Abstract
Rhodobacter sphaeroides is a photosynthetic bacterium which swims by rotating a single flagellum in one direction, periodically stopping, and reorienting during these stops. Free-swimming R. sphaeroides was examined by both differential interference contrast (DIC) microscopy, which allows the flagella of swimming cells to be seen in vivo, and tracking microscopy, which tracks swimming patterns in three dimensions. DIC microscopy showed that when rotation stopped, the helical flagellum relaxed into a high-amplitude, short-wavelength coiled form, confirming previous observations. However, DIC microscopy also revealed that the coiled filament could rotate slowly, reorienting the cell before a transition back to the functional helix. The time taken to reform a functional helix depended on the rate of rotation of the helix and the length of the filament. In addition to these coiled and helical forms, a third conformation was observed: a rapidly rotating, apparently straight form. This form took shape from the cell body out and was seen to form directly from flagella that were initially in either the coiled or the helical conformation. This form was always significantly longer than the coiled or helical form from which it was derived. The resolution of DIC microscopy made it impossible to identify whether this form was genuinely in a straight conformation or was a low-amplitude, long-wavelength helix. Examination of the three-dimensional swimming pattern showed that R. sphaeroides changed speed while swimming, sometimes doubling the swimming speed between stops. The rate of acceleration out of stops was also variable. The transformations in waveform are assumed to be torsionally driven and may be related to the changes in speed measured in free-swimming cells. The roles of and mechanisms that may be involved in the transformations of filament conformations and changes in swimming speed are discussed.
Bacteria swim by using the electrochemical ion gradient (usually the proton motive force [Δp]) across the cytoplasmic membrane to drive flagellar rotation. To change swimming direction, flagellar rotation can either stop or switch between counterclockwise (CCW) and clockwise (CW), depending on the species. A change in switching frequency biases the overall swimming direction of motile bacteria to an optimum environment for growth; for reviews, see references 1 to 3, 5, 15, and 20. Peritrichous species such as Escherichia coli swim smoothly when the majority of flagella are rotating CCW. The flagella rotate together as a bundle, which pushes the cell forward at about 20 μm s−1. Periodically, the flagella switch to rotate CW, and the bundle flies apart. This activity reorients the cell for its next period of smooth swimming. Rhodobacter sphaeroides, on the other hand, rotates its flagellum only CW and changes swimming direction by stopping that rotation, although the driving force, Δp, remains constant (4). The frequency of stopping was found to increase when the cells were moving down a gradient (16). High-intensity dark-field microscopy showed that a normal flagellar helix was formed during periods of swimming, but when the cells stopped, the filament relaxed into a short-wavelength, high-amplitude helix which coiled against the cell body. The helix reformed from the cell body out, and the cell usually swam off in a new direction. The “glare” around the cell body resulting from the high light intensity made it impossible to identify what was happening to the coiled form next to the cell (4).
The frequency of direction changing, which results from switching or stopping motor rotation, is controlled by environmental stimuli signalling through a phosphorelay system. Sensory receptors control the activity of a cytoplasmic histidine protein kinase, CheA, which in turn controls a small response regulator, CheY. Phosphorylated CheY can bind to the motor and cause switching. Interestingly, while the flagella of E. coli are controlled by one CheY protein, four CheY proteins control a single, unidirectional motor in R. sphaeroides. In addition, unlike enteric species, R. sphaeroides changes swimming speed independently of changes in Δp (10, 18). Any changes in E. coli flagellar rotation could be dampened by the flagella rotating together, while R. sphaeroides swimming reflects the behavior of a single flagellar motor (17).
In this study, we used video-enhanced differential interference contrast (DIC) microscopy to examine any changes that might occur in the structure of the R. sphaeroides flagellum during free swimming and direction changing. In addition, we tracked the swimming pattern of unstimulated cells by using a microscope which can accurately track behavior and swimming in three dimensions (6, 8). We found that R. sphaeroides does change speed while swimming. Using DIC microscopy, we found that the coiled form of the flagellum can rotate and probably contributes to direction changing. In addition, we also identified a new flagellar waveform which appears to be a rapidly rotating straight or low-amplitude conformation.
MATERIALS AND METHODS
Strains and growth conditions.
R. sphaeroides WS8 (a gift from W. Sistrom) was grown anaerobically in succinate medium under tungsten illumination at room temperature as previously described (14). Cells were either harvested and resuspended in sodium HEPES buffer (20 mM, pH 7.2) or examined directly in growth medium. Ficoll (2%) was added to increase viscosity and decrease the rate of rotation of the flagellum for DIC microscopy. E. coli AW405, a wild-type strain, was grown in tryptone broth to the mid-exponential phase, harvested, and resuspended at 107 per ml in 20 mM potassium phosphate buffer (pH 7.0). All measurements involving E. coli were made at 30°C, while all measurements for R. sphaeroides were made at room temperature.
Video-enhanced DIC microscopy.
Video-enhanced DIC microscopy was performed essentially by the method of Block et al. (9). Flagella were detected with a Nikon Diaphot microscope fitted with a ×100 DIC 1.4-numerical aperture (NA) objective and a 1.4-NA oil immersion condenser. The microscope was illuminated with a 100-W short-arc Hg lamp (Oriel, Stratford, Conn.). The light was passed through a water filter (to remove infrared light) and a 475-nm, long-pass filter (Shott Glass Technologies, Duryea, Pa.) before it was coupled to the microscope via a 0.48-NA multimode optical fiber (GeneralFiber Optics, Fairfield, N.J.). Light emitted from the fiber was collected by a ×20, 0.5-NA, 160-mm-tube-length DIC objective followed by a relay lens (100-mm focal length). The relay lens focused the back aperture of the objective onto the field iris, and the lens pair focused the end of the fiber onto the back focal plane of the condenser, the image of the fiber just filling that aperture. The rest of the optical train was standard Nikon configuration. Images were collected with a Nevicon video camera (Hamamatsu Corp., Tokyo, Japan). The background was subtracted, and two frames were averaged with an Argus-20 image processor (Hamamatsu). The data were stored on Hi-8 videotape.
Still images from the tape were digitized with a Raptor video board (Bitflow Inc., Woburn, Mass.). Images were deinterlaced and contrast stretched with Photoshop (Adobe Systems Inc., San Jose, Calif.).
Three-dimensional tracking.
A tracking microscope was used to obtain data on the swimming pattern of unstimulated R. sphaeroides, and these data were compared to the swimming pattern of E. coli. The microscope tracks individual bacteria as they move in three dimensions (6, 7). Growing, motile R. sphaeroides cells were diluted in growth medium to 107 cells per ml. This concentration was used to reduce the problems of cells visually interfering with each other during tracking (13). The bacteria were placed in a tracking chamber on the microscope, and individual cells were tracked in bulk solution, far from the surface. Cells remained motile at room temperature for several hours. E. coli cells were tracked similarly, except that the temperature of the stage was increased to 30°C.
The position of the bacterium being tracked was sampled every 1/12 s, and these data were collected and analyzed by use of a Macintosh Power PC 8100 running LabView 3.1 (National Instruments) with a 16-bit A-to-D converter (National Instruments NB-MIO-16XH). The data were analyzed to obtain the instantaneous velocity of a bacterium at any given point by use of the method of Berg and Brown (8). The velocities were then used to calculate the following: the average speed of each bacterium for the period during which it was tracked; the average speed of the population; the standard deviations of the average speed of the population; the maximum speed of each bacterium; the number of times each bacterium stopped or tumbled; the average bacterial run time; and the rate of acceleration out of a stop or during swimming.
A cell was considered to have “stopped” when its measured velocity fell below 10 μm s−1, a speed which correlated well with the actual behavior examined by eye and which was the speed measured for cells subjected only to Brownian motion. All cells were examined by eye to confirm that events considered stops did not look like slow translational movement. Bacterial accelerations were calculated by determining the average speed of a given bacterium and the standard deviation of that speed to give a measure of the extent to which speed changed during swimming. Any measured change in velocity greater than or equal to one standard deviation of the speed occurring over one time step was considered a “significant” acceleration. The accelerations were described with two categories: “stop to go,” in which the bacterium accelerated out of a stop, and “go to go,” in which the bacterium was measured as changing speed while already swimming.
RESULTS
Three-dimensional tracking.
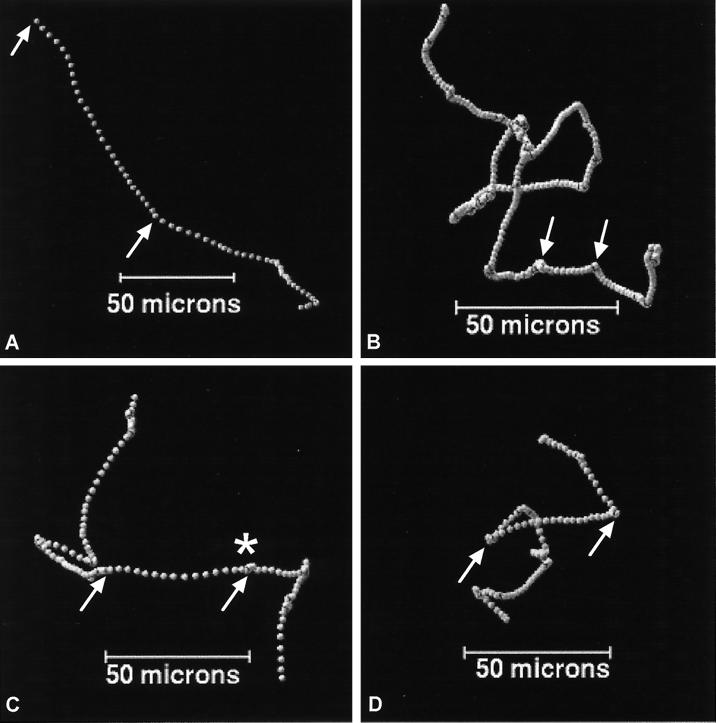
Figure 1 shows representative tracks from three different cells of R. sphaeroides and an E. coli cell. The direction in which each cell is swimming is indicated in the legend to Fig. 1. Each point represents 1/12 s; therefore, the closer the points are together when oriented within a two-dimensional plane, the slower the cell speed. Increases in speed are shown by increased distance between the points. In all cases, the images are printed so that the run between the two arrows is oriented in the plane of the paper. Away from this section, the cells are swimming in three dimensions, and it is difficult to decouple the changes in speed from the effects of projection onto the plane. The average swimming speed of all R. sphaeroides cells was 27 μm s−1; the average swimming speed measured for E. coli was 22 μm s−1. Both species showed a random three-dimensional swimming pattern; periods of smooth swimming in E. coli were interrupted by tumbles, and those in R. sphaeroides were interrupted by stops. The difference in the swimming behavior of the three R. sphaeroides cells is obvious. The cell in Fig. 1A swam very rapidly, with few changes in speed or direction, whereas the cell in Fig. 1B swam very slowly, stopped frequently, and changed direction during periods of slow swimming. The cell in Fig. 1A was tracked for 5 s before being lost, while that in Fig. 1B was tracked for well over 20 s. The cell in Fig. 1C was swimming parallel to the plane between the arrows and could be seen to accelerate out of a stop.
FIG. 1.
Three-dimensional swimming patterns of three individual R. sphaeroides cells (A, B, and C) and one E. coli cell (D). Between the arrows, the plots are oriented so that the cell is swimming parallel to the plane of view. Each spot represents 1/12 s. The closer together the spots, the slower the cell speed. The cells in panels A and D were swimming top to bottom, while those in panels B and C were swimming bottom to top. The asterisk marks a stop followed by acceleration out of that stop.
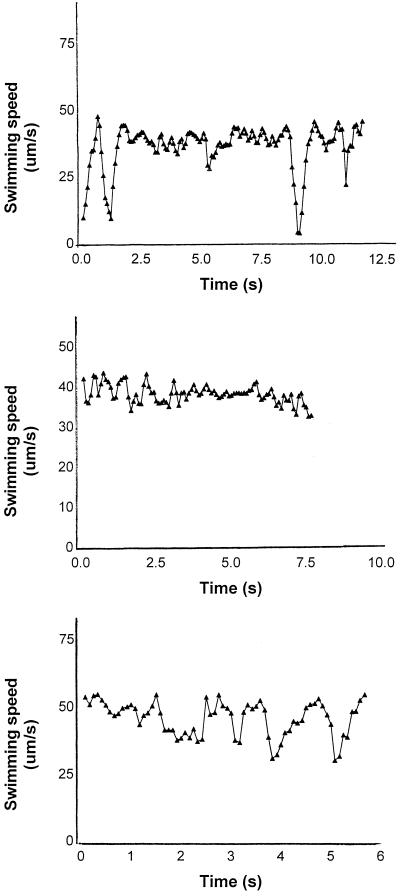
The speed of individual R. sphaeroides cells also could be seen to change between stops. This finding is shown more clearly in Fig. 2, where the swimming speeds of three different bacteria from the same population are plotted. Bacterium 1 showed a steady swimming speed of about 45 μm s−1 with short stops at 1.5 and 9 s. There was a brief reduction in speed at about 5 s. Bacterium 2 continued to swim at a steady average of about 38 μm s−1 throughout the 8 s tracked, but bacterium 3 showed a great variation in speed, between about 30 and 50 μm s−1.
FIG. 2.
Change in swimming speed of three individual R. sphaeroides cells tracked in three dimensions. When the speed falls below 10 μm s−1, the cells have probably stopped, and the measured speed probably represents Brownian motion, although in some cases there may be slow movement. The time scales are different, as the cells were tracked for different periods of time before being lost from tracking.
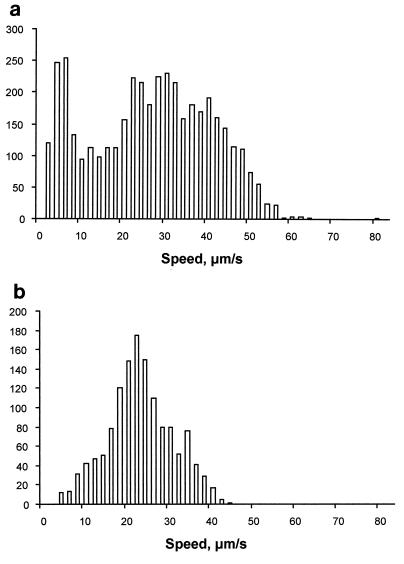
The average swimming speed within a run is plotted in Fig. 3 and shows that the speed of the individual cells within the population varied greatly. Below about 10 μm s−1, the cells were probably stopped, but there was slow nontranslational movement, probably as a result of both slow flagellar rotation and Brownian motion. Because the cells were incubated in high light and growth medium, the Δp of these photosynthetically grown cells remained at its maximum level, but there was still a great variation in the swimming speed measured between stops. The average velocity was 27.3 ± 10.4 μm s−1, and the average maximum speed was 46.3 ± 9.5 μm s−1 (the maximum speed measured was 62.3 μm s−1). In contrast, E. coli swam at an average velocity of 22.6 ± 3.5 μm s−1 and had an average maximum speed of 35.0 ± 5.4 μm s−1 (Table 1).
FIG. 3.
Distribution of swimming speed during periods of smooth swimming between stops, calculated from 32 tracked R. sphaeroides cells (a) and 11 tracked E. coli cells (b). Below 10 μm s−1, R. sphaeroides cells were considered stopped. The number of the y axis refers to the number of periods of smooth swimming, and the speed on the x axis is the average speed during a period of smooth swimming.
TABLE 1.
Average swimming behavior calculated for 32 individual R. sphaeroides cells and 11 individual E. coli cells by three-dimensional trackinga
| Cells | Avg
|
|||||
|---|---|---|---|---|---|---|
| Tracked time(s) | Number of stops | Time of stop(s) | Time of run between stop(s) | Velocity (μm s−1) | Maximum speed (μm s−1) | |
| R. sphaeroides | 10.7 ± 6.0 | 4.5 ± 4.8 | 0.34 ± 0.32 | 3.1 ± 2.2 | 27.3 ± 10.4 | 46.3 ± 9.5 |
| E. coli | 10.3 ± 5.5 | 4.7 ± 3.3 | 0.16 ± 0.06 | 3.2 ± 2.6 | 22.6 ± 3.5 | 35.0 ± 5.4 |
The data include standard deviations to show the greater variation in behavior shown by R. sphaeroides than by E. coli. The average time of a stop is the total time stopped divided by the number of stops for those cells.
The stopping frequency of individual cells varied greatly, with some cells stopping every second and others showing no stops over several minutes. The average number of stops in an average measuring period of 11 s was 4.5 ± 4.8. The time that cells remained stopped also varied between a fraction of a second and several seconds; the average time of a stop was 0.34 ± 0.32 s. In comparison, an average E. coli tumble lasted 0.16 ± 0.06 s (Table 1). In 137 measured periods, R. sphaeroides was found to stop and subsequently change direction 116 times, change direction without a measurable stop 19 times, and stop but not change direction 2 times. The mean turn angle was 76° ± 41°.
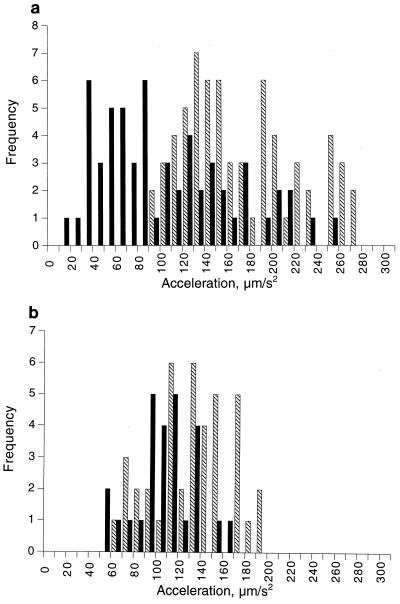
Fig. 1C and 2 show the rate at which a cell accelerates out of a stop. Each point is 1/12 s; in Fig. 1C, it takes about 0.3 s to reach a maximum swimming speed. The rates of acceleration from stop to go and from one speed to another (labelled go to go) during swims were calculated. The average rate of acceleration from stop to swim was about 175 ± 78 μm s−2, but accelerations ranged from 15 to a maximum of 520 μm s−2 (Fig. 4). Some cells therefore reach their initial swimming speed as rapidly as E. coli, which reaches 25 μm s−2 very rapidly after a tumble, usually within one time point (Fig. 4b), but others show distinctly slow rates of acceleration up to swimming speed.
FIG. 4.
Distribution of acceleration rates of 32 R. sphaeroides cells tracked from stop to swim (▧) and speed changes while free swimming (■) (a) and 11 tracked E. coli cells (b) (symbols are as in panel a). The frequency with which the individual acceleration rates were measured (as described in Materials and Methods) is shown on the y axis. Data show 95% confidence limits. The averages (mean ± standard deviation) of the behaviors measured over the entire histogram were as follows: ▧, 178 ± 18 and 122 ± 10 μm s−2 for R. sphaeroides and E. coli, respectively; ■, 95 ± 15 and 99 ± 10 μm s−2 for R. sphaeroides and E. coli, respectively.
Figure 4 plots the rates of acceleration for cells from a stop to smooth swimming and also within periods of smooth swimming. The rates are plotted as the frequency with which cells accelerate to beyond one standard deviation of the starting speed. Greater accelerations were seen for cells coming out of a stop than for cells already swimming. During a run, many cells accelerated to over one standard deviation of the initial swimming speed, an average of about 99 μm s−2, but accelerations out of stops averaged about 122 μm s−2. The ranges of speed changes and acceleration rates were seen not only between cells but also within the swimming patterns of individual cells. In contrast, in E. coli speed changes showed a much narrower range and the frequency with which cells changed speed while swimming was much lower.
DIC microscopy.
Video-enhanced DIC microscopy is able to reveal individual flagella in vivo. Ficoll was added to the medium to increase its viscosity and thus slow the rotation of the flagella; otherwise, rapidly rotating flagella would not be resolvable.
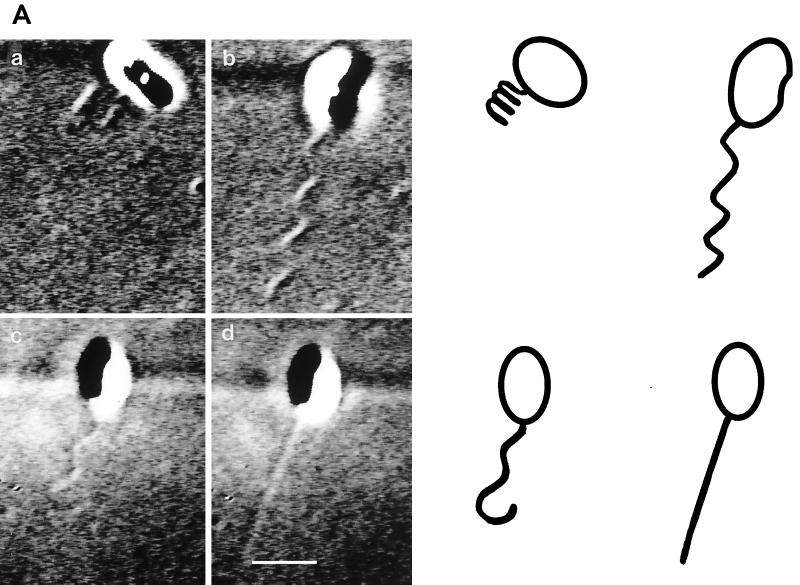
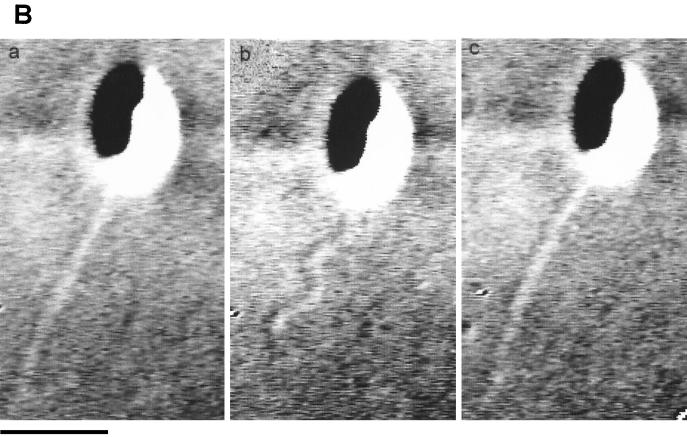
Figure 5 shows the changes in conformation seen for the R. sphaeroides flagellum. Figure 5A shows various flagellar conformations: a coiled form (a), a functional helix (b), a helix relaxing into a coil (c), and an apparently straight conformation (d). Figure 5B shows a straight filament forming a helix (b) and reforming the straight conformation (c). On occasion, coiled flagella were seen to rotate slowly before reforming a functional helix from the cell body out. Figure 5C shows the slow reforming of a coiled conformation to a functional helix; the shadow on the cell body shows the change in orientation during this period. The slow rotation reoriented the cell during the stop, so that the next period of swimming was in a different direction. The time taken for the transformation from the coiled to the helical waveform varied from about three frames to several seconds and was dependent on the rate of motor rotation and the length of the filament, which varied between 1 and 6 μm. The time taken also might have been longer because the medium contained Ficoll.
FIG. 5.
DIC images of the flagellar filament of R. sphaeroides. (A) Various flagellar conformations: a coiled form (a), a functional helix (b), a helix relaxing into a coil (c), and an apparently straight conformation (d). The cartoon shows the likely cell size and the flagellar shape. (B) Filament changing between straight and helical conformations. (C) Sequential formation of a functional helix from a coiled filament. The two images at the top show the coiled form rotating. Bars, 1 μm.
Frequently, the flagellum was seen to transform into an apparently straight form (Fig. 5A, panel d, and 5B, panels a and c). This transformation could occur within one video frame, and the flagellum could reform into a helix within one frame. The filament could transform directly from a coil to a straight form or from a helix to a straight form. The transformation to the straight form happened from the motor out, and the filament could be seen to rotate very rapidly, as evidenced by the movement of the filament in the medium. The straight form of the filament was clearly longer than the helical form of the filament (Fig. 5B), eliminating the possibility that it was a rapidly rotating, short-wavelength, low-amplitude conformation. When the normal helix reformed from the straight conformation, it did so rapidly from the distal end of the filament. The change from coiled to helical form or coiled to straight form happened from the cell body out, while the formation of the helix from the straight form or the helix from the coiled form happened from the distal end of the filament. The slow rotation of the coiled flagellum (upper two panels in Fig. 5C) and the reformation of a helix from a coiled flagellum could be seen clearly.
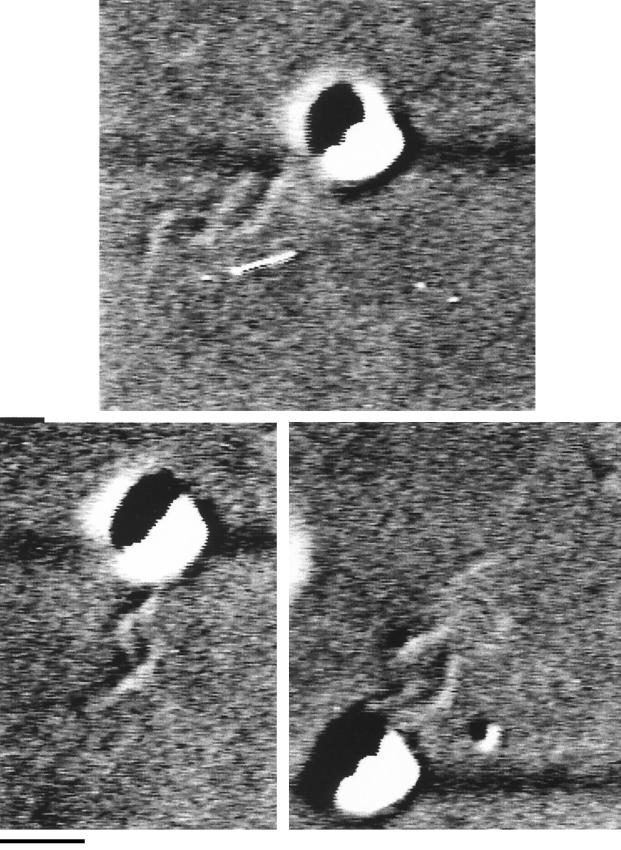
Several cells which were predivisional and had two flagella were seen. The flagella clearly rotated independently and were never seen to form the equivalent of a bundle; i.e., they never rotated in phase (Fig. 6). Periodically, one or the other filament would stop rotating and would coil against the cell body, while the other filament continued rotating and pushing the cell forward.
FIG. 6.
DIC images of a dividing cell with two independently rotating flagellar filaments. The two flagella rotated independently, even when close together. Bar, 1 μm.
DISCUSSION
The data presented here suggest that R. sphaeroides can change swimming speed under unstimulated conditions, possibly as the result of a change in either motor rotation rate or flagellar conformation. Increases in swimming speed were measured when free-swimming R. sphaeroides was given some metabolizable chemoattractants, and this sustained increase was independent of changes in Δp (18). Tethered R. sphaeroides cells have also been found to show a much greater variation in rotation rate than tethered enteric cells under nonstimulated conditions (17). Recent work with a quadrant photodiode, sampling at a rate much higher than that of video analysis, has suggested that the bias of the tethered R. sphaeroides motor, i.e., the stopping frequency, is significantly higher than the switching frequency of E. coli. This finding may indicate the occurrence of unresolved very short stops, which may affect both measured swimming speed and flagellar conformation (8a). Tethered cells are under much higher frictional loads than free-swimming cells. Changes in swimming speed may therefore reflect short stops or transient transformations in flagellar conformation.
Within a population, there was great variation in the behavior of individual cells and changes in the patterns of behavior of single cells during a period of swimming. There were also great variations in measured stopping frequency and length of stops. When swimming rapidly, the cells tended to swim a straighter course than when swimming slowly, when the cells swam in more obvious curves. Bacteria have been shown to swim in very curved paths when swimming close to surfaces (13), but the bacteria being tracked here were well away from the surface. There were also occasions when cells appeared to change direction after slowing down rather than after stopping. Changes in swimming speed (which may be the manifestation of a series of extremely short stops or flagellar transformation), as well as measurable stopping, may therefore be involved in behavioral responses. Recent data have shown that the related bacterium Sinorhizobium meliloti may change direction by altering the speed of individual flagella within a bundle (5, 19).
It had been assumed that Brownian motion caused the reorientation of R. sphaeroides during a stop, but calculations suggested that, in the relatively viscous environment in which the bacteria live, Brownian motion would be a slow and inefficient mechanism for redirecting swimming given the short average time of a stop. The data from the DIC microscopy showed that the coiled form of the flagellum did result from a stop in rotation but that, once coiled against the cell body, the flagellum often slowly rotated before the helix reformed from the cell body out. The Δp was still maximal under these conditions, and the reduced rate of rotation was presumably the result of the increased viscous drag on the high-amplitude helix. The slow rotation of the flagellum may also explain the movement of the cells measured during a stop.
The rapid rotation of the flagellar motor caused the helix to reform from the cell body out, producing a functional helix. It might be expected that, as thrust can be equated with the helical shape of the filament, the speed at which a cell swims might relate to the extent of functional helix reformation. Therefore, a long filament would take longer to form a functional helix, and the rate of acceleration from a stop to a smooth swimming speed would be slower than that measured for a cell with a short filament. The wide range of acceleration rates from stop to swim may therefore partly reflect the difference in time taken to go from the coiled form to the functional helix conformation. With DIC microscopy, a long flagellum was seen to take longer to reform a complete helix than a short filament, and this finding would be expected to affect speed. The R. sphaeroides flagellum probably grows continuously, unlike, for example, the Caulobacter crescentus single flagellum (11). Individual R. sphaeroides cells can have flagella between 1 and 9 μm in length (21).
On several occasions, cells that were clearly dividing and had two flagella were seen. These flagella were seen to rotate independently, sometimes one stopping while the other continued to rotate. They were not seen to rotate together as any type of bundle, even when apparently very close. This finding suggests that either a specific hook structure is required to form a bundle or a critical number of rotating filaments is needed.
A third waveform—a very rapidly rotating straight form—was apparent with DIC microscopy. It is tempting to speculate that the transformation among coiled, helical, and straight forms is involved in the change in swimming speed seen in the tracking experiments. The resolution of the DIC microscope is limited to about 200 nm as a result of the optical “blooms” caused by the techniques. If the filament formed a shallow waveform with less than a 200-nm amplitude, it would appear straight. We could not determine, therefore, whether the filament was really straight or was a low-amplitude helix. However, the very obvious increase in length compared to the length of the helical form showed that there had been a major change in conformation, and it was not to a short-wavelength, low-amplitude helix, such as that which forms during the normal-to-curly transformations of E. coli flagella when the motor switches rotational direction.
Why and how does this conformation form, and what effect does it have on the pattern of swimming? Mutations in the flgL gene of E. coli, coding for the hook-associated protein HAP3, which links the flexible hook and the more rigid filaments (12), result in flagella which go through the conformational changes seen in free-swimming R. sphaeroides. These so-called sag mutants show normal swimming behavior in liquid medium but poor swarming ability in soft agar, probably because the filaments are forced into a straight conformation by the viscous drag in agar. The transformations seen in sag mutants were similar to those seen in R. sphaeroides, with torsion-induced changes probably occurring with changes in torque, the straight conformation forming from the cell body out and reforming a helix from the distal end. A straight filament would naturally rotate faster than a helical form because of the reduction in drag; however, it would also produce less driving force. It was suggested that HAP3 is responsible for maintaining the E. coli flagellar filaments in a helical form by preventing torsion-induced transformations. It was also argued that the torque produced in E. coli would allow the formation of straight filaments but that this process is prevented by HAP3, which allows the hook subunits to move but restricts the movement of the flagellin subunits. Nothing is known about the HAP proteins from R. sphaeroides, but these data suggest that there may be less control over conformational changes in R. sphaeroides flagellar filaments than in those of enteric species. A change in rotation rate or transient stops may cause a switch in filament conformation in free-swimming cells, and this switch would cause changes in swimming speed. It may be worth noting that R. sphaeroides does not swarm quite as well as E. coli in soft agar, but it does swarm much better than sag mutants. R. sphaeroides has four different CheY proteins. In E. coli, the single CheY protein is responsible for flagellar switching. The reason for multiple copies in a monoflagellate species with a unidirectional motor is unknown, but it is tempting to speculate that one or more of these proteins may have a role in transient changes in torque and thus flagellar transformation and swimming speed.
ACKNOWLEDGMENTS
J.P.A. thanks the Rowland Institute of Science and Howard Berg for use of their facilities and Howard Berg and Richard Berry for useful discussion of the data.
J.P.A. is supported by the BBSRC, and H.L.P. is supported by NERC. T.P.P. is supported by the Rowland Institute of Science, and M.A.-S.V. is supported by IBM and NSF through the PIRCH program sponsorship.
REFERENCES
- 1.Adler J, Dahl L. A method for measuring the motility of bacteria and comparing random and non-random motility. J Gen Microbiol. 1967;46:161–173. doi: 10.1099/00221287-46-2-161. [DOI] [PubMed] [Google Scholar]
- 2.Amsler C D, Matsumura P. Chemotactic signal transduction in Escherichia coli and Salmonella typhimurium. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 89–103. [Google Scholar]
- 3.Armitage J P. Bacterial motility and chemotaxis. Sci Prog. 1992;76:451–477. [PubMed] [Google Scholar]
- 4.Armitage J P, Macnab R M. Unidirectional intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol. 1987;169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armitage J P, Schmitt R. Bacterial chemotaxis: variations on a theme: Rhodobacter sphaeroides and Sinorhizobium meliloti. Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 6.Berg H C. How to track bacteria. Rev Sci Instrum. 1972;42:868–871. doi: 10.1063/1.1685246. [DOI] [PubMed] [Google Scholar]
- 7.Berg H C. The tracking microscope. Adv Opt Electron Microsc. 1978;7:1–13. [Google Scholar]
- 8.Berg H C, Brown D A. Chemotaxis in Escherichia coli analyzed by three dimensional tracking. Antibiot Chemother. 1974;19:55–78. doi: 10.1159/000395424. [DOI] [PubMed] [Google Scholar]
- 8a.Berry, R. M., and J. P. Armitage. Unpublished data.
- 9.Block S M, Fahrner K A, Berg H C. Visualization of bacterial flagella by video-enhanced light microscopy. J Bacteriol. 1991;173:933–936. doi: 10.1128/jb.173.2.933-936.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown S, Poole P S, Jeziorska W, Armitage J P. Chemokinesis in Rhodobacter sphaeroides is the result of a long term increase in the rate of flagellar rotation. Biochim Biophys Acta Bioenerg. 1993;1141:309–312. [Google Scholar]
- 11.Dingwall A, Shapiro L, Ely B. Flagellar biosynthesis in Caulobacter. In: Armitage J P, Lackie J M, editors. Biology of the chemotactic response. Cambridge, United Kingdom: Cambridge University Press; 1990. pp. 155–176. [Google Scholar]
- 12.Fahrner K A, Block S M, Krishnaswamy S, Parkinson J S, Berg H C. A mutant hook-associated protein (HAP3) facilitates torsionally induced transformations of the flagellar filament of Escherichia coli. J Mol Biol. 1994;238:173–186. doi: 10.1006/jmbi.1994.1279. [DOI] [PubMed] [Google Scholar]
- 13.Frymier P D, Ford R M, Berg H C, Cummings P T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc Natl Acad Sci USA. 1995;92:6195–6199. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauden D E, Armitage J P. Electron transport-dependent taxis in Rhodobacter sphaeroides. J Bacteriol. 1995;177:5853–5859. doi: 10.1128/jb.177.20.5853-5859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones C J, Aizawa S. The bacterial flagellum and flagellar motor: structure, assembly and function. Adv Microb Physiol. 1991;32:109–172. doi: 10.1016/s0065-2911(08)60007-7. [DOI] [PubMed] [Google Scholar]
- 16.Packer H L, Gauden D E, Armitage J P. The behavioral response of anaerobic Rhodobacter sphaeroides to temporal stimuli. Microbiology. 1996;142:593–599. doi: 10.1099/13500872-142-3-593. [DOI] [PubMed] [Google Scholar]
- 17.Packer H L, Lawther H, Armitage J P. The Rhodobacter sphaeroides flagellar motor is a variable speed motor. FEBS Lett. 1997;409:37–40. doi: 10.1016/s0014-5793(97)00473-0. [DOI] [PubMed] [Google Scholar]
- 18.Poole P S, Brown S, Armitage J P. Swimming changes and chemotactic responses in Rhodobacter sphaeroides do not involve changes in the steady state membrane potential or respiratory electron transport. Arch Microbiol. 1990;153:614–618. [Google Scholar]
- 19.Sourjik V, Schmitt R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol. 1996;22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 20.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 21.Takahashi, N., and J. P. Armitage. Unpublished data.