Abstract
Objectives:
We examined whether molecular cluster membership was associated with public health identification of HIV transmission potential among named partners in Chicago.
Design:
Historical cohort study
Methods:
We matched and analyzed HIV surveillance and partner services data from HIV diagnoses (2012–2016) prior to implementation of cluster detection and response interventions. We constructed molecular clusters using HIV-TRACE at a pairwise genetic distance threshold of 0·5% and identified clusters exhibiting recent and rapid growth according to the CDC’s definition (3 new cases diagnosed in past year). Factors associated with identification of partners with HIV transmission potential were examined using multivariable Poisson regression.
Results:
There were 5,208 newly diagnosed index clients over this time period. Average age of index clients in clusters was 28; 47% were Black, 29% Latinx/Hispanic, 6% female and 89% men who have sex with men (MSM). Of the 537 named partners, 191 (35·6%) were linked to index cases in a cluster and of those 16% were either new diagnoses or viremic. There was no statistically significant difference in the probability of identifying partners with HIV transmission potential among index clients in a rapidly growing cluster versus those not in a cluster (adjusted Relative Risk 1·82, (0·81–4·06)).
Conclusion:
Partner services that were initiated from index clients in a molecular cluster yielded similar new HIV case finding or identification of those with viremia as did interviews with index clients not in clusters. It remains unclear whether these findings are due to temporal disconnects between diagnoses and cluster identification, unobserved cluster members, or challenges with partner services implementation.
Keywords: molecular surveillance, HIV, partner services, linkage to care, cluster membership
Cluster detection and response1 is a newer strategy where HIV molecular analysis is used to detect growing HIV transmission clusters and direct HIV prevention and care services where they can have the greatest impact. Cluster detection and response using such molecular HIV data is a promising approach to both improve HIV prevention and prioritize limited public health resources. Sexually transmitted infections, including HIV, diffuse through contact networks2, and the number of onward transmissions varies widely.3 With limited public health resources, it is important to focus and optimize enhanced prevention and care services among those most likely to transmit HIV and their sexual partners. Simulations also demonstrate that concentrating efforts towards people living with HIV centrally positioned in molecular clusters could be an efficient way to decrease onward transmissions.4,5 This approach has also been demonstrated to reduce HIV transmission in applied public health settings. For example, members of a rapidly expanding, phylogenetically-identified drug resistant HIV cluster in Canada were provided enhanced public health follow up to ensure linkage to care6 – resulting in reduced transmission of drug resistant HIV from this cluster.
Despite the interest and rapid development of molecular cluster-guided public health activities using HIV-TRACE7 (Transmission Cluster Engine) and other programs6, there are several important questions that have not been tested, and may not be feasibly tested, in experimental trials. The most pressing question is whether molecular cluster-based approaches are more effective in yielding those most likely to acquire or transmit HIV - the members of the risk environment8 - as compared with current surveillance and partner services-based approaches conducted by local health departments.9 The current standard is for partner services staff to interview as many persons newly diagnosed with HIV as feasible in order to elicit information about their sex or drug-using partners who are then confidentially notified of their possible exposure to HIV; there are major constraints for many jurisdictions in expeditiously reaching all those who are newly diagnosed, prompting this study evaluating prioritization based on molecular data. The goal is for health departments to identify people who are undiagnosed and offer treatment, or to offer prevention services to people who are HIV-negative and report exposure to an HIV case. We evaluated if aligning molecular cluster response and partner services efforts enables better identification of people newly diagnosed with HIV or viremic and at-risk people who are HIV-negative and part of the larger risk environment, relative to current real-world practice. The risk environment includes both molecular clusters, those in active HIV transmission networks, inclusive of people living without HIV but in close proximity to those with HIV transmission potential. We conducted a historical cohort study using linked partner services and HIV pol sequence data to determine ifthe likelihood of identifying partners with HIV transmission potential (newly diagnosed or previously diagnosed with viremic infection) was increased by molecular cluster membership. In this way, we can begin to understand the potential public health impact of cluster detection and potential challenges of implementing a real-world response with the goal of accelerating the end of the HIV epidemic.
Methods
Our analysis included health department data collected through routine HIV surveillance and partner services. We matched and analyzed HIV surveillance registry10, partner services data and HIV pol sequences for all Chicago residents newly diagnosed from 2012–2016. We used partner services data from 2011–2018 to allow for adequate follow-up and matching between the datasets. HIV registry data included10 age, sex at birth, race/ethnicity, transmission category (MSM, injection drug use), and HIV diagnostic, genotype and viral load results (VL of >200 copies). Partner services data abstracted from Chicago Health Information Management System (CHIMS) included newly diagnosed index clients, partners of index clients, and case investigation disposition. Data management and analysis followed ethics review from the University of Chicago and the Chicago Department of Public Health and adhered to local, state, and federal security and confidentiality guidelines. The institutional review boards from the Chicago Department of Public Health and the University of Chicago reviewed and approved all study procedures.
HIV-TRACE data and cluster growth definitions
HIV-TRACE11 constructs molecular clusters of HIV sequences by calculating all pairwise genetic distances between aligned sequences. Genetic distance is a proxy for epidemiological relatedness because it increases as a function of time since transmission (recipient’s virus diverging from source’s virus, with each changing). The molecular clock underlying this sequence divergence between source-recipient pairs of HIVs, however, is highly imprecise due to immune/drug selection pressure, viral latency, and other factors. Furthermore, the virus can evolve at different rates in the donor and recipient, so the genetic distance between source and recipient strains is not simply translatable to a standardized time since they diverged. Clusters include sequences that diverge less than a pre-specified threshold (usually 0·5–2%). This threshold is chosen because it is an average estimate of within patient evolution12, segregates well between the two distributions of distances seen in large sets of sequences13 and agrees with the genetic distance seen between named HIV risk partners.14 Clustered sequences can help focus public health intervention based on concern for their size, associated epidemiological or clinical features, or growth.
We constructed molecular clusters using HIV-TRACE at a pairwise genetic distance threshold of 0·5%. Our primary analysis to identify if cluster membership increases identification of those with HIV transmission potential uses CDC’s recent and rapid growing cluster (hereinafter referred to as RR3) metric which is defined as clusters at 0·5% genetic distance threshold formed in the most recent 3-years, where at least 3 cases in the cluster were diagnosed in the most recent year.14 As additional analyses we explored two additional growth metrics: 1) CDC defined recent and rapid cluster with at least 5 cases in the last year (RR5); and 2) Square root growth - the number of cases added to this cluster in the past 12 months divided by the square root of cluster size, defined at distance threshold 0·5%.4 To dynamically calculate these growth statistics, we performed a network graph depth-first search for each individual on the date of their diagnosis, ignoring links to partner nodes with a diagnosis date that postdated this individual’s diagnosis date. For this calculation, we considered nodes linked in the network with diagnoses as early as 2009 (i.e., 36 months prior to 2012 diagnoses).
Primary Outcome Variable
The primary outcome variable was identification of any named partners with HIV transmission potential19 vs. no partners with HIV transmission potential. All indexes who did not name a partner were excluded from this analysis. HIV transmission potential includes persons in three categories: 1) people previously undiagnosed and not yet on ART; 2) people previously diagnosed with HIV and with viremia (detectable viral load (>= 200 copies) according to their viral load sampled closest to their index’s partner services interview assignment date or initiation date within one year prior ); or 3) people previously diagnosed who did not have a viral load registered in eHARS within one year prior to, or after, the Index’s Partner Services interview assignment date or initiation date. Each of these are epidemiologically important because they pose transmission potential. Having no partners with HIV transmission potential includes: (1) named partners whose viral load was suppressed (<200 copies) according to their viral load sampled closest to their index’s partner services interview assignment date or initiation date (within one year prior); (2) named partners who were presumed HIV negative- (no match to eHARS and partner services’ outcome of HIV negative or suggesting not HIV+), and (3) other HIV dispositions inconclusive of HIV status.
This definition adds a focus on the previously diagnosed who are not suppressed through ART to the original intention of partner services in order to identify new diagnoses. In the US, people who are viremic as opposed to those newly diagnosed contribute the most to onwards HIV transmission events 15 thus, focusing on those who are viremic in robust (re-)engagement in care activities are critical for modern partner services efforts.16,17
Analysis
The prevalence of those with HIV transmission potential was compared by index participant characteristics (in molecular cluster or not) using chi-square tests. We used multivariable Poisson regression with robust error variance to examine associations between membership in rapidly growing clusters (RR3), other index patient characteristics, and the likelihood of identifying partners with HIV transmission potential. We computed relative risks and 95% confidence intervals to quantify the magnitude of associations. Poisson regression avoids convergence issues frequently encountered in log-binomial regression and relative risks were computed because the odds ratio over-estimates the relative risk in situations where the outcome is common, and the relative risk has a more intuitive interpretation. As sensitivity analyses, we also explored the use of two other clustering metrics, including RR5 and the square root growth16 using the same analytic approach. Analyses were conducted in SAS software version 9.4. (SAS Institute, Cary NC).
Role of Funder
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The lead authors had final responsibility for the decision to submit for publication.
Results
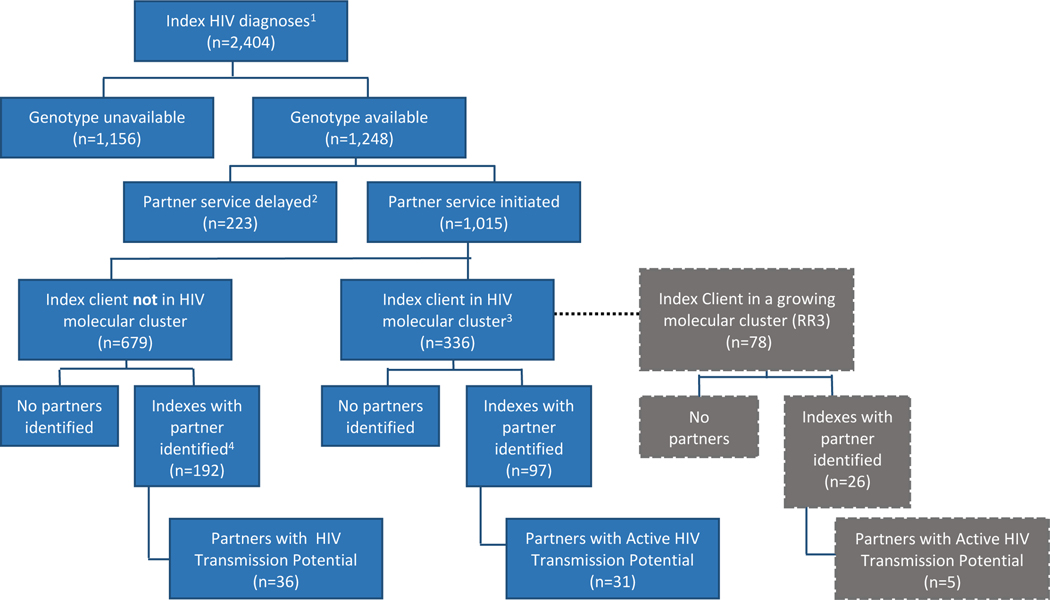
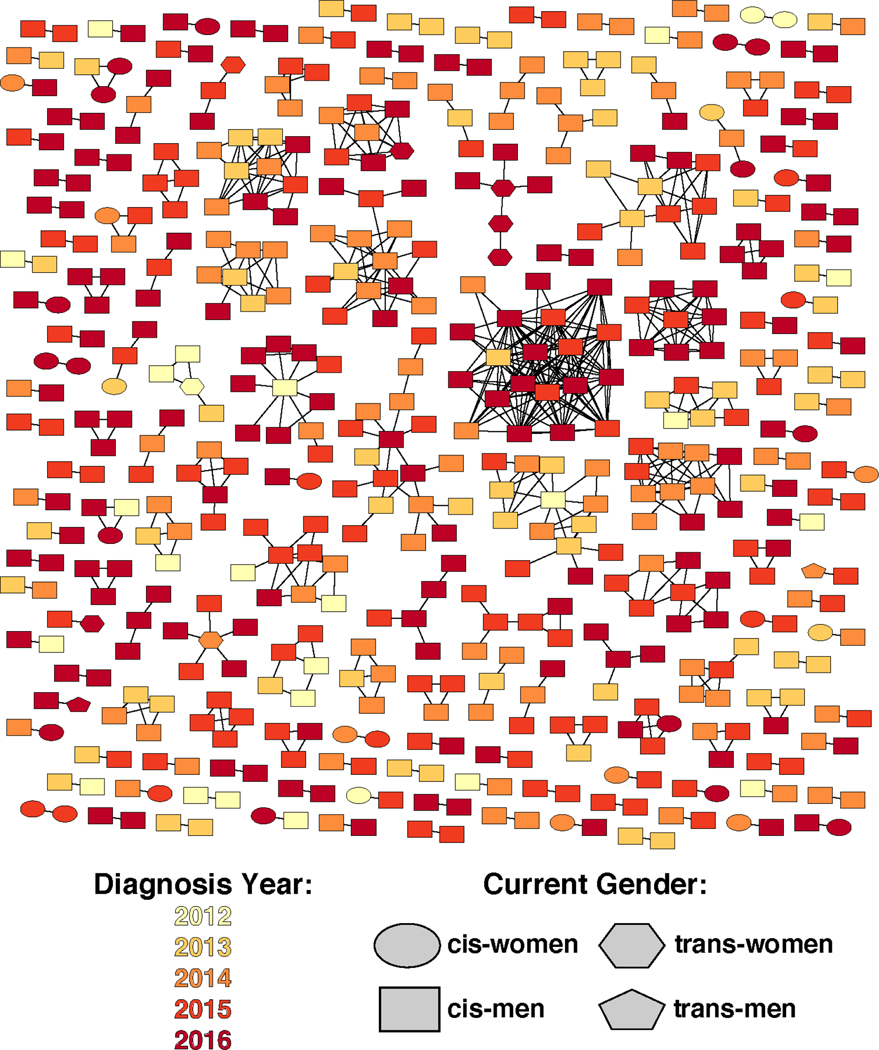
The sample was diverse and reflective of HIV epidemics in many urban epicenters (Table 1). A flow schematic of partner services’ index cases newly diagnosed with HIV between 2012–2016 is presented in (Figure 1). Of 5,208 newly diagnosed persons in 2012–2016, 1,015 (19%) had both HIV sequence data available and partner services initiated within 12 months of diagnosis. Of these, 336 (33%) had HIV pol sequences that molecularly clustered at 0.5% genetic distance and 97 (29%) of them named at least one risk partner. The entire network generated from diagnoses over this time period is visualized (Figure 2). There were 289 indexes identified for analysis, regardless of cluster membership, who had identified at least one partner. One index was dropped from our analysis because their partner record was missing from our partner services dataset. The majority of these index clients were aged 25–34 (39·9%), 52·1% were Black, 26·4% Latinx, 7·6% female and 84·0% MSM. Of these 288 indexes, 100 indexes had at least one HIV-negative partner named, with 36% originating from index clients in a cluster versus 64% from index clients not in a cluster (data not shown). Of the 191 partners named by index clients in a 0·5% molecular cluster, 46 were living with HIV and of those 67% were either viremic or had no labs reported.
Table 1.
Index and partner characteristics, 2012–2016, Chicago IL
| Index | Any named partners with HIV transmission potential1 | No named partners with HIV transmission potential2 | ||||
|---|---|---|---|---|---|---|
| Variable | N | col % | N | % | N | % |
| Total | 288 | 65 | 22·6 | 223 | 77·4 | |
| Age3 | ||||||
| 13–24 | 110 | 38·2 | 30 | 46·2 | 80 | 35·9 |
| 25–34 | 115 | 39·9 | 21 | 32·3 | 94 | 42·2 |
| 35–44 | 35 | 12·2 | 7 | 10·8 | 28 | 12·6 |
| 45+ | 28 | 9·7 | 7 | 10·8 | 21 | 9·4 |
| Sex at birth | ||||||
| Male | 266 | 92·4 | 59 | 90·8 | 207 | 92·8 |
| Female | 22 | 7·6 | 6 | 9·2 | 16 | 7·2 |
| Race/Ethnicity | ||||||
| Black | 150 | 52·1 | 38 | 58·5 | 112 | 50·2 |
| Latine | 76 | 26·4 | 16 | 24·6 | 60 | 26·9 |
| White | 46 | 16·0 | 7 | 10·8 | 39 | 17·5 |
| Other | 16 | 5·6 | <5 | 6·2 | 12 | 5·4 |
| MSM identified | ||||||
| Yes | 242 | 84·0 | 54 | 83·1 | 188 | 84·3 |
| No | 46 | 16·0 | 11 | 16·9 | 35 | 15·7 |
| Cluster growth I4 | ||||||
| Non-clustered | 191 | 66·3 | 36 | 55·4 | 155 | 69·5 |
| Clustered, not rapidly growing5 | 82 | 28·5 | 24 | 36·9 | 58 | 26·0 |
| Clustered and rapidly growing | 15 | 5·2 | 5 | 7·7 | 10 | 4·5 |
Newly diagnosed and/or unsuppressed virus identified through standard partner services
Virally suppressed or HIV negative
At diagnosis
CDC definition
>3 cases in the last year
Number of cases added to this cluster in the past 12 months divided by the square root of cluster size, defined at distance threshold 0·5% (continuous outcome dichotomized at 1)
Figure 1.
Flow schematic of index clients newly diagnosed with HIV, Chicago IL, 2012–2016
146·2% of 2012–2016 new HIV diagnosis assigned to partner services
2Partner services initiated after 12 months- Initiation after 12 months may be due to lag time in HIV case or lab reporting.
30·5% genetic distance
4 1 partner identified was dropped because their partner record was missing from our partner services dataset
Figure 2.
Network Visualization of molecular clusters and growth parameters, Chicago IL 2012–2016 Diagnoses.1 Node color indicates year of sampling and shape denotes gender.
10·5% genetic distance
For our primary outcome, adjusted models demonstrate there was not a statistically significant difference in the probability of identifying people with HIV transmission potential among partners of index clients in a three year rapidly growing cluster (RR3) versus partners of index clients not in a RR3 cluster (adjusted Relative Risk [aRR] 1·82; 95% confidence interval [CI]0·81–4·06) (Table 2). In a sensitivity analyses using a different cluster metric (RR5), there was no statistically significant difference in the yield of people with HIV transmission potential linked to an index client in RR5 clusters versus those not in a RR5 cluster (aRR 1·53; 95% CI 1·0–2·34). There also was no statistical difference in the probability of identifying people with HIV transmission potential among clustered versus unclustered index clients using the square root growth metric (aRR 1·45; 95% CI 0·84–2·48) to define index clusters.
Table 2.
Poisson regression of factors associated with identification of clients with HIV transmission potential through partner services linked to molecular cluster analysis in Chicago 2012–2018.
| Any partners with HIV transmission potential1 | aRR2 | 95% CI | ||
|---|---|---|---|---|
| Variable | ||||
| Age4 (n, %) | ||||
| 13–24 | 30 | 46·2 | Ref | |
| 25–34 | 21 | 32·3 | 0·71 | 0·44 – 1.16 |
| 35–44 | 7 | 10·8 | 0·89 | 0·42 – 1.88 |
| 45+ | 7 | 10·8 | 1·07 | 0·49 – 2.37 |
| Sex at birth | ||||
| Male | 59 | 90·8 | Ref | |
| Female | 6 | 9·2 | 1·25 | 0·42 – 3.73 |
| Race/Ethnicity | ||||
| Black | 38 | 58·5 | Ref | |
| Latine | 16 | 24·6 | 0·86 | 0·50 – 1.45 |
| White | 7 | 10·8 | 0·61 | 0·28 – 1.34 |
| Other | <5 | 6·2 | 1·06 | 0·44 – 2.56 |
| MSM identified | ||||
| No | 11 | 16·9 | Ref | |
| Yes | 54 | 83·1 | 1·16 | 0·50 – 2.66 |
| Cluster growth I5 | ||||
| Non-clustered | 36 | 55·4 | Ref | |
| Clustered, not rapidly growing6 | 24 | 36·9 | 1·51 | 0·97 – 2.35 |
| Clustered and rapidly growing | 5 | 7·7 | 1·82 | 0·81 – 4.06 |
Newly diagnosed and/or unsuppressed virus
Adjusted for all variables for which estimates are presented
At diagnosis
CDC definition
>3 cases in the last year
Discussion
Our findings suggest that deployment of partner services for index clients in a growing transmission cluster may not increase the identification of partners who have a new HIV diagnosis or are viremic. This finding suggests the need for a more robust implementation of partner services linked to molecular cluster interventions; or perhaps other network-based interventions that can be deployed through current public health systems. It remains unknown whether the yield of people with HIV transmission potential would increase if the time between identification of a new diagnosis and deployment of partner services, to detection of cluster growth, were decreased. In addition, it remains unknown whether the yield of HIV negative partners, potential PrEP candidates through partner services, would increase as a result of cluster response, potentially reducing onward transmission. Of course, better PrEP engagement than currently exists19 would need to also be actualized to achieve any benefit from engagement with people who are potential PrEP clients. Current public health data systems are unable to link HIV negative but at-risk persons from partner services data to those eligible for or engaged in PrEP care, thus we were unable to better classify those persons which might improve status neutral approaches to HIV elimination. Partner services clearly can increase clients in PrEP care,19,20 and further study could examine whether the yield of people who are HIV negative from partner services deployed for people in growing molecular clusters improves identification of those at higher risk of HIV acquisition.
Following molecular cluster growth over time by repeated HIV-TRACE analyses of accumulating sequences from additional new HIV cases is now recommended by CDC, rather than using phylogenetics.14 Phylogenetics is much more computationally- and time-intensive, especially if a time variable is added; the latter limits the feasible number of sequences that can be used. Another complication is that as more sequences are added to phylogenetic trees, formerly robust clusters can break down. When the goal is tracking molecular cluster growth over time while adding more individuals’ sequences, this is highly undesirable. Sequences clustered using HIV-TRACE will always be clustered using HIV-TRACE; adding more data can only increase the size of clusters, not break them apart. For these reasons, HIV-TRACE was adopted as the feasible implementation for “phylodynamics” to guide health department interventions. Cluster growth cut-offs can be modified through HIV-TRACE and are likely relevant at different stages of epidemics and what response can be generated (ie RR3 would generate more clusters than RR5). We use RR3 given the evolving epidemic and future work that focuses on low-burden epidemics or outbreaks as getting to zero efforts are realized.Our findings should be interpreted with some notable limitations. We classified people lacking a recent lab as having “HIV transmission potential”, although a lack of HIV care lab values may result when people move out of the public health jurisdiction (i.e., out of Chicago) and has been found elsewhere to substantially distort care status determination.21 Similarly, people who are deceased may be categorized as out of care and HIV TRACE was not utilized across jurisdictions in this analysis. Additionally, we only considered partners who had enough information to initiate partner services. Anonymous or semi-anonymous partners may have been excluded when there was not enough identifying information reported to the health department to attempt locating.
Given that the cluster detection and response strategy is relatively new and implementation is still in formation, there is little evidence of the impact of focused interventions for people in growing clusters and their associated transmission and risk networks. However, there are on-going evaluation efforts in the US in Houston, the state of Texas, state of New York and New York City in addition to a third in Vancouver, Canada that utilize molecular cluster data to identify and intervene within molecular clusters and their networks.6 One such CDC-funded demonstration project was implemented in Houston Texas, New York State, and New York City and focused efforts on cluster detection and response among Hispanic/Latino MSM and people in their networks.22,23 The project, named “Project Conéctate” by CDC, was initiated in 2017 with project close-out and results expected in 2021. Notably, Houston utilized a social network strategy which may have several strengths as a network approach when compared to traditional partner services that were utilized in this analysis.24 Despite lack of evidence of impact highlighted by our findings, cluster detection and response has shown promising results in identifying prevention and care service gaps and needs of specific populations.
There are other approaches that could be utilized with timely molecular cluster data. Indeed, this aspect of current partner services yield cannot be assessed unless interview data on named partners are collected from those lacking sequences as well as those with genotypes, as proposed here and not previously completed in other published reports.11 In addition, partner services currently ends with investigation of new diagnoses, while molecular cluster-informed investigations could re-initiate/repeat interviews with those in growing clusters who are out of care or unsuppressed in care. Optimizing partner services or social network strategies that move more progressively further away from cluster membership (network members of network members) irrespective of HIV status are worth further investigation. Will targeting these more intensive services to fewer persons in concerning clusters improve yield? Will these better reach HIV negative, at-risk individuals for linkage to effective prevention services (ie. PreP)? Further, examination of the optimal frequency of revisiting clusters and intervening is critically needed. Finally, mathematical modeling and computational simulation may provide guidance into optimization of partner services25 and other network-based intervention efforts that follow molecular cluster identification given the complexity in empirical testing of such intervention efficacy. Greater efficiency, however, of network and temporal based approaches to building upon molecular cluster analyses will likely also be highly susceptible to variations in epidemic phase and other contextual factors as we move closer to HIV elimination.
Acknowledgement:
Funding for this research was provided by the National Institute of Allergy and Infectious Disease, National Institute of Health (1R01AI136056) and through a supplemental grant from the Third Coast Center for AIDS Research (CFAR), an NIH funded center (P30AI117943). This work was supported through funding by the Centers for Disease Control and Prevention (cooperative agreement NU62PS924572). The content, findings, and views expressed are those of the authors and do not necessarily represent the official views of the Houston Health Department, the Centers of Disease Control and Prevention, the Department of Health and Human Services, or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interest: We have no competing interests to declare
References
- [1].Oster AM, Panneer N, Lyss SB, McClung RP, Watson M, Saduvala N, et al. Increasing Capacity to Detect Clusters of Rapid HIV Transmission in Varied Populations-United States. Viruses 2021;13(4):577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anderson RM & May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press; 1992. [Google Scholar]
- [3].Hamilton DT, Handcock MS, Morris M. Degree distributions in sexual networks: A framework for evaluating evidence. Sex Transm Dis 2008;35:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wertheim JO, Murrell B, Mehta SR, Forgoine LA, Kosakovsky Pond SL, Smith DM, Torian LV. Growth of HIV-1 Molecular Transmission Clusters in New York City. J Infect Dis 2018;218(12):1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, et al. Using HIV networks to inform real time prevention interventions. PLoS One 2014;9:e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Poon AF, Gustafson R, Daly P, Zerr L, Demlow SE, Wong J, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV 2016;3:e231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (TRAnsmission Cluster Engine): a Tool for Large Scale Molecular Epidemiology of HIV-1 and Other Rapidly Evolving Pathogens. Mol Biol Evol 2018;35(7):1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Morgan E, Skaathun B, Schneider JA. Sexual, Social, and Genetic Network Overlap: A Socio-Molecular Approach Toward Public Health Intervention of HIV. Am J Public Health 2018;108(11):1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gore D, Ferreira M, Khanna AS, Schneider J. Human Immunodeficiency Virus Partner Notification Services Among a Representative Sample of Young Black Men Who Have Sex with Men Demonstrates Limited Service Offering and Potential Benefits of Clinic Involvement. Sex Transm Dis 2018;45(9):636–641. doi: 10.1097/OLQ.0000000000000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cohen SM, Gray KM, Ocfemia MC, Johnson AS, Hall HI. The status of the National HIV Surveillance System, United States, 2013. Public Health Rep 2014;129(4):335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wertheim JO, Pond SLK, Forgione LA, Mehta SR, Murrell B, Shah S et al. Social and Genetic Networks of HIV-1 Transmission in New York City. Plos Pathog 2017;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004;18:719–28. [DOI] [PubMed] [Google Scholar]
- [13].Morgan E, Skaathun B, Nikolopoulos GK, Paraskevis D, Williams LD, Smyrnov P et al. A Network Intervention to Locate Newly HIV Infected Persons Within MSM Networks in Chicago. AIDS Behav 2019;23(1):15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Centers for Disease Control and Prevention. Detecting, and Responding to HIV Transmission Clusters: A guide for Health Departments. Version 2.0, June 2018. https://www.cdc.gov/hiv/pdf/funding/announcements/ps18-1802/CDC-HIV-PS18-1802-AttachmentE-Detecting-Investigating-and-Responding-to-HIV-Transmission-Clusters.pdf.
- [15].Li Z, Purcell DW, Sansom SL, Hayes D, Hall HI. Vital Signs: HIV Transmission Along the Continuum of Care — United States, 2016. MMWR Morb Mortal Wkly Rep 2019;68:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oster AM, France AM, Panneer N, Ocfemia MC, Campell E, Dasgupta S, et al. Identifying Clusters of Recent and Rapid HIV Transmission Through Analysis of Molecular Surveillance Data. J Acquir Immune Defic Syndr 2018;79(5):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DS et al. The global transmission network of HIV-1. J Infect Dis 2014;209:304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oster AM, Wertheim JO, Hernandez AL, Ocfemia MCB, Saduvala N, Hall HI. Using Molecular HIV Surveillance Data to Understand Transmission Between Subpopulations in the United States. Jaids-J Acq Imm Def 2015;70:444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Teixeira da Silva D, Bouris A, Ramachandran A, Blocker O, Davis B, Harris J, et al. Embedding a Linkage to Preexposure Prophylaxis Care Intervention in Social Network Strategy and Partner Notification Services: Results From a Pilot Randomized Controlled Trial. J Acquir Immune Defic Syndr 2021;86(2):191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bhatia R, Modali L, Lowther M, Glick N, Bell M, Rowan S, et al. Outcomes of Preexposure Prophylaxis Referrals From Public STI Clinics and Implications for the Preexposure Prophylaxis Continuum. Sex Transm Dis 2018;45(1):50–55. [DOI] [PubMed] [Google Scholar]
- [21].Buskin SE, Kent JB, Dombrowski JC, Golden MR. Migration distorts surveillance estimates of engagement in care: results of public health investigations of persons who appear to be out of HIV care. Sex Transm Dis 2014;41(1):35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Braunstein S. Fast-Track Cities: Opportunities for Implementation Science: Our data-driven approach to ending the epidemic in New York City [Lecture]. International Association of Providers in AIDS Care November 2020. [Google Scholar]
- [23].Oster A. Detecting HIV transmission clusters to better prioritize prevention efforts [Lecture]. National Latinx Conference May 2018. [Google Scholar]
- [24].Skaathyn B, Pho MT, Pollack HA, Friedman SR, McNulty MC, Friedman EE, et al. Comparison of effectiveness and cost for different HIV screening strategies implemented at large urban medical centre in the United States. J Int AIDS Soc 2020; 23(10): e25554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roberts ST, Khanna AS, Barnabas RV, Goodreau SM, Baeten JM, Celum C, et al. Estimating the impact of universal antiretroviral therapy for HIV serodiscordant couples through home HIV testing: insights from mathematical models. J Int AIDS Soc 2016;19(1):20864. [DOI] [PMC free article] [PubMed] [Google Scholar]