Abstract
Background
Quantitative structure–activity relationship (QSAR) modeling is one of the most important computational tools employed in drug discovery and development. The external validation of QSAR models is the main point to check the reliability of developed models for the prediction activity of not yet synthesized compounds. It was performed by different criteria in the literature.
Methods
In this study, 44 reported QSAR models for biologically active compounds reported in scientific papers were collected. Various statistical parameters of external validation of a QSAR model were calculated, and the results were discussed.
Results
The findings revealed that employing the coefficient of determination (r2) alone could not indicate the validity of a QSAR model. The established criteria for external validation have some advantages and disadvantages which should be considered in QSAR studies.
Conclusion
This study showed that these methods alone are not only enough to indicate the validity/invalidity of a QSAR model.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13065-022-00856-4.
Keywords: Biological activity, External validation, QSAR, Statistical parameters
Introduction
Quantitative structure–activity relationship (QSAR) is a numerical method for finding the relationships between chemical structure and drug properties i.e., biological activity in drug discovery processes [1]. Developing a QSAR model composed of different stages i.e., (1) collecting data from the literature, (2) calculation of parameters performed by different software packages such as Dragon software or image analysis (2D-QSAR), force field calculations based on three-dimensional structures (3D-QSAR) and etc., (3) developing the QSAR model by various statistical technique e.g. multiple linear regression, artificial neural network and partial least square, and (4) validation of the model by internal (leave one out and leave many out) and external validation [2]. There are various critical points in QSAR studies that should be considered by researchers [3]. However, the challenges on selecting appropriate parameters for external validation have been seen in the literature [4, 5].
In QSAR studies, training a model by linear and non-linear models is not enough to confirm the prediction capability. The developed model should be applied to other data sets which did not synthesize in virtual screening and designing new drug compounds. On the way, whenever we can say a QSAR model is acceptable that it could predict the activity of other compounds with reasonable accuracy. Therefore, external validation (splitting data into training and test sets) is one of the major challenges in QSAR studies [6–8]. Various types of cross validation analysis i.e., leave one out, leave many out and repeated double cross validation are recommended in QSAR studies especially when the available sample size is small [9, 10]. However, external validation is one of the most common criteria for evaluating the validity of a QSAR model [11–13].
Different criteria and rules were proposed for evaluating the validity of the QSAR models, which most of them focused on the external validation [13, 14]. Five criteria proposed in authentic journals were selected in this study and details have been described in method section. They are highly cited and several researchers were used them to evaluate validity of QSAR models [15–18]. Designers of each criterion have been shown advantages of them in comparison with others for external validation of QSAR models [5, 6, 19–21]. Some models have certain defects from the statistical viewpoint and various results are observed based on the applied software e.g. the correlation coefficient (r2) of regression through origin [5]. Nevertheless, there is no comprehensive comparison between them for the evaluation of the external validity of QSAR models. The aim of this study is the comparison of external validation of QSAR models by them to find advantages and disadvantages of each method.
Methods
Forty-four data sets (training and test sets) composed of experimental biological activity and corresponding calculated activity (re-substitution value for training data set) using QSAR models with various statistical approaches were collected from the published articles [22–48] indexed in Scopus database (see Additional file 1 and Table 1). The absolute error (AE) of each datum (absolute difference between experimental and calculated data) was calculated. External validation of these data set was assessed with the following methods:
Table 1.
The numerical values of statistical parameters which need to calculate mentioned criteria for external validation for 44 developed QSAR models
| No. | Number of compounds in training set | Number of compounds in test set | r2 > 0.6 | (Eq. 3) | (Eq. 4) | (Eq. 5) | AEE ± SD Training set |
AEE ± SD Test set |
Training set range | Refs. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 39 | 10 | 0.917 | 0.909 | 0.917 | 0.999 | 0.161 ± 0.114 | 0.221 ± 0.110 | 4.07 | [23] |
| 2 | 39 | 10 | 0.880 | 0.879 | 0.857 | 0.999 | 0.237 ± 0.234 | 0.318 ± 0.150 | 4.07 | [23] |
| 3 | 31 | 10 | 0.715 | 0.715 | 0.617 | 0.997 | 0.167 ± 0.171 | 0.266 ± 0.244 | 1.72 | [24] |
| 4 | 26 | 11 | 0.725 | 0.310 | 0.691 | 0.997 | 0.233 ± 0.167 | 0.354 ± 0.301 | 2.74 | [25] |
| 5 | 40 | 10 | 0.906 | 0.904 | 0.904 | 0.999 | 0.121 ± 0.091 | 0.206 ± 0.095 | 2.68 | [26] |
| 6 | 40 | 10 | 0.892 | 0.879 | 0.892 | 0.999 | 0.122 ± 0.087 | 0.195 ± 0.146 | 2.68 | [26] |
| 7 | 68 | 17 | 0.261 | 0.012 | 0.052 | 0.957 | 0.503 ± 0.435 | 1.165 ± 0.715 | 5.00 | [27] |
| 8 | 68 | 17 | 0.444 | 0.220 | 0.404 | 0.977 | 0.331 ± 0.674 | 0.435 ± 0.326 | 4.60 | [27] |
| 9 | 42 | 11 | 0.834 | 0.823 | 0.818 | 0.824 | 0.872 ± 0.678 | 1.630 ± 1.256 | 14.46 | [28] |
| 10 | 42 | 9 | 0.588 | 0.552 | 0.511 | 0.999 | 0.040 ± 0.035 | 0.169 ± 0.124 | 1.85 | [29] |
| 11 | 42 | 9 | 0.748 | 0.496 | 0.730 | 0.999 | 0.053 ± 0.043 | 0.133 ± 0.077 | 1.85 | [29] |
| 12 | 20 | 6 | 0.963 | 0.962 | 0.983 | 0.787 | 0.052 ± 0.043 | 0.035 ± 0.035 | 0.91 | [30] |
| 13 | 90 | 22 | 0.372 | 0.376 | -0.292 | 0.950 | 0.432 ± 0.648 | 0.538 ± 0.647 | 6.95 | [31] |
| 14 | 68 | 17 | 0.382 | 0.136 | 0.309 | 0.975 | 0.364 ± 0.324 | 0.457 ± 0.356 | 4.90 | [31] |
| 15 | 27 | 5 | 0.088 | − 2.263 | − 1.129 | 0.995 | 0.074 ± 0.094 | 0.315 ± 0.135 | 0.40 | [32] |
| 16 | 27 | 7 | 0.818 | − 1.721 | 0.563 | 0.993 | 0.412 ± 0.352 | 0.645 ± 0.489 | 3.76 | [33] |
| 17 | 27 | 7 | 0.763 | − 4.030 | 0.462 | 0.992 | 0.359 ± 0.290 | 0.729 ± 0.511 | 3.76 | [33] |
| 18 | 89 | 19 | 0.932 | 0.932 | 0.928 | 0.998 | 0.187 ± 0.151 | 0.246 ± 0.204 | 3.95 | [34] |
| 19 | 89 | 19 | 0.821 | 0.813 | 0.811 | 0.995 | 0.255 ± 0.186 | 0.339 ± 0.368 | 3.95 | [34] |
| 20 | 66 | 16 | 0.703 | 0.514 | 0.914 | 0.984 | 0.444 ± 0.338 | 0.678 ± 0.411 | 5.45 | [35] |
| 21 | 66 | 16 | 0.671 | 0.475 | 0.700 | 0.983 | 0.384 ± 0.324 | 0.706 ± 0.461 | 5.45 | [35] |
| 22 | 66 | 16 | 0.914 | 0.908 | 0.670 | 0.995 | 0.288 ± 0.232 | 0.297 ± 0.307 | 5.45 | [35] |
| 23 | 32 | 11 | 0.790 | 0.006 | 0.683 | 0.993 | 0.120 ± 0.094 | 0.501 ± 0.249 | 4.68 | [47] |
| 24 | 40 | 12 | 0.876 | 0.875 | 0.845 | 0.999 | 0.090 ± 0.079 | 0.238 ± 0.088 | 3.35 | [36] |
| 25 | 40 | 12 | 0.866 | 0.814 | 0.861 | 0.999 | 0.079 ± 0.084 | 0.205 ± 0.140 | 3.35 | [36] |
| 26 | 63 | 16 | 0.999 | 0.999 | 0.999 | 1.000 | 0.011 ± 0.006 | 0.011 ± 0.006 | 3.76 | [37] |
| 27 | 40 | 4 | 0.960 | 0.693 | 0.863 | 1.000 | 0.155 ± 0.118 | 0.178 ± 0.105 | 4.25 | [38] |
| 28 | 22 | 7 | 0.995 | 0.995 | 0.995 | 1.000 | 0.043 ± 0.064 | 0.046 ± 0.032 | 2.56 | [39] |
| 29 | 22 | 7 | 0.971 | 0.971 | 0.971 | 0.999 | 0.101 ± 0.127 | 0.097 ± 0.097 | 2.56 | [39] |
| 30 | 50 | 18 | 0.914 | 0.796 | 0.879 | 1.000 | 0.041 ± 0.038 | 0.068 ± 0.134 | 2.35 | [40] |
| 31 | 50 | 18 | 0.994 | 0.993 | 0.992 | 1.000 | 0.031 ± 0.028 | 0.026 ± 0.028 | 2.35 | [40] |
| 32 | 52 | 12 | 0.815 | 0.686 | 0.801 | 0.983 | 0.340 ± 0.269 | 0.297 ± 0.261 | 3.32 | [41] |
| 33 | 58 | 6 | 0.964 | 0.949 | 0.958 | 1.000 | 0.051 ± 0.048 | 0.127 ± 0.117 | 2.90 | [42] |
| 34 | 58 | 6 | 0.966 | 0.965 | 0.962 | 1.000 | 0.066 ± 0.052 | 0.105 ± 0.076 | 2.90 | [42] |
| 35 | 47 | 16 | 0.899 | 0.878 | 0.898 | 0.999 | 0.195 ± 0.117 | 0.186 ± 0.153 | 2.16 | [43] |
| 36 | 52 | 20 | 0.533 | 0.367 | 0.511 | 0.995 | 0.566 ± 0.378 | 0.383 ± 0.314 | 4.28 | [44] |
| 37 | 52 | 20 | 0.659 | 0.533 | 0.657 | 0.997 | 0.554 ± 0.521 | 0.327 ± 0.230 | 4.28 | [44] |
| 38 | 52 | 20 | 0.744 | 0.714 | 0.733 | 0.998 | 0.355 ± 0.343 | 0.282 ± 0.213 | 4.28 | [44] |
| 39 | 52 | 20 | 0.815 | 0.785 | 0.814 | 0.998 | 0.290 ± 0.358 | 0.246 ± 0.181 | 4.28 | [44] |
| 40 | 31 | 10 | 0.658 | 0.475 | 0.658 | 0.995 | 0.097 ± 0.064 | 0.272 ± 0.202 | 2.17 | [45] |
| 41 | 68 | 8 | 0.898 | 0.865 | 0.935 | 0.999 | 0.125 ± 0.110 | 0.204 ± 0.151 | 4.03 | [46] |
| 42 | 68 | 8 | 0.855 | 0.702 | 0.828 | 0.998 | 0.199 ± 0.115 | 0.270 ± 0.148 | 4.03 | [46] |
| 43 | 53 | 18 | 0.806 | 0.678 | 0.795 | 0.996 | 0.122 ± 0.118 | 0.279 ± 0.203 | 3.78 | [48] |
| 44 | 53 | 18 | 0.676 | 0.109 | 0.640 | 0.993 | 0.329 ± 0.271 | 0.362 ± 0.276 | 3.78 | [48] |
Proposed criteria by Golbraikh and Tropsha
I. r2 > 0:6, r2 is the coefficient of determination between the experimental activity and predicted values based on regression analysis.
II. 0.85 < K < 1.15 or 0.85 < K' < 1.15.
K and K' are slopes of regression lines through the origin between the experimental activity and predicted, and vice versa, respectively.
III.
r0.2 and is the coefficient of determination between the experimental activity and predicted values and predicted versus experimental activity, respectively, based on regression through origin analysis (linear regression by least square method without a constant term) [19].
Proposed criteria by Roy based on regression through origin (RTO)
Roy and coworkers suggested which calculated by Eq. 1, and it is one of the most famous equations which used by QSAR experts in literature [20, 49]:
| 1 |
In this equation, value computed using regression through origin (RTO) and RTO referred to linear regression by least square method without a constant term.
Concordance correlation coefficient (CCC)
Gramatica and coworker [4] suggested the concordance correlation coefficient (CCC) for external validation of a QSAR model:
| 2 |
Yi is the experimental value, is the average of experimental values, is the predicted value of activity and is the average of the predicted value of the activity. EXT is external prediction set or test set. CCC > 0.8 accounts as a valid model.
Statistical significant between deviation of experimental activity and calculated data
In 2014, our research group challenged the regression through origin and proposed the calculation of model errors for training and test sets and comparison of them as a reliable method to external validation of QSAR models [5].
Criteria based on training set range and the deviation between experimental and calculated data
Roy and coworkers [21] similar to our method (method 4) proposed new principles based on training set range and absolute average error (AAE) i.e., the difference between experimental and the predicted values of test set, and corresponding standard deviation (SD) for training and test sets as follows:
Good prediction: AAE ≤ 0.1 × training set range and AAE + 3 × SD ≤ 0.2 × traning set range
Bad prediction: AAE > 0.15 × training set range or AAE + 3 × SD > 0.25 × traning set range
A good model should be passed both above criteria. However, the predictions which fall into one of the conditions could be considered as of moderately acceptable model.
Results and discussion
Table 1 listed the numerical values of statistical parameters that need to calculate the mentioned criteria for external validation of 44 developed QSAR models.
The main factor in the validation of QSAR models from a statistical point is different equations even to calculate simple parameters such as r2 and r02 [22, 50]. These different equations will affect the comparison. The r2 in this work was calculated by SPSS software based correlation between experimental and calculated values. However, in the studied criteria in this work, there is a controversy in the calculation of r02. The following equations were applied to the calculation of r02 and in method 1, 2 and Excel software [21]
| 3 |
| 4 |
Instead, the alternative formula was proposed instead of the Eqs. 3 and 4 because of statistical defects to the calculation of r2 of RTO [5, 22] which recommended by statistical books in the literature [51, 52]:
| 5 |
In addition to statistical defects in Eq. (3) and (4) for the calculation of r02 and r0′2, QSAR researchers, may apply Eq. (5) which proposed as an appropriate equation for r02 and officinal statistical package such as SPSS, and do not give reasonable results. Calculation of based on computed by Eq. (5) (or SPSS software) is not possible because of r2 is commonly less than and therefore . This is the most defect of methods 1 and 2 for the external validation of QSAR models.
Seven of the studied models have r2 < 0.6 (Table 2). Therefore, they could not account as valid models. r2 is simple parameter to evaluate the correlation between experimental and predicted values in QSAR studies and for estimating the correlation between concentration and response in analytical chemistry. It is a primary criterion, and a QSAR model or a developed analytical method with a high r2 value does not necessarily have an acceptable validity [53, 54]. In addition, the squared factors e.g. r2, negatively affects the possibility to distinguish errors in one or in another direction: overpredicted or underpredicted values; these two kinds of errors have a huge different in toxicity and regulatory evaluation.
Table 2.
Values of the proposed criteria (method 1–5) for external validation of QSAR models
| Model | Method 1 | Method 2 | Method 3 | Method 4 | Method 5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 > 0.6 | 0.85 < K or K´ < 1.15 | > 0.5 | CCC > 0.8 | p-value | Ib | IIc | IIId | IVe | Vf | VIg | ||||
| 1 | 0.917 | 1.00 | 1.00 | 0.010 | 0.000 | 0.83 | 0.95 | 0.14 | 0.55 | 0.41 | 0.81 | 0.61 | 1.02 | G |
| 2 | 0.880 | 1.01 | 0.98 | 0.000 | 0.025 | 0.86 | 0.92 | 0.31 | 0.77 | 0.41 | 0.81 | 0.61 | 1.02 | G |
| 3 | 0.715 | 1.00 | 1.00 | 0.000 | 0.138 | 0.71 | 0.84 | 0.18 | 1.02 | 0.17 | 0.34 | 0.26 | 0.43 | B |
| 4 | 0.725 | 0.98 | 1.02 | 0.573 | 0.047 | 0.26 | 0.77 | 0.23 | 1.26 | 0.27 | 0.55 | 0.41 | 0.69 | B |
| 5 | 0.906 | 1.00 | 1.00 | 0.002 | 0.003 | 0.86 | 0.95 | 0.01 | 0.49 | 0.27 | 0.54 | 0.40 | 0.67 | G |
| 6 | 0.892 | 1.00 | 1.00 | 0.015 | 0.000 | 0.79 | 0.94 | 0.16 | 0.63 | 0.27 | 0.54 | 0.40 | 0.67 | M |
| 7 | 0.261 | 0.98 | 0.98 | 0.956 | 0.800 | 0.13 | 0.51 | < 0.01 | 3.31 | 0.50 | 1.00 | 0.75 | 1.25 | B |
| 8 | 0.444 | 0.97 | 1.01 | 0.506 | 0.090 | 0.23 | 0.66 | 0.543 | 1.41 | 0.46 | 0.92 | 0.69 | 1.15 | B |
| 9 | 0.834 | 0.74 | 1.11 | 0.014 | 0.020 | 0.75 | 0.89 | 0.08 | 5.40 | 1.35 | 2.70 | 2.02 | 3.37 | B |
| 10 | 0.588 | 1.02 | 0.98 | 0.062 | 0.131 | 0.48 | 0.68 | 0.01 | 0.54 | 0.19 | 0.37 | 0.28 | 0.46 | B |
| 11 | 0.748 | 0.98 | 1.02 | 0.336 | 0.024 | 0.37 | 0.75 | 0.01 | 0.36 | 0.19 | 0.37 | 0.28 | 0.46 | G |
| 12 | 0.963 | 1.05 | 0.92 | 0.001 | − 0.021 | 0.93 | 0.97 | 0.41 | 0.14 | 0.09 | 0.18 | 0.14 | 0.23 | G |
| 13 | 0.372 | 1.00 | 0.95 | − 0.012 | 1.786 | NDa | 0.57 | 0.49 | 2.48 | 0.70 | 1.40 | 1.05 | 1.74 | B |
| 14 | 0.382 | 1.01 | 0.97 | 0.644 | 0.189 | 0.19 | 0.61 | 0.30 | 1.53 | 0.49 | 0.98 | 0.74 | 1.23 | B |
| 15 | 0.088 | 1.02 | 0.97 | 26.745 | 13.844 | − 0.05 | − 0.25 | < 0.01 | 0.72 | 0.04 | 0.08 | 0.06 | 0.10 | B |
| 16 | 0.818 | 1.05 | 0.95 | 3.105 | 0.312 | − 0.49 | 0.55 | 0.16 | 2.11 | 0.38 | 0.75 | 0.56 | 0.94 | B |
| 17 | 0.763 | 1.05 | 0.94 | 6.282 | 0.394 | − 0.91 | 0.43 | 0.02 | 2.26 | 0.38 | 0.75 | 0.56 | 0.94 | B |
| 18 | 0.932 | 1.01 | 0.99 | 0.000 | 0.004 | 0.92 | 0.96 | 0.14 | 0.80 | 0.40 | 0.79 | 0.59 | 0.99 | M |
| 19 | 0.821 | 1.01 | 0.98 | 0.009 | 0.012 | 0.75 | 0.90 | 0.34 | 1.44 | 0.40 | 0.79 | 0.59 | 0.99 | B |
| 20 | 0.703 | 0.97 | 1.01 | 0.270 | − 0.300 | 0.40 | 0.81 | 0.02 | 1.91 | 0.55 | 1.09 | 0.82 | 1.36 | B |
| 21 | 0.671 | 0.96 | 1.02 | 0.292 | − 0.044 | 0.37 | 0.79 | 0.02 | 2.09 | 0.55 | 1.09 | 0.82 | 1.36 | B |
| 22 | 0.914 | 0.99 | 1.03 | 0.007 | 0.268 | 0.84 | 0.95 | 0.90 | 1.22 | 0.55 | 1.09 | 0.82 | 1.36 | M |
| 23 | 0.790 | 0.91 | 1.09 | 0.992 | 0.136 | 0.09 | 0.60 | < 0.01 | 1.25 | 0.47 | 0.94 | 0.70 | 1.17 | B |
| 24 | 0.876 | 1.00 | 1.00 | 0.002 | 0.035 | 0.84 | 0.93 | < 0.01 | 0.50 | 0.34 | 0.67 | 0.50 | 0.84 | G |
| 25 | 0.866 | 0.99 | 1.01 | 0.059 | 0.005 | 0.67 | 0.92 | 0.01 | 0.62 | 0.34 | 0.67 | 0.50 | 0.84 | G |
| 26 | 0.999 | 1.00 | 1.00 | 0.000 | 0.000 | 1.00 | 1.00 | 0.65 | 0.03 | 0.38 | 0.75 | 0.58 | 0.94 | G |
| 27 | 0.960 | 0.98 | 1.02 | 0.278 | 0.101 | 0.46 | 0.83 | 0.72 | 0.03 | 0.43 | 0.85 | 0.64 | 1.06 | G |
| 28 | 0.995 | 1.00 | 1.00 | 0.000 | 0.000 | 0.99 | 1.00 | 0.90 | 0.14 | 0.26 | 0.51 | 0.38 | 0.64 | G |
| 29 | 0.971 | 1.00 | 1.00 | 0.000 | 0.000 | 0.96 | 0.99 | 0.93 | 0.39 | 0.26 | 0.51 | 0.38 | 0.64 | G |
| 30 | 0.914 | 1.00 | 1.00 | 0.129 | 0.038 | 0.60 | 0.93 | 0.42 | 0.47 | 0.24 | 0.47 | 0.35 | 0.59 | M |
| 31 | 0.994 | 1.00 | 1.00 | 0.002 | 0.002 | 0.96 | 1.00 | 0.51 | 0.11 | 0.24 | 0.47 | 0.35 | 0.59 | G |
| 32 | 0.815 | 1.03 | 0.95 | 0.158 | 0.017 | 0.52 | 0.87 | 0.61 | 1.09 | 0.33 | 0.66 | 0.50 | 0.83 | B |
| 33 | 0.964 | 1.01 | 0.99 | 0.016 | 0.006 | 0.85 | 0.96 | 0.18 | 0.48 | 0.29 | 0.58 | 0.44 | 0.73 | G |
| 34 | 0.966 | 1.00 | 1.00 | 0.001 | 0.004 | 0.94 | 0.98 | 0.10 | 0.33 | 0.29 | 0.58 | 0.44 | 0.73 | G |
| 35 | 0.899 | 1.02 | 0.98 | 0.023 | 0.001 | 0.77 | 0.91 | 0.81 | 0.64 | 0.22 | 0.43 | 0.28 | 0.54 | B |
| 36 | 0.533 | 1.01 | 0.98 | 0.311 | 0.041 | 0.32 | 0.71 | 0.06 | 1.33 | 0.43 | 0.86 | 0.64 | 1.07 | B |
| 37 | 0.659 | 1.00 | 1.00 | 0.191 | 0.003 | 0.43 | 0.80 | 0.07 | 1.02 | 0.43 | 0.86 | 0.64 | 1.07 | M |
| 38 | 0.744 | 1.00 | 1.00 | 0.040 | 0.014 | 0.62 | 0.86 | 0.38 | 0.92 | 0.43 | 0.86 | 0.64 | 1.07 | M |
| 39 | 0.815 | 1.01 | 0.99 | 0.037 | 0.001 | 0.67 | 0.89 | 0.50 | 0.79 | 0.43 | 0.86 | 0.64 | 1.07 | G |
| 40 | 0.658 | 0.97 | 1.03 | 0.278 | 0.000 | 0.38 | 0.77 | 0.02 | 0.88 | 0.22 | 0.43 | 0.33 | 0.54 | B |
| 41 | 0.898 | 0.99 | 1.01 | 0.037 | − 0.041 | 0.73 | 0.94 | 0.03 | 0.66 | 0.40 | 0.81 | 0.60 | 1.01 | G |
| 42 | 0.855 | 1.00 | 1.00 | 0.179 | 0.032 | 0.52 | 0.89 | 0.06 | 0.71 | 0.40 | 0.81 | 0.60 | 1.01 | G |
| 43 | 0.806 | 1.00 | 0.99 | 0.159 | 0.014 | 0.52 | 0.87 | 0.01 | 0.89 | 0.38 | 0.76 | 0.55 | 0.95 | M |
| 44 | 0.676 | 0.99 | 1.00 | 0.838 | 0.053 | 0.17 | 0.74 | 0.66 | 1.19 | 0.38 | 0.76 | 0.55 | 0.95 | B |
a
bAAE + 3 × SD
c0.1 × training set range
d0.2 × training set range
e0.15 × training set range
f0.25 × training set range
G good, MG moderately good, B Bad
The numerical values of other proposed criteria in method 1 show that all models have K or K' between 0.85 and 1.15. The third rule () is only non-acceptable for 7 models which 3 of them have r2 < 0.6. Therefore, based on the suggested principles in method 1, 11 models are not valid.
Method 2 proposed based on RTO and r02 calculated by Eq. (3). Twenty-six models have > 0.5, and the results are similar to method 1 (both of them are based on RTO). The valid models based on method 1 with r2 > 0.75 have > 0.5 except model 27 with r02 = 0.101 (close to threshold, 0.1).
The third studied method was proposed by Gramatica and named CCC [4]. Twenty-nine models have CCC > 0.8. All of them are valid models based on method 1. The results of methods 2 and 3 are very similar. Two models (20 and 27) only have CCC > 0.8 while the defined values near to threshold i.e., 0.4 < < 0.5. Method 3 is comparable to developed methods based on RTO. However, it has not statistical defects and non-identical datum for r02 based on proposed equations (Eq. (3) and (4) or Eq. (5)) or software (e.g. Excel or SPSS).
Method 4 is based on the calculation of model errors for training and test sets and compares them as a possible reliable method to external validation for models with r2 > 0.6 for test set. The aim of developing a QSAR model is the prediction and elucidation of mechanisms of drug action. It is obvious that the prediction capability of training and test sets should be identical. Without considering the training set, it possible statistical parameters for external validation of test set could be acceptable but a significant difference (independent t-test) between prediction power of training and test set might be a weakness for the model. Twenty-six models have r2 > 0.6 and no significant difference between absolute error (AE) of training and test sets (p > 0.05). Twenty-three models of them have been selected by CCC as a valid model (CCC > 0.8 and p > 0.05). Model 16 has a CCC = 0.55, and AAE of training and test sets are 0.412 ± 0.352 and 0.645 ± 0.489 (p = 0.16), respectively. High values for SD because of outlier data, is the possible reason for non-significant difference between AEs and it could not account validity of the developed model. On the other hand, models 5, 24 and 25 have CCC > 0.9 and p < 0.01. The relative frequencies of AEs for models 5, 24 and 25 sorted in three subgroups, < 0.1, 0.1–0.2 and > 0.2 and illustrated in Figure 1. In these models, AAE values are low; however, there is 50–250% difference between AAE of training and test sets. On the other hand, in model 5, 48% of the training set and 10% of test sets have AE less than 0.1 while 15% of the training set and 60% of test set have AE more than 0.2. Similar patterns are observed in models 24 and 25. In addition, for those models, residual plots have been illustrated in Figure 2. These plots confirm that there is a significant difference between the prediction capability of developed models for training and test sets and it could not be acceptable for a QSAR model to approve prediction capability.
Fig. 1.
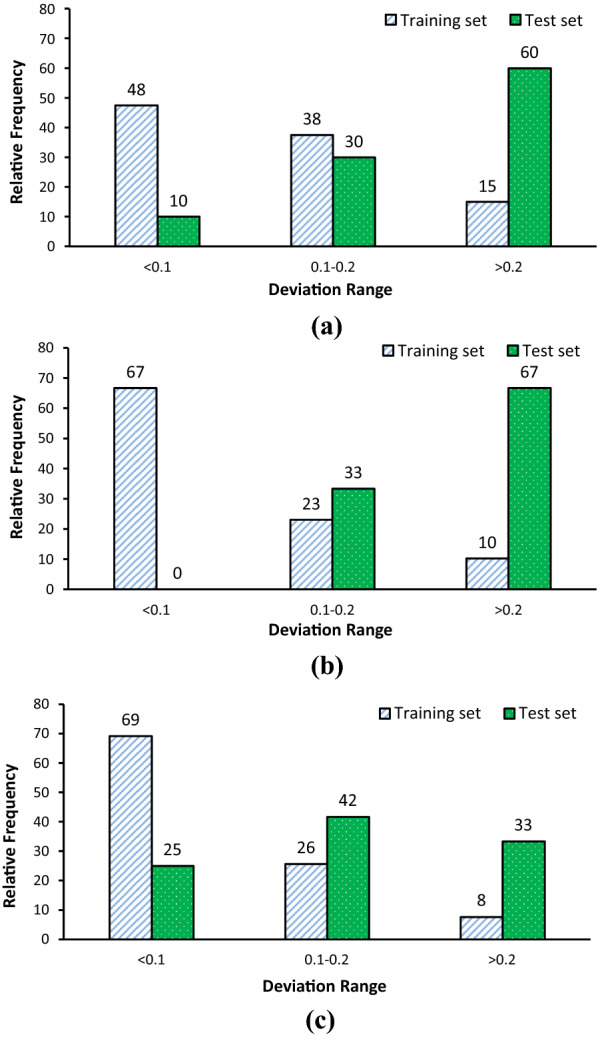
Relative frequency of individual deviation (absolute error) for model 5 (a), model 24 (b) and model 25 (c)
Fig. 2.
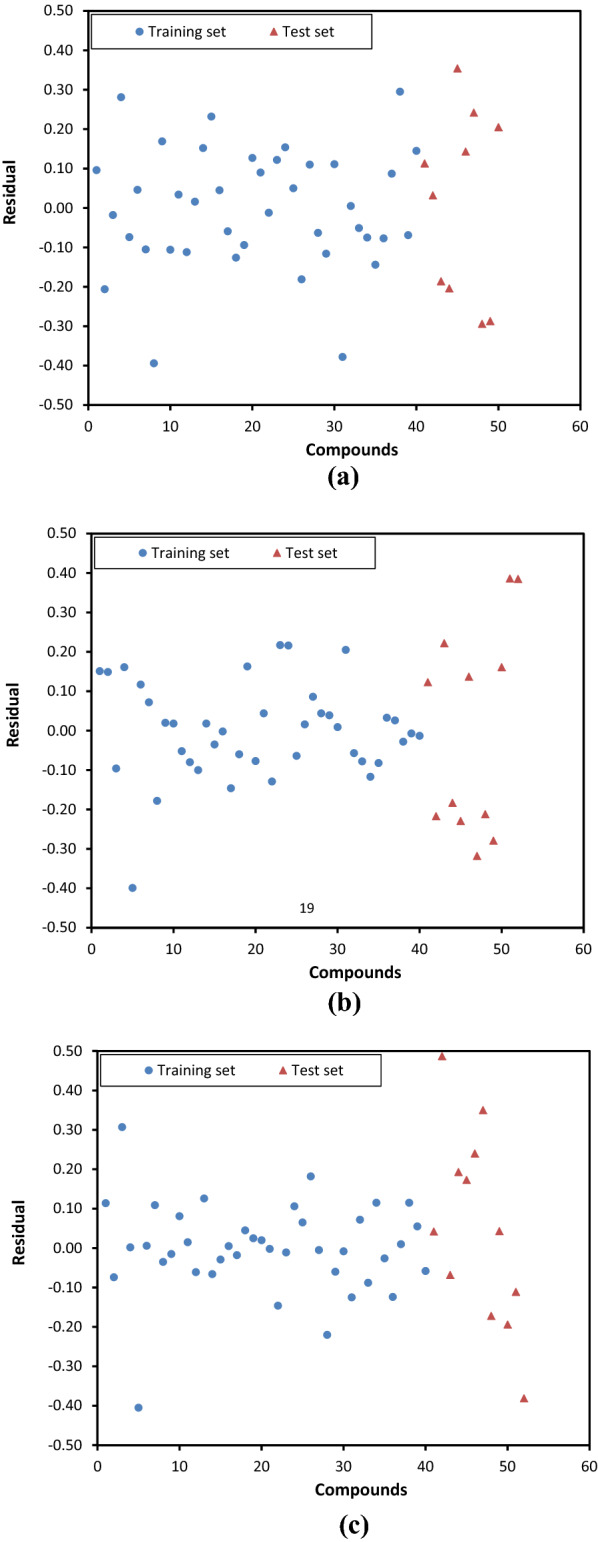
Residual plots for model 5 (a), model 24 (b) and model 25 (c)
The last method (method 5) proposed by Roy’s research group based on the training set range and mean and standard deviation of test set data [21]. The models could be classified as GOOD, MODERATELY GOOD and BAD according to their proposed parameters. Most of the models were categorized as BAD (45%) and GOOD (39%) and a few models were MODERATELY GOOD models (Table 2). The first point that should be considered is r2 > 0.6 as a necessary criterion. All models which have r2 < 0.6 classified as BAD model. Moreover, a good correlation is observed between CCC and GOOD model based on method 5. However, model 11 is a GOOD model while CCC = 0.75 and there is a significant difference between AE of training and test set (AAE of training and test sets are 0.05 and 0.13, respectively and p = 0.01). In comparison with method 4, models 5, 24 and 25 (GOOD models) have a vast difference between AAE of training and test set (Figure 1), although the proposed principles in method 5 could not detect it. A model with a statistically significant difference between the AE of training and test sets might not confirm developing a valid model.
Furthermore, model 3 is a BAD model while CCC = 0.84 and p-value for the difference between AE of training and test is 0.18. AAE of the training set is 0.167 ± 0.171 and 0.266 ± 0.244 (AE ± SD), respectively. High values for SD of training and test sets indicate that there are outlier data which could be considered using statistical parameters e.g. SD of mean errors, in the external validation of QSAR models.
Typographic errors and un-uniformity of applied data set for QSAR modeling or mistake in the determination of the biological activity of studied compounds are a common reason for outlier data, which can decrease the prediction capability of a model. Docking study of outlier cases and comparison with other compounds can help researchers to detect outlier data in developing a QSAR model [55].
These results confirm the results of previous studies which more than a single criterion is recommended to assess the real external predictivity of QSAR models [56]. Moreover, other recommended guidelines in developing QSAR models such as cross validation, appropriate splitting training and test sets variable allocation and correlation coefficients adjusted by degrees of freedom, are other important issues in QSAR studies which should be considered by researchers [10, 57–59]. In addition, cross (internal) validation analysis e.g., leave many out and leave one out are recommended in QSAR studies especially when the sample size is small [9, 10], and some reports showed its superiority in external validation [60]. Therefore, both internal and external validation analysis with considering various criteria are necessary to check the validity of a QSAR model.
Conclusion
The aim of developing a QSAR model is an acceptable prediction of activity of a compound before synthesis and biological evaluation. Therefore, external validation is necessary. All of the developed methods for external validation of a QSAR model are useful and a good correlation was observed between the studied methods for the selected models. However, some differences were detected between established methods. Methods 1 and 2 are valuable but they are some questionable points in the applied equation for calculation. CCC is a valuable parameter, though in some cases, it cannot detect outlier data. Similar to methods 1 and 2, training data set are not included in CCC. Method 4 and 5 established based on training and test sets. They detected most invalid models, but method 5 considered some model as a GOOD model while the difference between AE of training and test sets are substantial (p < 0.05). On the other way, high SD value in both of training and test sets may pass proposed criterion of method 4 while accounted as a invalid model because of outlier data in training and test sets. Finally, evaluation of a model with either established method is useful, but they did not necessarily mean validity/invalidity of a QSAR model. The results of this study show the importance of calculation error of training and test sets and detection of outliers for checking the validity of a model.
Supplementary Information
Additional file 1. Forty-four data sets (training and test sets) composed of experimental biological activity and corresponding calculated activity.
Acknowledgements
Not applicable.
Abbreviations
- QSAR
Quantitative structure–activity relationship
- AE
Absolute error
- RTO
Regression through origin
- CCC
Concordance correlation coefficient
- AAE
Absolute average error
- SD
Standard deviation
Author contributions
SS and AS performed data collecting, analysis and manuscript writing. All authors read and approved the final manuscript.
Funding
The authors would like to thanks from Tabriz University of Medical Sciences for the financial support (65369) of the project.
Availability of data and materials
All data is available as supplementary.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Norouzi S, Farahani M, Nejad Ebrahimi S. The Integration of pharmacophore-based 3D-QSAR modeling and virtual screening in identification of natural product inhibitors against SARS-CoV-2. Pharm Sci. 2021;27:S94–S108. [Google Scholar]
- 2.Dearden JC. Whither QSAR? Pharm Sci. 2017;23(2):82–83. doi: 10.15171/PS.2017.13. [DOI] [Google Scholar]
- 3.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, et al. QSAR modeling: Where have you been? Where are you going to? J Med Chem. 2014;57(12):4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirico N, Gramatica P. Real external predictivity of QSAR models: How to evaluate it? Comparison of different validation criteria and proposal of using the concordance correlation coefficient. J Chem Inf Model. 2011;51(9):2320–2335. doi: 10.1021/ci200211n. [DOI] [PubMed] [Google Scholar]
- 5.Shayanfar A, Shayanfar S. Is regression through origin useful in external validation of QSAR models? Eur J Pharm Sci. 2014;59(1):31–35. doi: 10.1016/j.ejps.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Gramatica P, Cassani S, Roy PP, Kovarich S, Yap CW, Papa E. QSAR modeling is not “push a button and find a correlation”: a case study of toxicity of (Benzo-)triazoles on Algae. Mol Informatics. 2012;31(11–12):817–835. doi: 10.1002/minf.201200075. [DOI] [PubMed] [Google Scholar]
- 7.Veselinović JB, Veselinović AM, Toropova AP, Toropov AA. The Monte Carlo technique as a tool to predict LOAEL. Eur J Med Chem. 2016;116:71–75. doi: 10.1016/j.ejmech.2016.03.075. [DOI] [PubMed] [Google Scholar]
- 8.Zivkovic M, Zlatanovic M, Zlatanovic N, Golubović M, Veselinović AM. The application of the combination of monte carlo optimization method based QSAR modeling and molecular docking in drug design and development. Mini-Rev Med Chem. 2020;20(14):1389–1402. doi: 10.2174/1389557520666200212111428. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins DM, Basak SC, Mills D. Assessing model fit by cross-validation. J Chem Inf Comput Sci. 2003;43(2):579–586. doi: 10.1021/ci025626i. [DOI] [PubMed] [Google Scholar]
- 10.Gütlein M, Helma C, Karwath A, Kramer S. A large-scale empirical evaluation of cross-validation and external test set validation in (Q)SAR. Mol Informatics. 2013;32(5–6):516–528. doi: 10.1002/minf.201200134. [DOI] [PubMed] [Google Scholar]
- 11.Filzmoser P, Liebmann B, Varmuza K. Repeated double cross validation. J Chemometr. 2009;23(4):160–171. doi: 10.1002/cem.1225. [DOI] [Google Scholar]
- 12.Esbensen KH, Geladi P. Principles of proper validation: use and abuse of re-sampling for validation. J Chemometr. 2010;24(3–4):168–187. doi: 10.1002/cem.1310. [DOI] [Google Scholar]
- 13.Gramatica P. External evaluation of QSAR models, in addition to cross-validation: verification of predictive capability on totally new chemicals. Mol Informatics. 2014;33(4):311–314. doi: 10.1002/minf.201400030. [DOI] [PubMed] [Google Scholar]
- 14.Muratov EN, Bajorath J, Sheridan RP, Tetko IV, Filimonov D, Poroikov V, Oprea TI, Baskin II, Varnek A, Roitberg A, et al. QSAR without borders. Chem Soc Rev. 2020;49(11):3525–3564. doi: 10.1039/D0CS00098A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Đorđević V, Pešić S, Živković J, Nikolić GM, Veselinović AM. Development of novel antipsychotic agents by inhibiting dopamine transporter: in silico approach. New J Chem. 2022;46(6):2687–2696. doi: 10.1039/D1NJ04759K. [DOI] [Google Scholar]
- 16.Perić V, Golubović M, Lazarević M, Marjanović V, Kostić T, Đorđević M, Milić D, Veselinović AM. Development of potential therapeutics for pain treatment by inducing Sigma 1 receptor antagonism: in silico approach. New J Chem. 2021;45(27):12286–12295. doi: 10.1039/D1NJ00883H. [DOI] [Google Scholar]
- 17.Živković JV, Trutić NV, Veselinović JB, Nikolić GM, Veselinović AM. Monte Carlo method based QSAR modeling of maleimide derivatives as glycogen synthase kinase-3β inhibitors. Comput Biol Med. 2015;64:276–282. doi: 10.1016/j.compbiomed.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Hamzeh-Mivehroud M, Khoshravan-Azar Z, Dastmalchi S. QSAR and molecular docking studies on non-imidazole-based histamine h3 receptor antagonists. Pharm Sci. 2020;26(2):165–174. doi: 10.34172/PS.2019.64. [DOI] [Google Scholar]
- 19.Golbraikh A, Tropsha A. Beware of q2! J Mol Graph Model. 2002;20(4):269–276. doi: 10.1016/S1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 20.Roy PP, Roy K. On some aspects of variable selection for partial least squares regression models. QSAR Comb Sci. 2008;27(3):302–313. doi: 10.1002/qsar.200710043. [DOI] [Google Scholar]
- 21.Roy K, Das RN, Ambure P, Aher RB. Be aware of error measures. Further studies on validation of predictive QSAR models. Chemometr Intell Lab Syst. 2016;152:18–33. doi: 10.1016/j.chemolab.2016.01.008. [DOI] [Google Scholar]
- 22.Eisenhauer JG. Regression through the origin. Teach Stat. 2003;25(3):76–80. doi: 10.1111/1467-9639.00136. [DOI] [Google Scholar]
- 23.Zhang X, Zhang H. 3D-QSAR studies on 1,2,4-triazolyl 5-azaspiro [2.4]-heptanes as D3R antagonists. Chem Phys Lett. 2018;704:11–20. doi: 10.1016/j.cplett.2018.05.022. [DOI] [Google Scholar]
- 24.Patil RB, Barbosa EG, Sangshetti JN, Sawant SD, Zambre VP. LQTA-R: A new 3D-QSAR methodology applied to a set of DGAT1 inhibitors. Comput Biol Chem. 2018;74:123–131. doi: 10.1016/j.compbiolchem.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Aouidate A, Ghaleb A, Ghamali M, Ousaa A, Choukrad M, Sbai A, Bouachrine M, Lakhlifi T. 3D QSAR studies, molecular docking and ADMET evaluation, using thiazolidine derivatives as template to obtain new inhibitors of PIM1 kinase. Comput Biol Chem. 2018;74:201–211. doi: 10.1016/j.compbiolchem.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Gao J, Sun J, Wang T, Sheng S, Huang T. Combined 3D-QSAR modeling and molecular docking study on spiro-derivatives as inhibitors of acetyl-CoA carboxylase. Med Chem Res. 2017;26(2):361–371. doi: 10.1007/s00044-016-1743-3. [DOI] [Google Scholar]
- 27.Arthur DE, Uzairu A, Mamza P, Abechi SE, Shallangwa G. Activity and toxicity modelling of some NCI selected compounds against leukemia P388ADR cell line using genetic algorithm-multiple linear regressions. J King Saud Univ Sci. 2020;32(1):324–331. doi: 10.1016/j.jksus.2018.05.023. [DOI] [Google Scholar]
- 28.González MP, Teran Moldes MDC, Fall Y, Dias LC, Helguera AM. A topological sub-structural approach to the mutagenic activity in dental monomers. 3. Heterogeneous set of compounds. Polymer. 2005;46(8):2783–2790. doi: 10.1016/j.polymer.2005.01.064. [DOI] [Google Scholar]
- 29.Xu F, Yang ZZ, Ke ZL, Xi LM, Yan QD, Yang WQ, Zhu LQ, Lin FL, Lv WK, Wu HG, et al. Synthesis, antitumor evaluation and 3D-QSAR studies of [1,2,4]triazolo[4,3-b][1,2,4,5]tetrazine derivatives. Bioorg Med Chem Lett. 2016;26(19):4580–4586. doi: 10.1016/j.bmcl.2016.08.078. [DOI] [PubMed] [Google Scholar]
- 30.Ugale VG, Patel HM, Surana SJ. Molecular modeling studies of quinoline derivatives as VEGFR-2 tyrosine kinase inhibitors using pharmacophore based 3D QSAR and docking approach. Arab J Chem. 2017;10:S1980–S2003. doi: 10.1016/j.arabjc.2013.07.026. [DOI] [Google Scholar]
- 31.Arthur DE, Uzairu A, Mamza P, Abechi SE, Shallangwa G. Insilco study on the toxicity of anti-cancer compounds tested against MOLT-4 and p388 cell lines using GA-MLR technique. BeniSuef Univ J Basic Appl Sci. 2016;5(4):320–333. [Google Scholar]
- 32.Bhatia MS, Pakhare KD, Choudhari PB, Jadhav SD, Dhavale RP, Bhatia NM. Pharmacophore modeling and 3D QSAR studies of aryl amine derivatives as potential lumazine synthase inhibitors. Arab J Chem. 2017;10:S100–S104. doi: 10.1016/j.arabjc.2012.05.008. [DOI] [Google Scholar]
- 33.Aouidate A, Ghaleb A, Ghamali M, Chtita S, Ousaa A, Choukrad M, Sbai A, Bouachrine M, Lakhlifi T. QSAR study and rustic ligand-based virtual screening in a search for aminooxadiazole derivatives as PIM1 inhibitors. Chem Cent J. 2018;12:32. doi: 10.1186/s13065-018-0401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma MC, Jain S, Sharma R. Trifluorophenyl-based inhibitors of dipeptidyl peptidase-IV as antidiabetic agents: 3D-QSAR COMFA, CoMSIA methodologies. Netw Model Anal Health Inform Bioinform. 2018;7:1. doi: 10.1007/s13721-017-0163-8. [DOI] [Google Scholar]
- 35.Tong J, Lei S, Qin S, Wang Y. QSAR studies of TIBO derivatives as HIV-1 reverse transcriptase inhibitors using HQSAR, CoMFA and CoMSIA. J Mol Struct. 2018;1168:56–64. doi: 10.1016/j.molstruc.2018.05.005. [DOI] [Google Scholar]
- 36.Liu G, Wang W, Wan Y, Ju X, Gu S. Application of 3D-QSAR, pharmacophore, and molecular docking in the molecular design of diarylpyrimidine derivatives as HIV-1 nonnucleoside reverse transcriptase inhibitors. Int J Mol Sci. 2018;19(5):1436. doi: 10.3390/ijms19051436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behgozin SM, Fatemi MH. 3D-QSAR modeling of maximum steady-state fluxes of some substituted benzenes and quinolone derivatives through polydimethylsiloxane membrane. J Iran Chem Soc. 2018;15(6):1293–1300. doi: 10.1007/s13738-018-1328-9. [DOI] [Google Scholar]
- 38.Kaczor AA, Żuk J, Matosiuk D. Comparative molecular field analysis and molecular dynamics studies of the dopamine D2 receptor antagonists without a protonatable nitrogen atom. Med Chem Res. 2018;27(4):1149–1166. doi: 10.1007/s00044-018-2137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ZZ, Ma CY, Yang J, Gao QB, Sun XD, Ding L, Liu HM. Investigating the binding mechanism of (4-Cyanophenyl)glycine derivatives as reversible LSD1 by 3D-QSAR, molecular docking and molecular dynamics simulations. J Mol Struct. 2019;1175:698–707. doi: 10.1016/j.molstruc.2018.08.029. [DOI] [Google Scholar]
- 40.Singh U, Gangwal RP, Dhoke GV, Prajapati R, Damre M, Sangamwar AT. 3D-QSAR and molecular docking analysis of (4-piperidinyl)-piperazines as acetyl-CoA carboxylases inhibitors. Arab J Chem. 2017;10:S617–S626. doi: 10.1016/j.arabjc.2012.10.023. [DOI] [Google Scholar]
- 41.Türkmenoğlu B, Güzel Y. Molecular docking and 4D-QSAR studies of metastatic cancer inhibitor thiazoles. Comput Biol Chem. 2018;76:327–337. doi: 10.1016/j.compbiolchem.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Chun-Zhi H, Shu-Wei X, Hu W, Jun X, Liangmin Y. Using 3D-QSAR and molecular docking insight into inhibitors binding with complex-associated kinases CDK8. J Mol Struct. 2018;1173:498–511. doi: 10.1016/j.molstruc.2018.05.072. [DOI] [Google Scholar]
- 43.Ajay Kumar TV, Athavan AAS, Loganathan C, Saravanan K, Kabilan S, Parthasathy V. Design, 3D QSAR modeling and docking of TGF-β type I inhibitors to target cancer. Comput Biol Chem. 2018;76:232–244. doi: 10.1016/j.compbiolchem.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Ounissi M, Kameli A, Tigrine C, Rachedi FZ. Computer-aided identification of natural lead compounds as cyclooxygenase-2 inhibitors using virtual screening and molecular dynamic simulation. Comput Biol Chem. 2018;77:1–16. doi: 10.1016/j.compbiolchem.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Ghasemi JB, Davoudian V. 3D-QSAR and docking studies of a series of β -carboline derivatives as antitumor agents of PLK1. J Chem. 2014;2014:10. doi: 10.1155/2014/323149. [DOI] [Google Scholar]
- 46.Zheng J, Kong H, Wilson JM, Guo J, Chang Y, Yang M, Xiao G, Sun P. Insight into the interactions between novel isoquinolin-1,3-dione derivatives and cyclin-dependent kinase 4 combining QSAR and molecular docking. PLoS ONE. 2014;9(4):e93704. doi: 10.1371/journal.pone.0093704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Ning J, Wang Y, Wang C, Sun C, Huo X, Yu Z, Feng L, Zhang B, Tian X, et al. Drug interaction study of flavonoids toward CYP3A4 and their quantitative structure activity relationship (QSAR) analysis for predicting potential effects. Toxicol Lett. 2018;294:27–36. doi: 10.1016/j.toxlet.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Hao M, Ren H, Luo F, Zhang S, Qiu J, Ji M, Si H, Li G. A computational study on thiourea analogs as potent MK-2 inhibitors. Int J Mol Sci. 2012;13(6):7057–7079. doi: 10.3390/ijms13067057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ojha PK, Mitra I, Das RN, Roy K. Further exploring rm 2 metrics for validation of QSPR models. Chemometr Intell Lab Syst. 2011;107(1):194–205. doi: 10.1016/j.chemolab.2011.03.011. [DOI] [Google Scholar]
- 50.Avdeef A. Do you know your r2? ADMET DMPK. 2021;9(1):69–74. doi: 10.5599/admet.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chattefuee S, Hadi AS. Regression analysis by example. 4. Hoboken: Wiley; 2006. [Google Scholar]
- 52.Hulsizer MR, Woolf LM. a guide to teaching statistics: innovations and best practices. Oxford: Wiley; 2009. [Google Scholar]
- 53.Kaneko H. Beware of r2 even for test datasets: using the latest measured y-values (r2 LM) in time series data analysis. J Chemometr. 2019;33(2):e3093. doi: 10.1002/cem.3093. [DOI] [Google Scholar]
- 54.Shayanfar A, Ershadi S. Developing new criteria for validity evaluation of analytical methods. J AOAC Int. 2019;102(6):1908–1916. doi: 10.5740/jaoacint.19-0007. [DOI] [PubMed] [Google Scholar]
- 55.Ghandadi M, Shayanfar A, Hamzeh-Mivehroud M, Jouyban A. Quantitative structure activity relationship and docking studies of imidazole-based derivatives as P-glycoprotein inhibitors. Med Chem Res. 2014;23(11):4700–4712. doi: 10.1007/s00044-014-1029-6. [DOI] [Google Scholar]
- 56.Gramatica P, Sangion A. A historical excursus on the statistical validation parameters for QSAR models: a clarification concerning metrics and terminology. J Chem Inf Model. 2016;56(6):1127–1131. doi: 10.1021/acs.jcim.6b00088. [DOI] [PubMed] [Google Scholar]
- 57.Rácz A, Bajusz D, Héberger K. Consistency of QSAR models: Correct split of training and test sets, ranking of models and performance parameters. SAR QSAR Environ Res. 2015;26(7–9):683–700. doi: 10.1080/1062936X.2015.1084647. [DOI] [PubMed] [Google Scholar]
- 58.Tóth G, Király P, Kovács D. Effect of variable allocation on validation and optimality parameters and on cross-optimization perspectives. Chemometr Intelligent Lab Syst. 2020;204:104106. doi: 10.1016/j.chemolab.2020.104106. [DOI] [Google Scholar]
- 59.Dearden JC, Cronin MTD, Kaiser KLE. How not to develop a quantitative structure-activity or structure-property relationship (QSAR/QSPR) SAR QSAR Environ Res. 2009;20(3–4):241–266. doi: 10.1080/10629360902949567. [DOI] [PubMed] [Google Scholar]
- 60.Majumdar S, Basak SC. Beware of external validation!-a comparative study of several validation techniques used in qsar modelling. Curr Comput-Aided Drug Des. 2018;14(4):284–291. doi: 10.2174/1573409914666180426144304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Forty-four data sets (training and test sets) composed of experimental biological activity and corresponding calculated activity.
Data Availability Statement
All data is available as supplementary.


