ABSTRACT
Macroautophagy/autophagy is an evolutionarily conserved intracellular degradation pathway that maintains cellular homeostasis. Over the past two decades, a series of scientific breakthroughs have helped explain autophagy-related molecular mechanisms and physiological functions. This tremendous progress continues to depend largely on powerful research methods, specifically, various autophagy marker Atg8–PE protein-based methods for studying membrane dynamics and monitoring autophagic activity. Recently, several biochemical approaches have been successfully developed to produce the lipidated protein Atg8–PE or its mimics in vitro, including enzyme-mediated reconstitution systems, chemically defined reconstitution systems, cell-free lipidation systems and protein chemical synthesis. These approaches have contributed important insights into the mechanisms underlying Atg8-mediated membrane dynamics and protein-protein interactions, creating a new perspective in autophagy studies. In this review, we comprehensively summarize Atg8–PE protein-based in vitro biochemical approaches and recent advances to facilitate a better understanding of autophagy mechanisms. In addition, we highlight the advantages and disadvantages of various Atg8–PE protein-based approaches to provide general guidance for their use in studying autophagy.
Abbreviations: ATG: autophagy related; ATP: adenosine triphosphate; COPII: coat protein complex II; DGS-NTA: 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt); DPPE: 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DSPE: 1,2-distearoyl-sn-glycero-3-phosphoethanolamine; E. coli: Escherichia coli; EPL: expressed protein ligation; ERGIC: ER-Golgi intermediate compartment; GABARAP: GABA type A receptor-associated protein; GABARAPL1: GABA type A receptor associated protein like 1; GABARAPL2: GABA type A receptor associated protein like 2; GFP: green fluorescent protein; GUVs: giant unilamellar vesicles; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MBP: maltose binding protein; MEFs: mouse embryonic fibroblasts; MESNa: 2-mercaptoethanesulfonic acid sodium salt; NCL: native chemical ligation; NTA: nitrilotriacetic acid; PE: phosphatidylethanolamine; PS: phosphatidylserine; PtdIns3K: class III phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; SPPS: solid-phase peptide synthesis; TEV: tobacco etch virus; WT: wild-type.
KEYWORDS: Atg8–PE, autophagy, biochemical approaches, cell-free lipidation system, chemically defined reconstitution systems, enzyme-mediated reconstitution system, LC3–PE, protein chemical synthesis
Introduction
Macroautophagy/autophagy is an intracellular degradation mechanism that is evolutionarily conserved from yeast to mammals [1,2]. During the early stages of autophagy, a membrane cisterna, called the phagophore, is formed upon amino acid starvation or exposure to other stimuli. Then, along with the nucleation, expansion, and sealing of the phagophore to form double-membrane vesicles, termed autophagosomes, the cargoes to be degraded are isolated and enclosed inside autophagosomes (Figure 1). The subsequent fusion of autophagosomes with lysosomes causes the degradation of autophagic cargoes by lysosomal hydrolyses [3,4]. Autophagy was initially considered a nonselective degradation pathway through which a random portion of the cytoplasm is degraded and recycled. However, it has been established that autophagy can be a highly selective process that serves to specifically degrade protein aggregates, various damaged organelles and intracellular pathogens [5–7]. Accumulating evidence shows that autophagy plays a very central role in maintaining cellular homeostasis by eliminating harmful or excessive cellular contents both under normal circumstances and in response to different environmental and cellular stresses [8,9]. Dysregulation of autophagy has been implicated in multiple human diseases, including cancers, cardiovascular diseases, neurodegenerative diseases and aging [10–14].
Figure 1.
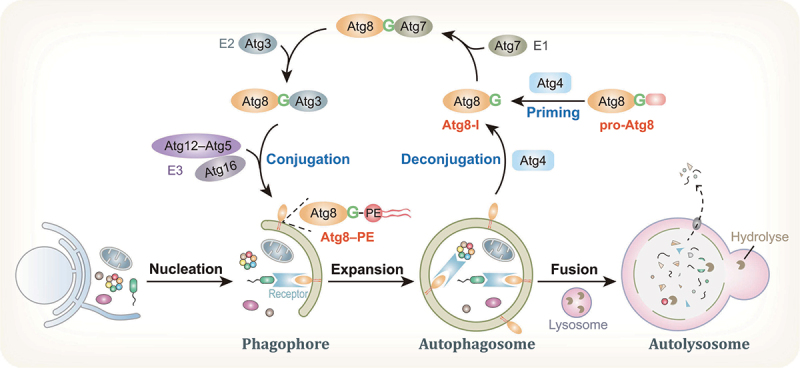
Overview of the autophagy pathway and Atg8 processing. The autophagosome is formed along with the nucleation, expansion, and sealing of the phagophore, upon autophagy induction by environmental stresses. After the subsequent fusion of autophagosomes with lysosomes, cargoes are degraded by lysosomal hydrolyses inside autolysosomes and recycled. In the autophagy process, the newly synthesized Atg8 protein is primed by the cysteine protease Atg4 to expose a C-terminal glycine residue and, thus, generate Atg8-I. Subsequently, a ubiquitin-like conjugation reaction catalyzed by E1-like Atg7, E2-like Atg3 and the E3-like Atg12–Atg5-Atg16 complex mediates Atg8 conjugation to PE on membranes. In addition, Atg4 is also responsible for Atg8–PE deconjugation from the outer surface of autophagosomes.
Over the past two decades, a series of breakthroughs in the autophagy field have been achieved because of continuous efforts of researchers and scientific communities [15]. The molecular mechanisms and physiological functions associated with autophagy have been largely revealed. Importantly, the fundamental importance of this process was recognized when, in 2016, the field was awarded the Nobel Prize for discoveries on the mechanisms of autophagy [16–18]. This tremendous progress continues to depend largely on powerful research approaches, including electron microscopy, fluorescence microscopy and various biochemical assays [19–22]. In particular, in vitro biochemical assays have provided important insights into the mechanistic details underlying autophagosome biogenesis and cargo recruitment, thereby accelerating the pace of autophagy research [23–27]. In vitro approaches represent reductionist strategies and are powerful for studying the complicated cellular biological process of autophagy because they enable control of multicomponent compositions and spatiotemporal arrangements, which cannot be achieved with in vivo approaches. Among these in vitro assays, autophagy marker Atg8 protein-based approaches have been widely used to study membrane dynamics in autophagosome biogenesis. Recently, several biochemical approaches have been developed, including enzyme-mediated reconstitution systems, chemically defined reconstitution systems, cell-free lipidation systems and protein chemical synthesis, to produce the lipidated protein Atg8–phosphatidylethanolamine (PE) or its mimics in vitro [28–33]. These biochemical approaches have been extensively applied and have contributed important insights into the mechanisms underlying Atg8-mediated membrane dynamics and protein-protein interactions, thereby creating a new perspective in autophagy studies. In this review, we focus on these Atg8–PE protein-based in vitro biochemical approaches and the biological questions they have been used to answer. In addition, the advantages and disadvantages of these approaches are highlighted to provide general guidance for their applications in autophagy studies.
The prequel to Atg8–PE serving as a bona fide autophagosome marker
The formation of autophagosomes is a critical event in autophagy and is governed by a distinctive set of autophagy related (Atg) proteins [34,35]. More than 40 Atg proteins have been identified in yeast to date, and many of these proteins are conserved in higher eukaryotes. A subset of these proteins, referred to as the autophagy core machinery, is required for selective and nonselective autophagy. Among these Atg proteins, a ubiquitin-like protein, Atg8, is crucial for autophagosome maturation (Figure 1). During the autophagic process, a key reaction involves the conjugation of Atg8 to PE, which is catalyzed by two interconnected ubiquitin-like conjugation systems called the Atg8 and Atg12 systems [36,37]. Similar to Atg8, Atg12 is a ubiquitin-like protein and is constitutively conjugated to Atg5 through a sequential cascade reaction involving E1 enzyme-like Atg7 and E2 enzyme-like Atg10 [38]. The Atg12–Atg5 conjugate exhibits E3-like ligase activity by facilitating Atg8 conjugation to PE in autophagic membranes [39,40]. In addition, Atg12–Atg5 interacts with the dimeric coiled-coil protein Atg16 (ATG16L1 in mammalian cells) to form a constitutive complex, Atg12–Atg5-Atg16, which is essential for autophagosome biogenesis [39,41–45].
Whereas yeast has a single Atg8 protein, mammals harbor two subfamilies of Atg8 proteins classified by sequence similarity: the MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) subfamily consisting of LC3A, LC3B, LC3B2 and LC3C (referred to as LC3 proteins) and the GABARAP (GABA type A receptor-associated protein) subfamily consisting of GABARAP, GABARAPL1 and GABARAPL2 (referred to as GABARAP proteins) [46–48]. LC3B was identified as the first and most extensively studied mammalian Atg8 homolog. During the autophagic process, Atg8-family proteins undergo a unique conjugation reaction to covalently link with PE on autophagic membranes [36,37]. In summary, a newly synthesized Atg8 protein (pro-Atg8) with an additional amino acid or sequence of amino acids at its C terminus is immediately primed by an endogenous cysteine protease, Atg4, to expose a C-terminal glycine residue and thus generate Atg8-I. Subsequently, the Atg8 conjugation reaction is mediated by a ubiquitin-like conjugation reaction catalyzed by E1-like Atg7, E2-like Atg3 and the E3-like Atg12–Atg5-Atg16 complex [40,49]. Remarkably, the target of Atg8 is the lipid PE, not a protein. Atg8 is covalently conjugated to PE through an amide bond that forms between the amino group of PE and the C-terminal glycine of processed Atg8, thereby generating the lipidated form, termed Atg8-II or Atg8–PE [36,50,51]. In addition, Atg4 is critical for Atg8–PE deconjugation from the outer surface of autophagosomes; deconjugated Atg8 can be recycled, while Atg8–PE inside autolysosomes is degraded [37,52,53]. Atg4-mediated Atg8–PE deconjugation is an important step that facilitates multiple key events, including Atg14 dissociation from complete autophagosomes and the fusion of autophagosomes with lysosomes [54]. Yeast has only one Atg4, whereas mammals have at least four conserved Atg4 homologs, including ATG4A, ATG4B, ATG4C and ATG4D [55].
Interestingly, Atg8–PE is the first identified protein that specifically associates with autophagy-related membrane structures, including phagophores and mature autophagosomes [56]. Atg8–PE levels on phagophores correlate with the size of autophagosomes, implying that Atg8–PE is directly involved in phagophore expansion [57]. Atg8–PE is not only involved in the biogenesis of autophagosomes but is also critical for the recruitment of autophagic cargoes in selective autophagy [58,59]. Phagophore-associated Atg8–PE recruits ubiquitinated cargoes into autophagosomes by binding with autophagy acceptors [60,61]. In addition, the level of autophagy activity can be estimated by monitoring the amount of Atg8–PE in cells. Therefore, the lipidated protein Atg8–PE, but not the cytosolic form, serves as a widely used bona fide marker of autophagosomes [62].
Due to the crucial importance of Atg8–PE, various methods based on Atg8–PE protein levels are among the most common tools used to study membrane dynamics and to monitor autophagic activity. Indeed, various fluorescent protein-tagged Atg8/LC3 proteins, such as GFP-Atg8, tandem GFP-mCherry-Atg8 and the autophagic flux probe GFP-LC3B-RFP-LC3BΔG, have been developed for capturing a dynamic autophagic process [63–68]. In living cells, these fluorescently tagged Atg8 probes are converted to the lipidated form, Atg8–PE, thereby reflecting autophagy progression and autophagic activity, which can be further monitored by fluorescence microscopy or western blot analysis. However, it is difficult to clearly demonstrate Atg8–PE protein-mediated membrane dynamics and protein-protein interactions in this dynamic autophagic process by employing these fluorescent protein-tagged Atg8 probes in vivo. Therefore, establishing in vitro approaches to compensate for the disadvantages of in vivo methods is important in the study of autophagy. Recently, several in vitro reductionist biochemical approaches have been developed to produce the Atg8–PE protein or its mimics (Table 1). In these cases, Atg8–PE is produced manually in vitro, not through the conversion of Atg8 in living cells. These Atg8–PE protein-based in vitro approaches have been powerful tools for elucidating the molecular mechanisms of Atg8 lipidation and Atg8–PE deconjugation, as well as Atg8–PE protein-mediated membrane dynamics. In addition, the produced Atg8–PE protein and its mimics allow us to gain insights into the biochemical mechanisms of Atg8–PE-mediated protein-protein interactions.
Table 1.
Summary of Atg8–PE protein-based in vitro biochemical approaches.
| Approach | Mechanism | Required components | Product | Pros | Cons | Applications for autophagy studies | Ref |
|---|---|---|---|---|---|---|---|
| Enzyme-mediated reconstitution system | Mimicking Atg8 lipidation using purified components and liposomes | Recombinant Atg8-I, Atg7 and Atg3, PE-containing liposomes or giant unilamellar vesicles (GUVs), ATP | Native Atg8–PE | 1. Standard and effective approach for in vitro Atg8 lipidation 2. Native Atg8–PE products |
Time-consuming and laborious | 1. Analyzing Atg8-mediated membrane dynamics 2. Determining influential factors involved in Atg8–PE conjugation 3. Investigating Atg8–PE deconjugation mediated by Atg4 |
[30,69] |
| Chemically defined reconstitution system | Maleimide-thiol coupling: Michael addition of the thiol group of cysteine and maleimide moiety | Recombinant LC3BG120C mutant, PE-maleimide-containing liposomes | LC3B–PE maleimide conjugate | 1. Simple and time-effective approach for in vitro LC3 lipidation 2. Bypassing the requirement for the conjugation machinery |
The product is non-native LC3–PE conjugate | Analyzing LC3-mediated membrane dynamics | [31] |
| PolyHis-NTA strategy: high-affinity binding of polyHis-tagged proteins to Ni-NTA | Recombinant LC3B-His12, DGS-NTA-containing liposomes | LC3B-DGS noncovalent complex | 1. The commercial DGS-NTA lipids are not native PE structures 2. The product is LC3-DGS noncovalent complex |
Analyzing LC3-mediated membrane dynamics | [70] | ||
| Cell-free lipidation system | Mimicking LC3 lipidation using purified substrates, fractionated cytosols, membranes and nucleotides. | Recombinant T7-LC3B-I, the cytosols from WT MEFs, the membranes from atg5 knockout MEFs, ATP | Native LC3B–PE | 1. Enabling the LC3–PE conjugation in the physiological lipid composition 2. Native LC3–PE products |
1. Time-consuming and laborious process, technical challenge 2. Difficult to separate target compartments |
1. Defining the origin source of autophagic membranes 2. Dissecting the molecular mechanisms of early autophagic membrane generation |
[32,33] |
| Protein chemical synthesis strategy | PolyArg-assisted solubilization strategy: EPL of LC3[1-114] protein thioester and PE-modified peptide with polyArg tag | LC3B[1-114] MESNa thioester, PE-modified peptide with polyArg tag, MPAA (catalyst) | Native LC3B–PE | Enabling functional LC3–PE in preparative amounts | 1. Time-consuming and laborious, technical challenge 2. Need to remove polyArg tag and to refold proteins after ligation |
Analyzing LC3-mediated membrane tethering and fusion | [29] |
| MBP-assisted solubilization strategy: EPL of MBP-LC3[1-114] protein thioester and PE-modified peptide | MBP-LC3B[1-114] MESNa thioester, PE-modified peptide, β-octylglucoside (detergent), MPAA (catalyst) | Native LC3B–PE | 1. Enabling functional LC3–PE with various mutants and modifications in preparative amounts 2. The ligation works under folding conditions |
Time-consuming and laborious process, technical challenge | 1. Analyzing LC3-mediated membrane tethering and fusion 2. Elucidating the biochemical mechanisms of Legionella RavZ-mediated LC3–PE deconjugation |
[28,71] |
Various biochemical approaches for producing Atg8–PE in vitro
Enzyme-mediated reconstitution systems
Atg8-family proteins are conjugated to PE through a sequential enzymatic cascade involving Atg7, Atg3 and the Atg12–Atg5-Atg16 complex in cells, as described above. Mimicking the in vivo lipidation system, the yeast Atg8–PE conjugation reaction is first reconstituted in vitro using a coexpression system or through enzymatic reactions between purified Atg proteins and liposomes in 2004 [30]. As an initial attempt to reconstitute yeast Atg8–PE, Escherichia coli (E. coli) BL21 cells carrying recombinant plasmids encoding Atg8-I, Atg7 and Atg3 are constructed and then induced to coexpress these proteins. Subsequent urea-SDS-PAGE and immunoblot analysis with an anti-Atg8 antibody show that the Atg8–PE protein is successfully produced. In this system, E. coli membrane phospholipids provide enough PE for Atg8 conjugation. Thus, this coexpression experiment suggests that Atg7 and Atg3 are sufficient for the lipidation of yeast Atg8.
Furthermore, the yeast Atg8–PE conjugation reaction in vitro can be reconstituted by using purified Atg proteins and liposomes containing PE (Figure 2) [30]. In this case, GST-fused yeast Atg8-I, Atg7 and Atg3 are expressed by a bacterial system and further purified using glutathione beads. PE-containing liposomes are generated from a mixture of 70% purified E. coli PE and 30% total E. coli GN10 lipids. GN10 possesses a null mutant of the pssA gene that encodes phosphatidylserine (PS) synthase; therefore, it completely lacks PE [72]. Subsequently, incubation of the purified components, including Atg8-I, Atg7 and Atg3, with PE-containing liposomes efficiently allows the production of Atg8–PE in the presence of an adenosine triphosphate (ATP) regeneration system. Therefore, the minimal essential factors necessary for yeast Atg8–PE conjugation are Atg8-I, Atg7, Atg3, PE-containing membranes and the energy donor ATP. In this reaction, ATP is an adenylate donor for the adenylation of the Atg8 C terminus in a reaction catalyzed by E1-like Atg7, and the resulting Atg8–acyl adenylate is then attacked by Atg7 to generate intermediate Atg8–Atg7 with a thioester bond [73,74]. In contrast to the Atg8 lipidation system in cells, Atg8–PE reconstitution in vitro does not require the E3-like Atg12–Atg5-Atg16 complex for the transfer of Atg8 to PE. In addition, the PE content of the membranes is an important factor governing the efficiency of Atg8–PE formation [30].
Figure 2.
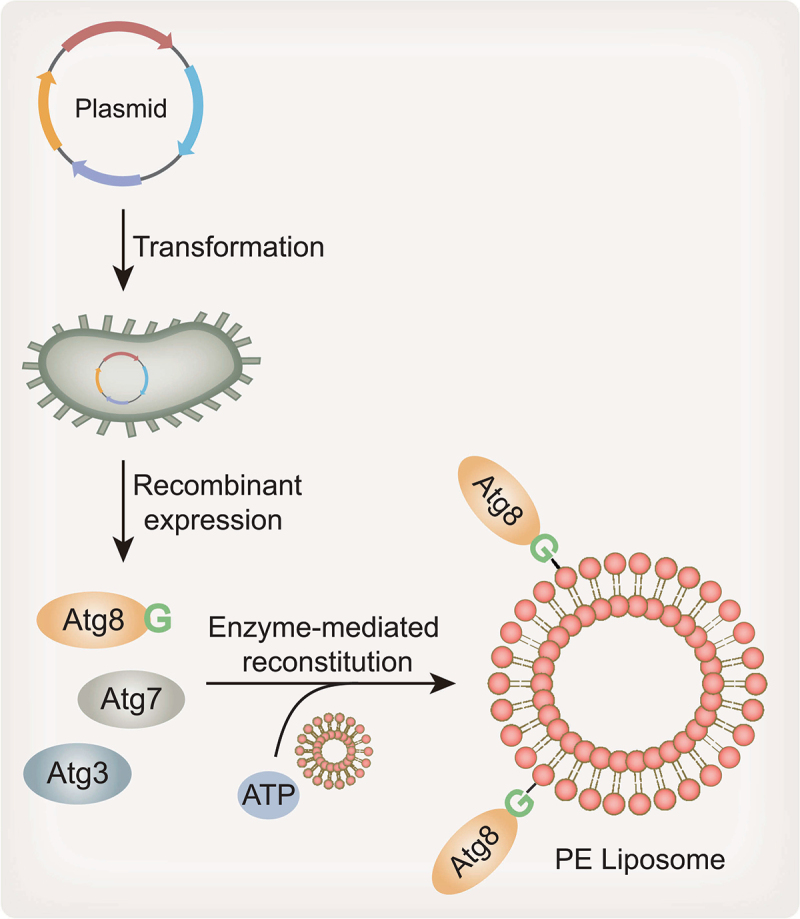
Enzyme-mediated Atg8–PE reconstitution system. Recombinant Atg8-I, Atg7 and Atg3 are obtained by bacterial expression and purification systems. Incubation of the purified components, including Atg8, Atg7 and Atg3, with PE-containing liposomes enables the production of Atg8–PE in the presence of ATP.
Similarly, through in vitro conjugation systems in which purified human ATG7, ATG3 and synthetic phospholipid liposomes are used in the presence of ATP, three Atg8 homologs, LC3B, GABARAP and GABARAPL2, are reconstituted [69]. The minimum components necessary for human Atg8-family protein conjugation to PE are human ATG7, ATG3, the respective Atg8 homolog (LC3B, GABARAP or GABARAPL2), PE-containing liposomes and ATP. Importantly, Atg8–PE conjugation is considered the predominant lipidation of autophagy in vivo [69], whereas Atg8-family proteins undergo alternative lipidation to PS at single membranes during LC3-associated phagocytosis or influenza A virus infection [75], suggesting that both PS and PE are targets of all human Atg8 homologs in vivo. The in vitro conjugation system can mediate the conjugation of these proteins with PS as efficiently as with PE. However, in contrast to PE conjugation, in vitro PS conjugation of yeast Atg8 is markedly suppressed at physiological pH levels [76]. Recently, an in vitro reconstitution strategy shows that the ATG5–ATG12-ATG16L1 complex in mammals also functions as an E3-like enzyme, which is required for efficient LC3 lipidation [44,77].
In addition, giant unilamellar vesicles (GUVs), which are 10–100 µm in diameter, can also serve as a platform for the conjugation of Atg8-family proteins in vitro [77–80]. GUVs containing PE are produced by electroformation using a hydrated lipid film [81]. Because of their large size, GUVs conjugated by fluorescent recombinant Atg8-family proteins allow imaging insights into Atg8 lipidation and membrane dynamics.
Overall, an enzyme-mediated reconstitution system can be used as a standard and effective approach for driving in vitro Atg8 lipidation.
Chemically defined reconstitution systems
The enzyme-mediated Atg8–PE reconstitution system requires various purified recombinant proteins, which is a time-consuming and laborious process. To bypass the need for conjugation machinery, two chemically defined reconstitution systems have been developed to mimic the lipidation process through which Atg8-family proteins are anchored to a membra
Since the first report of the thiol-Michael addition reaction in the 1960s [82], this reaction system quickly becomes an indispensable and powerful tool for protein chemical modification [83–86]. The chemoselective Michael addition of a sulfhydryl group to a maleimide group is a well-characterized conjugation reaction under neutral pH conditions, and it is commonly used for the coupling of fluorophores to biomolecules (proteins and peptides) with surface-exposed cysteine residues [85,87]. As a Michael acceptor, maleimide reacts with cysteine thiolate to form a covalent thiosuccinimide bond. Recently, a new liposome-based maleimide-thiol coupling strategy is developed for the preparation of LC3B–PE conjugates [31,88]. In this strategy, PE with a maleimide moiety in the head group (PE-maleimide) is incorporated into liposomes. LC3B does not contain a cysteine residue, and the C-terminal glycine residue in LC3B-I can be replaced by Cys (LC3BG120C) to enable a site-specific reaction between LC3B-I and maleimide-containing liposomes (Figure 3A) [31]. Recombinant LC3BG120C is prepared by an E. coli expression system and subsequent protein purification. PE-maleimide readily and directly interacts with LC3BG120C via a thiol-Michael addition reaction. This strategy allows the conjugation of LC3B-I to PE to generate LC3B–PE maleimide in vitro. Similar to endogenous LC3B–PE extracted from cells, this LC3B–PE conjugate is a fast-migrating band in SDS-PAGE. In contrast to an enzyme-mediated reconstitution system, which can be affected by several factors, such as the PE content in liposomes, reconstitution component ratio and conjugation reaction conditions, this maleimide-thiol coupling strategy enables efficient production of the LC3B–PE conjugate at the physiological PE level due to the high reactivity and selectivity of the thiol-Michael addition. Furthermore, short peptides corresponding to the first α-helix in LC3B and GABARAPL2 are synthesized through a C-terminal cysteine residue. Peptide–PE conjugates are obtained by incubating a synthetic α-helix peptide of LC3B or GABARAPL2 with liposomes containing PE-maleimide, which demonstrates that the first α-helix alone can mediate membrane lipid mixing [31]. Similarly, a GABARAP–PE conjugate in nanodiscs is prepared based on this strategy to investigate the function and structure of lipidated GABARAP protein [89].
Figure 3.
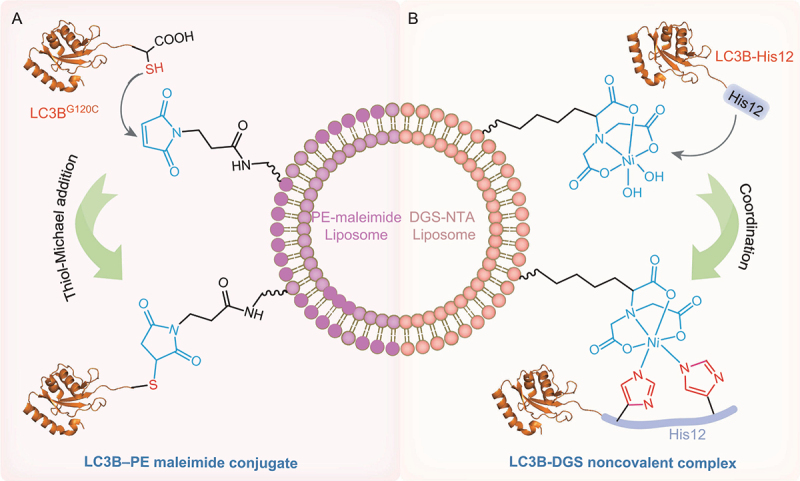
Chemically defined LC3B–PE reconstitution systems. (A) LC3B–PE reconstitution system based on maleimide-thiol coupling. PE with a maleimide moiety in the head group (PE-maleimide) is incorporated into liposomes. LC3BG120C directly interactes with PE-maleimide via a thiol-Michael addition reaction. (B) LC3B–PE reconstitution system based on a polyHis-NTA strategy. The PE mimic lipid DGS-NTA is included in the lipid composition used for preparing liposomes. DGS-NTA coordinates with His12-tagged LC3B to produce the LC3B-DGS noncovalent complex.
Alternatively, another chemically defined reconstitution system is based upon the high-affinity binding profile of polyHis-tagged protein to nickel-nitrilotriacetic acid (Ni-NTA) (Figure 3B) [70]. In this system, recombinant LC3B and GABARAPL2 proteins are purified with an additional modified polyHis tag (His12) added downstream of the C‐terminal Gly120 residue of LC3B and the Gly116 residue of GABARAPL2 (denoted as LC3B-His12 and GABARAPL2-His12, respectively). 1,2-Dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS-NTA), a synthetic diacyl lipid with a His-tag-binding head group, is included in the lipid composition used for preparing liposomes. The purified LC3B-His12 and GABARAPL2-His12 proteins can be stably and specifically attached to the membrane surface of liposomes through high‐affinity binding of the His-tag to DGS‐NTA lipids. The attachment of Atg8 proteins to the surface of liposomes mimics the membrane‐bound state of native Atg8 proteins [70]. However, mimicking the PE group using DGS‐NTA is a compromise solution since commercial DGS‐NTA lipids are not native PE structures. This DGS-NTA strategy has been widely used to mimic membrane anchoring of various proteins and peptides [90–95]. Notably, the link between the protein and DGS-NTA is noncovalent, which limits the application scope.
In summary, chemically defined reconstitution systems based on unilamellar liposomes, including systems based on a maleimide-thiol coupling or polyHis-NTA strategy, can be used to bypass the requirement for conjugation machinery to establish Atg8–PE, thereby providing simple and time-effective approaches to produce Atg8–PE mimics in vitro. However, notably, LC3B–PE-maleimide conjugates and the Atg8-DGS noncovalent complex are not native proteins and cannot mimic the physiological behaviors of native Atg8–PE.
Cell-free lipidation systems
The aforementioned enzyme-mediated and chemically defined reconstitution systems are based on unilamellar membranes with varying sizes and lipid compositions that mimic in vitro phagophore membranes. Actually, the lipid composition of phagophore membranes at the physiological level is complicated and still unclear, although some insights into the lipid composition of yeast autophagosomes and their precursors have been gained recently [80,96], thereby rendering it difficult to reconstitute phagophore membranes in vitro. To overcome this limitation, a novel cell-free lipidation system is recently developed (Figure 4) [32]. In this process, a cell-free reaction that mimics a certain cellular process is generated in vitro by combining cytosol and cellular membranes from cells lysed in a test tube. Cell-free systems have successfully evolved into several key platforms for synthetic biology applications [97,98].
Figure 4.
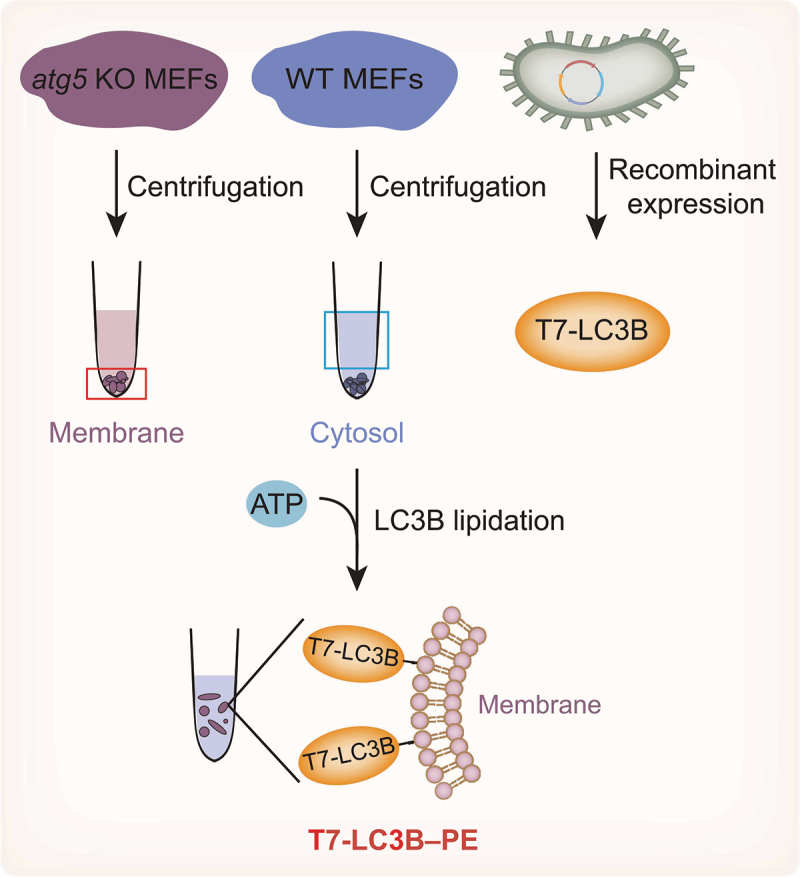
Cell-free LC3B–PE lipidation system. The cellular membrane fractionated from atg5 knockout MEFs donates the PE. The cytosol from WT MEFs provides the core components of LC3B lipidation, such as ATG proteins, regulatory proteins and ATP. Purified recombinant T7-tagged LC3B-I is used as a substrate for cell-free lipidation. Incubating T7-LC3B-I with the membrane and cytosol results in the production of T7-LC3B–PE.
In a cell-free LC3B lipidation system, mammalian cellular membranes are used instead of liposomes to mimic phagophore membranes. The key components of this cell-free lipidation system include the cytosol, the cellular membrane, nucleotides and the LC3B substrate [32,99,100]. The cytosol provides the core components for LC3B lipidation, including different ATG proteins and regulatory proteins. The cellular membrane donates PE. Nucleotides provide ATP for energy and guanosine triphosphate (GTP) for activating certain GTP-binding proteins, which are usually required for membrane remodeling events in cells. Moreover, LC3B-I is highly enriched in the cytosol, whereas LC3B–PE is precipitated with membranes derived from wild-type (WT) mouse embryonic fibroblasts (MEFs). In MEFs lacking Atg5, LC3B-I is present in both cytosolic and membrane fractions. In addition, the required membranes are obtained from fractionated atg5-knockout MEFs and, thus, mimic the physiological lipid compositions of MEF membranes. The fractionated membranes are incubated with the cytosol from WT MEFs containing LC3B-I and enzymes, resulting in the formation of LC3B–PE in a time- and ATP-dependent manner.
Furthermore, recombinant LC3B-I is used as a substrate in a cell-free lipidation system (Figure 4) [32]. To distinguish recombinant LC3B-I from endogenous LC3B-I, recombinant LC3B-I is tagged with T7 in the N-terminus and detected with a T7 antibody. The in vitro lipidation reaction is performed by incubating T7-LC3B with WT MEF cytosol and atg5-knockout MEF membranes. T7-LC3B–PE is observed in a cytosol and membrane concentration-dependent manner. In addition, the cytosol and membranes isolated from starved cells stimulate T7-LC3B lipidation in vitro. Similarly, the class III phosphatidylinositol 3-kinase (PtdIns3K) inhibitors 3-methyladenine (3-MA) and wortmannin and the phosphatidylinositol-3-phosphate (PtdIns3P)-blocking peptide FYVE can block in vitro T7-LC3B lipidation, suggesting that this cell-free lipidation system can reflect and respond to major regulatory pathways of autophagy in living cells to a great extent [32].
Therefore, compared with traditional in vitro liposome-based reconstitution systems, the cell-free lipidation system enables LC3 lipidation in an environment more reflective of the physiological state and is a general method that is useful for elucidating the molecular mechanisms of LC3 lipidation. However, in this approach, there is no full control of the molecular composition of the cell fractions, subsequently, it is difficult to separate the target fractions from other contaminants.
Protein chemical synthesis strategy
Proteins with various modifications can be obtained using a protein chemical synthesis strategy [101–104]. Native chemical ligation (NCL) is effective in near-neutral pH aqueous solutions and is the most widely used method for preparing modified proteins [105,106]. In NCL, the thiol group of an N-terminal cysteine residue of a peptide attacks the C-terminal thioester of another peptide to generate a thioester intermediate. Then, the intermediate undergoes rearrangement through spontaneous intramolecular N→S acyl shifting, resulting in the formation of a native amide bond between two peptides. The applications of NCL are largely limited by the length of the peptides that can be generated through solid-phase peptide synthesis (SPPS). To expand the synthetic scope of NCL, expressed protein ligation (EPL) based on intein protein splicing technology has been proposed [107]. EPL is a semisynthetic version of NCL in which synthetic and recombinant polypeptides are chemically ligated. The target protein fused to the N-terminus of an intein is obtained by recombinant protein expression and undergoes an N→S acyl shift, causing target protein transfer to the sulfhydryl group in cysteine. Subsequently, the engineered intein is cleaved by a thiol reagent, such as 2-mercaptoethanesulfonic acid sodium salt (MESNa), to form protein α-thioester, which is chemically ligated to the synthesized peptide to form a complete protein. To date, NCL and EPL have been employed to prepare a few hundred natural and chemically modified proteins [85,87,108–110]. Recently, LC3B–PE is produced directly using an EPL-based chemical synthesis strategy [28,29]. Protein chemical synthesis enables exogenously expressed truncated LC3B to form a covalent link with synthetic peptides harboring a cysteine mutation and PE modification. Given the poor solubility of LC3B–PE, two solubilization strategies involving polyArg and maltose binding protein (MBP) tags have been developed to overcome this problem.
Because the LC3B[1-114] MESNa thioester is produced at the highest level and has the best reactive efficiency, Ala114-Ser115 residues of LC3B are chosen as the ligation site in two strategies. In the light activatable solubilizing side chain strategy, a removable solubilizing side chain, polyArg (Arg4 tag), is employed to make a peptide–PE soluble in aqueous solution (Figure 5A) [29]. A fully protected hexapeptide is synthesized by standard SPPS. The polyArg tag capped with a Boc-Gly-OH residue on the N-terminus is linked to the amino group in a glutamine residue of the lipidated peptide via a designed photosensitive nitrobenzyl linker. The synthetically protected peptide is subsequently coupled with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE). After removing the protecting group, the EPL reaction between the lipidated peptide and the LC3B[1-114] MESNa thioester performed using an intein strategy is successful under detergent-free conditions. With UV irradiation, the polyArg tag and the photosensitive linker are removed, and LC3B–DSPE is then prepared after refolding (Figure 5A). In addition, the Cys115 residue of synthetic LC3B–DSPE can be labeled with a small fluorophore through maleimide coupling to provide a novel replacement for fluorescent LC3B–PE [29].
Figure 5.
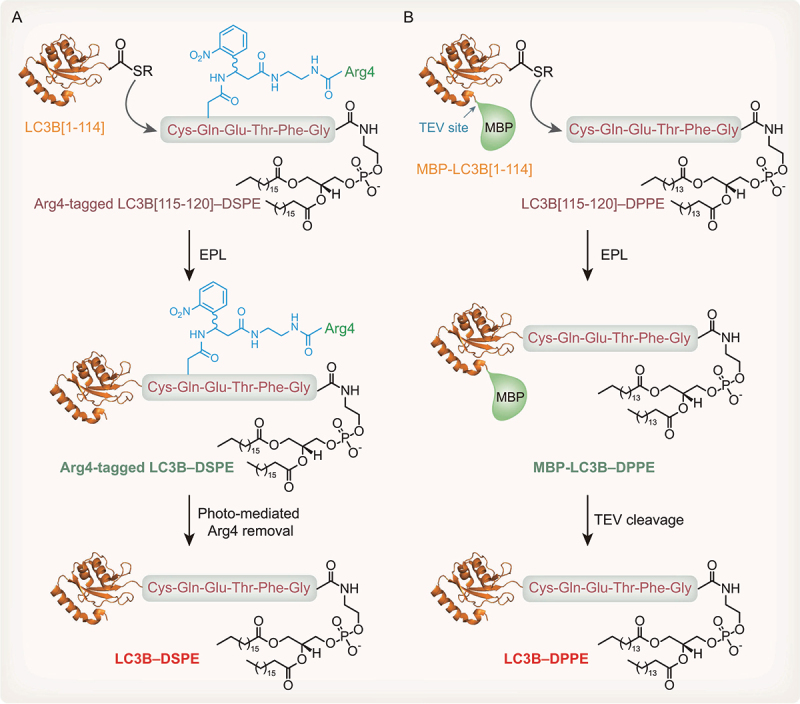
Protein chemical synthesis strategies to produce LC3B–PE in vitro. (A) PolyArg-assisted solubilization strategy to produce LC3B–PE. Recombinant LC3B[1-114] MESNa thioester is obtained by an intein strategy. LC3B[115-120] peptide CQETFG containing DSPE fused with a polyArg tag is obtained by SPPS. LC3B[1-114] MESNa thioester and Arg4-tagged peptide-DSPE are conjugated by EPL. Following photosensitive Arg4-tag removal, LC3B–DSPE proteins are produced. (B) MBP-assisted solubilization strategy to produce LC3B–PE. Recombinant LC3B[1-114] MESNa thioester fused with an MBP tag is obtained by an intein strategy. LC3B[115-120] peptide CQETFG containing DPPE is produced by SPPS. LC3B[1-114] MESNa thioester and peptide-DPPE are conjugated by EPL. Following TEV protease cleavage, LC3B–DPPE proteins are produced.
Alternatively, our group employs a facile tobacco etch virus (TEV) protease-cleavable MBP tag to facilitate solubilization of the lipidated proteins (Figure 5B) [28]. In summary, we fuse an LC3B[1-114] MESNa thioester with an MBP tag using an intein strategy. The protected hexapeptide is synthesized using an acid-sensitive chlorotrityl resin through standard SPPS and is subsequently activated as a pentafluorophenyl ester and coupled with 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) to produce a protected lipidated peptide. Treatment with high concentrations of trifluoroacetic acid (TFA) results in the removal of all acid-sensitive protection groups. Finally, the PE-modified peptide is ligated with MBP-LC3B[1-114] MESNa thioester under folding conditions in the presence of the detergent β-octylglucoside, with 4-mercaptophenylacetic acid (MPAA) serving as a catalyst. The resulting MBP-LC3B–DPPE protein is soluble in aqueous buffer without detergents, making work with the lipidated LC3B protein convenient. In addition, the MBP tag can be removed by TEV protease to obtain LC3B–DPPE [28].
Similar to endogenous LC3B–PE, chemically semisynthesized LC3B–PE proteins, including LC3B–DSPE and LC3B–DPPE, can be cleaved by the endogenous cysteine protease ATG4B, suggesting that semisynthetic LC3B–PE proteins are functional [28,29]. More importantly, semisynthetic LC3B–PE can be inserted into liposomes to study LC3B–PE protein-mediated membrane dynamics and protein-protein interactions. Indeed, the semisynthetic LC3B–PE proteins are successfully anchored to the liposome membranes, leading to the fusion of membranes to create larger liposome clusters in vitro. Remarkably, a protein chemical synthesis strategy enables LC3B proteins to undergo various modifications and mutations at any site. Using an EPL-based solubilizing MBP strategy, LC3B–PE with mutations in the C-terminal amino acid tail and several LC3B–lipid conjugates, such as LC3B–1,2-dihexanoyl-sn-glycero-3-phosphoethanolamine (LC3B–DHPE) and LC3B–1-hexadecanol (LC3B–C16), have been successfully prepared [71,111]. Therefore, functional LC3B–PE, mutants and LC3B–lipid conjugates are successfully prepared using this strategy, and these products are powerful tools for exploring the perplexing role of LC3B–PE in autophagy.
Applications of Atg8–PE protein-based in vitro approaches used in autophagy studies
Analyzing Atg8 protein-mediated membrane dynamics using a liposome-based reconstitution strategy
The Atg8 ubiquitin-like conjugation system plays a central role in autophagy. In vivo experiments have shown that yeast Atg8 controls phagophore expansion during autophagosome formation and that steady-state levels of Atg8 regulate autophagosome size [57]. Despite their likely importance during autophagosome formation, the precise functions of Atg8 proteins and the Atg8 mode of action are incompletely understood. To replicate features of autophagosome biogenesis in vitro, potentially flexible membrane substrates should be employed. For this purpose, liposome-based recombinant enzyme-mediated and chemically defined reconstitution approaches have been employed to anchor Atg8-family proteins to PE in unilamellar liposomes to analyze Atg8 protein-mediated membrane dynamics. Liposomes are desirable membrane substrates because they are both architecturally flexible and experimentally tractable and well suited for the purpose of developing autophagosome in vitro mimics [23,112]. Liposome-based assays allow for explicit testing of the capacity of different homologs to induce membrane fusion in vitro.
Ohsumi et al. [113] reports the first experimental evidence indicating that the yeast Atg8 protein directly drives the growth of phagophore membranes; their work is based on the use of a recombinant enzyme-mediated yeast Atg8–PE reconstitution strategy. Specifically, in vitro lipidation of yeast Atg8 causes membrane tethering and hemifusion, which is required for expansion of phagophore membranes. These data, however, relies on in vitro fusion reactions using liposomes containing 55% PE. Liposomes with physiological PE concentrations are not able to undergo Atg8–PE-dependent hemifusion, suggesting that Atg8–PE-mediated fusion is dependent on the high concentration of PE but not the physiological concentration [88]. In addition, a maleimide-thiol coupling-defined reconstitution system shows that LC3B and GABARAPL2 crosslinked to PE via their respective C-termini promote liposome tethering and membrane fusion in vitro [31]. These findings suggest that Atg8-family proteins have intrinsic and evolutionarily conserved membrane tethering and fusogenic functions.
Mammalian Atg8 homologs, including LC3 and GABARAP proteins, are all essential for the autophagic process. The LC3 subfamily is involved in elongation of the phagophore membrane, whereas the GABARAP subfamily is essential for later stage autophagosome maturation [114]. In vitro recombinant enzyme-mediated reconstitution systems are used to confirm that mammalian Atg8 homologs have different abilities to induce membrane tethering and fusion [115]. The lipidated forms of GABARAP and GABARAPL2 promote extensive membrane tethering and fusion, whereas lipidated LC3B promotes tethering and fusion to a profoundly lesser extent. In accordance with reported in vitro results, GABARAP and GABARAPL2 induce dramatic growth of vesicles, leading to roughly spherical structures in living cells, whereas LC3, a much less efficient fusogen, gives rise to elongated and peanut-shaped structures. In addition, a DGS‐NTA reconstitution assay reveals that GABARAPL2 is a more potent agent than LC3B for tethering the membranes of large flat vesicles (e.g., 200 and 400 nm in diameter). However, for highly curved small vesicles (e.g., 50 nm in diameter), LC3B can drive tethering more efficiently than GABARAPL2 [70]. These findings suggest that mammalian Atg8 homologs have different abilities to induce membrane tethering and fusion in different membrane curvature-inducing systems, shedding light on the roles of various mammalian Atg8-family proteins during autophagosome biogenesis.
More importantly, the lipidation of Atg8-family proteins can affect membrane curvature and morphology. The lipidation of yeast Atg8 stabilizes membrane curvature and produces profoundly curved membrane structures, such as nanotubes [116]. A structural analysis reveals that lipidated yeast Atg8 adopts a preferred orientation on the membrane that disrupts the membrane structure [117,118]. The membrane perturbation ability of Atg8–PE has been shown to be essential for efficient autophagosome biogenesis. In addition, lipidated Atg8-family proteins cooperate with cargo receptors to generate membrane curvature. In selective autophagy, the cargo-activated receptor Atg19 mediates tight apposition of the cargo and yeast Atg8-coated membranes by exposing multiple Atg8 binding sites, contributing to tight membrane bending of the phagophore membrane around the cargo [119,120]. The cargo receptor human SQSTM1/p62 stabilizes its interaction with LC3B and linear ubiquitin via oligomerization, which is sufficient to bend the membrane around the cargo [121]. Both cargo receptors, including Atg19 and SQSTM1/p62, control membrane bending via locally condensed Atg8–PE. Moreover, yeast Atg8 lipidation activates the Atg1 kinase, stimulating substrate phosphorylation along the growing phagophore membrane [122].
Overall, liposome-based reconstitution approaches have contributed important insights into the mechanisms underlying Atg8-family protein-mediated membrane dynamics.
Determining influential factors involved in Atg8–PE conjugation using an enzyme-mediated reconstitution system
Since the formation of Atg8–PE is central to autophagosome formation, understanding how the Atg8–PE conjugation reaction is regulated is very important. Indeed, the enzyme-mediated Atg8–PE reconstitution approach has provided unique insights into the influential factors involved in Atg8–PE conjugation. As previously mentioned, the minimal essential factors necessary for Atg8–PE conjugation are Atg7, Atg3, Atg8, PE-containing membranes and ATP [30]. The efficiency of in vitro Atg8–PE conjugation depends on the states of the enzymes, membrane lipid composition, membrane curvature and other important factors. The enzyme-mediated reconstitution system has been employed to yield important insights into the regulation of Atg8 lipidation.
First, the states of the enzymes Atg7 and Atg3 regulate the overall reaction efficiency. Atg8 proteins bind to Atg7 before being transferred to Atg3. A recent study shows that human ATG3 and ATG7 are susceptible to catalytic thiol oxidation, which affects the activities of the enzymes [123]. Treatment with oxidized glutathione (GSSG) decreases the amount of LC3B covalently bound to ATG3 and ATG7, resulting in defects in LC3B–PE formation in an enzyme-mediated lipidation reconstitution assay. In addition, acetylation promotes yeast Atg3 membrane binding and Atg8 lipidation [124]. Specifically, Lys19 and Lys48 residues of yeast Atg3 are acetylated by essential SAS2-related acetyltransferase 1 (Esa1) [125]. To further address the ability of Atg3 acetylation to regulate yeast Atg8 lipidation, homogeneous Lys19 and Lys48-diacetylated Atg3 is semisynthesized through sequential hydrazide-based native chemical ligation [124]. An enzyme-mediated lipidation reconstitution assay with semisynthetic proteins confirms that Atg3 acetylation could promote the lipidation of Atg8. The acetylation of Atg3 enhances its binding to physiological levels of PE in a liposome size-dependent manner, which in turn promotes the lipidation process [124].
Second, the membrane lipid composition affects the efficiency of the Atg8–PE lipidation reaction in vitro. The yield of yeast Atg8–PE increases in a dose-dependent manner with increasing PE content, constituting up to 70% of the total lipid content. The use of liposomes containing more than 90% PE, however, results in a remarkably diminished quantity of the yeast Atg8–PE conjugates [30]. PE has a small hydrophilic headgroup that is critical for its tendency to form nonbilayer structures. Liposomes with excess PE may possess an unfavorable structure for Atg8 lipidation. Thus, the PE content of membranes is an important factor governing the efficiency of Atg8–PE formation. In addition, the lipidation reaction is completely inhibited when 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) is replaced by DPPE and DSPE in the liposome [126]. DOPE contains 18 unsaturated carbon acyl chains and is a cone-shaped lipid, whereas DPPE and DSPE have 16 or 18 saturated carbon acyl chains, respectively, and are cylindrical lipids. The dependence of lipid formation on PE species indicates lipid packing constraints. Importantly, the yield of yeast Atg8–PE is enhanced by increases in either phosphatidylinositol (PI) or phosphatidylglycerophosphate (PG) or by the addition of either PS or phosphatidic acid (PA) [30]. Therefore, the Atg8–PE conjugation reaction appears to be sensitive to membrane lipid composition.
Third, membrane curvature influences Atg8–PE formation in vitro [116,126]. Lipidation reaction efficiency varies with the curvature of the underlying membrane. Smaller liposomes have a more highly curved surface than larger liposomes. In the enzyme-mediated Atg8 lipidation reconstitution assay, liposomes extruded through 800-nm filters are incompatible with lipidation, whereas 50-nm liposomes are quite compatible, suggesting that the extent of yeast Atg8 lipidation greatly depends on the curvature of the membrane in vitro [116]. Further experiments show that the E1-like enzyme Atg3 is a membrane curvature sensor and harbors a curvature-sensitive membrane-binding motif. Twenty-six amino-terminal amino acids in Atg3 ordinarily function as a weak amphipathic helix that dictates membrane curvature. Therefore, Atg3 is designed to function with highly curved membranes, possibly including the limiting edge of growing phagophores [126]. Atg3 affects PE‐like lipid dynamics and rearrangement, thereby promoting Atg8 lipidation [127].
In addition, other important factors have been identified to regulate Atg8 lipidation. Cargo receptors, including CALCOCO2/NDP52, TAX1BP1 (Tax1 binding protein 1) and OPTN (optineurin), are capable of driving LC3B lipidation [128]. The lipid PtdIns3P generated by PtdIns3K recruits WIPI2 (WD repeat domain, phosphoinositide interacting 2) interacting with ATG16L1, thereby activating LC3B lipidation on GUV membranes [77,79]. Human ATG12–ATG5 facilitates lipidation of human Atg8 proteins owing to its well-known ligase-like activity [78]. Moreover, Atg9 vesicles can serve as the seeds of the phagophore, recruiting Atg2 to transfer lipids for Atg8 lipidation in yeast [80].
In conclusion, using an enzyme-mediated reconstitution system, several influential factors involved in Atg8 lipidation have been clearly identified, expanding our knowledge of the Atg8 lipidation process.
Investigating Atg8–PE deconjugation mediated by Atg4 using an enzyme-mediated reconstitution system
Atg8–PE deconjugation is an important step required to facilitate multiple events during autophagy [54]. The inability to deconjugate Atg8–PE results in the mislocalization of this protein to the vacuolar membrane and the production of increasingly smaller autophagosomes in cells. Thus, it is quite important to clearly elucidate the mechanism of Atg8–PE deconjugation. In addition to converting pro-Atg8 to Atg8-I, Atg4 is critical for Atg8–PE deconjugation. Four mammalian Atg4 homologs exhibit distinctive functions and enzymatic activities with respect to soluble and lipidated forms of mammalian Atg8-family proteins. Mammalian Atg4 homologs are most active only on membrane-associated lipidated proteins, whereas ATG4B is capable of processing both soluble and lipidated forms of Atg8-family proteins. Numerous early studies on ATG4B soluble protein processing indicate that the peptide sequence of Atg8 downstream of the reactive glycine residue is unimportant. Similarly, the ATG4B crystal structure reveals only ATG4B-LC3B interactions on LC3B sequences upstream of glycine [129–131]. However, ATG4B cleaves soluble mammalian LC3 homologs very efficiently, whereas an intrinsically slow process is observed in the ATG4B release of lipidated proteins, suggesting that the mode of ATG4B-mediated LC3–PE deconjugation is different from that of ATG4B-mediated pro-LC3 cleavage [52].
Two recently reported models explain that Atg4 mediates the cleavage of soluble and lipidated forms of Atg8-family proteins in different ways. Yeast Atg4 is recruited to phagophore membranes by directly binding to Atg8 via two evolutionarily conserved Atg8 recognition sites, a classical LC3-interacting region (LIR) in the C-terminus of the protein and a novel motif in the N-terminus [132,133]. An in vitro deconjugation assay shows that a mutant N-terminal LIR causes a complete defect in yeast Atg8 release from its lipid anchor and exerts no effect on the cleavage of soluble Atg8, suggesting that the N-terminal LIR motif plays an important role in the Atg4 recognition of Atg8–PE [132]. In another report, an “interfacial inhibition” model of mammalian ATG4B is proposed to explain the effect of the membrane on ATG4B activity [52]. In an enzyme-mediated reconstitution assay, GABARAPL1–PE is proteolyzed by ATG4B much faster in the presence of detergent. The protease activity of ATG4B is interfacially inhibited on intact GABARAPL1–PE-containing bilayers. The degree of physical anchoring of Atg8-family proteins in the membrane strongly determines the rate of delipidation, suggesting that local cues augment or mitigate ATG4B activity. This mechanism suggests that a small fraction of a substrate pool is bioactivated and engaged in productive downstream events on the membrane, while a larger soluble pool is essentially unaffected by the same sets of downstream factors [52]. Overall, enzyme-mediated reconstitution assays have shown that Atg4 mediates cleavage of the soluble and lipidated forms of Atg8-family proteins in distinct ways.
Elucidating the molecular mechanism of early autophagic membrane generation using a cell-free lipidation system
A cell-free lipidation system allows LC3B to be lipidated in an environment more similar to the physiological environment. More importantly, this approach has enabled the discovery of autophagic membrane origins and the molecular mechanism of early autophagic membrane generation. In this approach, cellular membranes are fractionated through a sequential three-step membrane fractionation procedure involving differential centrifugation, a sucrose gradient and an OptiPrep gradient [32]. The lipidation activity of various membranes is monitored using a cell-free lipidation assay. Through this process, an ER-Golgi intermediate compartment (ERGIC)-enriched fraction is identified as the most active membrane source for triggering LC3 lipidation, suggesting that ERGIC is the membrane source for autophagosome formation [32,134]. Based on this strategy, the upstream pathway in ERGIC-regulating membrane sources has been identified. In addition, starvation induces the recruitment of coat protein complex II (COPII) to the ERGIC via activation of the PtdIns3K complex to cause budding of LC3 lipidation-active vesicles, which are potential membrane sources for autophagosome formation [33,135]. Moreover, RB1CC1/FIP200 (RB1 inducible coiled-coil 1) and MIA2/CTAGE5 (MIA SH3 domain ER export factor 2) facilitate starvation-induced remodeling of ER exit sites (ERESs), a prerequisite for COPII vesicles budding from the ERGIC, which contributes to autophagosome formation [136]. TMED9 (transmembrane p24 trafficking protein 9) is identified as an ERGIC determinant for autophagosome biogenesis [137]. In summary, the use of the cell-free lipidation system has led to unique insight into the molecular mechanism of early autophagic membrane generation.
Elucidating the biochemical mechanism of LC3–PE deconjugation mediated by the Legionella effector RavZ using semisynthetic LC3–PE
Legionella pneumophila is a gram-negative bacterium that invades alveolar macrophages and causes pneumonia in humans [138,139]. Specifically, Legionella manipulates membrane transport pathways to create a specialized vacuole that supports bacterial replication in host cells. Legionella can evade autophagy by delivering an effector protein, RavZ, into the host cytosol [140]. RavZ is a cysteine protease that irreversibly deconjugates all lipidated human Atg8-family proteins [140]. The deconjugation of Atg8–PE mediated by Legionella RavZ prevents the formation and maturation of autophagosomes, which in turn allows Legionella to escape host autophagy. In contrast to endogenous Atg4, Legionella RavZ cleaves only Atg8–PE, not pro-Atg8.
RavZ targets LC3B–PE to produce LC3B[1-119], which cannot remain conjugated with PE due to the lack of a glycine residue in the C-terminus of LC3B [140]. To address the mechanism of RavZ-LC3B–PE recognition, LC3B proteins with different C-terminal modifications are treated with RavZ in vitro [28,71,111,141]. RavZ cleaves these PE-modified LC3B proteins with a preference for long fatty acid chains, suggesting that the PE lipid moiety is essential for RavZ recognition and proteolysis. The N-terminal domain of RavZ has a fold similar to that of the yeast phospholipid transfer protein Sec14, implying that RavZ may have a lipid-binding site that accommodates the PE moiety [142]. Supporting this notion, microscale thermophoresis (MST) measurements show that RavZ binds to lipidated LC3B with a three-fold higher affinity than unlipidated LC3B [111,143]. In addition, the C-terminal residues of LC3B, Gln116, Phe119 and Gly120 are critical for RavZ recognition. The specific interactions of the LC3B C-terminal motif with RavZ may determine the specificity of the cleavage site of RavZ [144]. Therefore, various semisynthetic LC3B–PE proteins have been used as powerful tools with which to elucidate the biochemical mechanism of LC3B–PE deconjugation by Legionella RavZ.
Conclusions and outlook
Atg8–PE protein-based in vitro biochemical approaches have evolved sufficiently to constitute a robust toolkit for autophagy studies. As exemplified by the variety of studies covered in this review, these biochemical approaches are well poised to address numerous queries regarding the complexities of autophagy because they enable control of multicomponent compositions and spatiotemporal arrangements. Within the past two decades, these approaches have demonstrated their usefulness in analyses of the functions and mechanisms of Atg8-family proteins in autophagy. On the one hand, these in vitro Atg8–PE protein-based approaches have been employed to elucidate the molecular mechanisms of Atg8 lipidation and deconjugation processes. On the other hand, Atg8–PE and its mimics produced using these approaches have led to insights into the biochemical mechanisms of Atg8–PE protein-mediated membrane dynamics and protein-protein interactions. Therefore, these biochemical approaches have been extensively used and have contributed important insights into the mechanisms underlying Atg8 protein-mediated membrane dynamics, Atg8–PE conjugation and deconjugation, early autophagic membrane generation and RavZ-mediated irreversible Atg8–PE deconjugation. Overall, these biochemical approaches have compensated for defects of in vivo fluorescent protein-tagged Atg8 probes, thereby providing a new perspective in autophagy studies.
As described in the previous sections, although various in vitro biochemical approaches are available for the study of autophagy, the principal advantages and disadvantages of these methods should be considered when selecting an appropriate strategy for a given scientific purpose (Table 1). For example, an enzyme-mediated reconstitution system has been used as a standard and effective approach for in vitro Atg8 lipidation; however, this is a time- and laboratory resource-consuming process due to the need for expression and purification of recombinant enzymes and substrates. Although chemically defined reconstitution systems, including the maleimide-thiol coupling strategy and polyHis-NTA strategy, do not require the machinery for Atg8–PE conjugation to produce Atg8–PE mimics in vitro in a simple and time-effective manner, their products, Atg8–PE-maleimide conjugates and the Atg8-DGS complex, are not native proteins and cannot mimic the physiological behaviors of native Atg8–PE in some cases. Enzyme-mediated and chemically defined reconstitution systems are based on unilamellar liposomes and GUVs, which serve as in vitro mimics of phagophore membranes. However, the lipid composition of endogenous phagophore membranes at the physiological level is quite complicated and still unclear, rendering it difficult to reconstitute phagophore membranes in vitro. To better reconstitute LC3B lipidation in vitro, a cell-free lipidation system can be used. In this approach, the cellular membrane is fractionated and used as a membrane mimic, which enables LC3B lipidation in a physiological lipid environment. More importantly, the cell-free lipidation system can reflect and respond to the major regulatory pathways of autophagy in living cells. However, in the cell-free lipidation system, it should be noted that the target fractions are difficult to separate from other contaminants because of the lack of full control of the molecular composition of the cell fractions. The use of a chemical synthesis strategy enables the production of functional LC3B–PE with various mutants and modifications in preparative amounts for further analysis of protein–protein interactions. However, LC3B–PE synthesis using the EPL strategy remains a technical challenge because of SPPS and protein ligation, resulting in semisynthetic LC3B–PE protein production difficulties in a regular biological laboratory. Therefore, the advantages and disadvantages of various Atg8–PE protein-based in vitro biochemical approaches outlined in this paper provide general guidance for the use of in vitro biochemical approaches in the study of autophagy (Table 1).
Overall, these Atg8–PE protein-based in vitro biochemical approaches have played and will continue to play vital roles in advancing our understanding of Atg8–PE as a bona fide marker of autophagosomes. We envisage these in vitro biochemical approaches to contribute profoundly to the analysis of Atg8-mediated membrane dynamics and protein-protein interactions and to open a new door into the autophagy research field.
Funding Statement
This work was supported by the National Natural Science Foundation of China (91854101, 31801166, 32170185, 22011530161), the Natural Science Foundation of Chongqing, China (cstc2021jcyj-msxmX0030) and the Fundamental Research Funds for the Central Universities (2018CDQYSM0037, 2019CDCGSM303).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. DOI: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36(13):1811–1836. DOI: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhao YG, Codogno P, Zhang H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat Rev Mol Cell Biol. 2021;22(11):733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26(1):6–16. [DOI] [PubMed] [Google Scholar]
- [6].Sharma V, Verma S, Seranova E, et al. Selective autophagy and xenophagy in infection and disease. Front Cell Dev Biol. 2018;6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. [DOI] [PubMed] [Google Scholar]
- [8].Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–364. [DOI] [PubMed] [Google Scholar]
- [9].Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366(6467):818–822. [DOI] [PubMed] [Google Scholar]
- [10].Gatica D, Chiong M, Lavandero S, et al. The role of autophagy in cardiovascular pathology. Cardiovasc Res. 2021. DOI: 10.1093/cvr/cvab158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24(1):69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao YG, Zhang H. Core autophagy genes and human diseases. Curr Opin Cell Biol. 2019;61:117–125. [DOI] [PubMed] [Google Scholar]
- [13].Wen X, Yang Y, Klionsky DJ. Moments in autophagy and disease: past and present. Mol Aspects Med. 2021;82:100966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. 2020;383(16):1564–1576. [DOI] [PubMed] [Google Scholar]
- [15].Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 2021;17(1):1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tooze SA, Dikic I. Autophagy captures the Nobel Prize. Cell. 2016;167(6):1433–1435. [DOI] [PubMed] [Google Scholar]
- [17].Levine B, Klionsky DJ. Autophagy wins the 2016 Nobel Prize in physiology or medicine: breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A. 2017;114(2):201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mizushima N. The exponential growth of autophagy-related research: from the humble yeast to the Nobel Prize. FEBS Lett. 2017;591(5):681–689. [DOI] [PubMed] [Google Scholar]
- [19].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mizushima N, Murphy LO. Autophagy assays for biological discovery and therapeutic development. Trends Biochem Sci. 2020;45(12):1080–1093. [DOI] [PubMed] [Google Scholar]
- [21].Ueno T, Komatsu M. Monitoring autophagy flux and activity: principles and applications. Bioessays. 2020;42(11):e2000122. [DOI] [PubMed] [Google Scholar]
- [22].Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18(9):1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Turco E, Martens S. Insights into autophagosome biogenesis from in vitro reconstitutions. J Struct Biol. 2016;196(1):29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rao Y, Matscheko N, Wollert T. Autophagy in the test tube: in vitro reconstitution of aspects of autophagosome biogenesis. FEBS J. 2016;283(11):2034–2043. [DOI] [PubMed] [Google Scholar]
- [25].Alam JM, Noda NN. In vitro reconstitution of autophagic processes. Biochem Soc Trans. 2020;48(5):2003–2014. [DOI] [PubMed] [Google Scholar]
- [26].Moparthi SB, Wollert T. Reconstruction of destruction - in vitro reconstitution methods in autophagy research. J Cell Sci. 2018;132(4):jcs223792. [DOI] [PubMed] [Google Scholar]
- [27].Brier LW, Zhang M, Ge L. Mechanistically dissecting autophagy: insights from in vitro reconstitution. J Mol Biol. 2016;428(9):1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang A, Li Y, Pantoom S, et al. Semisynthetic lipidated LC3 protein mediates membrane fusion. ChemBioChem. 2013;14(11):1296–1300. DOI: 10.1002/cbic.201300344. [DOI] [PubMed] [Google Scholar]
- [29].Huang YC, Li YM, Chen Y, et al. Synthesis of autophagosomal marker protein LC3-II under detergent-free conditions. Angew Chem Int Edit. 2013;52(18):4858–4862. DOI: 10.1002/anie.201209523. [DOI] [PubMed] [Google Scholar]
- [30].Ichimura Y, Imamura Y, Emoto K, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279(39):40584–40592. DOI: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- [31].Weidberg H, Shpilka T, Shvets E, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20(4):444–454. DOI: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- [32].Ge L, Melville D, Zhang M, et al. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014;3:e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fujioka Y, Alam JM, Noshiro D, et al. Phase separation organizes the site of autophagosome formation. Nature. 2020;578(7794):301–305. DOI: 10.1038/s41586-020-1977-6. [DOI] [PubMed] [Google Scholar]
- [35].Nishimura T, Tooze SA. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020;6(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39–50. [DOI] [PubMed] [Google Scholar]
- [37].Yu ZQ, Ni T, Hong B, et al. Dual roles of Atg8–PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–892. DOI: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. DOI: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- [39].Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. DOI: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- [40].Otomo C, Metlagel Z, Takaesu G, et al. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20(1):59–66. DOI: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18(14):3888–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kuma A, Mizushima N, Ishihara N, et al. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277(21):18619–18625. DOI: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- [43].Romanov J, Walczak M, Ibiricu I, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–4317. DOI: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lystad AH, Carlsson SR, de La Ballina LR, et al. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat Cell Biol. 2019;21(3):372–383. DOI: 10.1038/s41556-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Popelka H, Reinhart EF, Metur SP, et al. Membrane binding and homodimerization of Atg16 via two distinct protein regions is essential for autophagy in yeast. J Mol Biol. 2021;433(5):166809. DOI: 10.1016/j.jmb.2021.166809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(9):859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schaaf MB, Keulers TG, Vooijs MA, et al. LC3/GABARAP family proteins: autophagy-(un)related functions. Faseb J. 2016;30(12):3961–3978. DOI: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- [48].Grunwald DS, Otto NM, Park JM, et al. GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction. Autophagy. 2020;16(4):600–614. DOI: 10.1080/15548627.2019.1632620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1–10. [DOI] [PubMed] [Google Scholar]
- [50].Kabeya Y, Mizushima N, Uero T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. DOI: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. DOI: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- [52].Kauffman KJ, Yu SL, Jin JX, et al. Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy. 2018;14(6):992–1010. DOI: 10.1080/15548627.2018.1437341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276. DOI: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nair U, Yen WL, Mari M, et al. A role for Atg8–PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8(5):780–793. DOI: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li M, Hou Y, Wang J, et al. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286(9):7327–7338. DOI: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Koyama-Honda I, Itakura E, Fujiwara TK, et al. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9(10):1491–1499. DOI: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- [57].Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gubas A, Dikic I. A guide to the regulation of selective autophagy receptors. FEBS J. 2022;289(1):75–89. [DOI] [PubMed] [Google Scholar]
- [59].Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432(1):80–103. [DOI] [PubMed] [Google Scholar]
- [60].Martens S, Behrends C. Molecular mechanisms of selective autophagy. J Mol Biol. 2020;432(1):1–2. [DOI] [PubMed] [Google Scholar]
- [61].Lamark T, Johansen T. Mechanisms of selective autophagy. Annu Rev Cell Dev Biol. 2021;37(1):143–169. [DOI] [PubMed] [Google Scholar]
- [62].Martens S, Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4(5):621–628. [DOI] [PubMed] [Google Scholar]
- [64].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. [DOI] [PubMed] [Google Scholar]
- [65].Morishita H, Kaizuka T, Hama Y, et al. A new probe to measure autophagic flux in vitro and in vivo. Autophagy. 2017;13(4):757–758. DOI: 10.1080/15548627.2016.1278094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kaizuka T, Morishita H, Hama Y, et al. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64(4):835–849. DOI: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- [67].Tornero-Écija A, Tábara L-C, Bueno-Arribas M, et al. A dictyostelium model for BPAN disease reveals a functional relationship between the WDR45/WIPI4 homolog Wdr45l and Vmp1 in the regulation of autophagy-associated PtdIns3P and ER stress. Autophagy. 2021;1–17. DOI: 10.1080/15548627.2021.1953262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Geng J, Klionsky DJ. Direct quantification of autophagic flux by a single molecule-based probe. Autophagy. 2017;13(4):639–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sou YS, Tanida I, Komatsu M, et al. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281(6):3017–3024. DOI: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- [70].Taniguchi S, Toyoshima M, Takamatsu T, et al. Curvature-sensitive trans-assembly of human Atg8-family proteins in autophagy-related membrane tethering. Protein Sci. 2020;29(6):1387–1400. DOI: 10.1002/pro.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yang A, Hacheney I, Wu YW. Semisynthesis of autophagy protein LC3 conjugates. Bioorg Med Chem. 2017;25(18):4971–4976. [DOI] [PubMed] [Google Scholar]
- [72].Saha SK, Nishijima S, Matsuzaki H, et al. A regulatory mechanism for the balanced synthesis of membrane phospholipid species in Escherichia coli. Biosci Biotechnol Biochem. 1996;60(1):111–116. DOI: 10.1271/bbb.60.111. [DOI] [PubMed] [Google Scholar]
- [73].Taherbhoy AM, Tait SW, Kaiser SE, et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol Cell. 2011;44(3):451–461. DOI: 10.1016/j.molcel.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Noda NN, Satoo K, Fujioka Y, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44(3):462–475. DOI: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- [75].Durgan J, Lystad AH, Sloan K, et al. Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Mol Cell. 2021;81(9):2031–2040. DOI: 10.1016/j.molcel.2021.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Oh-Oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. J Biol Chem. 2008;283(32):21847–21852. [DOI] [PubMed] [Google Scholar]
- [77].Fracchiolla D, Chang C, Hurley JH, et al. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol. 2020;219(7):e201912098. DOI: 10.1083/jcb.201912098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kaufmann A, Beier V, Franquelim HG, et al. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156(3):469–481. DOI: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- [79].Strong LM, Chang C, Riley JF, et al. Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy. eLife. 2021;10:e70372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sawa-Makarska J, Baumann V, Coudevylle N, et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 2020;369(6508):eaaz7714. DOI: 10.1126/science.aaz7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fracchiolla D, Zens B, Martens S. In vitro reconstitution of Atg8 conjugation and deconjugation. Methods Enzymol. 2017;587:377–390. [DOI] [PubMed] [Google Scholar]
- [82].Allen C, Fournier JO, Humphlett WJ. The thermal reversibility of the michael reaction: IV. Thiol adducts. Can J Chem. 2011;42(11):2616–2620. [Google Scholar]
- [83].Ravasco J, Faustino H, Trindade A, et al. Bioconjugation with maleimides: a useful tool for chemical biology. Chem Eur J. 2019;25(1):43–59. DOI: 10.1002/chem.201803174. [DOI] [PubMed] [Google Scholar]
- [84].Boutureira O, Bernardes GJ. Advances in chemical protein modification. Chem Rev. 2015;115(5):2174–2195. [DOI] [PubMed] [Google Scholar]
- [85].Luo Y, Jiang C, Yu L, et al. Chemical biology of autophagy-related proteins with posttranslational modifications: from chemical synthesis to biological applications. Front Chem. 2020;8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Gunnoo SB, Madder A. Chemical protein modification through cysteine. ChemBioChem. 2016;17(7):529–553. [DOI] [PubMed] [Google Scholar]
- [87].Yang A, Zhao L, Wu YW. Chemical synthesis and biological function of lipidated proteins. Top Curr Chem. 2015;362:137–182. [DOI] [PubMed] [Google Scholar]
- [88].Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. DOI: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ma P, Mohrluder J, Schwarten M, et al. Preparation of a functional GABARAP-lipid conjugate in nanodiscs and its investigation by solution NMR spectroscopy. ChemBioChem. 2010;11(14):1967–1970. DOI: 10.1002/cbic.201000354. [DOI] [PubMed] [Google Scholar]
- [90].Bhattacharya A, Niederholtmeyer H, Podolsky KA, et al. Lipid sponge droplets as programmable synthetic organelles. Proc Natl Acad Sci U S A. 2020;117(31):18206–18215. DOI: 10.1073/pnas.2004408117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chikh GG, Li WM, Schutze-Redelmeier MP, et al. Attaching histidine-tagged peptides and proteins to lipid-based carriers through use of metal-ion-chelating lipids. Biochim Biophys Acta-Biomembr. 2002;1567(1–2):204–212. DOI: 10.1016/S0005-2736(02)00618-1 [DOI] [PubMed] [Google Scholar]
- [92].Liu J, Zhu L, Zhang X, et al. Peptide-based NTA(Ni)-nanodiscs for studying membrane enhanced FGFR1 kinase activities. PeerJ. 2019;7:e7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Thomas FA, Visco I, Petrasek Z, et al. Introducing a fluorescence-based standard to quantify protein partitioning into membranes. Biochim Biophys Acta-Biomembr. 2015;1848(11 Pt A):2932–2941. DOI: 10.1016/j.bbamem.2015.09.001 [DOI] [PubMed] [Google Scholar]
- [94].Schafer J, Forster L, Mey I, et al. Neuroligin-2 dependent conformational activation of collybistin reconstituted in supported hybrid membranes. J Biol Chem. 2020;295(52):18604–18613. DOI: 10.1074/jbc.RA120.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Malecka KA, Szentpetery Z, Peterson JR. Synergistic activation of p21-activated kinase 1 by phosphatidylinositol 4,5-bisphosphate and Rho GTPases. J Biol Chem. 2013;288(13):8887–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Schutter M, Giavalisco P, Brodesser S, et al. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell. 2020;180(1):135–149. DOI: 10.1016/j.cell.2019.12.005. [DOI] [PubMed] [Google Scholar]
- [97].Noireaux V, Liu AP. The new age of cell-free biology. Annu Rev Biomed Eng. 2020;22:51–77. [DOI] [PubMed] [Google Scholar]
- [98].Lu Y. Cell-free synthetic biology: engineering in an open world. Synth Syst Biotechnol. 2017;2(1):23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang M, Ge L. Cell-free reconstitution of autophagic membrane formation. Methods Mol Biol. 2019;1880:135–148. [DOI] [PubMed] [Google Scholar]
- [100].Zhang M, Liu D, Ge L. In vitro dissection of autophagy. Curr Protoc Cell Biol. 2017;77:11.23.01–11.23.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Spicer CD, Davis BG. Selective chemical protein modification. Nat Commun. 2014;5(1):4740. [DOI] [PubMed] [Google Scholar]
- [102].Kent SBH. Novel protein science enabled by total chemical synthesis. Protein Sci. 2019;28(2):313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hackenberger CP, Schwarzer D. Chemoselective ligation and modification strategies for peptides and proteins. Angew Chem Int Edit. 2008;47(52):10030–10074. [DOI] [PubMed] [Google Scholar]
- [104].Bondalapati S, Jbara M, Brik A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat Chem. 2016;8(5):407–418. [DOI] [PubMed] [Google Scholar]
- [105].Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69(1):923–960. [DOI] [PubMed] [Google Scholar]
- [106].Dawson PE, Muir TW, Clark-Lewis I, et al. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–779. DOI: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- [107].Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95(12):6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Flavell RR, Muir TW. Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc Chem Res. 2009;42(1):107–116. [DOI] [PubMed] [Google Scholar]
- [109].Conibear AC, Watson EE, Payne RJ, et al. Native chemical ligation in protein synthesis and semi-synthesis. Chem Soc Rev. 2018;47(24):9046–9068. DOI: 10.1039/C8CS00573G. [DOI] [PubMed] [Google Scholar]
- [110].Kulkarni SS, Sayers J, Premdjee B, et al. Rapid and efficient protein synthesis through expansion of the native chemical ligation concept. Nat Rev Chem. 2018;2(4):0122. DOI: 10.1038/s41570-018-0122. [DOI] [Google Scholar]
- [111].Yang A, Pantoom S, Wu YW. Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. eLife. 2017;6:e23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jotwani A, Richerson DN, Motta I, et al. Approaches to the study of Atg8-mediated membrane dynamics in vitro. Methods Cell Biol. 2012;108:93–116. [DOI] [PubMed] [Google Scholar]
- [113].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. [DOI] [PubMed] [Google Scholar]
- [114].Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29(11):1792–1802. DOI: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Landajuela A, Hervas JH, Anton Z, et al. Lipid geometry and bilayer curvature modulate LC3/GABARAP-mediated model autophagosomal elongation. Biophys J. 2016;110(2):411–422. DOI: 10.1016/j.bpj.2015.11.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Knorr RL, Nakatogawa H, Ohsumi Y, et al. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PLoS One. 2014;9(12):e115357. DOI: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Maruyama T, Noda NN. Delineating the lipidated Atg8 structure for unveiling its hidden activity in autophagy. Autophagy. 2021;17(10):3271–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Maruyama T, Alam JM, Fukuda T, et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat Struct Mol Biol. 2021;28(7):583–593. DOI: 10.1038/s41594-021-00614-5. [DOI] [PubMed] [Google Scholar]
- [119].Sawa-Makarska J, Abert C, Romanov J, et al. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16(5):425–433. DOI: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yamasaki A, Alam JM, Noshiro D, et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell. 2020;77(6):1163–1175. DOI: 10.1016/j.molcel.2019.12.026. [DOI] [PubMed] [Google Scholar]
- [121].Wurzer B, Zaffagnini G, Fracchiolla D, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife. 2015;4:e08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Schreiber A, Collins BC, Davis C, et al. Multilayered regulation of autophagy by the Atg1 kinase orchestrates spatial and temporal control of autophagosome formation. Mol Cell. 2021;81(24):5066–5081.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Frudd K, Burgoyne T, Burgoyne JR. Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat Commun. 2018;9(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Li YT, Yi C, Chen CC, et al. A semisynthetic Atg3 reveals that acetylation promotes Atg3 membrane binding and Atg8 lipidation. Nat Commun. 2017;8(1):14846. DOI: 10.1038/ncomms14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336(6080):474–477. DOI: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- [126].Nath S, Dancourt J, Shteyn V, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2014;16(5):415–424. DOI: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang S, Li Y, Ma C. Atg3 promotes Atg8 lipidation via altering lipid diffusion and rearrangement. Protein Sci. 2020;29(6):1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chang C, Shi X, Jensen LE, et al. Reconstitution of cargo-induced LC3 lipidation in mammalian selective autophagy. Sci Adv. 2021;7(17):eabg4922. DOI: 10.1126/sciadv.abg4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Satoo K, Noda NN, Kumeta H, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28(9):1341–1350. DOI: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Kumanomidou T, Mizushima T, Komatsu M, et al. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol. 2006;355(4):612–618. DOI: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- [131].Sugawara K, Suzuki NN, Fujioka Y, et al. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280(48):40058–40065. DOI: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- [132].Abreu S, Kriegenburg F, Gomez-Sanchez R, et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2018;18(5):765–780. DOI: 10.15252/embr.201643146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kriegenburg F, Reggiori F. LIR and APEAR, two distinct Atg8-binding features within Atg4. Oncotarget. 2017;8(47):81717–81718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ge L, Schekman R. The ER-Golgi intermediate compartment feeds the phagophore membrane. Autophagy. 2014;10(1):170–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ge L, Wilz L, Schekman R. Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy. 2015;11(12):2372–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ge L, Zhang M, Kenny SJ, et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017;18(9):1586–1603. DOI: 10.15252/embr.201744559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Li SL, Yan R, Xu JL, et al. A new type of ERGIC-ERES membrane contact mediated by TMED9 and SEC12 is required for autophagosome biogenesis. Cell Res. 2021. DOI: 10.1038/s41422-021-00563-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Gomez-Valero L, Rusniok C, Carson D, et al. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci U S A. 2019;116(6):2265–2273. DOI: 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Qiu J, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol. 2017;15(10):591–605. [DOI] [PubMed] [Google Scholar]
- [140].Choy A, Dancourt J, Mugo B, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338(6110):1072–1076. DOI: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Mei L, Qiu X, Jiang C, et al. Host delipidation mediated by bacterial effectors. Trends Microbiol. 2021;29(3):238–250. DOI: 10.1016/j.tim.2020.09.012. [DOI] [PubMed] [Google Scholar]
- [142].Schaaf G, Ortlund EA, Tyeryar KR, et al. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29(2):191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Pantoom S, Yang A, Wu YW. Lift and cut: anti-host autophagy mechanism of Legionella pneumophila. Autophagy. 2017;13(8):1467–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Yang A, Pantoom S, Wu YW. Distinct mechanisms for processing autophagy protein LC3-PE by RavZ and ATG4B. ChemBioChem. 2020;21(23):3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]


