebrafish embryos. We identified DDcAMP as a protein sensor that is specifically stabilized by cAMP, and we created transgenic fish that ubiquitously express DDcAMP.
DDcAMP specifically responds to the production of cAMP in developing muscle cells. Our analyses suggest that DDcAMP turnover is regulated by cAMP concentration and that this sensor produces a strong GFP signal specifically in response to cAMP. These features enable in vivo time lapse imaging without phototoxicity and bleaching; however DDcAMP is relatively slow to respond to changes in cAMP levels. There is a 3-fold increase in DDcAMP GFP intensity after 4 h of exposure to 20 μM FSK. This time lag presumably reflects the time required for newly translated DDcAMP to be stabilized by cAMP. Thus, DDcAMP is suitable for imaging endogenous cAMP levels in living zebrafish embryos, but it may not be ideal for monitoring changes in cAMP levels that occur over rapid time scales or in subcellular compartments with restricted access to the ubiquitin-proteasome system. DDcAMP has complementary features to other recently developed cAMP sensors, and it will enable new approaches to analyze cAMP in different experimental settings, cell types, and model organisms.
Acknowledgments
We thank J. Chen at Stanford for the Tg(gli:mCherry-NLS) transgene and J. Reiter at UCSF for the Arl13b construct. We are grateful to T. Reyes and C. Hill for fish care, to the Stanford Neuroscience Macroscopy Service for support with image acquisition, and to the Data Studio Office at Stanford for advice on statistical analyses. We thank J. Rodrigues for help in modeling the CNBD structure using PyMOL. This work was supported by grant R35 NS111584 from the National Institutes of Health to W.S.T. and a Stanford Discovery Innovation Award to T.J.W. M.S. was supported by postdoctoral fellowships from the Wu Tsai Neuroscience Institute (formerly Stanford Neuroscience Institute) and the National MS Society (FG-1807-31636). W.S.T. is a Kennedy-Grossman Fellow in Human Biology. Some diagrams in the figures were designed using Biorender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.2c00333.
Cloning and genotyping primers and sgRNA sequences (XLSX)
Model organisms and strains, recombinant DNA resources, and software and algorithms (XLSX)
Confocal time lapse image of Tg(ubi:DDcAMP) embryo starting at 15 hpf (t = 0′). The embryo is oriented dorsally, with the anterior developing head on top. Developing adaxial cells gradually increase GFP intensity in the somites. The images were taken every 8 minutes for over 7 hours of time lapse (MP4)
Experimental information on CNBD structure, transgene, and pharmacological and genetic validation, details on the experimental procedures for the CNBD selection, transgene generation, and image acquisition (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Willoughby D.; Cooper D. M. F. Organization and Ca2+ Regulation of Adenylyl Cyclases in CAMP Microdomains. Physiol Rev. 2007, 87 (3), 965–1010. 10.1152/physrev.00049.2006. [DOI] [PubMed] [Google Scholar]
- Morozov A.; Muzzio I. A.; Bourtchouladze R.; Van-Strien N.; Lapidus K.; Yin D.; Winder D. G.; Adams J. P.; Sweatt J. D.; Kandel E. R. Rap1 Couples CAMP Signaling to a Distinct Pool of P42/44MAPK Regulating Excitability, Synaptic Plasticity, Learning, and Memory. Neuron 2003, 39 (2), 309–325. 10.1016/S0896-6273(03)00404-5. [DOI] [PubMed] [Google Scholar]
- Lee D. Global and Local Missions of CAMP Signaling in Neural Plasticity, Learning, and Memory. Front Pharmacol 2015, 6, 161. 10.3389/fphar.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdo N. C.; Berrera M.; Koschinski A.; Brescia M.; Machado M. R.; Carr C.; Wright P.; Gorelik J.; Morotti S.; Grandi E. M.; et al. FRET Biosensor Uncovers CAMP Nano-Domains at β-Adrenergic Targets That Dictate Precise Tuning of Cardiac Contractility. Nat. Commun. 2017, 8 (1), 15031. 10.1038/ncomms15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschinski A.; Zaccolo M. Activation of PKA in Cell Requires Higher Concentration of CAMP than in Vitro : Implications for Compartmentalization of CAMP Signalling. Sci. Rep 2017, 7 (1), 1–12. 10.1038/s41598-017-13021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.-L. F.; Leroux M. R. cAMP and cGMP Signaling: Sensory Systems with Prokaryotic Roots Adopted by Eukaryotic Cilia. Trends Cell Biol. 2010, 20 (8), 435–444. 10.1016/j.tcb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Zaccolo M.; Zerio A.; Lobo M. J. Subcellular Organization of the CAMP Signaling Pathway. Pharmacol Rev. 2021, 73 (1), 278–309. 10.1124/pharmrev.120.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger J. U.; Nikolaev V. O. Biophysical Techniques for Detection of CAMP and CGMP in Living Cells. Int. J. Mol. Sci. 2013, 14 (4), 8025–8046. 10.3390/ijms14048025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.; Wan X.; Li Y.; Zhou W.; Peng L. Multiplexed 3D FRET Imaging in Deep Tissue of Live Embryos. Sci. Rep 2015, 5 (1), 13991. 10.1038/srep13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.; Shin S.; Bae S. W. CAMP Biosensors Based on Genetically Encoded Fluorescent/Luminescent Proteins. Biosensors (Basel) 2021, 11 (2), 39. 10.3390/bios11020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka H.; Arai S.; Inoue T.; Kitaguchi T. Genetically-Encoded Yellow Fluorescent CAMP Indicator with an Expanded Dynamic Range for Dual-Color Imaging. PLoS One 2014, 9 (6), e100252. 10.1371/journal.pone.0100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley C. R.; Mazzoni E. O.; Blau J. cAMPr: A Single-Wavelength Fluorescent Sensor for Cyclic AMP. Sci. Signal. 2018, 11 (520), eaah3738. 10.1126/scisignal.aah3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y.; Furuta T.; Nagai T.; Horikawa K. Red Fluorescent CAMP Indicator with Increased Affinity and Expanded Dynamic Range. Sci. Rep 2018, 8 (1), 1866. 10.1038/s41598-018-20251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K.; Ito M.; Wang X.; Tanaka M.; Wongso D.; Konno A.; Hirai H.; Hirase H.; Tsuboi T.; Kitaguchi T. Red Fluorescent Protein-Based CAMP Indicator Applicable to Optogenetics and in Vivo Imaging. Sci. Rep 2017, 7 (1), 7351. 10.1038/s41598-017-07820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oe Y.; Wang X.; Patriarchi T.; Konno A.; Ozawa K.; Yahagi K.; Hirai H.; Tsuboi T.; Kitaguchi T.; Tian L.; et al. Distinct Temporal Integration of Noradrenaline Signaling by Astrocytic Second Messengers during Vigilance. Nat. Commun. 2020, 11 (1), 471. 10.1038/s41467-020-14378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Sami A.; Noristani H. N.; Slattery K.; Qiu J.; Groves T.; Wang S.; Veerasammy K.; Chen Y. X.; Morales J.; et al. Glial Metabolic Rewiring Promotes Axon Regeneration and Functional Recovery in the Central Nervous System. Cell Metab 2020, 32 (5), 767–785.e7. 10.1016/j.cmet.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.; Jester B. W.; Tinberg C. E.; Mandell D. J.; Antunes M. S.; Chari R.; Morey K. J.; Rios X.; Medford J. I.; Church G. M.; et al. A General Strategy to Construct Small Molecule Biosensors in Eukaryotes. eLife Sciences 2015, 4, e10606. 10.7554/eLife.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L. A.; Chen L.-C.; Maynard-Smith L. A.; Ooi A. G. L.; Wandless T. J. A Rapid, Reversible, and Tunable Method to Regulate Protein Function in Living Cells Using Synthetic Small Molecules. Cell 2006, 126 (5), 995–1004. 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M.; Björklund T.; Lundberg C.; Kirik D.; Wandless T. J. A General Chemical Method to Regulate Protein Stability in the Mammalian Central Nervous System. Chem. Biol. 2010, 17 (9), 981–988. 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro R.; Chen L.-C.; Rakhit R.; Wandless T. J. A Novel Destabilizing Domain Based on a Small-Molecule Dependent Fluorophore. ACS Chem. Biol. 2016, 11 (8), 2101–2104. 10.1021/acschembio.6b00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton G. M.; Silverman W. R.; Heginbotham L.; Morais-Cabral J. H. Structural Basis of Ligand Activation in a Cyclic Nucleotide Regulated Potassium Channel. Cell 2004, 119 (5), 615–627. 10.1016/j.cell.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Mukherjee S.; Jansen V.; Jikeli J. F.; Hamzeh H.; Alvarez L.; Dombrowski M.; Balbach M.; Strünker T.; Seifert R.; Kaupp U. B.; et al. A Novel Biosensor to Study CAMP Dynamics in Cilia and Flagella. eLife Sciences 2016, 5, e14052. 10.7554/eLife.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünke S.; Stoldt M.; Lecher J.; Kaupp U. B.; Willbold D. Structural Insights into Conformational Changes of a Cyclic Nucleotide-Binding Domain in Solution from Mesorhizobium Loti K1 Channel. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (15), 6121–6126. 10.1073/pnas.1015890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C.; Kaufman C. K.; Li P.; Pugach E. K.; Tamplin O. J.; Zon L. I. Ubiquitous Transgene Expression and Cre-Based Recombination Driven by the Ubiquitin Promoter in Zebrafish. Development 2011, 138 (1), 169–177. 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S. J.; Devoto S. H.; Westerfield M.; Moon R. T. Positive and Negative Regulation of Muscle Cell Identity by Members of the Hedgehog and TGF-Beta Gene Families. J. Cell Biol. 1997, 139 (1), 145–156. 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi M. J.; Stickney H. L.; Devoto S. H. The Zebrafish Slow-Muscle-Omitted Gene Product Is Required for Hedgehog Signal Transduction and the Development of Slow Muscle Identity. Development 2000, 127 (10), 2189–2199. 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- Truong M. E.; Bilekova S.; Choksi S. P.; Li W.; Bugaj L. J.; Xu K.; Reiter J. F. Vertebrate Cells Differentially Interpret Ciliary and Extraciliary CAMP. Cell 2021, 184 (11), 2911–2926.e18. 10.1016/j.cell.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard J. B.; Malhotra D.; Gvozdenovic-Jeremic J.; Josey M.; Chen M.; Weinstein L. S.; Lu J.; Shore E. M.; Kaplan F. S.; Yang Y. Activation of Hedgehog Signaling by Loss of GNAS Causes Heterotopic Ossification. Nat. Med. 2013, 19 (11), 1505–1512. 10.1038/nm.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. T. H.; Zhao Z.; Ingham P. W. Hedgehog Signalling. Development 2016, 143 (3), 367–372. 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- Hirsinger E.; Stellabotte F.; Devoto S. H.; Westerfield M. Hedgehog Signaling Is Required for Commitment but Not Initial Induction of Slow Muscle Precursors. Dev. Biol. 2004, 275 (1), 143–157. 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Ingham P. W.; Kim H. R. Hedgehog Signalling and the Specification of Muscle Cell Identity in the Zebrafish Embryo. Exp. Cell Res. 2005, 306 (2), 336–342. 10.1016/j.yexcr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Huang P.; Schier A. F. Dampened Hedgehog Signaling but Normal Wnt Signaling in Zebrafish without Cilia. Development 2009, 136 (18), 3089–3098. 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich J. K.; Payumo A. Y.; Rack P. G.; Chen J. K. In Vivo Imaging of Hedgehog Pathway Activation with a Nuclear Fluorescent Reporter. PLoS One 2014, 9 (7), e103661. 10.1371/journal.pone.0103661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S.; Wen X.; Ratti N.; Loktev A.; Rangell L.; Scales S. J.; Jackson P. K. The Ciliary G-Protein-Coupled Receptor Gpr161 Negatively Regulates the Sonic Hedgehog Pathway via CAMP Signaling. Cell 2013, 152 (1–2), 210–223. 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Bachmann V. A.; Mayrhofer J. E.; Ilouz R.; Tschaikner P.; Raffeiner P.; Röck R.; Courcelles M.; Apelt F.; Lu T.-W.; Baillie; et al. Gpr161 Anchoring of PKA Consolidates GPCR and CAMP Signaling. Proc. Natl. Acad. Sci. U.S.A. 2016, 113 (28), 7786–7791. 10.1073/pnas.1608061113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaikner P. M.; Regele D.; Röck R.; Salvenmoser W.; Meyer D.; Bouvier M.; Geley S.; Stefan E.; Aanstad P. Feedback Control of the Gpr161-Gαs-PKA Axis Contributes to Basal Hedgehog Repression in Zebrafish. Development 2021, 148 (4), dev192443. 10.1242/dev.192443. [DOI] [PubMed] [Google Scholar]
- Wang M.; Li P.; Wang H.; Dong L.; Wu C.; Zhao Z. Identification and Spatiotemporal Expression of Gpr161 Genes in Zebrafish. Gene 2020, 730, 144303. 10.1016/j.gene.2019.144303. [DOI] [PubMed] [Google Scholar]
- Hwang S.-H.; Somatilaka B. N.; White K.; Mukhopadhyay S. Ciliary and Extraciliary Gpr161 Pools Repress Hedgehog Signaling in a Tissue-Specific Manner. eLife 2021, 10, e67121. 10.7554/eLife.67121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeler E. L.; Urner L. M.; Rakhit R.; Liu C. W.; Wandless T. J. Ligand-Switchable Substrates for a Ubiquitin-Proteasome System. J. Biol. Chem. 2011, 286 (36), 31328–31336. 10.1074/jbc.M111.264101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y.; Imoto H.; Chen L.; Wandless T. J. Destabilizing Domains Derived from the Human Estrogen Receptor. J. Am. Chem. Soc. 2012, 134 (9), 3942–3945. 10.1021/ja209933r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.