Abstract
Objectives
We aimed to explore the shared and specific signalling pathways involved in diabetic retinopathy (DR), diabetic peripheral neuropathy (DPN) and diabetic nephropathy (DN).
Methods
Differentially expressed mRNAs and lncRNAs were identified by high-throughput sequencing. Subsequently, functional enrichment analysis, protein-protein interaction (PPI) analysis and lncRNAs-mRNAs networks were conducted to determine the pathogenic mechanisms underlying DR, DPN and DN.
Results
Twenty-six biological pathways were shared among DR, DPN and DN groups compared to the type 2 diabetes mellitus (T2DM) group without complications, and most of the shared pathways and core proteins were involved in immune and inflammatory responses of microvascular damage. Cytokine‒cytokine receptor interactions and chemokine signalling pathway were the most significant and specific pathways for DR, and the lncRNA‒mRNA regulatory networks affected DR by targeting these pathways. Sphingolipid metabolism and neuroactive ligand-receptor pathways were found to be specific for the pathogenesis of DPN. Moreover, multiple amino acid metabolic pathways were involved in the occurrence and progression of DN.
Conclusions
Diabetic retinopathy, DPN and DN exhibited commonality and heterogeneity simultaneously. The shared pathologic mechanisms underlying these diabetic complications are involved in diabetic microvascular damage via immune and inflammatory pathways. Our findings predict several biomarkers and therapeutic targets for these diabetic complications.
Keywords: Diabetic retinopathy, diabetic peripheral neuropathy, diabetic nephropathy, mRNA, long noncoding RNA, high-throughput sequencing
Introduction
Diabetic retinopathy (DR), diabetic peripheral neuropathy (DPN) and diabetic nephropathy (DN) are common in patients with longstanding or uncontrolled diabetes, and are generally identified as “diabetic triopathy”. 1 Transcriptome sequencing is widely used to detect crucial protein-encoding genes and long noncoding RNAs (lncRNAs) that act as key regulators in diabetes and its complications.2-4 Many previous studies are limited to transcriptomic profiles of single diabetic complications, and studies to determine the common and distinct mechanisms underlying different diabetic complications are warranted. The current study aimed to determine the commonalities and differences in the expression of mRNAs and lncRNAs in type 2 diabetes mellitus (T2DM), DR, DPN and DN patients, and further elucidate the biological mechanisms of these diabetic microvascular complications.
Methods
A total of five healthy control individuals, five T2DM patients without complications , five DR patients, five DPN patients and five DN patients were recruited for the RNA sequencing analysis (Supplementary materials). Clinical information from electronic medical records, morningness-eveningness questionnaire (MEQ) and blood samples were subsequently collected.
The detailed process for RNA preparation, library construction, sequencing quality control and quantitative analysis of mRNAs and lncRNAs is described in the Supplementary materials. The current RNA-seq data are available in the Gene Expression Omnibus database (GEO, accession number GSE185011).
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to predict the biological functions of differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 and KEGG Orthology Based Annotation System (KOBAS) 3.0. An adjusted p value < .05 was considered to denote statistical significance. The results of the enrichment analysis were visualized using the R software (version 4.1.0) package ggplot2. Protein-protein interaction (PPI) and lncRNA-mRNA networks were constructed by CytoHubba of Cytoscape3.8.0.
The study adhered to the tenets of the Declaration of Helsinki and was approved by the Medical Ethics Committee of Ningbo University. Informed consent was obtained from all patients.
Results
The fasting glucose levels in the T2DM, DR, DPN, and DN groups were significantly higher than those in the control group, but there was no significant difference between the T2DM, DR, DPN, and DN groups (Table S1). The MEQ results revealed that the majority of control individuals (80%) were intermediate type, while patients with diabetes or its associated complications were mostly moderately morning type (82.3%). The distribution of sleep chronotypes was significantly different between control individuals and T2DM, DR, DPN, and DN patients (p = .017).
Compared to healthy control individuals, patients with T2DM, DR, DPN and DN exhibited 2776, 2697, 3641 and 1606 DEmRNAs, respectively. When compared to T2DM patients, DR, DPN and DN patients presented 2926, 614 and 1265 DEmRNAs, respectively (Table S2). Additionally, a total of 1537, 1490, 1925 and 882 DElncRNAs were identified in the T2DM, DR, DPN and DN groups, respectively (Table S2). The Venn diagrams further show the shared and distinct DEmRNAs and DElncRNAs in the DR, DPN and DN groups (Figure S1 and Figure S2).
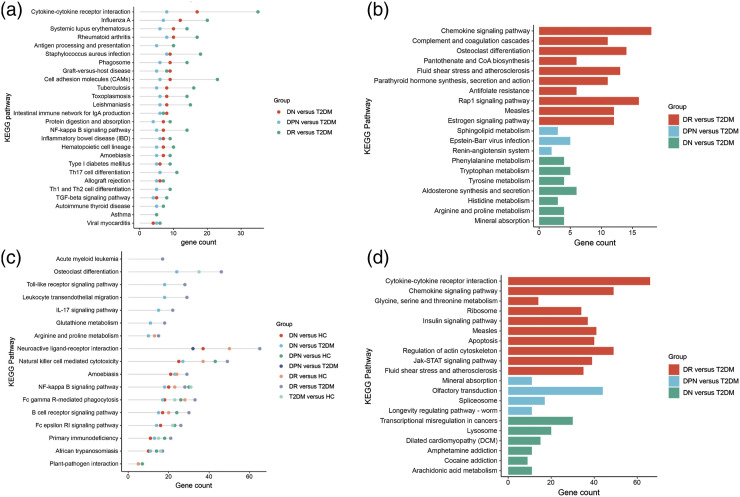
The shared biological process (BP) GO terms of T2DM, DR, DPN and DN mainly included complement activation classical pathway, immune response, receptor-mediated endocytosis, complement activation and regulation of immune response (Figure S3 and Table S3). In addition, the specific GO terms enriched by the DEmRNAs of DR, DPN and DN were shown in Figure S4. According to the KEGG analysis, the T2DM, DR, DPN and DN groups shared 26 pathways (Table S4). The identified shared pathways included cell adhesion molecules (hsa04514), cytokine-cytokine receptor interactions (hsa04060), TNF signalling pathway (hsa04668), PI3K-Akt signalling pathway (hsa04151), and IL-17 signalling pathway (hsa04657), all of which are involved in immune-regulatory function. The most enriched pathway in DR group was cytokine-cytokine receptor interactions (hsa04060), and this was followed by cell adhesion molecules (hsa04514) and TNF signalling pathway (hsa04668) (hsa05323) (Figure S5(A)). The top 10 enriched pathways in the DPN and DN groups were displayed in Figure S5(B)-5(C). Twenty-six KEGG pathways were shared between the DR, DPN and DN groups compared with the T2DM group. Most of these common pathways were also associated with immune and inflammatory responses, such as cytokine-cytokine receptor interactions (hsa04060), cell adhesion molecules (hsa04514), NF-kappa B signalling pathway (hsa04064), Th17 cell differentiation (hsa04659), and Th1 and Th2 cell differentiation (hsa04658) (Figure 1(A)).
Figure 1.
Shared and Specific KEGG Pathways for DR, DPN and DN. (A) Shared pathways of DR, DPN and DN groups enriched by DEmRNAs between diabetic complications and T2DM, (B) Specific pathways of DR, DPN and DN groups enriched by DEmRNAs between diabetic complications and T2DM, (C) Shared pathways of DR, DPN and DN groups enriched by lncRNA-targeted genes, (D) Specific pathways of DR, DPN and DN groups enriched by lncRNA-targeted genes. Scatterplots of different colors in Figure 1(a) and (c) presented different comparison groups. The horizontal axis was gene number. The length of the bars in Figure 1(b) and (d) indicated the number of genes enriched in the pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEmRNAs differentially expressed mRNAs; T2DM, type 2 diabetes mellitus; DR, diabetic retinopathy; DPN, diabetic peripheral neuropathy; DN, diabetic nephropathy.
Overall, enrichment analysis determined several specific pathways of DR, DPN and DN (Figure 1(B)). The results revealed that the most significantly specific pathway in DR was the chemokine signalling pathway (hsa04062). The specific pathways of DPN and DN mainly focused on amino acid metabolism pathways and mineral absorption pathways, respectively.
Identification of Hub mRNAs from the PPI Networks
Protein-protein interaction networks were established to investigate the functions of the coding genes. The key nodes for DR (CXCL8, CXCL1, CXCR4, CXCL6, C3, CXCR1, CXCL3, CXCL2, C5AR1 and CCL20), DPN (CXCL1, CXCL8, CAMP, LTF, DEFA4, CTSG, ADORA3, HLA-DQA1, HLA-DRA and DEFA3) and DN (CXCL8, CXCL10, CXCL1, CXCR4, CCL4L1, HCAR2, CCR8, ADORA3, MCHR1, and GPR183) were identified (Figure S6). IL6, IL1B, CXCL8, IL10, CCL2, TNF, IL1A, IFNG, CSF2 and CXCL1 were the top 10 key genes in the shared pathways of T2DM, DR, DPN and DN (Figure S7(A)). Furthermore, CXCL8, IL10, CCL2, CCL3, CXCL1, IL1B, CXCL10, BCXCL2, IFNG and IL1R1 were the crucial nodes in the shared pathways of these diabetic complications compared with T2DM (Figure S7(B)).
Pathway analysis of the LncRNA-targeted genes
KEGG analyses of the lncRNA target genes showed that the Fc epsilon RI signalling pathway (hsa04664) and Fc gamma R-mediated phagocytosis (hsa04666) were shared among T2DM, DR, DPN and DN patients (Figure 1(C)). Relative to T2DM, the remarkably enriched pathways in DR and DN were mostly involved in the immune system; these pathways include the B-cell receptor signalling pathway (hsa04662), Fc gamma R-mediated phagocytosis (hsa04666), osteoclast differentiation (hsa04380), natural killer cell-mediated cytotoxicity (hsa04650) and primary immunodeficiency (hsa05340) (Figure 1(C)). Moreover, DR and DPN simultaneously exhibited neuroactive ligand-receptor interactions (hsa04080) (Figure 1(C)). Analysis also presented several distinct pathways of DR, DPN and DN compared with T2DM (Figure 1(D)). The detailed KEGG pathways were presented in Table S5.
Construction of LncRNA-mRNA networks
The enriched KEGG pathways of lncRNA target genes, including cytokine-cytokine receptor interactions, chemokine signalling pathway and neuroactive ligand-receptor interactions, were relatively specific for diabetic DR or DPN. Thus, lncRNA-mRNA networks were constructed to investigate the regulatory relationship between the lncRNAs and target genes for these two diabetic complications. We found that 40 DElncRNAs targeted 40 genes enriched in cytokine-cytokine receptor interactions and chemokine signalling pathway (Figure S8), and 48 DElncRNAs could be paired with 38 target genes of the neuroactive ligand-receptor interaction pathway (Figure S9). The networks indicated that the IFNA (IFNA4, IFNA13, and IFNA14), CCL (CCL4, CCL15, CCL16, CCL15-CCL14, CCL23, and CCL18) and MAPK3 genes in the cytokine/chemokine signalling pathway were regulated by two or more lncRNAs, and the CHRM1, GABRR2, GABRQ, GABRA5, ADRB3, CHRNA4, HTR5A and NPB genes in the neuroactive ligand-receptor interaction pathway were targeted cores. These results indicate that the above lncRNA-mRNA networks might be involved in the specific mechanisms of diabetes complications.
Discussion
To the best of our knowledge, this study is the first to evaluate the shared and specific biological pathways of DR, DPN and DN by high-throughput sequencing. Notably, most of the shared biological pathways enriched by DEmRNA or DElncRNA-target genes were simultaneously involved in inflammation immunity-related biological processes. The key genes enriched in these shared pathways included CXCL8, IL10, CCL2, CCL3, CXCL1, IL1β, CXCL10, BCXCL2, IFNG and IL1R1. In particular, CXCL8, 5 MCP-1/CCL2, 6 CCL3/MIP-a 7 and IL1β 8 are potent proangiogenic mediators that elevate vascular endothelial growth factor (VEGF) levels in endothelial cells via NF-kappaβ signalling pathway, thereby resulting in an angiogenic response to inflammatory processes. In addition, the most specific pathway in DR was the chemokine signalling pathway. Recent studies have suggested that neurodegeneration is preceded by clinically visible vascular lesions in DR.9,10 Inflammation-based diabetic microvasculopathy is well established in DR,11,12 but whether the chemokine signalling pathway is implicated in retinal neurodegeneration has not been clarified. The amino acids metabolism and sphingolipid metabolism pathways were enriched in DN and DPN patients, respectively, which were rarely reported in previous studies. Collectively, our findings were consistent with previous research that reported the critical role of the chronic inflammatory status in diabetic microvascular damage. Moreover, we further explored the distinct pathogenic mechanisms underlying these diabetic complications.
However, the current study was limited to preliminary exploration via high-throughput sequencing and bioinformatics analysis, and the shared and specific biological signalling pathways in DR, DPN and DN merit further verification in functional studies.
In conclusion, the pathogenic mechanisms underlying DR, DPN and DN simultaneously exhibited commonality and heterogeneity in T2DM patients. The shared pathologic mechanisms are involved in diabetic microvascular damage via immune and inflammatory pathways and core proinflammatory cytokines. Our findings might provide novel insight into the specific molecular mechanisms of DR, DPN and DN, and identify predictive biomarkers and therapeutic targets for these diabetic complications.
Supplemental Material
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Acknowledgements
We thank Dr Yue-xi Wang for necessary help in blood sample collecting.
Author contributions: Jin Xu and Lin-dan Ji designed the study. Hui Zhu and Yan-ming Chen analyzed the data and drafted the manuscript. Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu contributed to the acquisition of subjects and data. Jin Xu and Lin-dan Ji have primary responsibility for the final content. All the authors have read and approved the final manuscript.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Zhejiang Public Welfare Technology Application Research Program [grant number LGF20H260009, LGF20H040005]; Ningbo Nonprofit Science and Technology Project [grant number 2019C50085, 2021S132]; Ningbo Major Science and Technology Task Project [grant number 2021Z054]; and Zhejiang University Students Science and Technology Innovation Activity Plan (New Seedling Talent Plan) [2021R405042].
Data availability statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplemental Material: Supplemental material for this article is available online
ORCID iD
References
- 1.Eid S, Sas KM, Abcouwer SF, et al. New insights into the mechanisms of diabetic complications: role of lipids and lipid metabolism. Diabetologia. 2019; 62: 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu B, Cong C, Ma Y, et al. Potential value of lncRNAs as a biomarker for proliferative diabetic retinopathy. Eye (Lond) 2021.36, 575, 584, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo L, Zhou WH, Cai JJ, et al. Gene expression profiling identifies downregulation of the neurotrophin-MAPK signaling pathway in female diabetic peripheral neuropathy patients. J Diabetes Res. 2017; 2017: 8103904–8103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodgin JB, Nair V, Zhang H, et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013; 62: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem. 2009; 284: 6038–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong KH, Ryu J, Han KH. Monocyte chemoattractant protein-1-induced angiogenesis is mediated by vascular endothelial growth factor-A. Blood. 2005; 105: 1405–1407. [DOI] [PubMed] [Google Scholar]
- 7.Shehadeh N, Pollack S, Wildbaum G, et al. Selective autoantibody production against CCL3 is associated with human type 1 diabetes mellitus and serves as a novel biomarker for its diagnosis. J Immunol. 2009; 182: 8104–8109. [DOI] [PubMed] [Google Scholar]
- 8.Chang CK, LoCicero J, 3rd. Overexpressed nuclear factor kappaB correlates with enhanced expression of interleukin-1beta and inducible nitric oxide synthase in aged murine lungs to endotoxic stress. Ann Thorac Surg. 2004; 77: 1222–1227; discussion 7. [DOI] [PubMed] [Google Scholar]
- 9.Barber AJ, Lieth E, Khin SA, et al. Neural apoptosis in the retina during experimental and human diabetes. Early onset and effect of insulin. J Clin Invest. 1998; 102: 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altmann C, Schmidt MHH. The role of microglia in diabetic retinopathy: inflammation, microvasculature defects and neurodegeneration. Int J Mol Sci. 2018; 19, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubsam A, Parikh S, Fort PE. Role of inflammation in diabetic retinopathy. Int J Mol Sci. 2018; 19, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nawaz MI, Van Raemdonck K, Mohammad G, et al. Autocrine CCL2, CXCL4, CXCL9 and CXCL10 signal in retinal endothelial cells and are enhanced in diabetic retinopathy. Exp Eye Res. 2013; 109: 67–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research
Supplemental Material for Shared and specific biological signalling pathways for diabetic retinopathy, peripheral neuropathy and nephropathy by high-throughput sequencing analysis by Zhu Hui, Yan-ming Chen, Wei-kun Gong, Jing-bo Lai, Bin-bin Yao, Zhi-jia Zhao, Qin-kang Lu, Ke Ye, Lin-dan Ji, and Jin Xu in Diabetes and Vascular Disease Research