Abstract
A wide range of perfusable microvessel models have been developed, exploiting advances in microfabrication, microfluidics, biomaterials, stem cell technology, and tissue engineering. These models vary in complexity and physiological relevance, but provide a diverse tool kit for the study of vascular phenomena and methods to vascularize artificial organs. Here we review the state-of-the-art in perfusable microvessel models, summarizing the different fabrication methods and highlighting advantages and limitations.
Introduction
The endothelium is an organ system that comprises over 60 trillion cells that form 100 000 km of interconnected vessels with a surface area of 4000 m2.1–3 The diameter of blood vessels in humans spans more than four orders of magnitude, from about 8 μm in capillaries to more than 1 cm in large elastic arteries.1 In larger vessels there are hundreds of cells around the perimeter, whereas in a capillary a single endothelial cell (EC) may wrap around to form a junction with itself as well as its upstream and downstream neighbors.4 In addition to the ultrastructural diversity across arteries, veins, and capillaries, ECs also exhibit broad molecular heterogeneity.1–3 For example, endothelial permeability in different vascular beds is modulated by the expression of different junctional proteins.5,6 The endothelium performs multiple functions, including regulating permeability, vasomotor tone, leukocyte trafficking, hemostasis, and angiogenesis.1–3 Endothelial cells respond to a wide range of input stimuli including biochemical (e.g. small molecules, hormones, proteins, and cells) and physical cues (e.g. hemodynamic shear stress, oxygen, and curvature).7,8
In vitro microvascular models provide new tools for fundamental and translational studies. In basic science these models can be used to study the structure and function of the endothelium in response to a wide range of biochemical stimuli (e.g. vasomodulators, pro-angiogenic factors) and physical perturbations (e.g. flow rate, pressure), and the mechanisms of angiogenesis and vessel formation. In vitro models can also be used to study the endothelial dysfunction and provide insight into the molecular mechanisms of disease. Endothelial dysfunction is associated with cardiovascular disease (e.g. coronary artery disease, peripheral artery disease, and hypertension), the leading cause of death in the US.9 These models can be used to study drug transport, uptake, and efficacy. In regenerative medicine, in vitro models can be used to develop design rules for vascularizing tissues and organs. Overall, in vitro models allow a reductive approach to addressing scientific questions with control over all experimental variables, and hence are complementary to animal models which have greater physiological relevance but where it is more difficult to independently control experimental variables.
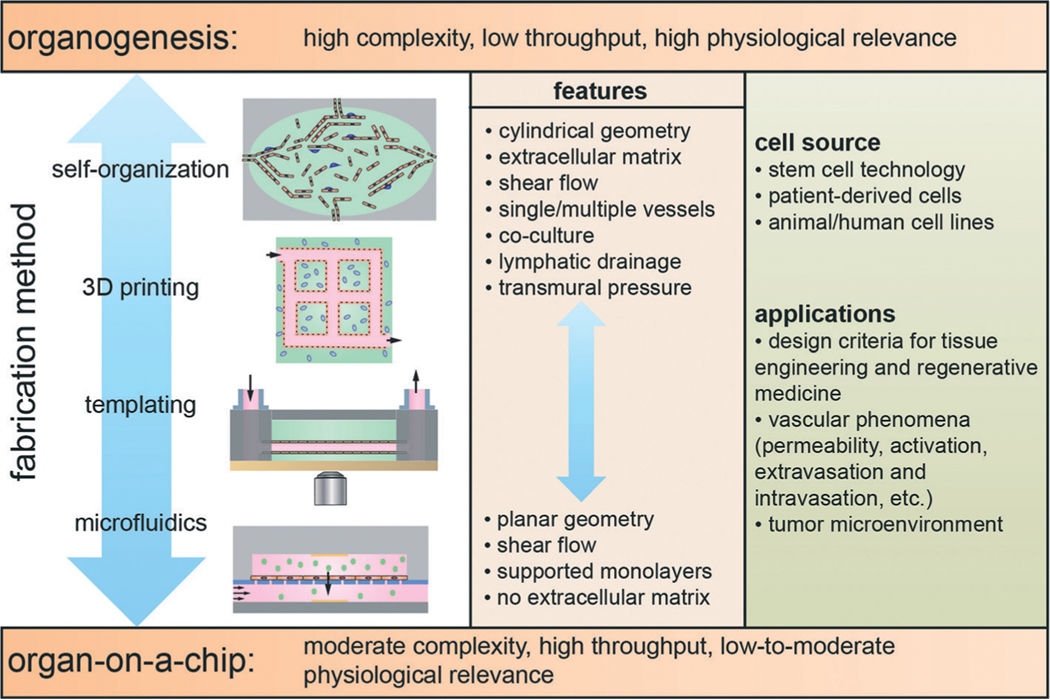
In vitro microvessel models can be broadly categorized as organ-on-a-chip platforms or organogenesis-based models (Fig. 1). Organ-on-a-chip platforms exploit microfabrication and microfluidics technologies to recapitulate specific aspects of vessel structure and function. In general these platforms have moderate complexity and are high throughput since they are relatively easy to fabricate and the endothelium can be formed in 2–4 days.10,11 In contrast, organogenesis involves the self-organization of stem cells and/or organ specific stem cells into a structure that recapitulates specific functions of the organ.12 These models are usually characterized by high complexity and low throughput since culturing stem cells is generally challenging and time consuming, and microvessel formation typically takes 1–2 weeks. While organ-on-a-chip platforms and organogenesis models represent very different approaches, there are many hybrid models that use elements of both. These models may use cell lines, primary cells, or patient derived pluripotent stem cells. Many models take advantage of advances in 3D cell culture, stem cell technology, and the development of extra-cellular matrix (ECM) models.13–20
Fig. 1.
Schematic illustration of the fabrication methods and features of in vitro microvessel models. Models can be categorized as organ-on-a-chip platforms, that have moderate complexity and are high throughput, and organogenesis-based models that are characterized by high complexity and low throughput. Organogenesis refers to the self-organization of stem cells and/or organ-specific progenitor cells into tissue that resembles a specific organ. Platforms exploiting self-organization are generally challenging due to difficulties in differentiating and culturing stem cells, and time consuming since endothelium formation takes 1–2 weeks. Organ-on-a-chip platforms are devices that use microfabrication and microfluidics technologies to recapitulate specific aspects of organ structure and function. In general, organ-on-a-chip platforms are relatively easy to fabricate and endothelial layers can be formed in 2–4 days. The fabrication methods include: microfluidics, templating, 3D printing, and self-organization.
Here we review the state-of-the-art in perfusable in vitro microvascular models. These models incorporate elements of microfabrication, tissue engineering, stem cell technology, biomaterials, and cell biology. We consider four broad fabrication methods: microfluidics, templating, 3D printing, and self-organization. In each case we discuss the capabilities and the features of the endothelium that can be recapitulated.
Microfluidic-based devices
Microfluidic devices have been exploited in microvascular research, primarily for their ability to impose laminar flow on monolayers of vascular ECs in a 2D planar geometry. Microfluidic devices are also easily integrated into live cell chambers, enabling real time imaging of the response of endothelial monolayers to shear stress or other external biochemical or mechanical perturbations. Research using these devices has elucidated the role of shear stress in regulating endothelial cell morphology, alignment, cytoskeleton reorganization, and protein/gene expression.7,21–27
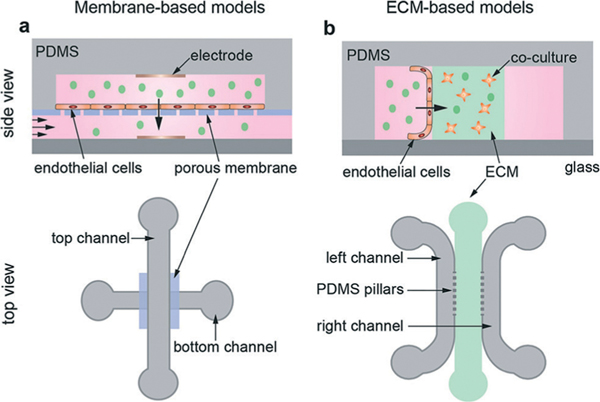
More complex devices, incorporating multiple microfluidic channels, generally fall into two categories depending on whether the endothelial monolayer is cultured on a membrane or on an extracellular matrix material (ECM) (Fig. 2 and Table 1). In membrane-based devices, endothelial monolayers are cultured horizontally on a porous membrane that separates an upper channel from a lower channel (Fig. 2a). In ECM-containing devices, endothelial monolayers are cultured onto the sidewall of an extracellular matrix material that separates two microfluidic channels (Fig. 2b). The devices are usually several centimeters in length and several millimetres in width, with channels that are typically 100 μm to 500 μm in height. The region containing the ECM is typically 1–5 mm in width.
Fig. 2.
Schematic illustration of membrane-based and ECM-containing microfluidic devices. (a) A membrane device with endothelial cells cultured on a porous membrane sandwiched between two orthogonal polydimethylsiloxane (PDMS) channels. Electrodes for transendothelial electrical resistance (TEER) measurements can be embedded in the top and bottom channels. These platforms are similar to transwell devices with the addition of shear flow. (b) ECM device with ECM separating two parallel channels. Endothelial cells are seeded onto the vertical sidewall of one of the channels. In addition, other cell types can be co-cultured in the ECM.
Table 1.
Features of membrane-based microfluidic models and selected measurements. P – permeability, TEER – transendothelial electrical resistance. PAEC – pulmonary aortic endothelial cells, HPMVEC – human pulmonary microvascular endothelial cells, b.End3 – immortalized mouse brain endothelial cell line
| Cells | Co-culture | Shear stress (dyne cm −2) | Molecule | P (cm s−1) | TEER (Ω cm2) | Ref. |
|---|---|---|---|---|---|---|
|
| ||||||
| b.end3 mouse brain ECs | n/a | n/a | n/a | n/a | 200−250 | 28 |
| PAECs | n/a | 1−100 | FITC−BSA | 10−5 | n/a | 29 |
| b.end3 mouse brain ECs | Astrocyte | Not reported | 4, 20, 70 kDa dextrans | 10−5–10−6 | 300 | 30 |
| HPMVEC | Alveolar epithelial cells | 15 | FITC−BSA | Not reported | 800 | 33 |
Membrane-based devices.
The membrane-based device models (Table 1) represent miniaturized versions of the standard transwell device used to measure the barrier properties of endothelial monolayers, but with the key difference that laminar flow can be introduced into one or both channels (Fig. 2a). Incorporation of electrodes into each channel allows measurement of the transendothelial electrical resistance (TEER).28 The permeability of the endothelial monolayer can be assessed by introducing a molecule of interest in the upper chamber and measuring transport to the lower chamber, for example by using a fluorescent tag.29–31 By culturing different cell lines, these devices have been used to model the gut, lung, and the blood-brain barrier.28–33
In a further modification of these membrane-based devices, upper and lower channels are aligned in parallel and large channels on either side of the stack are used to stretch the membrane by decreasing the air pressure.32,33 In a lung-on-a-chip model, alveolar epithelial cells are cultured on the top of the porous membrane and endothelial cells are cultured on the bottom. Simulating lung expansion during air inhalation by stretching the endothelial and epithelial monolayers was found to reduce permeability, increase TEER, and increase the susceptibility of epithelial cells to the cytotoxic effects of silica nanoparticles.33
ECM-containing devices.
In ECM-containing microfluidic devices (Table 2), narrowly spaced PDMS pillars are used to confine an extracellular matrix material between the two channels (Fig. 2b). A pillar spacing of less than 200 μm is required to contain the ECM material within its respective channel. Endothelial cells are then cultured on the vertical face of the ECM between the pillars in one or both of the channels. The introduction of ECM allows live cell imaging of interactions between the endothelium and the microenvironment. Growth factors or other chemicals can be introduced into the empty channel or into the ECM to establish gradients across the endothelium. This technique has been used to demonstrate that VEGF gradients, as well as interstitial flow and shear stress, play a significant role in sprouting and angiogenesis of HUVEC monolayers into the ECM.34 Cancer cells can be introduced into the microfluidic channels to observe extravasation into the ECM, or they can be embedded in the ECM in order to observe intravasation from the ECM across the endothelial monolayer into the microfluidic channel. ECM-containing models have been used to quantify the enhancement of cancer cell extravasation in the presence of CXCL5 and CXCL12 gradients35,36 by measuring the fraction of cancer cells extravasated and the distance of cancer cell migration into the ECM. In studies of intravasation, cancer cells in the ECM exhibited enhanced intravasation in the presence of TNF-α and EGF gradients.37
Table 2.
Features of ECM-containing microfluidic models and selected measurements. HUVEC – human umbilical vein endothelial cells, MDA-MB-231 – metastatic breast cancer cell line, hBM-MSCs – human bone marrow mesenchymal stem cells, ACC-M – metastatic adenoid cystic carcinoma cell line, HT1080 – fibrosarcoma cell line
| ECM, (cancer cell type) | Co-culture | Chemical gradients | Measurement | Values | Ref. |
|---|---|---|---|---|---|
|
| |||||
| Collagen | n/a | VEGF | % area of HUVEC invasion (3 days) | 0–80% | 34 |
| Matrigel (MDA-MB-231) | hBM-MSCs | CXCL5 | % extravasated distance migrated (5 days) | 40–80% 20–60 μm | 35 |
| Basement membrane extract (ACC-M) | n/a | CXCL-12 | Distance migrated (2 days) | 5–40 μm | 36 |
| Collagen (HT1080) | RAW264.7 Macro- phages | EGF, TNF-α | % cells intravasated migration speed P (70 kDa Dextran) | 0–8% 0–30 μm h−1 10−5 cm s−1 | 37 |
Advantages and limitations.
The membrane-based microfluidic devices have relatively high throughput with moderate complexity. These models are attractive for organs with barrier function such as gut and lung, where the cylindrical vessel geometry and tissue microenvironment are not thought to play a significant role in establishing barrier properties. The ability to introduce laminar flow is a significant advantage over conventional transwell devices where shear stress is important in establishing cell morphology and barrier function. A disadvantage of these platforms is the difficulty in live cell imaging due to the presence of the porous membrane.
The ECM-containing devices have increased complexity but do allow live cell imaging. The design of the PDMS pillars is crucial to allow the ECM precursor to flow into the central channel without leaking into the microfluidic channels on either side. The incorporation of ECM allows elements of the tissue microenvironment, such as co-culture with different cell types, to be incorporated into the platform. Similar to the membrane-based devices, these models do not recapitulate the cylindrical geometry and continuous lumen of blood vessels. Since cells can sense changes in matrix stiffness up to 100–200 μm away, devices with an ECM height less than around 500 μm would be considered quasi-2D.
Templating 3D microvessel models
Fabrication.
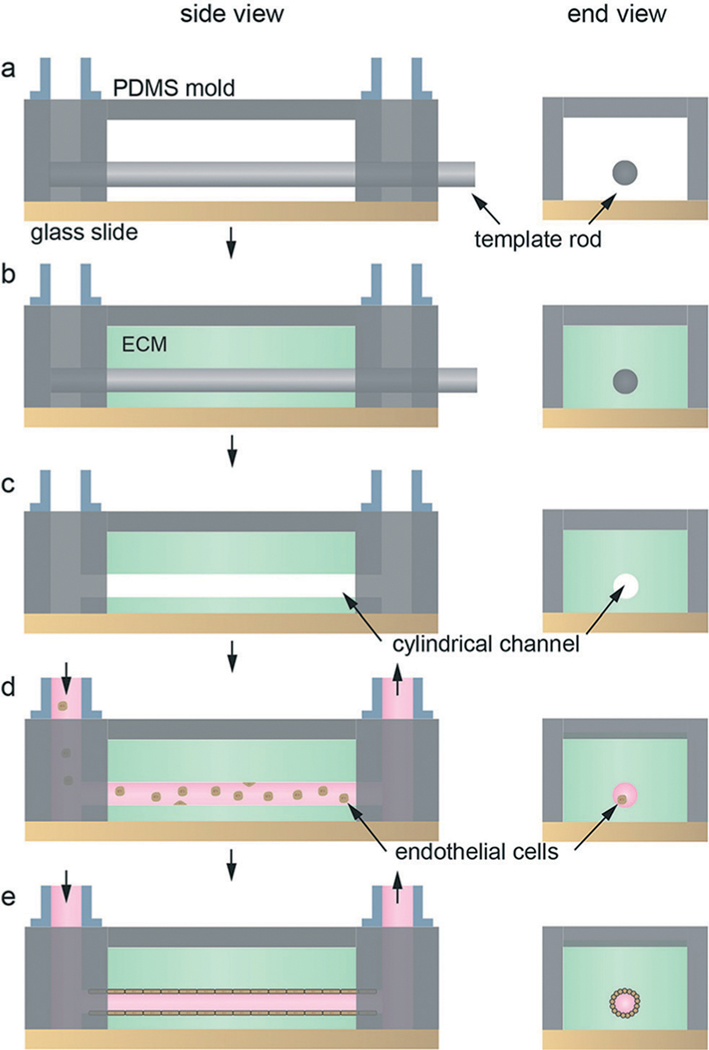
Microvessel templating methods involve casting an ECM material around a removable template and seeding ECs within the resulting empty channel or network (Fig. 3). The inlet and outlet of the channel or network is then connected to a flow loop to allow perfusion. Cylindrical channels can be fabricated using a needle or rod as a template that is physically removed by pulling the needle out of the surrounding ECM material, typically collagen type I or fibrin. The ECM is required to be sufficiently stiff to support the formation of a well-defined endothelium, inhibit EC invasion into the matrix, and resist shear and elastic deformation from channel perfusion. To fulfill these requirements, ECM protein concentrations are typically greater than 6 mg ml−1 (Table 3).38,39 Typical diameters of the template rod are 60–200 μm.40,41 While this technique produces geometrically relevant cylindrical channels, the template removal process limits the formation of vessels to simple linear structures.
Fig. 3.
Microvessel fabrication with cylindrical template. (a) A template rod inserted into a PDMS mold defines the location of the vessel. (b) A solution of the ECM, often collagen type I or fibrin, containing cells is introduced around the cylindrical template within the PDMS housing. (c) After gelation/cross-linking, the template rod is removed. (d) The platform is connected to a flow loop and endothelial cells are seeded into the cylindrical channel. (e) Adhesion and spreading of the endothelial cells on the internal surface of the ECM form the vessel lumen.
Table 3.
Features of templated perfusable microvessel models and selected results. P − permeability, EC – endothelial cell, Cn – collagen type I, Fn – fibrin. HDMEC – human dermal microvascular endothelial cells, HUVEC – human umbilical vein endothelial cells, LEC – lymphatic endothelial cells, HMVEC – human microvascular endothelial cells, TIME-RFP – telomerase immortalized microvascular endothelial cells – red fluorescent protein, HBVPC – human brain vascular pericytes, HUASMC – human umbilical arterial smooth muscle cells
| Template | Diameter (μm) | Shear stress (dyne cm−2) | P (cm s−1) sodium fluorescein | P (cm s−1) BSA | P (cm s−1) 70 kDa Dextran | ECM/density (mg ml−1) | EC type | Other cells | Ref. |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Needle | 55−150 | 1−30 | 0.1−7.9 × 10−6 | Cn/7, Fn/10 | HDMEC, HUVEC, LEC | Pericyte, leukocyte | 38, 40, 57, 69 | ||
| Needle | 700 | 1−10 | Cn/8 | TIME−RFP | MDA−MB−231 | 73 | |||
| Needle | 400 | Cn/2.5 | HUVEC, HMVEC | 39 | |||||
| Needle | 150−200 | 1−10 | 0.43−2.7 × 10−6 | Cn/7 | HUVEC, HMVEC | MDA−MB−231 | 54 | ||
| Lithographic pattern | 150 | 0.1−30 | 7.0 × 10−6 | 4.1 × 10−6 | Cn/6−10 | HUVEC | HBVPC, HUASMC | 43 | |
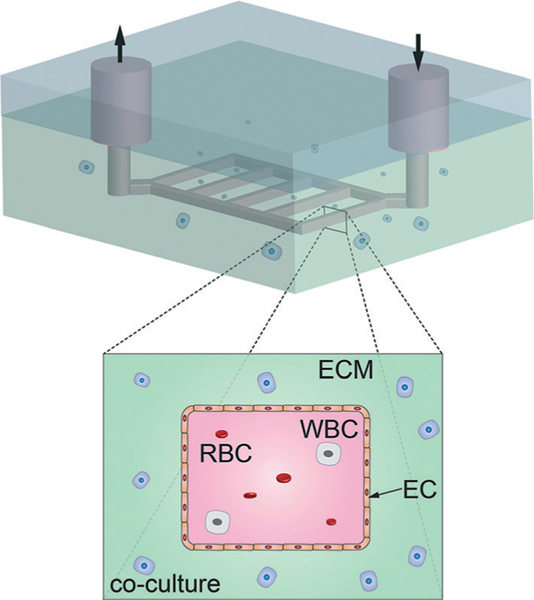
More complex vascular networks can be formed using lithographic techniques to produce molds or removable templates with interconnected rectangular channels (Fig. 4).42,43 Although templates produced by lithographic methods inherently have a rectangular cross-section, it has been shown that ECs seeded within the channels are able to form approximately cylindrical vessels despite the rigid corners.43 Templated networks of rectangular channels permit the study of branched vessels and thus vascular phenomena associated with bifurcations; however, these techniques are limited to 2D planar networks and similarly have been used to produce vessels with diameters ranging from 60–200 μm.42,43
Fig. 4.
2D microvessel array fabrication by lithographic patterning. Standard lithographic patterning is used to create a 2D array of rectangular channels in a matrix material. Following seeding with endothelial cells, the microvessels have rounded corners and display the versatility of co-culture with multiple cell types. RBC - red blood cells, WBC – white blood cells, EC – endothelial cells, and other relevant cells within the extracellular matrix (ECM).
The endothelium in templated microvessels is formed by introducing a suspension of ECs into the channel or network and allowing the cells to adhere and spread on the internal surface of the ECM (Fig. 3). This method of seeding generally limits vessel diameters to greater than 50 μm due to difficulties in distributing and achieving sufficient endothelial cell densities within small diameter templated channels; endothelial coverage of capillary-scale templated channels relies on enhancing endothelial migration from larger diameter portions.44 Another approach to obtaining perfused small diameter vessels involves guiding angiogenesis or vasculogenesis, from established larger vessels or dispersed endothelial cells embedded within the ECM, to form capillaries 10–20 μm in diameter.39,45,46 Perfusion through these self-organized capillaries is achieved by directing microvessel formation between both an inlet and outlet source of flow. Although these capillary networks have been established between perfusable cylindrical channels and separate PDMS compartments over significant distances (0.3 to 1 mm), they ultimately produce random and unpredictable networks and require multiple days to weeks to form.39,45,46 Other techniques that allow the design of capillary networks involve a combination of immobilized soluble or insoluble biological gradients (e.g. VEGF, RGD) or mechanical guidance to direct ECs laden within the ECM material to assemble into tubules and vascular networks.47,48
Vessel characterization.
The quality and functionality of templated 3D microvessel models is typically assessed by measuring their permeability to fluorescent solutes, such as dextran molecules of varying molecular weights, or other biologically relevant molecules, such as bovine serum albumin (BSA).49–51 Albumin is the most common protein in blood at a concentration of approximately 0.3–0.5 mg ml−1 and a molecular weight of about 65–70 kDa. In vivo vascular permeability ranges from 10−6 to 10−7 cm s−1 for both BSA and 70 kDa dextran, depending on vessel origin and location (e.g. brain, mesentery, tumor, etc.).49,50,52,53 In vitro artificial vessels typically achieve permeability values as low as 10−6 cm s−1 for BSA and 70 kDa dextran (Table 3),38,43,54 which is comparable to in vivo values in post-capillary venules and tumor vasculature (Table 4) and in vitro values for HUVEC monolayers cultured on transwell membranes.55,56 Lower permeability values for BSA (about 10−7 cm s−1) have been achieved by increasing shear stress and transmural pressure on artificial vessels as well as supplementing perfusion media with a cyclic AMP analog.40,57 Although 70 kDa dextran has similar molecular weight to BSA, it exhibits higher permeability values in vivo, suggesting that electrostatic and biological interactions decrease the apparent permeability of BSA.50
Table 4.
In vivo vessel permeability to fluorescent solutes
| Organ | Vessel | Host animal | Method | P (cm s−1) × 10−6 sodium fluorescein | P (cm s−1) × 106 α−lactalbumin | P (cm s−1) × 10−6 albumin | P (cm s−1) × 10−6 70 kDa dextran | Ref. |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Brain pial microvessels | Post−capillary venule | Rat | Intravital injection | 2.71 | 0.15 | 49 | ||
| Brain cerebral microvessels | Post−capillary venule | Rat | Intravital injection | 1.3−1.5 | 0.11−0.13 | 51 | ||
| Brain pial microvessels | Venule | Rat | Intravital injection | 0.1 | 75 | |||
| Cremaster muscle | Venule | Rat | Intravital injection | 0.6 | 75 | |||
| Skin | Mouse | In vivo injection | 0.163 | 76 | ||||
| Mesentery | Post−capillary venule | Rat | In vivo cannulation | 26 | 6.9 | 0.82 | 52 | |
| Mesentery | Post−capillary venule | Rat | In vivo cannulation | 0.8−0.9 | 77 | |||
| Mesentery | Venular | Frog | In vivo cannulation | 1.7 | 78 | |||
| Mesentery | Capillary | Frog | In vivo cannulation | 2.1−4.0 | 79 | |||
| Mesentery | Capillary | Frog | In vivo cannulation | 50 | 80 | |||
| Coronary | Venule | Pig | Ex vivo cannulation | 3.9−6.8 | 81 | |||
| Mammary carcinoma in brain | Rat | Intravital injection | 0.17 | 53 | ||||
| LS174T tumor in dorsal skin | Mouse | Intravital injection | 0.16 | 82 | ||||
| Squamous carcinoma | Mouse | Intravital | 0.49 | 0.98 | 50 | |||
Another measure of endothelial functionality is hydraulic conductivity which characterizes the flux of water across a vessel wall. While hydraulic conductivity has not been directly measured in artificial 3D microvessels, the resistance of the microvessel to water flux may determine the optimal transmural pressure required to prevent endothelial delamination from scaffold walls, a common challenge for increasing the lifespan of engineered microvessels.40,43,58 In vitro measurements of hydraulic conductivity have been performed in the transwell apparatus and reported to be on the order of 10−7 cm s−1 cm H2O−1 for bEnd3 and BAECs.59,60 This is comparable to in vivo measurements of frog mesentery venules and approximately two orders of magnitude higher than those obtained from frog pial microvessels.61,62 Similar to permeability, hydraulic conductivity will vary across different vascular beds and in vitro endothelial monolayers may be optimized to produce tighter or leakier vessels to water to model different vascular tissues.60
TEER is another measure of vessel barrier function and intactness by characterizing electrical impedance across an endothelium. TEER provides a relatively simple and fast measurement of monolayer integrity compared to permeability and hydraulic conductivity, but has not been utilized for 3D microvessels on templated ECM scaffolds. TEER has been most widely used in the transwell assay and has been adapted to membrane-based microfluidic devices (Fig. 2) and 3D cylindrical porous scaffolds.28,63,64 TEER values for primary HUVEC monolayers typically range from 10–100 Ω cm2 but may be as high as several hundred to thousands for treated or derived human brain endothelial cells, which is comparable to that of in vivo venules and arterioles.63,65–68 Both TEER and hydraulic conductivity may provide useful comparisons of endothelial function for more advanced in vitro microvessels.
Immunofluorescence staining of vessels can also be used to assess the expression and quality of continuous junctional networks (e.g. VE-cadherin, PECAM) as well as the deposition of a de novo basement membrane comprised of laminin and collagen type IV.39,40 Live cell imaging enables the measurement of EC speed as well as the presence of focal leaks.54,69,70 Fewer focal leaks has been associated with lower vessel permeability values and increased barrier properties.40,69 Functional quiescent vessels exhibit low levels of leukocyte adhesion or platelet aggregation,38,43 and have low rates of EC proliferation and apoptosis.57
Applications of templated 3D microvessel models.
Templated microvessel models have been used to study tissue engineering, vascular phenomena, and the tumor microenvironment (Table 5). The development of 3D microvessel models has shown the influence of several factors on improving vessel stability and decreasing permeability to in vivo levels (Table 4). Mechanical forces such as shear stress (due to flow) and transmural pressure (the pressure drop across the endothelium), as well as bioactive molecules added to the perfusion media, are able to decrease vessel permeability by two orders of magnitude and increase vessel lifespan to longer than 2 weeks.40,57 The fabrication and maintenance of microvessels has established tissue engineering design principles for creating vascularized tissues.71,72
Table 5.
Features and applications of templated microvessel models
| Template | Purpose/discoveries | Ref | |
|---|---|---|---|
|
| |||
| Monolayer formation | Needle/rod | Pioneered design criteria for engineering vascularized organs; understanding the roles of shear stress, transmural pressure, plasma expanders, cyclic AMP, and lymphatic drainage on vessel stability and permeability | 38, 40, 42 |
| Needle/rod | Explored the influence of shear stress on paracrine signaling between microvessels and breast cancer cells | 41, 73 | |
| Needle/rod | Mechanisms of invasion and intravasation | 54 | |
| Needle/rod | Capillary formation bridging angiogenic source to sink, anti-angiogenic therapies tested | 39 | |
| Lithographic patterning | Blood-endothelium interactions (e.g. thrombosis), pericyte-endothelium interactions, vessel angiogenesis | 43 | |
| Guided self-organization | Lithographic confinement | Implanted cords anastomose, in vivo regenerative medicine | 83–85 |
| Lithographic patterning | Method for creating microvascular networks for tissue engineering | 48, 86 | |
| Lithographic patterning | Method for immobilizing biomolecules in hydrogel scaffolds to direct endothelial tubulogenesis and vascular network formation | 47 | |
The ability to co-culture relevant cell types within the surrounding ECM of microvessels has permitted the study of vessel paracrine signaling with smooth muscle cells, pericytes, and cancer cells.38,43,73 Pericytes have been implicated in modulating the response of the vessel endothelium to proangiogenic factors.43 Extraction of tumor cells from the surrounding ECM and analysis of their gene expression has shown that tumor cell invasiveness is mediated by the presence of microvessels and the level of vessel shear stress.73 Live cell imaging of co-cultured microvessels with tumor cells in the ECM has recapitulated interactions thought to occur during cancer metastasis such as invasion and intravasation.54
Templated 3D microvessels have been used to explore a variety of vascular phenomena, such as inflammation and response to vascular mediators. By introducing whole blood into the microvessels, blood–endothelium interactions have shown that vessel activation during thrombosis exhibits greater platelet aggregation associated with bifurcations and junctions.43 Leukocyte adhesion and changes in vessel permeability in response to vascular mediators have been used to demonstrate the functional response of the vessel endothelium.38
Advantages and limitations.
Templated 3D microvessel models recapitulate the cylindrical geometry and surrounding ECM associated with vessels in vivo. These platforms also allow control of shear stress and transmural pressure, important for regulating interstitial flow, and multiple cell types can be seeded in the ECM. The geometry of these models is also convenient for live cell imaging of a wide range of processes including endothelium structure and function, solute transport, angiogenesis, cell intravasation and extravasation, and drug delivery.38,39,43,54,74 The single rod template models are limited to single straight microvessel segments. Microfabricated ECM templates can produce 2D microvessel networks, and although the template cross-section is rectangular, after seeding with endothelial cells, the vessel has rounded corners close to a cylindrical geometry. The main disadvantage of the templating methods is that the endothelium is formed by perfusing ECs into the lumen of the vessel, and hence the vessel diameter is limited to values larger than about 50 μm.
3D printing
The adaptation of 3D printing technology to print cells and ECM proteins has the potential for printing organs and tissues.87–92 In 3D bioprinting, liquid droplets containing hydrogels, ECM proteins, biochemical cues, and cells are dispensed from an array of one or more nozzles.90 The resolution for printing from aqueous solutions is about 100 μm, although printing more viscous solutions of ECM materials result in somewhat larger values.93 Since perfusion with oxygen and nutrients, and removal of metabolic waste are important for tissue survival, the ability to print vascular networks is critical for the future success of bioprinting.94–96 3D printing of microvessels can be divided into two main categories: direct printing and templating. Research in this field has largely focused on the technological challenges associated with 3D printing of vascular structures for tissue engineering and regenerative medicine.
Direct bioprinting of microvessels in ECM.
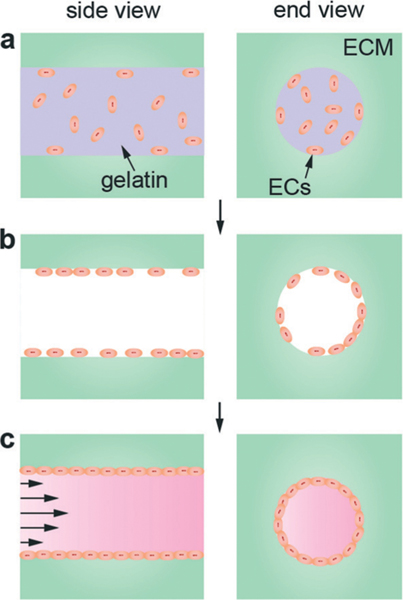
In the simplest case, 3D structures are printed from two components: a suspension of ECs in a dissolvable matrix precursor (e.g. gelatin) and a solution of an ECM material (e.g. collagen I) (Fig. 5). The two components are printed layer-by-layer such that the EC/dissolvable matrix component forms a continuous cylinder through the 3D structure. Following printing and gelation of the ECM, the matrix containing the ECs is dissolved. Adhesion and spread of the ECs results in the formation of the vessel lumen which is then connected to a flow loop. This technique is similar to the templating method except that the endothelial cells are seeded into the template. Due to the resolution of the droplets, vessel diameters are typically greater than 500 μm. This approach has been used to fabricate a single HUVEC microvessel in a collagen matrix, following dissolution of a gelatin template.97 The characteristics of microvessel models fabricated by direct bioprinting of ECM and ECs are summarized in Table 6.
Fig. 5.
Direct bioprinting of ECM and ECs in a dissolvable matrix. (a) Gelatin containing HUVECs printed as a cylinder embedded in a collagen ECM. (b) Following printing and gelation, the gelatin is dissolved by heating to 37 °C. During this step, the device is rotated to enhance adhesion of the HUVECs along the internal walls of the cylinder. (c) Proliferation and spreading of endothelial cells results in the formation of a vessel lumen, and the microvessel is connected to a flow loop for perfusion.
Table 6.
Features of microvessel models fabricated by direct bioprinting of ECM and endothelial cells. EC – endothelial cell, HUVEC – human umbilical vein endothelial cells, NHLF – normal human lung fibroblasts, CHO – Chinese hamster ovarian cells, HUVSMC – human umbilical vein smooth muscle cells, HSF – human skin fibroblasts
3D printing has the capability of producing complex vascular networks with multiple cell types, however, direct printing of small microvessels and capillaries is challenging due to the size of the droplets in the printing process. This limitation has been overcome by stimulating angiogenic sprouting and microvessel growth between two larger vessels.93 A fibringel embedded with ECs and fibroblasts is printed between two parallel 1 mm diameter printed vessels located a few millimeters apart. By applying a low shear stress to maintain viability of the larger vessels while avoiding suppression of sprouting at higher shear stresses, proliferation and recruitment of ECs results in the formation of small microvessels with diameters of 10–25 μm, similar to the diameters of arterioles or post-capillary venules.1 The connection between the two larger vessels was confirmed by perfusion of one of the larger vessels with 10 kDa fluorescently labeled dextran.
A variation of direct printing has been used to produce 1–2 mm diameter suspended tubes of smooth muscle cells and fibroblasts with no endothelium. In this method tubes are formed by printing droplets of large multicellular spheroids from one nozzle and extruding agarose from a second nozzle.98 By defining the regions where the two components are printed in each layer, the spheroids containing smooth muscle cells and fibroblasts formed a tubular structure embedded in agarose. Maturation of the structures over 2–4 days and dissolution of the agaraose led to the formation of robust, well-defined tubes.
Direct printing of suspended vessels.
An approach for producing suspended microvessels is to print droplets containing cells and an initiator (e.g. calcium chloride) from a single print head into a neutral buoyancy bath containing a hydrogel precursor (e.g. alginate).99 By printing droplets in a repeating circular pattern, a tube of cells embedded in a gel is extruded by gravity as printing continues and the construct sinks in the bath. Diameters as small as 200–300 μm can be achieved using this method. Printed suspended vessels could be used for high-throughput studies of transport properties, similar to experiments performed on resected vessels and capillaries isolated from different organs.100
Template printing with post-fabrication cell-seeding.
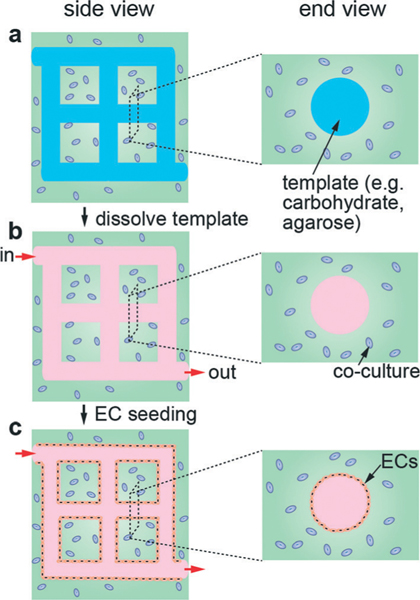
A hybrid strategy to fabricate perfusable vascular networks utilizes conventional 3D printing to produce a dissolvable template network that is then embedded in a matrix material. After the template is dissolved, ECs are seeded into the channels (Fig. 6). A wide range of template materials has been tested in combination with different ECM materials and cell types (Table 7). For example, carbohydrate glass templates with diameters as small as 200 μm have been prepared by 3D thermal extrusion printing.101 Carbohydrate glass provides both sufficient mechanical stiffness to support its own weight in an open lattice and can be dissolved in a biocompatible manner. The self-supporting lattice can then be encapsulated into an ECM containing cells. After cross-linking of ECM, the lattice is dissolved in cell media to yield a perfusable network. Coating the filaments with polyIJD-lactide-co-glycolide) prior to the encapsulation prevents carbohydrate diffusion into the ECM. Endothelial cells in suspension are then seeded into the empty channels to form a vascular network.
Fig. 6.
Schematic illustration of microvessel models formed by 3D template printing. (a) A printed 3D network of carbohydrate glass filaments is embedded in a hydrogel matrix. Other cell types, such as fibroblasts or smooth muscle cells can be embedded in the matrix. (b) The template is dissolved to form a perfusable network of cylindrical channels in the ECM. (c) Endothelial cells in suspension are introduced into the network of channels and allowed to adhere and spread to form the endothelium.
Table 7.
Features of microvessel models formed by 3D printing of templates. EC – endothelial cell, PEG – polyIJethylene glycol), GelMA – methacrylated gelatin, SPELA – star polyIJethylene glycol-co-lactide) acrylate, PEGDMA – polyIJethylene glycol) dimethactrylate, PEGDA – polyIJethylene glycol) diactrylate, HUVEC – human umbilical vein endothelial cells, HEK – human embryonic kidney cells, HNDF – human neonatal dermal fibroblasts, and MC3T3 – mouse calvarial pre-osteoblast cells
Other materials used to produce templates include Pluronic F127 (F127) and agarose.88,102 F127 is a triblock copolymer and forms gel above the critical micelle concentration at about 21 w/w%.,103,104 and can be removed by lowering the temperature below its critical micelle temperature of about 10 °C, when it undergoes a gel-to-fluid transition.105 Agarose is a naturally derived polysaccharide and can be printed as fibers in 3D networks, and is easily removed after embedding in an ECM material.102
Advantages and limitations.
Direct printing of matrix materials and cells in a dissolvable matrix allows the fabrication of 3D vascular networks in a single printing run, followed by dissolution of the vessel matrix and connection to the flow loop. These methods have the potential for the fabrication of complex 3D vascular structures, but are time intensive and limited to larger microvessels (>100 μm). At low resolution (≈500 μm–1 mm) a 1 cm long vessel can be printed in a few hours. Increasing the resolution to print smaller vessels would take considerably longer with current technologies. Following printing, a well-defined endothelial monolayer is formed in 3–5 days. The combination of this technology with self-organization allows the formation of hierarchical networks with microvessel diameters less than 100 μm and spacing necessary for perfusion of healthy tissues. The selforganization of capillary or microvessel networks between two large (≥500 μm) vessels typically takes 8–10 days. The use of 3D printed templates allows the extension of 2D template printing, with post-fabrication cell-seeding methods described in the previous section, to 3D networks.
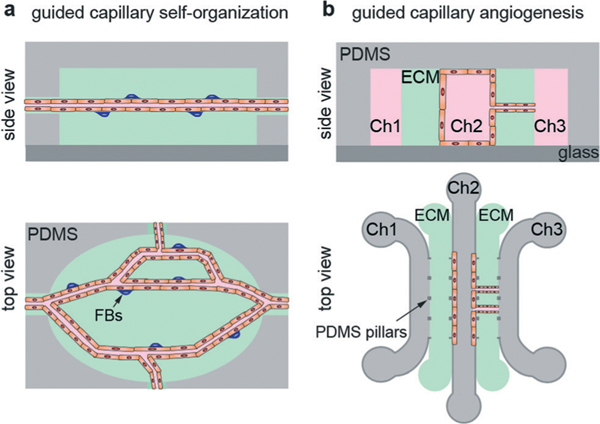
Self-organization
There are two general strategies for exploiting angiogenesis and tubulogenesis in the formation of perfusable microvessel models: guided capillary self-organization45,106 and guided capillary angiogenesis107–110 (Fig. 7). Both methods produce microvessel networks within an ECM but use different approaches, and hence have different constraints and benefits. In most cases the channel height is 100–500 μm and hence the microvessel array would be considered quasi-2D rather than a fully 3D network. The fabrication a 2D network ensures that all of the microvessels are within the focal plane for live cell imaging. Studies of the dynamics of angiogenic sprouts, tubulogenesis, or the invasion of non-perfusable vessel segments are beyond the scope of this review.43,111
Fig. 7.
Schematic illustration of microvessel models formed by self-organization. (a) Guided capillary self-organization and (b) guided capillary angiogenesis.
Guided capillary self-organization.
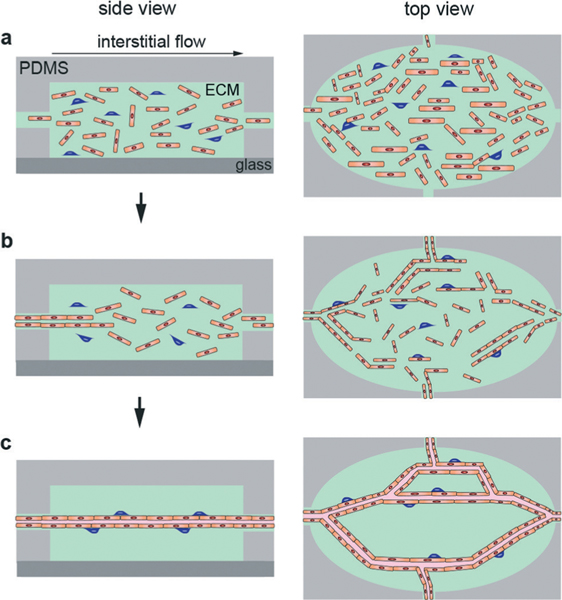
Guided capillary self-organization is used to create a network of capillaries/microvessels within a microfluidic chamber filled with an ECM (Fig. 8).45,106 This method employs a series of diamond shaped chambers (typically 1 mm × 2 mm × 0.1 mm) connected to each other and a series of channels (100 μm × 100 μm) to establish chemical and pressure gradients. The design of the microfluidic device allows for a pressure difference between source and sink channels that establishes interstitial flow through the ECM. Endothelial colony forming cell-derived endothelial cells (ECFC-ECs) and normal human lung fibroblasts (NHLFs) are mixed in a fibrinogen and thrombin ECM. The matrix containing cells is then pipetted into the chambers and allowed to gel (Fig. 8a). Each well is then subjected to a constant pressure to establish an interstitial flow that initiates self-organization (Fig. 8b), finally resulting in the formation of continuous microvessel networks after about 3 weeks (Fig. 8c).106
Fig. 8.
Schematic illustration of the steps in guided capillary self-organization of microvessels. (a) Cells are seeded into an ECM and introduced into the PDMS housing. (b) Interstitial flow drives self-organization. (c) Cells organize into a network of perfusable capillaries/microvessels.
To promote organization and anastomosis, the cells were grown under alternating interstitial flow in the absence of VEGF and bFGF for two weeks. Flow patterns and barrier properties were assessed using fluorescently-labeled polystyrene beads and fluorescently-labeled dextrans, respectively.45 This platform has been used to assess the efficacy and cytotoxicity of anti-cancer drugs by seeding cardiac and tumor tissue within the ECM (Table 8).112
Table 8.
Features of microvessel models fabricated by guided capillary self-organization and selected measurements. EC – endothelial cells
| Purpose | Flow speed | Cells/co-culture | Permeability | Diameter | Ref. |
|---|---|---|---|---|---|
|
| |||||
| Development of 3D vascularized matrix | 0–4000μm s−1 | ECs, lung fibroblasts | N/A | 15–50 μm | 106 |
| Determine perfusion, flow, and shear rate in vessels | 0–4000 μm s−1 | ECs, lung fibroblasts | Perfused with 70 kDa dextran | 15–50 μm | 45 |
| Co-culture tumor and heart tissue in matrix | 0–4000 μm s−1 | ECs, lung fibroblasts, cardiomyocytes, cancer cells (SW620) | Perfused with 70 kDa dextran | 15–50 μm | 112 |
Guided capillary angiogenesis.
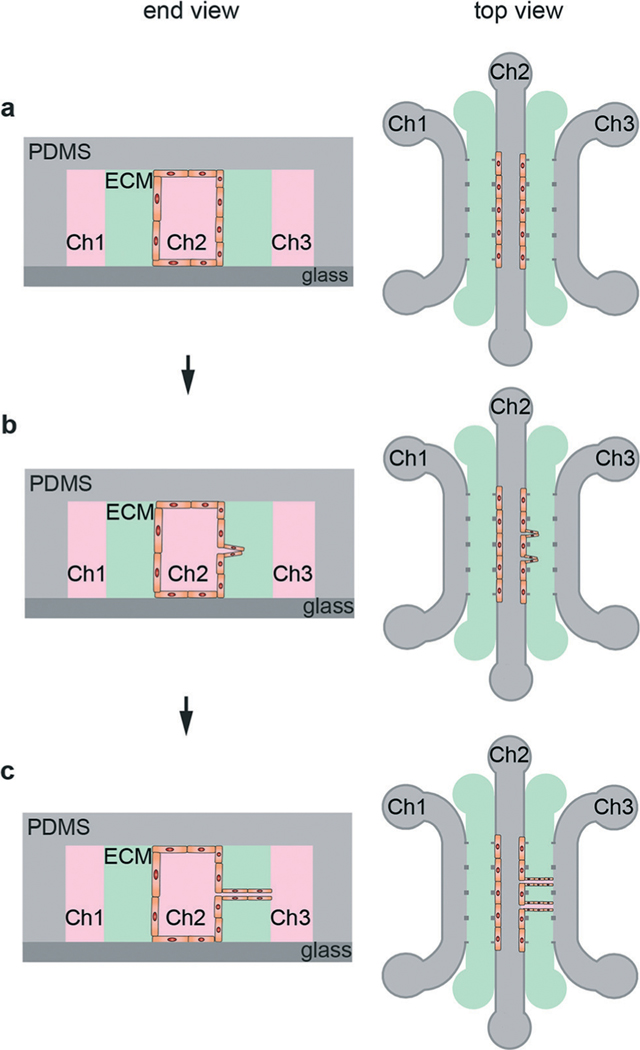
Guided capillary angiogenesis is used to create a network of microvessels across a microfluidic channel filled with an ECM material.107,108,110 This method is an extension of the ECM-based microfluidic models described previously (Fig. 2b). A microfluidic device is fabricated with two or more microfluidic channels, typically 100 μm in height, separated by a channel filled with ECM (Fig. 7b).108 Using three parallel microfluidic channels allows media perfusion on both outer channels, providing improved gas exchange and nutrient supply as well as allowing for the establishment of chemical or pressure gradients (Table 9). Endothelial cells are seeded into the source channel, resulting in the formation of a monolayer on the ECM wall. Chemical and/or pressure gradients can be used to stimulate the formation and growth of angiogenic sprouts that propagate from the source channel to the adjacent sink channel, forming a microvessel network. This process that takes about a week to form a network across a 1 mm wide ECM channel.107,108 By careful selection of the spacing of the pillars that confine the ECM during fabrication, the spacing of the angiogenic sprouts can be controlled.110
Table 9.
Features of microvessel models fabricated by guided capillary angiogenesis
| Purpose | Measurement | Co–culture | Diameter | ECM | Ref. |
|---|---|---|---|---|---|
|
| |||||
| Design of a microfluidic platform to model angiogenesis | Interaction between hepatocytes and ECs | HUVEC, fibroblasts, hepatocytes | 10–100 μm | Collagen | 108 |
| Control over perfusable angiogenesis platform using varying co-culture and growth factors | Perfusable segments (fluorescent beads), area coverage, branch number, and diameter | Stromal cells, ECs | 10–100 μm | Fibrin | 109 |
| Cell migration into matrix as a function of different co-cultures | EC migration into matrix | HMVEC, MTLn2/U87MG, 10T1/2 | 10–100 μm | Collagen | 107 |
The guided capillary angiogenesis model can be extended by seeding other cell types into the ECM. For example, with the incorporation of endothelial cells into the ECM, microvessel networks are formed by a combination of angiogenesis and self-organization. Networks of HUVEC microvessels 10–100 μm in diameter have been formed in devices with HUVECs and NHLFs encapsulated in a fibrin ECM in about 4 days (Fig. 9).109 Perfusion of fluorescent beads into one of the channels has been used to verify perfusion and measure flow rates within the microvessels.109 Various factors, including co-culture, cell density within the matrix, and growth factors influence matrix invasion and vascularization.107,108
Fig. 9.
Schematic illustration of the steps in guided capillary angiogenesis. (a) Endothelial cells are seeded into one of the microfluidic channels (Ch2), forming a monolayer on the side-wall of the ECM. Endothelial cells, fibroblasts, and other cell types can also be seeded into the ECM. (b) Chemical and/or pressure gradients between Ch2 and Ch3 promote formation and growth of angiogenic sprouts from the source channel (Ch2) towards the sink channel (Ch3). With the addition of endothelial cells in the ECM, both angiogenesis and self-organization contribute to the formation of a microvessel network. (c) A perfused microvessel network is formed between the source and sink channels.
Advantages and limitations.
Both guided capillary self-organization and guided capillary angiogenesis generate a perfusable, vascularized ECM, which can be used to study endothelial phenotype in vitro. Guided capillary self-organization generates an interconnected 2D network microvessels within a bulk ECM. The network of microfluidic channels connecting the ECM regions allows control over chemical gradients and interstitial flow, which can be used to model different circulatory systems, such as the lung, brain, or kidney. The flexibility is achieved with the drawback of the relatively long time (about three weeks) needed to establish the microvessel networks. Guided capillary angiogenesis creates a network of microvessels in the ECM between a source channel with an endothelial monolayer and a sink channel by directing the formation and growth of angiogenic sprouts from the source channel. This process takes about 7 days to traverse a 1 mm ECM channel. Incorporation of endothelial cells in the ECM results in the formation of a microvessel network through a combination of angiogenesis and self-organization. Chemical and interstitial flow gradients can be established using the three-channel platform, and microvessels can be formed in about four days. Different cell types can be incorporated into the ECM and into the source and sink channels. The first generation of perfusable models exploiting self-organization and/or angiogenesis has been based on conventional microfluidics technologies to produce quasi-2D networks. As these methods evolve, more complex geometries and physiological systems will be developed. Understanding how to exploit angiogenesis and self-organization will be key to future developments in the field.
Endothelial cell source
ECs are the main cellular component of blood vessels and are responsible for multiple functions including vasomotion (dilation and contraction), leukocyte trafficking, hemostasis (wound healing), and trafficking of small molecules, proteins, and hormones. An issue for all in vitro models is that the local microenvironment may alter the phenotype or genotype of the cells. The EC phenotype includes expression of Weibel–Palade bodies, secretion of von Willebrand factor, expression of ICAM, VCAM, and E-selectin, and VE-cadherin at cell–cell junctions.113 Depending on the location in the body, ECs exhibit significant differences in structure, protein/gene expression, and function. Therefore, the source of ECs may be important depending on the application and objectives of the in vitro microvessel model. Arterial, venous, and capillary endothelial cell lines are widely available, and may recapitulate specific functions and protein/gene expression profiles. Human umbilical vein endothelial cells (HUVECs) and bovine aortic endothelial cells (BAEs) are widely used in vascular and bioengineering research.113–115 Patient-derived cells are increasingly used in animal models (e.g. patient-derived xenografts, PDX) to study the mechanisms of disease and to identify patient-specific therapies,116–118 and may become more accessible for in vitro models. Stem cell-derived ECs represent a relatively new source of human cells for specific applications.119–122 For example, brain microvascular endothelial cells (BMECs) are highly specialized with tight junctions that almost completely prevent paracellular transport.31 Recent work has shown that induced pluripotent stem cells can be differentiated into hBMECs,121 overcoming the lack of cell lines with a blood-brain barrier phenotype.123
Summary
A wide range of perfusable microvessel models have been developed. Perfusable microvessel models can be classified by the fabrication methods used: microfluidics, templating, 3D printing, and self-organization. These models vary in complexity and physiological relevance, but provide a diverse tool kit for the study of vascular phenomena and methods to vascularize artificial organs (Table 10). Current models primarily use cell lines, however, advances in stem cell technology and access to patient derived cells will improve physiological relevance and will contribute to the development of precision medicine. The advances in the development of perfusable microvessel models summarized here will enable advances in basic science and the translation of vascular engineering to the clinic.
Table 10.
Overall comparison of in vitro microvessel models
| In vitro model | Features | Measurements/applications |
|---|---|---|
|
| ||
| Microfluidics | ||
| Membrane-based models | 2D endothelium, shear stress | Barrier function (TEER permeability) |
| ECM-containing models | 2D endothelium, ECM, shear stress, co-culture, chemical gradients | Barrier function (permeability), invasion, intravasation, extravasation |
| Templating | Single microvessel or 2D network, cylindrical geometry (d ≥ 50 μm), ECM, shear stress, co-culture, interstitial flow, transmural pressure | Endothelium structure and function, inflammation, invasion, intravasation, extravasation, drug transport, barrier function (permeability) |
| 3D printing | ||
| Direct bioprinting | Single microvessel or networks, cylindrical geometry (d ≥ 200 μm), capillary networks by anastomosis (d ≥ 10 μm), multiple ECM components, shear stress, co-culture, interstitial flow, transmural pressure | Endothelium structure and function, inflammation, invasion, intravasation, extravasation, drug transport, barrier function (permeability) |
| Template printing | 3D network of microvessels, cylindrical geometry (d ≥ 100 μm), multiple ECM components, shear stress, co-culture, interstitial flow, transmural pressure | Endothelium structure and function, inflammation, invasion, intravasation, extravasation, drug transport, barrier function (permeability) |
| Self-organization | ||
| Guided capillary self-organization | Interconnected microvessel/capillary networks (quasi-2D), cylindrical geometry (d = 10–50 μm), ECM, shear stress, co-culture, interstitial flow, transmural pressure | Angiogenesis, endothelium structure and function, barrier function (permeability) |
| Guided capillary angiogenesis | Parallel (quasi-2D) capillary array, (d = 10–50 μm), ECM, shear stress, co-culture, interstitial flow, transmural pressure | Angiogenesis, endothelium structure and function, barrier function (permeability) |
References
- 1.Aird WC, Thromb J. Haemostasis, 2005, 3, 1392–1406. [DOI] [PubMed] [Google Scholar]
- 2.Aird WC, Circ. Res, 2007, 100, 158–173. [DOI] [PubMed] [Google Scholar]
- 3.Aird WC, Circ. Res, 2007, 100, 174–190. [DOI] [PubMed] [Google Scholar]
- 4.Brightman MW, Exp. Eye Res, 1977, 25, 1–25. [DOI] [PubMed] [Google Scholar]
- 5.Dejana E, Nat. Rev. Mol. Cell Biol, 2004, 5, 261–270. [DOI] [PubMed] [Google Scholar]
- 6.Taddei A, Giampietro C, Conti A, Orsenigo F, Breviario F, Pirazzoli V, Potente M, Daly C, Dimmeler S. and Dejana E, Nat. Cell Biol, 2008, 10, 923–934. [DOI] [PubMed] [Google Scholar]
- 7.Chien S, Am. J. Physiol, 2007, 292, H1209–H1224. [DOI] [PubMed] [Google Scholar]
- 8.Ye M, Sanchez HM, Hultz M, Yang Z, Bogorad M, Wong AD and Searson PC, Sci. Rep, 2014, 4, 4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heron M, Natl. Vital Stat. Rep, 2013, 62, 1–96. [PubMed] [Google Scholar]
- 10.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR and Khademhosseini A, Expert Opin. Drug Discovery, 2014, 9, 335–352. [DOI] [PubMed] [Google Scholar]
- 11.Sei Y, Justus K, LeDuc P. and Kim Y, Microfluid. Nanofluid, 2014, 16, 907–920. [Google Scholar]
- 12.Lancaster MA and Knoblich JA, Science, 2014, 345, 1247125. [DOI] [PubMed] [Google Scholar]
- 13.Knight E. and Przyborski S, J. Anat, 2014, DOI: 10.1111/joa.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher DE, Mooney DJ and Zandstra PW, Science, 2009, 324, 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibbitt MW and Anseth KS, Sci. Transl. Med, 2012, 4, 160ps24. [DOI] [PubMed] [Google Scholar]
- 16.Griffith LG and Swartz MA, Nat. Rev. Mol. Cell Biol, 2006, 7, 211–224. [DOI] [PubMed] [Google Scholar]
- 17.Moon JJ, Saik JE, Poche RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME and West JL, Biomaterials, 2010, 31, 3840–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watt FM and Huck WT, Nat. Rev. Mol. Cell Biol, 2013, 14, 467–473. [DOI] [PubMed] [Google Scholar]
- 19.Burdick JA and Vunjak-Novakovic G, Tissue Eng., Part A, 2009, 15, 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S. and Engler AJ, Nat. Mater, 2014, 13, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies PF, Physiol. Rev, 1995, 75, 519–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song JW, Gu W, Futai N, Warner KA, Nor JE and Takayama S, Anal. Chem, 2005, 77, 3993–3999. [DOI] [PubMed] [Google Scholar]
- 23.Reinitz A, DeStefano J, Ye M, Wong AD and Searson PC, Microvasc. Res, 2015, 99, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eskin SG, Ives CL, McIntire LV and Navarro LT, Microvasc. Res, 1984, 28, 87–94. [DOI] [PubMed] [Google Scholar]
- 25.Malek AM and Izumo S, J. Cell Sci, 1996, 109(Pt 4), 713–726. [DOI] [PubMed] [Google Scholar]
- 26.Levesque MJ and Nerem RM, Biorheology, 1989, 26, 345–357. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Baker BM, Chen CS and Schwartz MA, Arterioscler., Thromb., Vasc. Biol, 2013, 33, 2130–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douville NJ, Tung Y-C, Li R, Wang JD, El-Sayed ME and Takayama S, Anal. Chem, 2010, 82, 2505–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young EW, Watson MW, Srigunapalan S, Wheeler AR and Simmons CA, Anal. Chem, 2010, 82, 808–816. [DOI] [PubMed] [Google Scholar]
- 30.Booth R. and Kim H, Lab Chip, 2012, 12, 1784–1792. [DOI] [PubMed] [Google Scholar]
- 31.Wong AD, Ye M, Levy AF, Rothstein JD, Bergles DE and Searson PC, Front. Neuroeng, 2013, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HJ, Huh D, Hamilton G. and Ingber DE, Lab Chip, 2012, 12, 2165–2174. [DOI] [PubMed] [Google Scholar]
- 33.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY and Ingber DE, Science, 2010, 328, 1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song JW and Munn LL, Proc. Natl. Acad. Sci. U. S. A, 2011, 108, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M. and Kamm RD, Biomaterials, 2014, 35, 2454–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Liu T. and Qin J, Lab Chip, 2012, 12, 2837–2842. [DOI] [PubMed] [Google Scholar]
- 37.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB and Kamm RD, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 13515–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chrobak KM, Potter DR and Tien J, Microvasc. Res, 2006, 71, 185–196. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen DH, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA and Chen CS, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 6712–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price GM, Wong KH, Truslow JG, Leung AD, Acharya C. and Tien J, Biomaterials, 2010, 31, 6182–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchanan CF, Voigt EE, Szot CS, Freeman JW, Vlachos PP and Rylander MN, Tissue Eng., Part C, 2014, 20, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golden AP and Tien J, Lab Chip, 2007, 7, 720–725. [DOI] [PubMed] [Google Scholar]
- 43.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA and Stroock AD, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tien J, Wong KHK and Truslow JG, Vascularization of microfluidic hydrogels, in Microfluidic Cell Culture Systems, ed. Bettinger C, Borenstein JT and Tao SL, Elsevier, Oxford, 2013, pp. 205–221, DOI: 10.1016/B978-1-4377-3459-1.00008-9. [DOI] [Google Scholar]
- 45.Moya ML, Hsu YH, Lee AP, Hughes CCW and George SC, Tissue Eng., Part C, 2013, 19, 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M. and Kamm RD, Proc. Natl. Acad. Sci. U. S. A, 2015, 112, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Culver JC, Hoffmann JC, Poche RA, Slater JH, West JL and Dickinson ME, Adv. Mater, 2012, 24, 2344–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikkhah M, Eshak N, Zorlutuna P, Annabi N, Castello M, Kim K, Dolatshahi-Pirouz A, Edalat F, Bae H, Yang Y. and Khademhosseini A, Biomaterials, 2012, 33, 9009–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan W, Lv Y, Zeng M. and Fu BM, Microvasc. Res, 2009, 77, 166–173. [DOI] [PubMed] [Google Scholar]
- 50.Dreher MR, Liu W, Michelich CR, Dewhirst MW, Yuan F. and Chilkoti A, J. Natl. Cancer Inst, 2006, 98, 335–344. [DOI] [PubMed] [Google Scholar]
- 51.Shi L, Zeng M, Sun Y. and Fu BM, J. Biomech. Eng, 2014, 136, 031005. [DOI] [PubMed] [Google Scholar]
- 52.Fu BM and Shen S, Microvasc. Res, 2004, 68, 51–62. [DOI] [PubMed] [Google Scholar]
- 53.Yuan F, Salehi HA, Boucher Y, Vasthare US, Tuma RF and Jain RK, Cancer Res., 1994, 54, 4564–4568. [PubMed] [Google Scholar]
- 54.Wong AD and Searson PC, Cancer Res., 2014, 74, 4937–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casnocha SA, Eskin SG, Hall ER and McIntire LV, J. Appl. Physiol, 1989, 67, 1997–2005. [DOI] [PubMed] [Google Scholar]
- 56.Ehringer WD, Edwards MJ and Miller FN, J. Cell. Physiol, 1996, 167, 562–569. [DOI] [PubMed] [Google Scholar]
- 57.Wong KH, Truslow JG and Tien J, Biomaterials, 2010, 31, 4706–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong KH, Truslow JG, Khankhel AH, Chan KL and Tien J, J. Biomed. Mater. Res., Part A, 2013, 101, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G, Simon MJ, Cancel LM, Shi ZD, Ji X, Tarbell JM, Morrison B 3rd and Fu BM, Ann. Biomed. Eng, 2010, 38, 2499–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sill HW, Chang YS, Artman JR, Frangos JA, Hollis TM and Tarbell JM, Am. J. Physiol, 1995, 268, H535–543. [DOI] [PubMed] [Google Scholar]
- 61.Clough G. and Michel CC, J. Physiol, 1988, 405, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fraser PA, Dallas AD and Davies S, J. Physiol, 1990, 423, 343–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wojciak-Stothard B, Potempa S, Eichholtz T. and Ridley AJ, J. Cell Sci, 2001, 114, 1343–1355. [DOI] [PubMed] [Google Scholar]
- 64.Cucullo L, Couraud PO, Weksler B, Romero IA, Hossain M, Rapp E. and Janigro D, J. Cereb. Blood Flow Metab, 2008, 28, 312–328. [DOI] [PubMed] [Google Scholar]
- 65.Dewi BE, Takasaki T. and Kurane I, J. Virol. Methods, 2004, 121, 171–180. [DOI] [PubMed] [Google Scholar]
- 66.Man S, Ubogu EE, Williams KA, Tucky B, Callahan MK and Ransohoff RM, Clin. Dev. Immunol, 2008, 2008, 384982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crone C. and Olesen SP, Brain research, 1982, 241, 49–55. [DOI] [PubMed] [Google Scholar]
- 68.Butt AM and Jones HC, Brain Res., 1992, 569, 100–105. [DOI] [PubMed] [Google Scholar]
- 69.Price GM, Chrobak KM and Tien J, Microvasc. Res, 2008, 76, 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morgan JP, Delnero PF, Zheng Y, Verbridge SS, Chen J, Craven M, Choi NW, Diaz-Santana A, Kermani P, Hempstead B, Lopez JA, Corso TN, Fischbach C. and Stroock AD, Nat. Protoc, 2013, 8, 1820–1836. [DOI] [PubMed] [Google Scholar]
- 71.Tien J, Curr. Opin. Chem. Eng, 2014, 3, 36–41. [Google Scholar]
- 72.Wong KH, Truslow JG, Khankhel AH and Tien J, Vascularization: Regenerative Medicine and Tissue Engineering, 2014, p. 109. [Google Scholar]
- 73.Buchanan CF, Verbridge SS, Vlachos PP and Rylander MN, Cell Adh. Migr, 2014, 8, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang XY, Pei Y, Xie M, Jin ZH, Xiao YS, Wang Y, Zhang LN, Li Y and Huang WH, Lab Chip, 2015, 15, 1178–1187. [DOI] [PubMed] [Google Scholar]
- 75.Gaber MW, Yuan H, Killmar JT, Naimark MD, Kiani MF and Merchant TE, Brain Res. Brain Res. Protoc, 2004, 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 76.Kim MH, Curry FR and Simon SI, Am. J. Physiol, 2009, 296, C848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai B, Fan J, Zeng M, Zhang L. and Fu BM, J. Appl. Physiol, 2012, 113, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu BM, Adamson RH and Curry FR, J. Biomech. Eng, 2005, 127, 270–278. [DOI] [PubMed] [Google Scholar]
- 79.Huxley VH, Curry FE and Adamson RH, Am. J. Physiol, 1987, 252, H188–197. [DOI] [PubMed] [Google Scholar]
- 80.Adamson RH, Lenz JF and Curry FE, Microcirculation, 1994, 1, 251–265. [DOI] [PubMed] [Google Scholar]
- 81.Yuan Y, Chilian WM, Granger HJ and Zawieja DC, Am. J. Physiol, 1993, 265, H543–552. [DOI] [PubMed] [Google Scholar]
- 82.Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP and Jain RK, Cancer Res., 1995, 55, 3752–3756. [PubMed] [Google Scholar]
- 83.Chaturvedi RR, Stevens KR, Solorzano RD, Schwartz RE, Eyckmans J, Baranski JD, Stapleton SC, Bhatia SN and Chen CS, Tissue Eng., Part C, 2015, 21, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raghavan S, Nelson CM, Baranski JD, Lim E. and Chen CS, Tissue Eng., Part A, 2010, 16, 2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN and Chen CS, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 7586–7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S. and Khademhosseini A, Biomaterials, 2010, 31, 5536–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy WL, McDevitt TC and Engler AJ, Nat. Mater, 2014, 13, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA and Lewis JA, Adv. Mater, 2014, 26, 3124–3130. [DOI] [PubMed] [Google Scholar]
- 89.Boland T, Xu T, Damon B. and Cui X, Biotechnol. J, 2006, 1, 910–917. [DOI] [PubMed] [Google Scholar]
- 90.Tasoglu S. and Demirci U, Trends Biotechnol., 2013, 31, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guillotin B. and Guillemot F, Trends Biotechnol., 2011, 29, 183–190. [DOI] [PubMed] [Google Scholar]
- 92.Pataky K, Braschler T, Negro A, Renaud P, Lutolf MP and Brugger J, Adv. Mater, 2012, 24, 391–396. [DOI] [PubMed] [Google Scholar]
- 93.Lee VK, Lanzi AM, Ngo H, Yoo S-S, Vincent PA and Dai G, Cell. Mol. Bioeng, 2014, 7, 460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Folkman J, Nat. Med, 1995, 1, 27–31. [DOI] [PubMed] [Google Scholar]
- 95.Radisic M, Yang L, Boublik J, Cohen RJ, Langer R, Freed LE and Vunjak-Novakovic G, Am. J. Physiol, 2004, 286, H507–516. [DOI] [PubMed] [Google Scholar]
- 96.Carmeliet P. and Jain RK, Nature, 2000, 407, 249–257. [DOI] [PubMed] [Google Scholar]
- 97.Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo S-S, Vincent PA and Dai G, Biomaterials, 2014, 35, 8092–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Norotte C, Marga FS, Niklason LE and Forgacs G, Biomaterials, 2009, 30, 5910–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishiyama Y, Nakamura M, Henmi C, Yamaguchi K, Mochizuki S, Nakagawa H. and Takiura K, J. Biomech. Eng, 2009, 131, 035001. [DOI] [PubMed] [Google Scholar]
- 100.Hartz AMS, Bauer B, Fricker G. and Miller DS, Mol. Pharmacol, 2004, 66, 387–394. [DOI] [PubMed] [Google Scholar]
- 101.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN and Chen CS, Nat. Mater, 2012, 11, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y. and Khademhosseini A, Lab Chip, 2014, 14, 2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bohorquez M, Koch C, Trygstad T. and Pandit N, J. Colloid Interface Sci, 1999, 216, 34–40. [DOI] [PubMed] [Google Scholar]
- 104.Linse P. and Malmsten M, Macromolecules, 1992, 25, 5434–5439. [Google Scholar]
- 105.Wu W, DeConinck A. and Lewis JA, Adv. Mater, 2011, 23, H178–H183. [DOI] [PubMed] [Google Scholar]
- 106.Hsu Y-H, Moya ML, Hughes CC, George SC and Lee AP, Lab Chip, 2013, 13, 2990–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V. and Kamm RD, Lab Chip, 2009, 9, 269–275. [DOI] [PubMed] [Google Scholar]
- 108.Chung S, Sudo R, Vickerman V, Zervantonakis IK and Kamm RD, Ann. Biomed. Eng, 2010, 38, 1164–1177. [DOI] [PubMed] [Google Scholar]
- 109.Whisler JA, Chen MB and Kamm RD, Tissue Eng., Part C, 2014, 20, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee H, Kim S, Chung M, Kim JH and Jeon NL, Microvasc. Res, 2014, 91, 90–98. [DOI] [PubMed] [Google Scholar]
- 111.Tourovskaia A, Fauver M, Kramer G, Simonson S. and Neumann T, Exp. Biol. Med, 2014, 239, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moya M, Tran D. and George SC, Stem Cell Res. Ther, 2013, 4, S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bouis D, Hospers GA, Meijer C, Molema G. and Mulder NH, Angiogenesis, 2001, 4, 91–102. [DOI] [PubMed] [Google Scholar]
- 114.Jaffe EA, Nachman RL, Becker CG and Minick CR, J. Clin. Invest, 1973, 52, 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paez A, Mendez-Cruz AR, Varela E, Rodriguez E, Guevara J, Flores-Romo L, Montano LF and Masso FA, Clin. Exp. Immunol, 2005, 141, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Edwards AM, Arrowsmith CH, Bountra C, Bunnage ME, Feldmann M, Knight JC, Patel DD, Prinos P, Taylor MD, Sundstrom M, Barker P, Barsyte D, Bengtson MH, Bell C, Bowness P, Boycott KM, Buser-Doepner C, Carpenter CL, Carr AJ, Clark K, Das AM, Dhanak D, Dirks P, Ellis J, Fantin VR, Flores C, Fon EA, Frail DE, Gileadi O, O'Hagan RC, Howe T, Isaac JTR, Jabado N, Jakobsson P-J, Klareskog L, Knapp S, Lee WH, Lima-Fernandes E, Lundberg IE, Marshall J, Massirer KB, MacKenzie AE, Maruyama T, Mueller-Fahrnow A, Muthuswamy S, Nanchahal J, O'Brien C, Oppermann U, Ostermann N, Petrecca K, Pollock BG, Poupon V, Prinjha RK, Rosenberg SH, Rouleau G, Skingle M, Slutsky AS, Smith GAM, Verhelle D, Widmer H and Young LT, Nat. Rev. Drug Discovery, 2015, 14, 149–150. [DOI] [PubMed] [Google Scholar]
- 117.Shaw SY and Brettman AD, Circulation, 2011, 124, 2444–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stroncek JD, Ren LC, Klitzman B. and Reichert WM, Acta Biomater., 2012, 8, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Adams WJ, Zhang Y, Cloutier J, Kuchimanchi P, Newton G, Sehrawat S, Aird WC, Mayadas TN, Luscinskas FW and Garcia-Cardena G, Stem Cell Rep., 2013, 1, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rufaihah AJ, Huang NF, Kim J, Herold J, Volz KS, Park TS, Lee JC, Zambidis ET, Reijo-Pera R. and Cooke JP, Am. J. Transl. Res, 2013, 5, 21–U122. [PMC free article] [PubMed] [Google Scholar]
- 121.Lippmann ES, Azarin SM, Kay JE, Nessler RA, Wilson HK, Al-Ahmad A, Palecek SP and Shusta EV, Nat. Biotechnol, 2012, 30, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.James D, Nam HS, Seandel M, Nolan D, Janovitz T, Tomishima M, Studer L, Lee G, Lyden D, Benezra R, Zaninovic N, Rosenwaks Z, Rabbany SY and Rafii S, Nat. Biotechnol, 2010, 28, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neuwelt E, Abbott N, Abrey L, Banks WA, Blakley B, Davis T, Engelhardt B, Grammas P, Nedergaard M, Nutt J, Pardridge W, Rosenberg GA, Smith Q. and Drewes LR, Lancet Neurol., 2008, 7, 84–96. [DOI] [PubMed] [Google Scholar]