Abstract
Antigen 43 (Ag43), the product of the flu gene, is a surface-displayed autotransporter protein of Escherichia coli. Ag43 is responsible for the autoaggregation and flocculation of static liquid cultures of many E. coli strains. The expression of Ag43 has been reported to be phase variable and controlled by the product of the oxyR gene. Type 1 fimbriae are thin adhesive thread-like surface organelles responsible for bacterial receptor recognition and tissue colonization. Like that of Ag43, the expression of type 1 fimbriae is phase variable. Interestingly, previous results have suggested that the expression of type 1 fimbriae and the expression of Ag43 are mutually exclusive. In the present report, we show, by use of well-defined mutants, that fimbriation abolishes Ag43-mediated autoaggregation but does not affect Ag43 expression. Autoaggregation is shown to require an intercellular Ag43-Ag43 interaction, and the physical presence of fimbriae on the cells seems to abrogate this interaction. The Ag43 or OxyR status does not appear to influence fimbria expression, and our results suggest that the expression of Ag43 and the expression of fimbriae are independent processes.
Many Escherichia coli strains have the ability to autoaggregate, observed as characteristic flocculation and settling of cells from liquid cultures that are left standing. This phenomenon was first reported by Diderichsen (13), who defined a locus, flu, mapping at 43 min on the E. coli K-12 chromosome. The flu locus appeared to control several surface properties. A number of strains exhibited two distinct but interconverting forms. Form 1 was characterized by large, flat, “frizzy,” and irregular colonies and was able to aggregate in static liquid medium. The other form gave rise to smaller, glossy colonies and did not autoaggregate in static liquid medium (13). Additionally, Diderichsen observed that certain strains with deletions in the 89-min region were fixed in form 1 (13). This region was later reported to harbor oxyR or mor (11, 15, 39). In separate studies, it was found that the product of the flu locus was identical to an outer membrane protein termed antigen 43 (Ag43) (16, 33). Ag43 was reported to consist of two equimolar protein subunits, α and β, with apparent molecular masses of 50 to 60 kDa and 53 kDa, respectively (9). The α protein is attached to the cell surface through an interaction with the β component, which is an integral outer membrane protein. Ag43 expression has been proposed to be negatively controlled by OxyR and positively through Dam methylation (33). Although Ag43 expression has been convincingly established to be correlated with bacterial autoaggregation, the underlying mechanism is still unknown.
Another prominent surface feature of many E. coli strains is type 1 fimbriae, thin, 7-nm-wide and approximately 1-μm-long, rod-shaped surface organelles. A typical type 1-fimbriated cell has 200 to 500 such organelles uniformly arranged on the surface. Type 1 fimbriae are widespread among members of the Enterobacteriaceae; they are adhesins involved in specific receptor recognition and tissue colonization (24). The expression of type 1 fimbriae is phase variable; i.e., bacterial cells with the potential to express these organelles fluctuate between two phenotypes, either fimbriated or bald. Phase variation of type 1 fimbriae is due to the inversion of a 314-bp DNA fragment (called the fim switch) located immediately upstream of the structural fim genes (1). A promoter residing in this phase switch drives the expression of the fim genes (31) when the switch is in the “on” orientation but not when it is in the “off” orientation. Two recombinases, FimB and FimE (14, 20), mediate the inversion of the phase switch.
Interestingly, it was noted that the expression of Ag43 and type 1 fimbriae might be coregulated. Autoaggregating cells were observed to be nonfimbriated, whereas nonaggregating cells were observed to be fimbriated (13). It is also noteworthy that both systems are subject to phase variation and that the phase variation frequencies seem to be similar, i.e., about 10−3 per cell per generation (4, 5, 33). In this study, we have explored the relationships among Ag43 expression, type 1 fimbria expression, and related phenotypes with a view to finding possible intersystem cross talk.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are described in Table 1. Cells were grown on solid medium or in liquid broth supplemented with the appropriate antibiotics unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Reference or construction |
|---|---|---|
| E. coli K-12 strains | ||
| BD1302 | ΔoxyR; autoaggregating | 13 |
| CC118λpir | λpir integrated on the chromosome | 19 |
| HEHA3 | BD1302 Δfim::kan | This study |
| HEHA10 | BD1302 flu::tet | This study |
| MS7 | PC31 Δfim::kan | 37 |
| PC31 | fim+ | 23 |
| Plasmids | ||
| pACYC184 | Camr Tetr | 10 |
| pBR322 | Ampr Tetr | 7 |
| pGP704 | Ampr; R6K-based origin (pir) | 19 |
| pHHA130 | oxyR+ | A 2,040-bp PCR fragment (primers 5 and 6) containing the oxyR gene from PC31 inserted into the HindIII site of pACYC184 |
| pHHA145 | StyI site deleted | pBR322 cut with StyI, made blunt with Klenow polymerase, and religated |
| pHHA146 | flu+ | A 3,550-bp PCR fragment containing the flu gene from PC31 inserted into the EcoRI/BamHI site of pBR322 |
| pHHA154 | flu+ | A 3,550-bp PCR fragment containing the flu gene from PC31 inserted into the EcoRI/BamHI site of pHHA145 |
| pHHA159 | flu::tet | A 1,724-bp BsaAI/SspI fragment containing the tet gene inserted into the (blunted) StyI site of pHHA154 |
| pHHA165 | flu::tet | A 5,450-bp EcoRI fragment containing the flu::tet construct inserted into the EcoRI site of pGP704 |
| pLBJ311 | Δfim::kan | 37 |
| pLIH14 | Has all fim genes except fimA | 25 |
| pMAS32 | lacUV5::fimACDFGH | Part of the fim gene cluster (fimA to fimH) inserted behind the isopropyl-β-d-thiogalactopyranoside-inducible lacUV5 promoter |
| pPAP5 | Contains the pap gene cluster | 27 |
| pPKL4 | Contains the fim gene cluster | 23 |
| pPKL5 | Contains the fimBEAICD genes | 21 |
| pPKL9 | fimB+ | 20 |
| pPKL143 | Contains the foc gene cluster | 22 |
DNA manipulations.
Isolation of plasmid DNA was carried out with a QIAprep Spin Miniprep Kit (Qiagen). Restriction endonucleases were used according to the manufacturer’s specifications (Biolabs). Chromosomal DNA was purified with a GenomicPrep Cell and Tissue DNA Isolation Kit (Amersham Pharmacia Biotech Inc.).
PCR methodology.
PCR was done as previously described (37). The primers used are listed in Table 2.
TABLE 2.
Primers
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| 1 | CCCGCGGCCGCGATATCCTTTGTCAGTAACATGC |
| 2 | CCCGCGGCCGCGGATCCTGTGGCGTTGAAGATCCG |
| 3 | CGCTGAGCAATGACATCCG |
| 4 | AATGTCACCCTGAAGCAGG |
| 5 | GGGAAGCTTGCGGCCGCTTAGCAGGCTGGCTGGG |
| 6 | GGGAAGCTTGCGGCCGCAAAGGTGGCGGCAACAC |
| 7 | GGGAAGCTTGCGGCCGCTTAGCAGCTGGCTGGG |
| 8 | GGGAAGCTTGCGGCCGCAAAGGTGGCGGCAACAC |
Nucleotide sequencing.
The nucleotide sequences of PCR products and flanking regions in the genetic constructs were determined by use of an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). Samples were electrophoresed on a Perkin-Elmer ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems) as described in the manufacturer’s specifications.
Construction of an flu::tetR mutant.
The flu gene was amplified by PCR from chromosomal DNA of strain PC31 with primers 1 and 2. The resulting fragment was inserted into the EcoRI/BamHI site of pHHA145 to generate pHHA154. This plasmid was then cut with StyI (1,138 bp inside the flu gene) and made blunt, and an SspI/BsaAI fragment from pACYC184 containing the tetR gene and its promoter was inserted to generate plasmid pHHA159. Plasmid pHHA159 was subsequently cut with EcoRI, and the fragment containing the flu::tetR construct was inserted into plasmid pGP704 and amplified in strain CC118λpir. After amplification, the plasmid was transformed into strain BD1302, and single-crossover mutants were selected on plates containing 8 μg of tetracycline per μl. Double-crossover mutants were then screened by replica plating on plates containing 8 μg of tetracycline and 100 μg of ampicillin per μl, and Tetr Amps colonies were picked for further work. Correct insertion of the flu::tetR construct on the chromosome was tested by PCR with primers 3 and 4, flanking the insertion point in flu. Colonies in which PCR patterns showed a shift in fragment size corresponding to the insertion were selected and tested for the loss of autoaggregation ability. One representative strain with this phenotype was designated HEHA10 and was used in this study (Fig. 1A).
FIG. 1.
(A) PCR analysis of pHHA159 (lane 2), HEHA10 (lane 3), and BD1302 (lane 4) with primers 3 and 4. Lane 1, size marker with the sizes indicated on the left (in kilobases). The sizes of PCR bands are indicated on the right. Above are a schematic representation of the two genetic variants and the positions of the primers. (B) Southern blot hybridization with a fim probe (lanes 1 to 4) and a kan probe (lanes 5 to 8) of chromosomal DNA cut with PvuII of strains MS7 (lanes 1 and 5), HEHA3 (lanes 2 and 6), PC31 (lanes 3 and 7), and BD1302 (lanes 4 and 8). Fragment sizes (in kilobases) are indicated on the left. Above are schematic representations of the two genetic variants; short vertical lines indicate the positions of the PvuII sites.
Construction of Δfim strains.
A Δfim variant of BD1302 was constructed by use of the λpir-dependent plasmid pLBJ311 containing the type 1 fim gene cluster with an npt gene (Kanr) inserted between truncated fimB and fimH, thus deleting all the fim genes. Insertion on the chromosome was done basically as described above and as described previously (37). Correct insertion was verified by PCR and Southern blotting (Fig. 1B) as previously described (36). A representative clone carrying the correct insertion was designated HEHA3 and was tested for the loss of mannose-sensitive yeast agglutination.
Cloning of the oxyR gene.
The oxyR gene was amplified by PCR from chromosomal DNA of strain PC31 with primers 7 and 8. The resulting fragment was inserted directly into the HindIII site of pACYC184 to generate pHHA130.
Autoaggregation assay.
Overnight cultures of the strains were adjusted to approximately the same optical density at 600 nm (OD600), and 10 ml of each culture was placed in a sterile 20-ml tube. At the beginning of each experiment, all cultures were vigorously shaken for 10 s. Two 100-μl samples were taken from each tube, approximately 1 cm from the top, and transferred to two new tubes, each containing 1 ml of 0.9% NaCl. The OD600 was then measured.
Detection of type 1 fimbriae.
The capacity of bacteria to express a d-mannose-binding phenotype was assayed by their ability to agglutinate yeast cells on glass slides. Aliquots of liquid cultures grown at an OD600 of 4.0 and a 5% (wt/vol) suspension of yeast cells were mixed, and the time until agglutination occurred was measured.
Immunofluorescence microscopy.
Surface presentation of type 1 fimbriae or Ag43 was assessed by immunofluorescence microscopy with a monoclonal antibody directed against FimA (37) or a polyclonal serum that recognizes the α subunit of Ag43 (a kind gift from Peter Owen), respectively. Cell fixation, immunolabeling, and microscopy were carried out as previously described (34) with a fluorescein isothiocyanate (FITC)-labeled secondary antibody.
RESULTS
The flu locus and Ag43.
The published N-terminal sequences of the α and β fragments of Ag43 (33) are encoded by an open reading frame of 1,039 codons (ORF1039) located at map position 43.6 min of the E. coli K-12 chromosome (3) and therefore coinciding with the flu locus. A strong consensus ribosomal binding site precedes the start codon, suggesting a theoretical molecular mass of 106.9 kDa for the primary product of the flu gene. N-terminal sequencing indicates that the mature α fragment starts another 52 amino acids into this sequence, predicting a mature Ag43 protein of 987 amino acids (101.6 kDa). The reported N-terminal sequence of the β fragment (33) indicates that this protein is further processed into an α fragment of 499 amino acids (49.8 kDa) and a β fragment of 488 amino acids (51.5 kDa), in reasonable agreement with the reported apparent molecular masses of 50 to 60 kDa (α fragment) and 53 kDa (β fragment) given in the literature (9).
Cloning of the flu gene and construction of a defined knockout mutant strain.
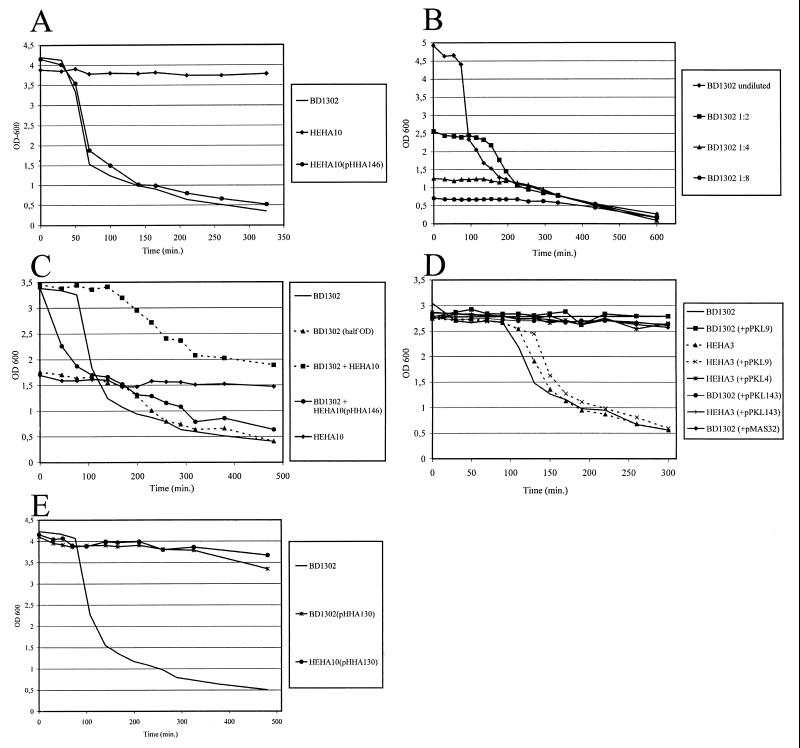
Two primers were made with sequences corresponding to positions 322 bp upstream and 76 bp downstream of ORF1039 and were used to amplify the flu gene from E. coli K-12 PC31. The 3.5-kb PCR fragment was cloned into pBR322, resulting in plasmid pHHA146. Subsequently, a tet cassette was inserted into the flu gene, and the flu::tet construct was inserted into the suicide vector pGP704 to yield pHHA165. E. coli BD1302 has a large chromosomal deletion in the 89-min region encompassing the oxyR gene (13, 39), resulting in the constitutive expression of Ag43. Plasmid pHHA165 was used to make a defined knockout mutation of the flu gene in this strain by homologous recombination with the flu::tet cassette, resulting in strain HEHA10 (Fig. 1A). When liquid cultures of strain BD1302 are left standing, the cells readily autoaggregate and settle (Fig. 2A). In contrast, cells from liquid cultures of the isogenic flu::tet strain HEHA10 stay in solution under similar conditions (Fig. 2A). When the Ag43-expressing plasmid pHHA146 was introduced into HEHA10, an autoaggregating phenotype identical to that of BD1302 could be reestablished (Fig. 2A). In order to gain further insight into Ag43 expression in these strains, immunofluorescence microscopy with specific anti-Ag43 serum was performed (Fig. 3B and D). It became clear from the results that the autoaggregation phenotype was concomitant with the ability to express Ag43 and that Ag43 must be directly responsible for this property.
FIG. 2.
Autoaggregation assays performed with BD1302, HEHA10, and HEHA10(pHHA146) (A); serial dilutions of BD1302 (B); BD1302, BD1302 (half OD600), HEHA10, and mixtures of BD1302 and HEHA10 and of BD1302 and HEHA10(pHHA146) (C); BD1302, BD1302(pPKL9), HEHA3, HEHA3(pPKL9), HEHA3(pPKL4), BD1302(pPKL143), HEHA3(pPKL143), and BD1302(pMAS32) (D); and BD1302, BD1302(pHHA130), and HEHA10(pHHA130) (E).
FIG. 3.
Phase-contrast microscopy (left panels on each side) and fluorescence microscopy (right panels on each side) of E. coli K-12 hosts. To detect the presence of type 1 fimbriae on the surface of the bacteria (left side), a monoclonal antibody directed against FimA was used, and this was detected with FITC-labeled rabbit anti-mouse serum. To detect the presence of Ag43 on the surface of the bacteria (right side), FITC-labeled pig anti-rabbit serum was used. The strains tested were BD1302 (A and B), HEHA10 (C and D), HEHA3 (E and F), BD1302(pPKL9) (G and H), HEHA3(pPKL9) (I and J), and BD1302(pHHA130) (K and L). Arrows indicate cells expressing a phenotype different from that of the majority.
Ag43-mediated autoaggregation kinetics.
To gain more insight into the autoaggregation phenomenon, we investigated the kinetics of Ag43-mediated autoaggregation, i.e., whether this process was dependent on cell density. For this purpose, the settling profiles of different concentrations of BD1302 were analyzed. It was apparent (Fig. 2B) that BD1302 cells settled regardless of the cell density used; i.e., virtually all cells had settled by the end of the experiment. Nevertheless, the time that passed before aggregation was initiated was highly dependent on the cell density of the initial suspension. When the cell density was halved, the time until the initiation of settling was approximately doubled. This result is in good agreement with a model following first-order kinetics, where the chance of two bacteria colliding at a given time interval is proportional to the cell density.
The Ag43-Ag43 interaction is responsible for bacterial autoaggregation.
In order to investigate whether Ag43-mediated autoaggregation was caused by an Ag43-Ag43 interaction between aggregating cells or, alternatively, whether Ag43 on one cell interacted with a non-Ag43 target on another cell, the following experiments were carried out. A model featuring an Ag43-Ag43 interaction as being responsible for autoaggregation would suggest that mixing equal portions of isogenic Ag43-positive and Ag43-negative cells would result in the precipitation of only half of the cells; i.e., Ag43-negative cells would not participate. This prediction was exactly what was observed in settling experiments with equal amounts of BD1302 and HEHA10 cells (Fig. 2C). Furthermore, careful sampling and plating of cells revealed that virtually all the cells remaining in suspension were HEHA10 and that the precipitated cells were BD1302. The introduction of plasmid pHHA146 into HEHA10 cells and mixing such cells with equal amounts of BD1302 cells caused settling identical to that seen in a monoculture of BD1302 cells (Fig. 2C). This result strongly suggests that an intercellular Ag43-Ag43 interaction is responsible for the autoaggregation phenomenon.
Ag43-mediated autoaggregation is abolished by fimbriation.
As previously noted, several observations have hinted that the expression of Ag43 and type 1 fimbriation might be mutually exclusive phenotypes and that the responsible genes might be reciprocally regulated (13). The support for this tenet was based on the observation that cells from glossy colonies are Ag43 negative, do not form autoaggregates, and agglutinate yeast cells in a d-mannose-sensitive manner (indicative of type 1 fimbriation), whereas cells from frizzy colonies are Ag43 positive, autoaggregate, and do not agglutinate yeast cells. E. coli BD1302 is the classic Ag43 reference strain. In order to investigate potential intersystem coregulation of Ag43 and type 1 fimbriae, derivatives of BD1302 which were forced to produce type 1 fimbriae were made. This was done in two ways, either by the introduction of a high-copy-number plasmid, pPKL4, carrying the fim gene cluster, or by the activation of the resident fim gene cluster of BD1302 by the introduction of a plasmid, pPKL9, encoding the fimB recombinase. Strain BD1302 readily autoaggregates; however, both BD1302(pPKL4) and BD1302(pPKL9) lost this faculty (Fig. 2D).
In order to examine whether FimB caused this effect by activation of the resident fim gene cluster, with ensuing fimbriation, or whether the abolition of autoaggregation was due to a yet-unknown effect of the recombinase, a derivative of BD1302, HEHA3, in which the fim gene cluster was deleted, was made (see Materials and Methods). Both HEHA3 and HEHA3(pPKL9) autoaggregated exactly like BD1302 (Fig. 2D). However, a control strain, HEHA3(pPKL4), did not. As a further control, we introduced into BD1302 a plasmid (pMAS32) containing a fim gene cluster in which the recombinase genes and phase switch had been replaced by a lacUV5 promoter. BD1302(pMAS32) cells did not autoaggregate (Fig. 2D). These experiments indicated that it was the physical presence of fimbriae on the cells that abolished autoaggregation.
To further examine this hypothesis, two plasmids, pLIH14, carrying the entire fim gene cluster with a truncated fimA gene, and pPKL5, which harbors a fim gene cluster missing the minor component containing the fimF, fimG, and fimH genes, were used. Host cells containing pLIH14 show a low level of d-mannose-specific adhesion but are bald due to the absence of the major organelle subunit (25), whereas host cells containing pPKL5 produce few, nonadhesive and abnormally long fimbriae due to the lack of minor components (21). The introduction of plasmid pLIH14 into HEHA3 did not interfere with the ability of this host to autoaggregate (data not shown). However, HEHA3(pPKL5) cells were unable to autoaggregate (data not shown), suggesting that the presence of even a few, albeit very long fimbriae, is sufficient to block the Ag43-Ag43 interaction.
Finally, with a view to examining whether fimbriation in general blocked Ag43-mediated autoaggregation, two plasmids encoding fimbriae distinct from type 1 fimbriae were introduced. These were pPKL143, carrying the foc gene cluster, and pPAP5, carrying the pap gene cluster, responsible for the production of F1C and P fimbriae, respectively. The plasmids were introduced into BD1302 and HEHA3. All four resulting strains, BD1302(pPKL143), BD1302(pPAP5), HEHA3(pPKL143), and HEHA3(pPAP5), were unable to autoaggregate (see, for example, Fig. 2D).
Ag43 expression is not affected by fimbriation.
The above results could be interpreted in two ways: either the physical presence of the fimbriae on the bacterial surface prevented the Ag43-expressing bacteria from establishing the required contact for an Ag43-Ag43 interaction to take place, thus abrogating autoaggregation or, alternatively, fimbrial expression somehow excluded Ag43 expression. To examine these possibilities, selected strains were submitted to immunofluorescence microscopy with specific antisera raised against type 1 fimbriae or Ag43. Not surprisingly, BD1302 cells were observed to react strongly with Ag43-specific antibodies (Fig. 3B); also, a small percentage of the cells reacted with fimbria-specific serum (Fig. 3A). Deletion of the Ag43-encoding gene on the chromosome (strain HEHA10) did not change the percentage of fimbriated cells (Fig. 3C) but, not unexpectedly, resulted in the disappearance of Ag43 from the cells (Fig. 3D). Likewise, HEHA3 cells in which the fim gene cluster was deleted showed no reaction with the fimbriae-specific serum, whereas the Ag43 level was identical to that in parent strain BD1302 (Fig. 3E and F). The introduction of plasmid pPKL9 into BD1302 resulted in a population where virtually all cells expressed both fimbriae and Ag43 (Fig. 3G and H). On the contrary, the introduction of plasmid pPKL9 into strain HEHA3 (Δfim) did not affect Ag43 production—it was indistinguishable from that of parent strain BD1302—whereas, as expected, no cells produced fimbriae (Fig. 3I and J). In light of these results, we therefore concluded that Ag43 production is unaffected by the level of fimbriation and that fimbrial neutralization of Ag43-mediated autoaggregation seems to be a physical rather than a coregulatory phenomenon.
Influence of oxyR.
Strain BD1302 has a large deletion in the 89-min region encompassing the oxyR locus and a number of flanking genes. It is a constitutive Ag43 producer, and this phenotype has been assumed, with some justification, to be due to the lack of OxyR in the cells, although this assumption has never been stringently proven. In order to prove this notion and to investigate the influence of OxyR on fimbrial expression, the oxyR locus in E. coli PC31 was amplified and cloned into plasmid pACYC184, resulting in plasmid pHHA130 (see Materials and Methods). The introduction of plasmid pHHA130 into BD1302 virtually abolished the capacity to autoaggregate (Fig. 2E). In keeping with this result, immunofluorescence microscopy revealed a dramatic decline in the number of Ag43-producing cells (Fig. 3L). However, the number of fimbriated cells seemed to be the same as in the parent strain (Fig. 3K). This result would suggest that OxyR does indeed repress Ag43 production but seems to have no influence on fimbriation.
DISCUSSION
Ag43 belongs to the growing family of autotransporter proteins from gram-negative bacteria. All the sequence information required to mediate transport and secretion through the outer membrane is contained within the protein itself (18). Members of the autotransporter family include important or putative virulence factors in many pathogens. Some, like the immunoglobulin A1 protease of Neisseria gonorrhoeae, are proteases (28); others, like the AIDA-I protein of diarrheagenic E. coli, are adhesins (2). Type 1 fimbriae are adhesins associated with many E. coli strains and have recently been shown to be critical for the ability of E. coli to colonize the urinary tract (12, 26, 30). The assembly and surface presentation of type 1 fimbriae follow a pathway different from that of Ag43, and these fimbriae, like other fimbriae, require chaperone- and usher-assisted assembly to cross the outer membrane.
Evidence accumulating in the literature indicated the possible coregulation of type 1 fimbriae and Ag43 and prompted us to examine this notion in greater detail. Work on Ag43 and on potential Ag43-type 1 fimbrial cross talk has been hampered by the lack of defined mutant strains. In this study, we focused on reference strain BD1302, originally characterized as a constitutive expressor of Ag43 (13). Suspensions of BD1302 cells readily autoaggregate and settle; however, replacement of the flu gene in BD1302 with a flu::tet cassette abolished this property (Fig. 2A). Complementation of the mutant with a plasmid carrying the flu gene reestablished the parental phenotype. Having established Ag43 as the causative agent of autoaggregation in BD1302, we proceeded to investigate the underlying mechanism for the autoaggregation, i.e., whether it was Ag43-Ag43 based or whether Ag43 recognized some other feature on the surface of E. coli. It turned out that the Ag43-Ag43 interaction indeed seemed to be responsible for cell aggregation, because in settling experiments with mixtures of isogenic strains BD1302 and HEHA10(flu::tet), the latter did not participate in the aggregation process. Also, the results from the settling experiments suggested that Ag43-mediated autoaggregation followed first-order kinetics.
BD1302 has a functional fim gene cluster, but only a small percentage of the cells at any given time were actually seen to express organelles, due to phase variation (Fig. 3). We have previously observed that the introduction of the fimB recombinase gene on a high-copy-number plasmid results in a population where virtually all cells are fimbriated (29). This finding was also obtained with BD1302(pPKL9) cells (Fig. 3). The introduction of a plasmid carrying the entire fim gene cluster had the same effect. In contrast to the BD1302 parent, such bacteria were unable to autoaggregate (Fig. 2D). The abolition of autoaggregation could be due to cross talk between the Ag43 and type 1 fimbrial systems; however, neutralization of autoaggregation was not seen when pPKL9 was introduced into a derivative of BD1302 from which the chromosomal fim genes had been deleted, i.e., in HEHA3. Furthermore, it was found, by immunofluorescence microscopy, that Ag43 production was not affected by concomitant type 1 fimbriation. This finding suggested that the physical presence of fimbriae on the cell surface negated the Ag43-Ag43 contact between cells and thereby prevented the autoaggregation phenotype (Fig. 4). This conclusion was further corroborated by the fact that the introduction of plasmids carrying the foc and pap gene clusters into BD1302 also neutralized autoaggregation. In this regard, it should be emphasized that the expression control systems of F1C and P fimbriae differ fundamentally from that of type 1 fimbriae (6, 8, 32, 35).
FIG. 4.
Schematic model of the relationship between autoaggregation (A) and fimbrial production (B).
Ag43 expression has been suggested to be negatively controlled by OxyR and positively controlled by Dam methylation. Henderson et al. (17) proposed an elegant model for the regulation of Ag43 expression in which OxyR acts as a repressor by binding to unmethylated Dam sites in the regulatory region of the flu gene. Also, according to this model, methylation prevents OxyR binding. Analysis of the region upstream of the flu gene identified a potential sigma-70 promoter located about 240 bp upstream of the start codon. Three GATC sites overlapping a motif similar to the proposed consensus sequence for OxyR-DNA binding (38) were found in this intergenic region (Fig. 5). It has not been stringently shown that OxyR regulates Ag43 expression. For this purpose, we amplified the oxyR gene from E. coli K-12 strain PC31 and cloned it on a pACYC184 vector (plasmid pHHA130). The introduction of plasmid pHHA130 into BD1302 virtually abrogated the autoaggregation phenotype (Fig. 2), and the number of Ag43-producing cells dropped dramatically (Fig. 3K and L). The residual Ag43-producing cells could be accounted for by the inability of OxyR to bind to methylated GATC sites, in accordance with the model of Henderson et al. (17). Interestingly, the fractions of cells expressing type 1 fimbriae were observed to be the same in BD1302 and BD1302(pHHA130) (Fig. 3). It therefore seems that neither the Ag43 status nor the OxyR status of the cells affects type 1 fimbriation. In fact, our results suggest that the expression of Ag43 and the expression of fimbriae are independent processes.
FIG. 5.
Schematic presentation of the promoter region of the flu gene showing the locations of the suggested promoter, Dam methylation sites, suggested OxyR-binding site, ribosomal binding site (RBS), and translation initiation site.
The biological reason for Ag43 production is somewhat controversial. Ag43 has been reported to confer a low level of adhesion to certain mammalian cells (33) but not in a manner that seems compatible with its widespread occurrence in E. coli strains. Since Ag43 is a self-recognizing protein which causes bacterial aggregation, this faculty might have a function in a mammalian host. However, the strength of binding of Ag43-Ag43-mediated cell interactions is very low compared to that of binding mediated by, for example, fimbrial adhesins, and cells that have autoaggregated as a result of Ag43 mediation are easily dispersed. Clearly, autoaggregating bacterial clumps may confer increased survival to individual bacteria under different conditions in many environments, and perhaps the mission of Ag43 is outside a mammalian host. The role of Ag43 indeed may be to cause autoaggregation of cells with ensuing settling under static liquid conditions. In this respect, it is interesting to speculate that when E. coli is shed by defecating animals in stagnant pools of water, it might be advantageous for the bacteria to become bottom dwellers in order to be in a nutrient-rich environment (feces) and perhaps to avoid UV radiation in the surface layer.
ACKNOWLEDGMENTS
We thank Peter Owen for generously supplying the Ag43 antibodies.
This work was supported by The Danish Natural Sciences Research Council (grant 9601682).
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benz I, Schmidt M A. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27) Infect Immun. 1992;60:13–18. doi: 10.1128/iai.60.1.13-18.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, III, Bloch C A, Perna T N, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield I C, McClain M S, Eisenstein B I. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol Microbiol. 1991;5:1439–1445. doi: 10.1111/j.1365-2958.1991.tb00790.x. [DOI] [PubMed] [Google Scholar]
- 6.Blyn L B, Braaten B A, Low D A. Regulation of pap pilin phase variation by a mechanism involving differential Dam methylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 8.Braaten B A, Nou X, Kaltenbach L S, Low D A. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 9.Caffrey P, Owen P. Purification and N-terminal sequence of the α subunit of antigen 43, a unique protein complex associated with the outer membrane of Escherichia coli. J Bacteriol. 1989;171:3634–3640. doi: 10.1128/jb.171.7.3634-3640.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christman M F, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connell H, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diderichsen B. flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol. 1980;141:858–867. doi: 10.1128/jb.141.2.858-867.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 15.Henderson I, Owen P. The autoregulatory protein Mor and OxyR are identical. Microbiology. 1997;143:1482. doi: 10.1099/00221287-143-5-1482. [DOI] [PubMed] [Google Scholar]
- 16.Henderson I R, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and auto-aggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 17.Henderson I R, Meehan M, Owen P. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv Exp Med Biol. 1997;412:349–355. doi: 10.1007/978-1-4899-1828-4_56. [DOI] [PubMed] [Google Scholar]
- 18.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 19.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 22.Klemm P, Christiansen G, Kreft B, Marre R, Bergmans H. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J Bacteriol. 1994;176:2227–2234. doi: 10.1128/jb.176.8.2227-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemm P, Jorgensen B J, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 24.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 9–26. [Google Scholar]
- 25.Klemm P, Krogfelt K A, Hedegaard L, Christiansen G. The major subunit of Escherichia coli type 1 fimbriae is not required for d-mannose-specific adhesion. Mol Microbiol. 1990;4:553–559. doi: 10.1111/j.1365-2958.1990.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 26.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 27.Lindberg F P, Lund B, Normark S. Genes of pyelonephritogenic E. coli required for digalactoside-specific agglutination of human cells. EMBO J. 1984;3:1167–1173. doi: 10.1002/j.1460-2075.1984.tb01946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomholt H, Poulsen K, Kilian M. Comparative characterization of the iga gene encoding IgA1 protease in Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae. Mol Microbiol. 1995;15:495–506. doi: 10.1111/j.1365-2958.1995.tb02263.x. [DOI] [PubMed] [Google Scholar]
- 29.McCormick B A, Klemm P, Krogfelt K A, Burghoff R L, Pallesen L, Laux D C, Cohen P S. Escherichia coli F-18 phase locked ‘on’ for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb Pathog. 1993;14:33–43. doi: 10.1006/mpat.1993.1004. [DOI] [PubMed] [Google Scholar]
- 30.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 31.Olsen P B, Klemm P. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol Lett. 1994;116:95–100. doi: 10.1111/j.1574-6968.1994.tb06681.x. [DOI] [PubMed] [Google Scholar]
- 32.Ott M, Hoschutzky H, Jann K, Van Die I, Hacker J. Gene clusters for S fimbrial adhesin (sfa) and F1C fimbriae (foc) of Escherichia coli: comparative aspects of structure and function. J Bacteriol. 1988;170:3983–3990. doi: 10.1128/jb.170.9.3983-3990.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen P, Meehan M, de Loughry-Doherty H, Henderson I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol. 1996;16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 34.Pallesen L, Poulsen L K, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial surface display system for heterologous sequences. Microbiology. 1995;141:2839–2848. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 35.Riegman N, Kusters R, Van Veggel H, Bergmans H, Van Bergen E, Hacker J, Van Die I. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J Bacteriol. 1990;172:1114–1120. doi: 10.1128/jb.172.2.1114-1120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schembri M A, Pallesen L, Connell H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 37.Stentebjerg-Olesen B, Pallesen L, Jensen L B, Christiansen G, Klemm P. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology. 1997;143:2027–2038. doi: 10.1099/00221287-143-6-2027. [DOI] [PubMed] [Google Scholar]
- 38.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 39.Warne S R, Varley J M, Boulnois G J, Norton M G. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J Gen Microbiol. 1990;136:455–462. doi: 10.1099/00221287-136-3-455. [DOI] [PubMed] [Google Scholar]