Abstract
The serotype 5 capsule gene cluster of Staphylococcus aureus comprises 16 genes (cap5A through cap5P), but little is known about how the putative gene products function in capsule biosynthesis. We propose that the N-acetylmannosaminuronic acid (ManNAcA) component of the S. aureus serotype 5 capsular polysaccharide (CP5) is synthesized from a UDP-N-acetylglucosamine (UDP-GlcNAc) precursor that is epimerized to UDP-N-acetylmannosamine (UDP-ManNAc) and then oxidized to UDP-ManNAcA. We report the purification and biochemical characterization of a recombinant UDP-GlcNAc 2-epimerase encoded by S. aureus cap5P. Purified Cap5P converted ∼10% of UDP-GlcNAc to UDP-ManNAc as detected by gas chromatography-mass spectrometry. The epimerization of UDP-GlcNAc to UDP-ManNAc occurred over a wide pH range and was unaffected by divalent cations. Surprisingly, CP5 expression in S. aureus was unaffected by insertional inactivation of cap5P. Sequence homology searches of the public S. aureus genomic databases revealed the presence of another putative UDP-GlcNAc 2-epimerase on the S. aureus chromosome that showed 61% identity to Cap5P. Redundancy of UDP-GlcNAc 2-epimerase function in S. aureus was demonstrated by cloning the cap5P homologue from strain Newman and complementing an Escherichia coli rffE mutant defective in UDP-GlcNAc 2-epimerase activity. Our results confirm the putative function of the S. aureus cap5P gene product and demonstrate the presence of a second gene on the staphylococcal chromosome with a similar function.
The serotype 5 capsular polysaccharide (CP5) produced by Staphylococcus aureus has a trisaccharide repeating unit structure of (→4)-3-O-Ac-β-d-ManNAcAp-(1→4)-α-l-FucNAcp-(1→3)-β-d-FucNAcp-(1→). Sixteen genes (cap5A through cap5P), clustered on the bacterial chromosome, are involved in CP5 biosynthesis (23). However, the function of only one of these gene products has been demonstrated; Cap5H O acetylates the N-acetylmannosaminuronic acid (ManNAcA) moiety of CP5 (2). The present work was initiated to ascribe biochemical function to the cap5P gene product and confirm its role in CP5 biosynthesis. Sequence analysis of Cap5P showed a putative protein of 391 amino acids with a high degree of homology to functionally characterized UDP-N-acetylglucosamine (UDP-GlcNAc) 2-epimerases, including RffE of Escherichia coli, Cps19fK of Streptococcus pneumoniae, and RfbC of Salmonella enterica (23). RffE is a UDP-GlcNAc 2-epimerase that catalyzes the conversion of UDP-GlcNAc to UDP-N-acetyl d-mannosamine (ManNAc) (17, 21). The latter serves as an intermediate in the biosynthesis of enterobacterial common antigen (ECA), a surface-associated glycolipid common to all members of the Enterobacteriaceae family (11). Both cps19fK and rfbC are involved in the synthesis of ManNAc-containing extracellular polysaccharides, and both gene products have been shown to complement rffE mutants of E. coli (8, 18). Similarly, we showed that S. aureus cap5P can functionally complement an ECA-negative rffE mutant of E. coli, suggesting that Cap5P also has UDP-GlcNAc 2-epimerase activity (9). Cap5O shows homology to RffD from E. coli, a protein with UDP-ManNAc dehydrogenase activity (23). S. aureus cap5O was able to complement an ECA-negative rffD mutant of E. coli (9). Thus, we propose that the ManNAcA residue of CP5 is synthesized as follows:
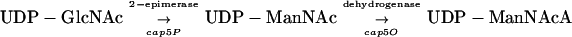 |
This paper confirms the function of Cap5P by demonstrating its enzymatic activity in vitro. The observation that a mutation in cap5P led to no observable phenotype directed us to the identification of a second staphylococcal gene with functional homology to cap5P.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. E. coli strains were propagated in Luria-Bertani medium. S. aureus strains were grown in tryptic soy broth (TSB) or tryptic soy agar (TSA) or on Columbia agar (Difco Laboratories, Detroit, Mich.) supplemented with 2% NaCl. Appropriate antibiotics were added to the culture medium as follows: chloramphenicol (Cm) at 10 μg/ml, erythromycin (Em) at 5 μg/ml, or kanamycin (Km) at 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F−ompT lon hsdSB(rB− mB−) DE3 | Novagen |
| DB11 | F−met thi gal nal rif hsdR Ems | 16 |
| JM110 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/rpsL (Strr) thr leu thi lacY galK galT ara fhuA dam dcm supE44 Δ(lac-proAB) | American Type Culture Collection |
| 21566 | K-12 thr-1 leuB6 Δ(gpt-proA)66 his G4 argE3 thi-1 rfbD1 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 supE44 rff::Tn10-66 | 17 |
| S. aureus | ||
| Newman | CP5 positive | NCTC 8178 |
| Newman P15-1 | ermB-inactivated cap5P gene | This study |
| RN4220 | Capsule negative, restriction negative | 20 |
| Plasmids | ||
| pERMB | 2.2-kb HpaI-SalI fragment from pRN8078 in pGEM-7Zf+ | This study |
| pET-24a+ | E. coli expression vector (Kmr) | Novagen |
| pGEM-7Zf+ | E. coli cloning vector (Apr) | Promega |
| pJCL84 | 6.2-kb HindIII fragment containing 3′ end of cap5 sequence in pGEM-7Zf+ | 9 |
| pKBK7 | 1.2-kb PCR amplicon carrying cap5P in pET-24a+ | This study |
| pKBK10 | 1.9-kb HindIII-EcoRI fragment carrying cap5P in pUC19 | 9 |
| pKBK13 | 1.3-kb BamHI-BclI fragment from pERMB inserted into cap5P in pKBK10 | This study |
| pKBK15 | 3.3-kb PCR amplicon from pKBK13 in pTS1 | This study |
| pKBK25 | 1.2-kb PCR amplicon carrying mnaA in pUC19 | This study |
| pRN8078 | Tn551 (ermB) in temperature-sensitive plasmid | 13 |
| pTS1 | Temperature-sensitive shuttle vector (Apr Cmr) | 19 |
| pUC19 | E. coli cloning vector (Apr) | New England Biolabs, Inc. |
Chemicals.
UDP-GlcNAc, GlcNAc, and ManNAc were obtained from Sigma Chemical Co. (St. Louis, Mo.). Ultrapure reagents used in sugar derivatizations were obtained from J. T. Baker, Inc. (Phillipsburg, N.J.). Restriction endonucleases and other DNA modification enzymes were obtained from Life Technologies, Inc. (Gaithersburg, Md.) or New England Biolabs, Inc. (Beverly, Mass.).
DNA manipulations.
Plasmid DNA was isolated with the QIAprep spin miniprep kit 250 (Qiagen, Inc., Santa Clarita, Calif.). Standard molecular cloning procedures were followed as detailed by Sambrook et al. (22). Both strands of the PCR-amplified cap5P product were sequenced by the Taq dideoxy terminator cycle method with an automated sequencer (model 373A; Applied Biosystems). The isoelectric point, hydropathy plot, and sequence alignment of Cap5P were determined with the University of Wisconsin Genetics Computer Group software package.
Subcloning cap5P into expression vector pET-24a+.
The entire cap5P open reading frame (ORF) from the first ATG codon, as identified by Sau et al. (23) (GenBank accession no. U81973), was amplified by PCR (25 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 7 min) with UlTma DNA polymerase (Perkin-Elmer, Foster City, Calif.), primers KK7 (5′-GATAAGCTagCATGTGTTTGAACTTCAGAGAGG-3′) and KK9 (5′-ATTAcTcGaGACGTAAAGGTACGAATTCATCCGG-3′), and plasmid pJCL84 as the template. (In primer KK7, the NheI site is underlined, and the first ATG start codon of the cap5P ORF is in bold print. In primer KK9, the XhoI site is underlined, and the sequence complementary to the TAA stop codon is in bold. Bases not complementary to the cap5 sequence are shown in lowercase letters.) The 1.2-kb amplicon was digested with NheI and XhoI and then ligated into pET-24a+ (Novagen, Inc., Madison, Wis.) to create plasmid pKBK7. Subcloning into the XhoI site of pET-24a+ resulted in an in-frame fusion to the His-Tag sequence of the vector.
Purification of Cap5P.
A Novagen kit for rapid affinity purification with pET His-Tag systems was used according to manufacturer’s specifications, except for a few modifications. A 100-ml culture of E. coli BL21(DE3) carrying pKBK7 was grown at 37°C to an absorbance of 0.6 at 600 nm. Isopropyl-β-d-thiogalactopyranoside (IPTG) (U.S. Biochemicals Corp., Cleveland, Ohio) was added at a final concentration of 1 mM, and the culture was incubated for an additional 3 h at 30°C. Bacterial cells were lysed in a French pressure cell (three cycles at 800 lb/in2). After centrifugation of the lysate, 1.25 ml of the supernatant was diluted with an equal volume of binding buffer and applied to the Ni2+ affinity column. Recombinant Cap5P was eluted in Tris-HCl buffer (pH 7.9) containing 1 M NaCl and 330 mM imidazole. The purified protein was dialyzed against 100 mM phosphate buffer (pH 7.0) and frozen in aliquots at −70°C. Protein content was determined by the Bradford dye-binding method (Bio-Rad Laboratories, Hercules, Calif.) (3), with bovine gamma globulin as the standard. A 100-pmol sample of the purified Cap5P protein was used to determine the N-terminal sequence by automated Edman degradation (4) on an Applied Biosystems protein sequencer (model 477A).
UDP-GlcNAc 2-epimerase assay.
The activity of UDP-GlcNAc 2-epimerase was assayed by measurement of the conversion of UDP-GlcNAc to UDP-ManNAc. The assay mixtures contained 0.5 mM UDP-GlcNAc, 100 mM phosphate buffer (pH 7.0), and 1.6 μg of purified Cap5P in a total volume of 100 μl. Control assay mixtures contained enzyme that was boiled for 10 min. Incubations were typically carried out for 2 h at 37°C. The reaction was stopped by the addition of 1 ml of 99% ethanol, followed by heating at 70°C for 10 min. The samples were hydrolyzed with 0.5 M trifluoroacetic acid at 100°C for 30 min to remove the UDP moieties. Alditol acetate derivatives of the samples were prepared (25) and identified by combined gas chromatography-mass spectrometry (GC-MS) with a model HP 6890 series gas chromatograph and a model HP 5973 mass selective detector (Hewlett-Packard). The column used for separation was a DB-17 (J & W Scientific) 30-m capillary column containing (50% phenyl) methylpolysiloxane, with a constant flow rate of 1 ml/min. The temperature program used for alditol acetate derivatives began at 150°C (delay, 2 min), with a subsequent 4°C/min rise to 200°C, then a 1°C/min rise to 225°C, and finally, a 4°C/min rise to 280°C (held for 6 min). The injector temperature was 230°C.
Enzymatic activity was expressed as a percent ratio of UDP-ManNAc to UDP-GlcNAc in the assay mixtures. UDP-GlcNAc and UDP-ManNAc were quantitated by integration of the area under the peaks separated by GC. Levels of enzymatic activity achieved under different experimental conditions were compared by the alternate Welch t test.
Insertional inactivation of cap5P.
A DNA fragment harboring the Tn551 ermB gene, which encodes Em resistance, was used to insertionally inactivate the cap5P gene. The 2.2-kb HpaI-SalI fragment of plasmid pRN8078, a temperature-sensitive vector carrying Tn551, was subcloned into pGEM-7Zf+, previously digested with SmaI and XhoI. The resulting plasmid, pERMB, was passaged through E. coli JM110 (dam mutant strain) and then digested with BamHI (vector site) and BclI (insert site), releasing a 1.3-kb fragment carrying the ermB gene flanked by Tn551 sequences. The cap5P subclone pKBK10, passaged through JM110, was digested with BclI, releasing a 93-bp fragment 72 bases downstream from the second ATG of the cap5P ORF. The 1.3-kb BamHI-BclI fragment of pERMB was ligated into the BclI site of pKBK10 to create plasmid pKBK13. Plasmids carrying the ermB cassette were isolated in Em-sensitive E. coli DB11 by selection on media containing Em at a concentration of 10 μg/ml.
A 3.3-kb segment of pKBK13 was amplified by PCR (25 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 4 min) with primers KK16 (5′-GAAGCGGGaAttCGTGTCTATCGC-3′; cap5O specific) and M13-40 (GGTTTTCCCAGTCACGACG-3′; vector specific) and eLONGase enzyme mix (Life Technologies). (In primer KK16, the EcoRI site is underlined, and bases not complementary to the cap5 sequence are shown in lowercase letters.) The amplicon was digested with EcoRI and ligated into pTS1, a shuttle vector with an E. coli origin of replication and an S. aureus temperature-sensitive origin of replication. In the resulting plasmid pKBK15, the ermB gene was inserted in the same orientation as the cap5P gene, as determined by restriction digest analysis.
Plasmid pKBK15 was electroporated (12) into the restriction-negative S. aureus strain RN4220 and then transduced with phage 80α (5) into strain Newman, in both instances with selection for Emr colonies at 30°C. Plasmid integrants in the chromosome were selected by plating on TSA + Em at 42°C. Single colonies were passaged three times at 30°C in TSB without antibiotics to allow for plasmid excision and then were plated at 42°C on TSA + Em. Colonies that were Emr and sensitive to Cm were determined by replica plating on TSA + Em and TSA + Cm. The identity of the cap5P mutant, Newman P15-1, was confirmed by Southern blot analysis. Briefly, chromosomal DNA from S. aureus was digested with EcoRI and EcoRV and electrophoresed in a 0.8% agarose gel. DNA was transferred to a GeneScreenPlus membrane (NEN Research Products, Boston, Mass.) with a Posiblot pressure blotter (Stratagene, La Jolla, Calif.). The blot was probed with PCR-amplified cap5P labeled with the AlkPhos labeling kit (Amersham Life Science, Arlington Heights, Ill.) and subjected to autoradiography.
Quantitation of CP5 expression.
CP5 expression by the parent strain Newman and the cap5P mutant was quantitated by rocket immunoelectrophoresis (15) and enzyme-linked immunosorbent assays (ELISA) (1, 15).
Cloning and characterization of the S. aureus cap5P homologue.
Preliminary S. aureus sequence data was obtained from The Institute of Genomic Research website (4a). Primers KK27 (5′-acaactgcAGAATCGGAGATAAGTAG-3′) and KK28 (5′-acaaggaTCCTCAGAAATCGCTTGG-3′) for the amplification of the gene encoding the cap5P homologue were designed to anneal to 5′ and 3′ ends of the gene that show no homology to the flanking sequences of cap5P. (The PstI and BamHI sites engineered in primers KK27 and KK28, respectively, are underlined, and bases not complementary to the S. aureus sequence are shown in lowercase letters.) The gene was amplified by PCR (94°C for 5 min; 4 cycles of 94°C for 1 min, 45°C for 1 min, and 68°C for 5 min; 21 cycles of 94°C for 30 s, 62°C for 30 s, and 68°C for 3 min) from S. aureus Newman chromosomal DNA with eLONGase enzyme mix. The 1.2-kb amplicon was digested with PstI and BamHI and ligated into pUC19 to create plasmid pKBK25.
RESULTS
Purification and characterization of Cap5P.
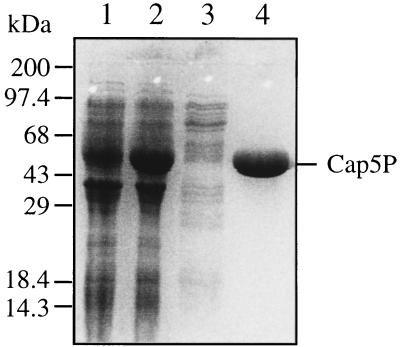
Cap5P was overexpressed from the clone pKBK7 in E. coli BL21(DE3). The hydropathy plot (data not shown) of the deduced amino acid sequence of Cap5P indicated that the protein was hydrophilic in nature. Accordingly, the majority of the overexpressed protein was present in the supernatant after centrifugation of the cell lysate at 39,000 × g. The 6-histidine-residue tag at the C-terminal end of Cap5P allowed purification of the protein over a Ni2+ affinity column. The final eluate from the column showed a single band by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1). At a concentration of 0.2 mg/ml, Cap5P retained solubility and activity at −70°C for at least 7 months.
FIG. 1.
SDS-PAGE analysis of S. aureus Cap5P expression and purification. Cell lysates were made from E. coli BL21(DE3) carrying pKBK7. Cap5P was overexpressed in the presence of 1 mM IPTG, and the soluble protein was purified over a nickel column. Lane 1, cell lysate from uninduced cells; lane 2, cell lysate from IPTG-induced cells; lane 3, column effluent reflecting unbound proteins; lane 4, purified S. aureus Cap5P. Molecular mass markers in kilodaltons are indicated on the left.
Examination of the cap5 sequence revealed two potential start sites for cap5P at nucleotides 15769 and 15799 and a termination codon at nucleotide 16942. Sau et al. tentatively identified the cap5P start site as the first ATG codon and, as a result, predicted a protein of 391 amino acids (23). N-terminal protein sequencing of Cap5P purified from E. coli BL21(DE3) yielded the sequence MKKIMVIFGT; this result indicated that the second ATG codon at position 15799 is the authentic translation start site for cap5P. This codon was preceded by a probable Shine-Dalgarno sequence (GAGAGG) at position 15785. The 1,143-nucleotide cap5P gene encoded 381 amino acids with a predicted polypeptide of 41.2 kDa, similar to that obtained with the histidine-tagged protein (42 kDa on SDS-PAGE). The predicted isoelectric point of Cap5P was 5.9.
Biochemical activity of purified Cap5P.
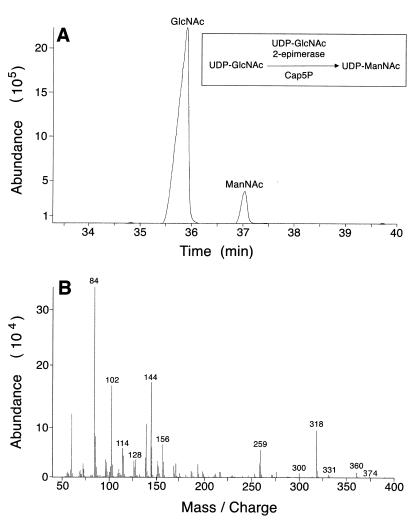
The purified Cap5P protein was incubated with UDP-GlcNAc as the substrate, and the products of the reaction mixture were hydrolyzed and derivatized before analysis by GC-MS. Gas chromatograms obtained with samples containing Cap5P showed two acetylhexosamine peaks with retention times of approximately 36 and 37 min (Fig. 2A). The peak at 36 min represents the hydrolyzed and derivatized product of authentic UDP-GlcNAc. The peak at 37 min showed a mass spectrum of m/z 84, 102, 114, 128, 144, 156, 259, 300, 318, 331, 360, and 374 (Fig. 2B), representing the amino sugar alditol 1,3,4,5,6-penta-O-acetyl-2-acetamino-2-deoxy-d-mannitol. This peak represents the hydrolyzed and derivatized product of UDP-ManNAc and elutes with the same retention time as the derivatized product of authentic ManNAc. Integration of the GC peaks yielded a 10:1 ratio of GlcNAc to ManNAc. Control reaction mixtures containing boiled Cap5P showed no peak at 37 min. Likewise, no conversion to UDP-ManNAc was observed when UDP-N-acetylgalactosamine was used as the substrate (data not shown).
FIG. 2.
GC-MS analysis of hydrolyzed and derivatized assay mixtures containing UDP-GlcNAc and S. aureus Cap5P. Elution products GlcNAc and ManNAc were identified by GC (A), in which peaks with retention times of 36 and 37 min corresponded to those of authentic GlcNAc and ManNAc standards, respectively, and MS (B), in which the peak at 37 min was shown to be an alditol acetate derivative of acetylhexosamine.
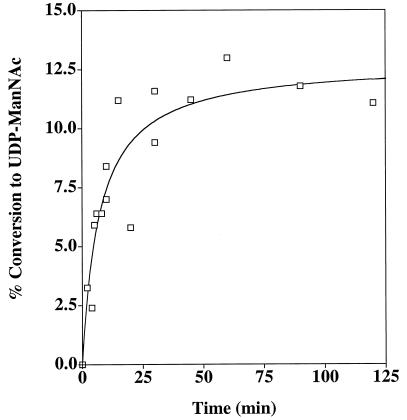
Under the standard Cap5P assay conditions (100 mM phosphate buffer [pH 7.0], 0.5 mM UDP-GlcNAc), the time of incubation at 37°C was varied from 2 min to 2 h. As shown in Fig. 3, the conversion of UDP-GlcNAc to UDP-ManNAc reached a plateau after ∼30 min of incubation. In six separate experiments catalyzed by two different preparations of Cap5P, ∼10% (range, 6 to 12%) of UDP-GlcNAc was converted to UDP-ManNAc. Doubling of the Cap5P enzyme concentration in a 2-h assay showed no effect on the net synthesis of UDP-ManNAc from UDP-GlcNAc (data not shown). Likewise, variation of the substrate concentration from 0.5 to 1 mM did not change the final ratio of GlcNAc to ManNAc in the assay mixture after 2 h at 37°C (data not shown).
FIG. 3.
Effect of incubation time on the activity of S. aureus cap5P-encoded UDP-GlcNAc 2-epimerase. Purified Cap5P was incubated with UDP-GlcNAc under standard conditions. Conversion to UDP-ManNAc was analyzed by GC-MS after alditol acetate derivatization of the samples. Results presented are pooled data from two experiments.
The effect of pH on the enzymatic activity of Cap5P was examined in three separate experiments. The results of a typical experiment (shown in Table 2) indicate that the activity of Cap5P was stable over a pH range of 6.6 to 9.0. The presence of divalent cations including Ca2+, Mn2+, or Mg2+ at concentrations of 10 mM had no significant effect on enzyme activity (data not shown). In addition, the epimerization of UDP-GlcNAc to UDP-ManNAc was unaffected by the presence of 5 mM EDTA or 4 mM ATP (data not shown) in the enzyme assay mixture.
TABLE 2.
Effect of pH on the activity of S. aureus cap5P-encoded UDP-GlcNAc 2 epimerase
| pH | % Conversiona (GlcNAc to ManNAc) |
|---|---|
| 6.0 | 3.5 |
| 6.6 | 6.5 |
| 7.0 | 6.8 |
| 7.6 | 5.3 |
| 8.0 | 6.1 |
| 8.5 | 5.3 |
| 9.0 | 6.8 |
Percent conversion of UDP-GlcNAc to UDP-ManNAc was calculated by integration of the GlcNAc and ManNAc peaks obtained on gas chromatograms.
Effect of cap5P mutation on CP5 expression.
S. aureus cap5P was inactivated by insertion of an ermB cassette into the 5′ end of the gene. In contrast to the wild-type cap5P gene in pKBK10 (9), the insertionally inactivated cap5P gene in pKBK13 was unable to complement an rffE mutation in E. coli 21566 (data not shown). This experimental result confirmed the loss of UDP-GlcNAc 2-epimerase activity in the ermB disrupted gene product. Therefore, the ermB-inactivated cap5P gene was subcloned into a temperature-sensitive plasmid and introduced by allelic exchange into the chromosome of S. aureus capsule type 5 strain Newman. To confirm the insertional disruption of cap5P in the chromosome of the mutant strain, genomic DNA from strain Newman and mutant P15-1 were digested with EcoRI and EcoRV, electrophoresed in an agarose gel, and analyzed by Southern blotting. The AlkPhos-labelled cap5P gene (amplified by PCR) hybridized only to 4- and 5.2-kb DNA bands from strain Newman and mutant P15-1, respectively. This size difference reflects a 72-bp deletion in the cap5P gene of the mutant concomitant with a 1.3-kb ermB insertion. Despite insertional inactivation of cap5P, CP5 was still produced by the cap5P mutant P15-1. Quantitative analysis by ELISA inhibition and rocket immunoelectrophoresis revealed that there was no significant decrease in CP5 production by the mutant compared with that of the parental strain.
Identification of a second UDP-GlcNAc 2-epimerase in S. aureus.
Based on the phenotype of the cap5P mutant, we suspected that a second UDP-GlcNAc 2-epimerase existed in S. aureus that could complement the function of Cap5P in the CP5 biosynthetic pathway. A BLAST search of the incomplete S. aureus strain COL genome sequence database (4a) revealed a gene product with striking similarity to Cap5P. The 1,125-bp ORF that shall be referred to as mnaA (UDP-ManNAc pathway) encodes a putative protein of 375 amino acids that is 60.8% identical and 71.2% similar to the Cap5P amino acid sequence (Fig. 4). The S. aureus mnaA gene was amplified from strain Newman and was shown to complement ECA expression in an E. coli rffE mutant (data not shown). These data support the presence of a second UDP-GlcNAc 2-epimerase in S. aureus that is capable of maintaining CP5 expression in the absence of a functional cap5P gene.
FIG. 4.
Sequence alignment of S. aureus Cap5P and MnaA. The complete amino acid sequences of Cap5P (23) and MnaA (4a) were aligned with the BestFit program (Wisconsin Package, Genetics Computer Group). Identical and similar residues between the two sequences are connected by vertical lines and dots, respectively.
DISCUSSION
Although capsular polysaccharides are ubiquitous among S. aureus strains, little is known about their biosynthesis. CP5 and CP8 synthesis in S. aureus is directed by allelic chromosomal loci, designated cap5 and cap8, that contain common genes flanking a central serotype-specific region (capH through capK) (23). Among these genes, only cap5H has been functionally characterized; its translational product O acetylates the third carbon of ManNAcA in CP5 (2). Cap5P, the 3′-terminal gene product of the cap5 gene cluster, has 99.2% amino acid identity with S. aureus Cap8P and is 67% similar to RffE, a UDP-GlcNAc 2-epimerase from E. coli. As in the ECA biosynthetic pathway of E. coli, the biosynthesis of ManNAcA (a component of both CP5 and CP8) in S. aureus is likely initiated by the action of a UDP-GlcNAc 2-epimerase. This study constitutes the first report of an in vitro demonstration of enzyme activity from the capsular polysaccharide biosynthetic pathway of S. aureus.
S. aureus Cap5P converted UDP-GlcNAc to UDP-ManNAc with an efficiency of ∼10%. This conversion level is similar to those observed with E. coli and Bacillus cereus enzymes. RffE from E. coli showed reversible UDP-GlcNAc 2-epimerase activity, attaining an equilibrium ratio of 10:1 in favor of UDP-GlcNAc (21). Similarly, UDP-GlcNAc 2-epimerase in a crude enzyme preparation from B. cereus was shown to convert UDP-GlcNAc to UDP-ManNAc in a reversible fashion with an efficiency of 10% (6). Increasing the amount of Cap5P or UDP-GlcNAc in the assay mixture did not improve the efficiency of conversion, a result suggesting that the epimerization of UDP-GlcNAc to UDP-ManNAc in S. aureus is also reversible. The epimerization reaction may be driven towards the synthesis of UDP-ManNAc within the bacterial cell, since this is the substrate for UDP-ManNAc dehydrogenase, the putative product of the S. aureus cap5O gene (9).
The UDP-GlcNAc 2-epimerase encoded by the cap5P gene appears to catalyze an early and critical step in the CP5 biosynthetic pathway. We predicted that inactivation of cap5P would interrupt UDP-ManNAcA synthesis and abrogate CP5 expression. However, the S. aureus cap5P mutant still produced CP5 at levels similar to those produced by the parent strain, Newman. Similarly, Sau et al. showed that a mutation in cap8P did not eliminate CP8 production by S. aureus Becker (24). We have identified a gene (mnaA) located outside of the cap5 gene cluster that encodes a protein highly homologous to Cap5P and functionally complements an rffE mutation in E. coli. We hypothesize that the mnaA gene compensates for the cap5P mutation in Newman P15-1, allowing for CP5 expression. We were also able to amplify this gene from S. aureus Becker, which explains why the cap8P mutation does not abrogate CP8 expression (24).
The Bacillus subtilis genome also encodes a putative UDP-GlcNAc 2-epimerase, YvyH, that has 61.4% identity and 71.6% similarity with S. aureus MnaA, as well as homology with S. aureus Cap5P (Table 3). The yvyH gene is located within a region containing genes involved in teichoic acid biosynthesis and is essential for B. subtilis cell growth (26). ManNAc is a component of the linkage unit between glycerol teichoic acid and peptidoglycan in cell walls of B. subtilis (7), and cell lysates from B. subtilis show UDP-GlcNAc 2-epimerase activity (27). Thus, B. subtilis YvyH is likely involved in the synthesis of the ManNAc-containing linkage unit between peptidoglycan and glycerol teichoic acid. By analogy, we predict that the S. aureus mnaA gene product is involved in teichoic acid biosynthesis since the linkage unit between ribitol teichoic acid and peptidoglycan in the cell walls of S. aureus also contains ManNAc (10). However, the genes encoding teichoic acid biosynthesis in S. aureus have not yet been characterized, so the definitive role of mnaA remains to be elucidated. Completion of the S. aureus genome sequence may provide additional clues to the functional role of the mnaA gene.
TABLE 3.
Bacterial gene products with established or putative UDP-GlcNAc 2-epimerase activity
| Organism | Gene product | Size (aa)d | % Identity/similarity (aa stretch/gaps)a | Polysaccharide | Polysaccharide constituentsc | Demonstration of function (reference) |
|---|---|---|---|---|---|---|
| Staphylococcus aureus | Cap5P | 381 | 100/100 (381/0) | CP5 | ManNAcA, d-FucNAc, l-FucNAc | Complementation (9), in vitro enzymatic activity (this study) |
| Staphylococcus aureus | Cap8P | 381 | 99.2/99.2 (381/0) | CP8 | ManNAcA, d-FucNAc, l-FucNAc | NDe |
| Staphylococcus aureus | MnaA | 375 | 60.8/71.2 (375/0) | Teichoic acid linker? | ManNAc, GlcNAc | Complementation (this study) |
| Bacillus subtilis | YvyH/OrfXf | 380 | 59.0/70.0 (373/0) | Teichoic acid linker? | ManNAc, GlcNAc | ND |
| Streptococcus pneumoniae | Cps19fKb | 362 | 54.4/66.5 (357/1) | Type 19f capsule | ManNAc, glucose, l-rhamnose | Complementation (8) |
| Escherichia coli | RffE/NfrC | 376 | 51.9/60.8 (369/1) | ECA | ManNAcA, Fuc4NAc, GlcNAc | In vitro enzymatic activity (21) |
| Pseudomonas solanacearum | EpsC | 396 | 49.5/62.3 (374/1) | EPS I | GalNAc, GalNAcA, N-acetyl-2,4,6-trideoxygalactose | ND |
| Neisseria meningitidis | SacA | 372 | 48.7/61.8 (367/1) | Serogroup A capsule | ManNAc-1-P | ND |
| Salmonella enterica | WecB/RfbC | 378 | 47.9/59.9 (369/2) | LPS O:54 antigen | ManNAc | Complementation (18) |
| Pseudomonas aeruginosa | WbpI | 362 | 30.7/43.2 (328/4) | Serotype O5, band B LPS | 2,3-diNAcManA Man(2NAc3N)A FucNAc | ND |
| Bordatella pertussis | WlbD/BplD | 362 | 30.7/41.2 (365/6) | Band A LPS | 2,3-diNAcManA FucNAcMe GlcNAc | ND |
| Staphylococcus aureus | Cap5(8)G | 374 | 30.2/42.1 (367/9) | CP5(8) | ManNAcA, d-FucNAc, l-FucNAc | ND |
Sequences were aligned and compared with S. aureus Cap5P with the BestFit program (Genetics Computer Group).
Cps19fK is nearly identical to Cps19aK, Cps19bK, and Cps19cK.
2,3-diNAcManA, 2,3-dideoxy-2,3-di-N-acetyl-d-mannosaminuronic acid (2,3-diacetamido-d-mannuronic acid); Fuc4NAc, 4-acetamido-4,6-dideoxy-d-galactose; FucNAc, 2-acetamido-2,6-dideoxy-d-galactose (N-acetyl fucosamine); FucNAcMe, N-acetyl-N-methylfucosamine; GalNAc, N-acetyl-d-galactosamine; GalNAcA, N-acetyl-d-galactosaminuronic acid; Man(2NAc3N)A, 2-acetamido-3-acetamidino-2,3-dideoxy-d-mannuronic acid.
aa, amino acid.
ND, not determined (function based on sequence homology and putative biosynthetic pathway).
Alternative names for one gene product are separated by the slash (names given by different researchers or names that have been changed).
Functional redundancy of the capsule biosynthetic genes of S. aureus appears not to be restricted to cap5(8)P. Sau et al. showed that mutations in cap8A, cap8B, and cap8C did not abrogate CP8 production by strain Becker (24). The S. aureus COL genome (4a), which contains the full cap5A-P locus, also possesses a genetic cluster homologous to capABC physically separated from the cap5 genes on the chromosome. These findings suggest that multiple genes within the S. aureus cap loci may be functionally redundant. Sau et al. reported that only 8 of the 16 S. aureus cap8 genes are required for capsule biosynthesis (24).
Genes involved in the biosynthesis of bacterial lipopolysaccharides (LPS) and capsular polysaccharides have been cloned from scores of microbes. Sequence analysis of an individual gene within a biosynthetic cluster may reveal that the putative gene product has homology with gene products from other microorganisms. However, only a few of these putative proteins have been purified and characterized biochemically. Thus, there is a risk of attributing the wrong function to a gene on the basis of protein sequence homologies alone, since the functions of a vast majority of these genes have been determined in silico, i.e., by homology to other genes in the databases. For example, S. aureus Cap5(8)G shows homology to Cap5(8)P and E. coli RffE (23). However, cap5G was unable to complement an rffE mutant of E. coli lacking UDP-GlcNAc 2-epimerase activity (9). Although we cannot rule out the possibility that Cap5(8)G has UDP-GlcNAc 2-epimerase activity in S. aureus, we believe that it more likely serves as a UDP-GlcNAc 3-epimerase in the biosynthesis of UDP-l-N-acetylfucosamine, the donor of l-FucNAc residues in the putative pathway for CP5(8) biosynthesis (14).
A number of other bacterial gene products have been assigned UDP-GlcNAc 2-epimerase function based on sequence homology alone (Table 3). Other than the characterization of S. aureus Cap5P and MnaA performed by our group, only E. coli RffE, S. enterica WecB, and S. pneumoniae Cps19fK have been shown to possess UDP-GlcNAc 2-epimerase activity (8, 18, 21). The organisms listed in Table 3 encoding a putative UDP-GlcNAc 2-epimerase all produce a ManNAc(A)-containing polysaccharide except for Pseudomonas solanacearum. Exopolysaccharide I (EPS I) of P. solanacearum consists primarily of GalNAc and GalNAcA, which indicates that EpsC may have an alternative function or that EPS I does contain ManNAc(A) that has been misidentified as GalNAc(A). Pseudomonas aeruginosa WbpI and B. pertussis WlbD are putative UDP-GlcNAc 2-epimerases that are proposed to play a role in the biosynthesis of a mannosaminuronic acid variant (2,3-diNacManA) in their LPS; however, their homology to Cap5P (30.7%) is considerably lower than that of the other established and putative UDP-GlcNAc 2-epimerases (≥47.9%) and is more similar to that of Cap5(8)G (30.2%). Like S. aureus CP5(8), both P. aeruginosa and B. pertussis LPS contain FucNAc. Therefore, WbpI and WlbD may represent functional homologues of Cap5(8)G (putative UDP-GlcNAc 3-epimerase) rather than of Cap5(8)P (UDP-GlcNAc 2-epimerase). Biochemical characterization and genetic complementation should be performed to delineate these two enzymatic activities. Our studies have confirmed both genetically and biochemically that S. aureus cap5P encodes a UDP-GlcNAc 2-epimerase.
ACKNOWLEDGMENTS
Kevin B. Kiser and Navneet Bhasin made equal contributions to this paper.
This study was supported by Public Health Service grants AI 29040 (to J. C. Lee) and T32-AI 07410 and T32-AI 07061 from the National Institute of Allergy and Infectious Diseases.
We thank John S. Anderson for useful discussions and Derek Frederickson, Adam Reitz, and Jessica Lam for their technical assistance. Preliminary sequence data were obtained from The Institute of Genomic Research website (4a). Sequencing of S. aureus was performed at The Institute for Genomic Research, with support from the National Institute of Allergy and Infectious Diseases and the Merck Genome Research Institute.
REFERENCES
- 1.Albus A, Arbeit R D, Lee J C. Virulence of Staphylococcus aureus mutants altered in type 5 capsule production. Infect Immun. 1991;59:1008–1014. doi: 10.1128/iai.59.3.1008-1014.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhasin N, Albus A, Michon F, Livolsi P J, Park J-S, Lee J C. Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol Microbiol. 1998;27:9–21. doi: 10.1046/j.1365-2958.1998.00646.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Edman P. Method for determination of the amino acid sequence in peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- 4a.Institute of Genomic Research Website. 1999, copyright date. 7 April 1999, revision date. Sequences. [Online.] The Institute for Genomic Research. http://www.tigr.org. [5 July 1999, last date accessed.]
- 5.Kasatiya S S, Baldwin J N. Nature of the determinant of tetracycline resistance in Staphylococcus aureus. Can J Microbiol. 1967;13:1079–1086. doi: 10.1139/m67-144. [DOI] [PubMed] [Google Scholar]
- 6.Kawamura T, Kimura M, Yamamori S, Ito E. Enzymatic formation of uridine diphosphate N-acetyl-d-mannosamine. J Biol Chem. 1978;253:3595–3601. [PubMed] [Google Scholar]
- 7.Kaya S, Yokoyama K, Araki Y, Ito E. N-Acetylmannosaminyl (1→4) N-acetylglucosamine, a linkage unit between glycerol teichoic acid and peptidoglycan in cell walls of several Bacillus strains. J Bacteriol. 1984;158:990–996. doi: 10.1128/jb.158.3.990-996.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 9.Kiser K B, Lee J C. Staphylococcus aureus cap5O and cap5P genes functionally complement mutations affecting enterobacterial common-antigen biosynthesis in Escherichia coli. J Bacteriol. 1998;180:403–406. doi: 10.1128/jb.180.2.403-406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojima N, Araki Y, Ito E. Structure of linkage region between ribitol teichoic acid and peptidoglycan in cell walls of Staphylococcus aureus H. J Biol Chem. 1983;258:9043–9045. [PubMed] [Google Scholar]
- 11.Kuhn H-M, Meier-Dieter U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee J C. Electrotransformation of staphylococci. Methods Mol Biol. 1995;47:209–216. doi: 10.1385/0-89603-310-4:209. [DOI] [PubMed] [Google Scholar]
- 13.Lee J C, Betley M J, Hopkins C A, Perez N E, Pier G B. Virulence studies, in mice, of transposon-induced mutants of Staphylococcus aureus differing in capsule size. J Infect Dis. 1987;156:741–750. doi: 10.1093/infdis/156.5.741. [DOI] [PubMed] [Google Scholar]
- 14.Lee J C, Lee C Y. Capsular polysaccharides of Staphylococcus aureus. In: Goldberg J B, editor. Genetics of bacterial polysaccharides. Boca Raton, Fla: CRC Press, Inc.; 1999. pp. 185–205. [Google Scholar]
- 15.Lee J C, Takeda S, Livolsi P J, Paoletti L C. Effects of in vitro and in vivo growth conditions on expression of type-8 capsular polysaccharide by Staphylococcus aureus. Infect Immun. 1993;61:1853–1858. doi: 10.1128/iai.61.5.1853-1858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macrina F L, Evans R P, Tobian J A, Hartley D L, Clewell D B, Jones K R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 17.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 18.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 19.O’Connell C, Pattee P A, Foster T J. Sequence and mapping of the aroA gene of Staphylococcus aureus 8325-4. J Gen Microbiol. 1993;139:1449–1460. doi: 10.1099/00221287-139-7-1449. [DOI] [PubMed] [Google Scholar]
- 20.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sala R F, Morgan P M, Tanner M E. Enzymatic formation and release of a stable glycal intermediate: the mechanism of the reaction catalyzed by UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc. 1996;118:3033–3034. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 24.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawardeker J S, Sloneker J H, Jeanes A. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal Chem. 1965;37:1602–1604. [Google Scholar]
- 26.Soldo B, Lazarevic V, Margot P, Karamata D. Sequencing and analysis of the divergon comprising gtaB, the structural gene of UDP-glucose pyrophosphorylase of Bacillus subtilis 168. J Gen Microbiol. 1993;139:3185–3195. doi: 10.1099/00221287-139-12-3185. [DOI] [PubMed] [Google Scholar]
- 27.Yoneyama T, Koike Y, Arakawa H, Yokoyama K, Sasaki Y, Kawamura T, Araki Y, Ito E, Takao S. Distribution of mannosamine and mannosaminuronic acid among cell walls of Bacillus species. J Bacteriol. 1982;149:15–21. doi: 10.1128/jb.149.1.15-21.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]