Abstract
In this issue of Molecular Cell, Weith et al. (2018) demonstrate that p97, together with a SEP adaptor, can catalyze ordered subunit exchange to facilitate the biogenesis of protein phosphatase-1 (PP1) holoenzyme, establishing a novel ubiquitin-independent “segregase” function for this versatile ATPase.
Protein phosphatase 1 (PP1) is an essential serine/threonine phosphatase that catalyzes a substantial fraction of protein dephosphorylation events in eukaryotic cells. It functions as a collection of obligate holoenzyme complexes, each consisting of a distinct regulatory adaptor that confers substrate specificity and a catalytic subunit, also named PP1. Intriguingly, during the biogenesis of PP1 holoenzymes, newly synthesized PP1 is first held by two cofactors—SDS22 and inhibitor-3 (I3)—forming a stable trimeric complex with no activity. These inhibitory partners are later exchanged with one of the many substrate-specific adaptors to produce active PP1 holoenzymes (Peti et al., 2013). How subunit exchange is achieved during PP1 biogenesis has been unclear. In this issue of Molecular Cell, Weith et al. uncovered an energy-dependent protein-disassembling reaction as an important step in PP1 biogenesis. This process is catalyzed by the type II AAA (ATPase-associated with diverse cellular activities) ATPase p97, which disrupts the inhibitory PP1 complex to facilitate subunit exchange and maturation of PP1 holoenzymes (Weith et al., 2018).
p97 (also named VCP in mammals or CDC48 in yeast) is an abundant, highly conserved hexameric ATPase whose major function is to separate polypeptides from protein complexes, chromatin DNA, or organelle membranes in various cellular compartments (Ye et al., 2017). Like other type II AAA ATPases, p97 contains two similar AAA ATPase domains (termed D1 and D2), which oligomerize into two stacked rings with a central pore. It also has an N-terminal regulatory domain that interacts with a variety of cofactors. These cofactors promote substrate recruitment or processing and thus define a complex functional repertoire for p97/CDC48 (Ye et al., 2017).
The best understood function of p97/CDC48 is in endoplasmic reticulum-associated degradation (ERAD), in which it acts as an ER-associated, ubiquitin-dependent protein “unfoldase.” This function is assisted by the Ufd1-Npl4 dimeric cofactor complex, which uses several ubiquitin binding motifs for substrate recruitment. Once ubiquitinated polypeptides engage p97, it hydrolyzes ATP, threading substrates through its central pore to cause their unfolding and separation from the ER membrane (Figure 1A) (Bodnar and Rapoport, 2017).
Figure 1. Ubiquitin-Dependent and -Independent “Segregase” Activity of p97.
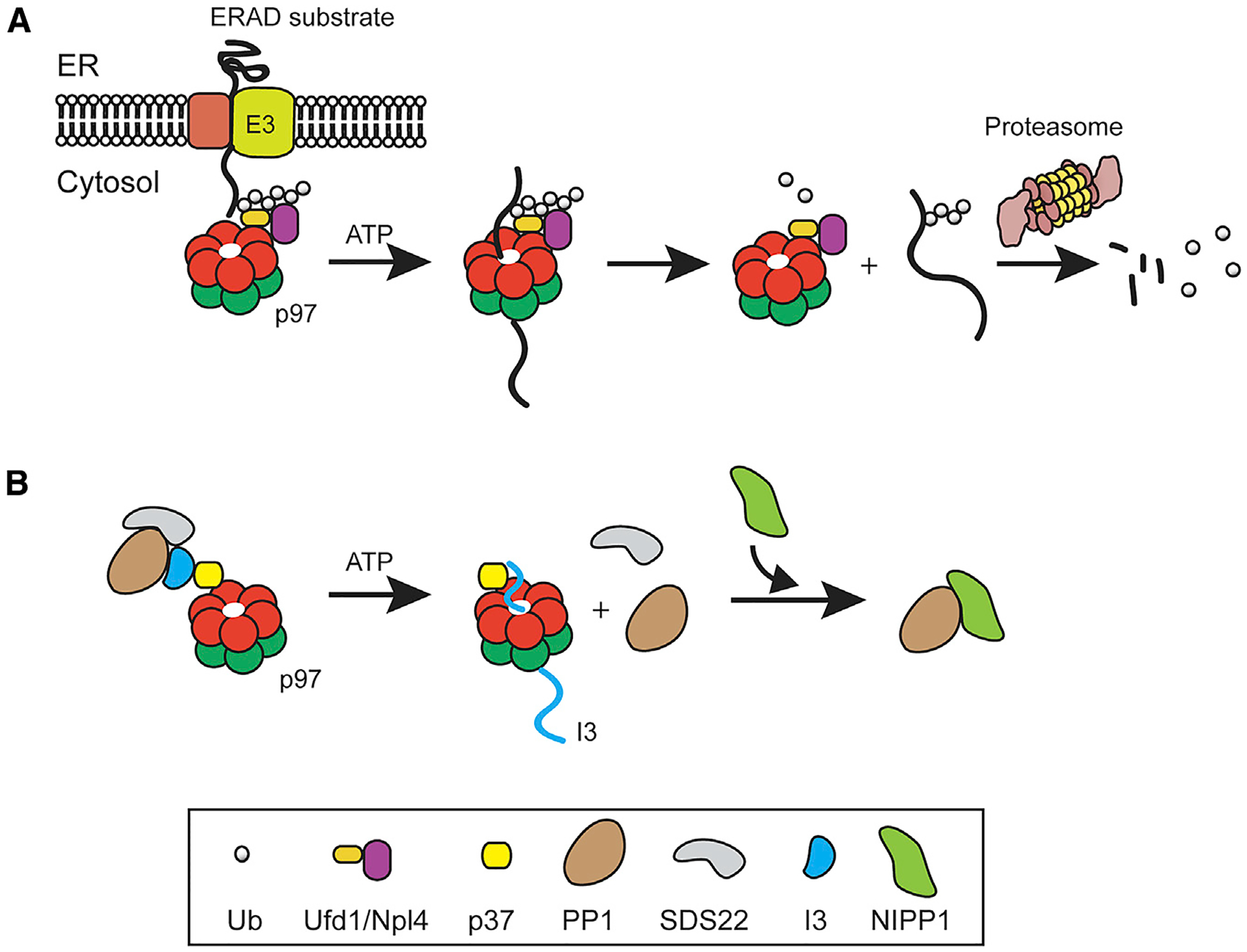
(A) In the ERAD pathway, substrates ubiquitinated during retrotranslocation by ER-associated ubiquitin ligase (E3) are recognized by the heterodimeric adaptor Ufd1-Npl4, which activates p97 to extract substrate from the ER membrane. p97 hydrolyzes ATP to move substrate through its central channel, thereby unfolding the substrate for proteasome degradation.
(B) In PP1 biogenesis, p97 associates with I3 in the PP1-SDS22-I3 complex via a SEP adaptor (e.g., p37). It then uses the energy from ATP hydrolysis to translocate and unfold I3, which results in the disassembly of the inhibitory PP1 complex and subsequent binding of a substrate-specific cofactor (e.g., NIPP1) to PP1.
Given the well-established “segregase” function, Weith et al. (2018) hypothesized that p97 may facilitate PP1 biogenesis by destabilizing the initial inhibitory complex. Using a pulse-chase approach, they first confirmed that newly synthesized PP1, SDS22, and I3 indeed form a transient complex whose disassembly allows PP1 to interact with a functional substrate specifier such as NIPP1. Strikingly, this highly coordinated subunit exchange process is prohibited when p97 is inactivated by either siRNA-mediated gene silencing or p97-specific inhibitors.
To elucidate the molecular mechanism of this p97-dependent process, Weith et al. (2018) focused on a class of p97 adaptors named SEP (for Shp1, eyes-closed, and p47) domain-containing adaptors because genetic and biochemical studies in yeast have implicated the SEP-containing protein Shp1 in PP1 biogenesis (Zhang et al., 1995). They first showed, by co-immunoprecipitation, that among the four mammalian SEP domain adaptors, p37, UBXN2A, and p47 all interact with the SDS22-PP1-I3 complex. Photo-crosslinking combined with mutagenesis defined a molecular model in which these adaptors bind SDS22-PP1-I3 via a direct interaction between their SEP domain and I3. Functionally, combined loss of these SEP adaptors reduces the recruitment of p97 to SDS22-PP1-I3 and delays its disassembly and the formation of a subset of PP1 holoenzyme complexes, which correlates with diminished cell viability and cell-cycle deregulation. Collectively, these findings suggested that SEP adaptors recruit p97 to the PP1 inhibitory complex and regulate the exchange of SDS22 and I3 with a functional substrate adaptor (Figure 1B). This model was further tested by an in vitro biochemical reconstitution assay in which pre-assembled SDS22-PP1-I3 complex could indeed be dismantled by purified p97 and p37, concomitant with the formation of the PP1-NIPP1 holoenzyme complex. Mechanistically, disassembly of SDS22-PP1-I3 is driven by ATP-dependent translocation of I3 into the central cavity of the p97 hexamer, which causes its unfolding (Figure 1B).
In contrast to the previously characterized role of p97 in ERAD, this study provides compelling biochemical evidence that p97, together with a SEP adaptor, is capable of catalyzing a protein disassembly reaction independently of ubiquitin. This conclusion resonates with earlier studies on VAT, a homolog of p97 in archaebacteria, which revealed a similar unfoldase activity (Gerega et al., 2005). Since archaebacteria do not have ubiquitin or adaptors for VAT, it seems that the original ubiquitin-independent unfolding activity is re-sculpted by later evolved adaptors, allowing p97/CDC48 to accommodate both ubiquitinated and non-ubiquitinated substrates in eukaryotes. Interestingly, to unfold polypeptides in ERAD, the ATPase activity of p97 needs to be upregulated by ubiquitin in the presence of Ufd1-Npl4 (Bodnar and Rapoport, 2017). How p97 is activated in ubiquitin-independent unfolding reactions remains to be elucidated because SEP adaptors can regulate p97 in both positive (p37) and negative (p47) manners in vitro (Ye et al., 2017; Zhang et al., 2015).
The study by Weith et al. (2018) also illustrates an interesting twist in protein complex biogenesis: an intermediate complex is “destructed” by an energy-dependent process to allow the formation of a functional holoenzyme. In protein complex biogenesis, it is prevalent that individual subunits form a transient complex with a stabilizing unit while waiting for their functional partner(s). For instance, analogous to PP1 biogenesis, the formation of the hemoglobin complex, which in adult animals is a tetramer of two hemoglobin-α and -β chains each, involves association of hemoglobin-α with a non-canonical chaperone named α-Hemoglobin stabilizing protein (AHSP). This interaction prevents hemoglobin-α self-aggregation and therefore promotes its assembly with the β-chain (Kihm et al., 2002). Likewise, the assembly of the 26S proteasome also involves transient complexes between certain proteasomal subunits and proteasome-specific chaperones (Madura, 2009). A recent ribosome profiling study suggested that ~30% of nascent complexes in yeast are assembled post-translationally via highly regulated and dynamic processes that involve exchange of a folding assistant with a functional subunit(s) (Shiber et al., 2018). Subunit exchange may also occur when cells need to remodel existing complexes to generate new functional entities. Whether resolving an existing complex in these subunit exchange processes is spontaneous in general or driven by energy as demonstrated for PP1 biogenesis in this study is unclear. Given the abundance and the powerful “segregase” activity of p97, it is conceivable that the molecular mechanism uncovered by Weith et al. (2018) may be applicable to other examples of protein complex biogenesis.
REFERENCES
- Bodnar NO, and Rapoport TA (2017). Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerega A, Rockel B, Peters J, Tamura T, Baumeister W, and Zwickl P (2005). VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem 280, 42856–42862. [DOI] [PubMed] [Google Scholar]
- Kihm AJ, Kong Y, Hong W, Russell JE, Rouda S, Adachi K, Simon MC, Blobel GA, and Weiss MJ (2002). An abundant erythroid protein that stabilizes free alpha-haemoglobin. Nature 417, 758–763. [DOI] [PubMed] [Google Scholar]
- Madura K (2009). Cell biology: the proteasome assembly line. Nature 459, 787–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, Nairn AC, and Page R (2013). Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 280, 596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiber A, Döring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, and Bukau B (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weith M, Seiler J, van den Boom J, Kracht M, Hülsmann J, Primorac I, Del Pino Garcia J, Kaschani F, Kaiser M, Musacchio A, et al. (2018). Ubiquitin-independent disassembly by a p97 AAA-ATPase complex drives PP1 holoenzyme formation. Mol. Cell 72, this issue, 766–777. [DOI] [PubMed] [Google Scholar]
- Ye Y, Tang WK, Zhang T, and Xia D (2017). A mighty “protein extractor” of the cell: structure and function of the p97/CDC48 ATPase. Front. Mol. Biosci 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Guha S, and Volkert FC (1995). The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol 15, 2037–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gui L, Zhang X, Bulfer SL, Sanghez V, Wong DE, Lee Y, Lehmann L, Lee JS, Shih PY, et al. (2015). Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. USA 112, E1705–E1714. [DOI] [PMC free article] [PubMed] [Google Scholar]


