Abstract
Pueraria lobata var. montana (P. montana) belongs to the genus Pueraria and originated in Asia. Compared with its sister P. thomsonii, P. montana has stronger growth vigour and cold-adaption but contains less bioactive metabolites such as puerarin. To promote the investigation of metabolic regulation and genetic improvement of Pueraria, the present study reports a chromosome-level genome of P. montana with length of 978.59 Mb and scaffold N50 of 80.18 Mb. Comparative genomics analysis showed that P. montana possesses smaller genome size than that of P. thomsonii owing to less repeat sequences and duplicated genes. A total of 6,548 and 4,675 variety-specific gene families were identified in P. montana and P. thomsonii, respectively. The identified variety-specific and expanded/contracted gene families related to biosynthesis of bioactive metabolites and microtubules are likely the causes for the different characteristics of metabolism and cold-adaption of P. montana and P. thomsonii. Moreover, a graphic genome was constructed based on 11 P. montana accessions. Total 92 structural variants were identified and most of which are related to stimulus-response. In conclusion, the chromosome-level and graphic genomes of P. montana will not only facilitate the studies of evolution and metabolic regulation, but also promote the breeding of Pueraria.
Keywords: Pueraria lobata var. montana, chromosomal-level genome, graphic genome, comparative genomics, structural variants
1. Introduction
The genus Pueraria belonging to the Leguminosae family was originated in Asia and comprises of more than 20 species. Thereinto, Pueraria lobata (P. lobata) is an important species with abundant health-beneficial metabolites such as puerarin and has been used as medicinal plant.1 The main medicinal compounds in the root of P. lobata are flavonoids and isoflavones including daidzein, genistein and puerarin (8-C-glycoside of daidzein), which possess various biological activities such as anti-alcohol, antioxidant, antipyretic and anticancer.2,3
Pueraria lobata var. montana (P. montana) and P. lobata var. thomsonii (P. thomsonii) are the two varieties of P. lobata, while their usages are completely different.4Pueraria thomsonii is traditionally used as an edible and medicinal plant owing to its high contents of starch and medicinal metabolites.5,6 The expanded root is the major edible part of P. thomsonii. However, the root of P. montana is not expanded. According to the Chinese Pharmacopoeia, P. lobata with high puerarin content (about 2.4%) is called as ‘Gegen’, while P. thomsonii with comparatively low puerarin content (about 0.3%) is regarded as ‘Fenge’. Nevertheless, P. montana barely has puerarin, resulting in less nutritional and medicinal values.7 It was reported the contents of amino acids and sugars contributing to nutritional values and flavonoids, isoflavones and phenolic acids contributing to medicinal values, are significantly higher in P. thomsonii than that in P. montana.8
Except for the significant differences in nutritional and medicinal values, P. montana and P. thomsonii also exhibit great differences in growth habits. Pueraria montana is tolerant to cold and can survive even under −10°C.9 Whereas, P. thomsonii is more susceptible to low temperature and mainly distributes in the temperate regions of eastern and southeast Asia.10 Morphologically, P. montana has smaller and almond-shaped leaves, thinner vines and more elongated roots than that of P. thomsonii, which possibly makes P. montana more malleable and stronger vigour. As a variety without artificial domestication, P. montana possibly contains some special resistance genes which were lost in P. thomsonii. The genome of P. thomsonii was recently published, which provided valuable resource for clarifying the metabolic pathways of medicinal compounds.11 However, the unavailability of P. montana genome greatly hinder the identification and utilization of excellent gene resources in this variety.
To address this problem, a high-quality and chromosome-level genome of P. montana was de novo assembled and annotated in this study, which was further used for comparative genomics analysis and construction of a graphic genome based on 11 P. montana accessions. The results showed that P. montana possesses smaller genome size than that of P. thomsonii, which is caused by less repeat sequences and gene duplication events. Besides, graphic genome analysis identified a total of 12 genes containing structural variants (SVs), and most of them are involved in the response to stimulus. Taken together, the high-quality genome of P. montana will provide valuable genomic resources for the evolutionary study of Pueraria genus and utilization of excellent gene resources in P. montana.
2. Materials and methods
2.1. Plant materials
Eleven representative accessions of Pueraria lobata var. montana (P. montana) were collected and used as the materials, including five accessions from Guangxi Province, four accessions from Yunnan Province, one accession from Jiangxi Province and one accession from Hunan Province. The detailed information is provided in Supplementary Table S1.
2.2. Sequencing
PM12 accession was selected for de novo genome assembly and the other 10 accessions were used for graphic genome construction. DNA was extracted from the young leaves. PCR-free single-molecule real-time library was constructed for PM12 and then sequenced on the PacBio Sequel platform (Pacific Biosciences, CA, USA) with CLR pattern according to the manufacturer’s instruction. Hi-C sequencing library was constructed via six steps: (i) cells were treated with paraformaldehyde to fix the conformation of the DNA; (ii) restriction endonuclease (Mbo I) processed cross-linked DNA to produce sticky ends; (iii) DNA terminal was repaired flattening and biotin was introduced to label the oligonucleotide terminal; (iv) DNA ligase-binded DNA fragments; (v) protease digested and removes cross-linking with DNA, and the purified DNA randomly interrupted into 300–500 bp fragments; (vi) the labeled DNA was captured by avidin magnetic beads. The Hi-C library was sequenced on MGISEQ-2000 PE150 platform. For gene annotation, RNA was extracted from the root, stem and leaves of PM12 and their full-length transcriptomes were sequenced on PacBio Sequel platform. Library construction and whole genome re-sequencing of the other 10 accessions were performed on BGIseq 500 platform (BGI, Shenzhen, China) according to the protocol provided by the manufacture.
2.3. De novo genome assembly
For de novo assembly of P. montana (PM12), the raw PacBio subreads were error-corrected and assembled into contigs using NextDenovo (seed_cutoff = 23826, read_cutoff = 1000) (https://github.com/Nextomics/NextDenovo, 5 October 2021, date last accessed). Subsequently, primary contigs were polished by the BGI short-reads with Pilon12 and the subreads with Arrow (https://github.com/PacificBiosciences/GenomicConsensus, 10 October 2021, date last accessed), respectively. The corrected contigs were further scaffolded into chromosomal-level genome via Hi-C.13 The raw Hi-C reads were filtered to remove adapter and low-quality sequences, and then aligned to the contigs with bwa-mem214 (v2.2.1). Next, Lachesis was used to remove the sequences beyond 500 bp from the restriction site.15 The obtained data were used for constructing chromosomal-level genome through clustering, sorting and orienting by juicer (v1.6.2).16 Visualization and manual correction were performed by JucieBox (https://github.com/aidenlab/juicebox/, 12 October 2021, date last accessed). BUSCO (v5.2.2) was applied to evaluate the completeness of assembly by mapping the genome sequence to the embryophyta_odb10 database.17 Quast (v5.0.2) was employed to calculate the statistics on the assembled genome.18
2.4. Reference-guided genome assembly
For reference-guided assembly of the other 10 accessions, the adapter and low-quality sequences were removed using fastp (v0.12.4) to get the clean reads.19 The clean reads were firstly corrected by CARE20 and then assembled into contigs using ALGA.21 The contigs were further scaffolded into chromosomal-level genomes by Ragtag (v2.0.1) using the de novo assembled genome of PM12 as the reference.22 Genome synteny analysis between the reference genome and the reference-guided genomes was performed by minimap2 with default parameters.23
2.5. Genome annotation
Repeat sequences were annotated with combination of homology and de novo predictions. RepeatMasker (v4.0.9) and RepeatProteinMask (http://www.repeatmasker.org, 20 October 2021, date last accessed) were applied to perform homology prediction based on RepBase database. Furthermore, RepeatModeler (v2.0.3) (http://repeatmasker.org/RepeatModeler/, 20 October 2021, date last accessed) and LTR-FINDER were used to perform de novo prediction based on self-sequence alignment and features of repeat sequences.24 TRF (v4.09.1) was used to annotate Tandem Repeat.25
tRNAscan-SE (v2.0.9) was applied to annotate transfer RNA (tRNA) sequences based on the structural features of the tRNA.26 Ribosomal RNA (rRNA) sequences were identified by applying BLASTN (v2.12) against rRNA sequences of closely related species. In addition, microRNAs (miRNAs) and small nuclear RNAs (snRNAs) were annotated with application of INFERNAL (v1.1.4) of Rfam.27
The gene structures were determined by combining the results of homology, de novo and transcriptome evidence-based predictions. Homology prediction was performed based on four closely related species (A. thaliana, G. max, M. truncatula and L. japonicus) by Exonerate (https://github.com/nathanweeks/exonerate), 20 October 2021, date last accessed (v2.4.0). De novo prediction was executed by application of Augustus (v3.4.0)28 and GeneMark (v4.33).29 RNA-seq and Iso-seq were passed to Trinity (https://github.com/trinityrnaseq/trinityrnaseq, 20 October 2021, date last accessed) (v2.13.2) and GMAP30 to identify transcript sequences which would be used to predict gene structure by applying PASA31 (v2.4.1). The EVM was subsequently used to integrate the gene sets predicted by various methods into a non-redundant and more complete gene set.32 Finally, the gene structures were updated by PASA using the transcriptome data. Gene functions were annotated by application of BLASTP (e-value <= 1e − 5) searches against multiple protein databases, including KEGG (http://www.genome.jp/kegg/, 3 November 2021, date last accessed), SwissProt (http://www.UniProt.org/, 3 November 2021, date last accessed), InterPro (https://www.ebi.ac.uk/interpro/, 3 November 2021, date last accessed), NR (http://www.ncbi.nlm.nih.gov, 3 November 2021, date last accessed) and TrEMBL (http://www.bioinfo.pte.hu/more/TrEMBL.htm, 3 November 2021, date last accessed).
2.6. Genome synteny analysis between P. montana and P. thomsonii
The sequence synteny between P. montana and P. thomsonii was performed by nucmer (–mum –mincluster 500) of mummer33 (v4). The gene collinearity was analyzed using DIAMOND34 (v2.0.14.152) for alignment and McscanX35 for identification of gene blocks (e-value cutoff of 1e−5), respectively. Finally, Ks values of pair genes were calculated using TBtools.36
2.7. Comparative genomics analysis
Individual gene sets from 15 species, including Abrus precatorius (A. precatorius), Arachis duranensis (A. duranensis), Cajanus cajan (C. cajan), Cicer arietinum (C. arietinum), Glycine max (G. max), Lupinus angustifolius (L. angustifolius), Mucuna pruriens (M. pruriens), Medicago truncatula (M. truncatula), P. thomsonii, P. montana, Senna tora (S. tora), Spatholobus suberectus (S. suberectus) and Vigna unguiculata (V. unguiculata), were filtered by keeping the longest transcript for genes with more than one isoforms. Gffread (v0.12.7) (https://github.com/gpertea/gffread, 24 February 2022, date last accessed) was used to extract protein sequences. Subsequently, orthoFinder (v2.2.7) was applied to perform cluster analysis and obtain all gene families.37
Single-copy gene families were used to construct phylogenetic trees. Muscle38 (v5.1) was applied to perform multiple sequence alignment and then protest39 (v3.4) (-all-distributions -F -AIC -BIC -tc 0.5) was applied to predict the best substitution model of amino acid. Using A. thaliana and O. sativa as the outgroups, phylogenetic tree was constructed by RaxML40 (v8.2.12) (-m PROTGAMMAIJTTF) with the maximum likelihood method.41 Thereafter, the constructed phylogenetic tree, combined with the TimeTree (http://timetree.org/, 27 February 2022, date last accessed), was delivered to the mcmctree (seqtype = 2) of PAML (v4.9) to estimate divergence time.42 Gene family expansion and contraction were analyzed using CAFE5 (v5.0, -k = 5).43 All contents of significantly expanded or contracted gene families (P-value < 0.05) were extracted and subjected to GO and KEGG functional enrichment analyses.
2.8. Functional enrichment analyses
GO and KEGG enrichment analyses for gene members of variety-specific or significantly expanded/contracted gene families were performed by clusterProfiler (v4.2.2).44 The significant GO terms and KEGG pathways were obtained by enrichment using hypergeometric test to test significance and Benjamini and Hochberg to correct the P values (parameter: pvalueCutoff = 0.05, pAdjustMethod = BH, qvalueCutoff = 0.2).
2.9. Graphic genome construction and identification of variants
Graphic genome of P. montana was constructed by minigraph45 (-xggs) using the de novo assembly genome of PM12 and the other 10 reference-guided assembly genomes. SVs (InDels or substitutions > 50 bp) were identified using gfatools (https://github.com/lh3/gfatools, 10 March 2022, date last accessed).
2.10. Cluster and principal component analysis
The clean reads of the other 10 accessions were aligned to PM12 genome by bwa-mem2 (v2.2.1) (mem -M -Y).14 Picard.jar (https://broadinstitute.github.io/picard/, 25 March 2022, date last accessed) was used to mark the PCR duplicates. SNPs were called by GATK (4.2.3). After hard-filter, Iqtree246 (2.2.0) was used to construct the phylogenetic tree (–seqtype DNA -m MFP -B 1000 -alrt 1000 -T AUTO). Principal component analysis (PCA) was performed using plink2 (v2.00a2.3LM, –max-alleles 2) (https://www.cog-genomics.org/plink/2.0/, 25 March 2022, date last accessed).
3. Results
3.1. De novo assembly and annotation of P. montana genome
For de novo assembly of P. montana genome, PM12, an accession of P. montana widely distributing in Guangxi Province, was selected as the material. Genome survey was firstly performed, which produced 161.9 M paired-end reads. K-mer analysis showed the estimated genome is 1,257.56 Mb in size with 0.55% of heterozygosity, 79.77% of repeated sequences and 37.65% of GC content. Then, sequencing using the PacBio long-read in CLR pattern and Hi-C technology generated 139.0 and 72.7 Gb data, respectively (Supplementary Table S2). The PacBio subreads were assembled into contigs using NextDenovo, which were further polished by the paired-end reads and CLR subreads. The polished contigs were subsequently clustered, ordered and oriented using the Hi-C reads. The heatmap of the interaction matrix showed strong interactions within each chromosome (Supplementary Fig. S1). After manual correction, 91.11% of the contigs were anchored into 11 pseudochromosomes, resulting in a chromosome-scale genome with size of 978.59 Mb (Supplementary Table S3). The lengths of contig N50, scaffold N50 and the longest scaffold are 1.61, 80.18, and 100.14 Mb, respectively, indicating high continuity of the assembled genome (Supplementary Table S4). 95.78% of the paired-end reads were successfully mapped onto the assembled genome with 99.85% genome coverage and 0.0041% of single base error rate, demonstrating high accuracy of the genome. The high completeness of the assembly was further verified by 99.3% of complete BUSCOs (Supplementary Table S3) with the OrthoDB database (embryophyta_odb10).
An integrated method based on ab initio prediction, homologous prediction using A. thaliana, G. max, M. truncatula and L. japonicus as the related species, and evidenced prediction (Iso-seq and RNA-seq) was employed to perform structural annotation. In total, 38,812 gene models and 54,476 proteins were annotated. The average lengths of genes and CDSs are 4,251.96 and 1,082.52 bp, respectively. The genes are mainly distributed on the arm of chromosome rather than in the middle where is principally composed of repeat sequences (Fig. 1a); 94.0% of the complete BUSCOs were found in the annotations, suggesting high completeness of the gene structural annotation (Supplementary Table S5). Out of all the predicted proteins, 49,656 proteins (91.15%) were functionally annotated by alignment to several protein databases including SwissProt, TrEMBL, KEGG, InterPro, GO and NR (Supplementary Table S6).
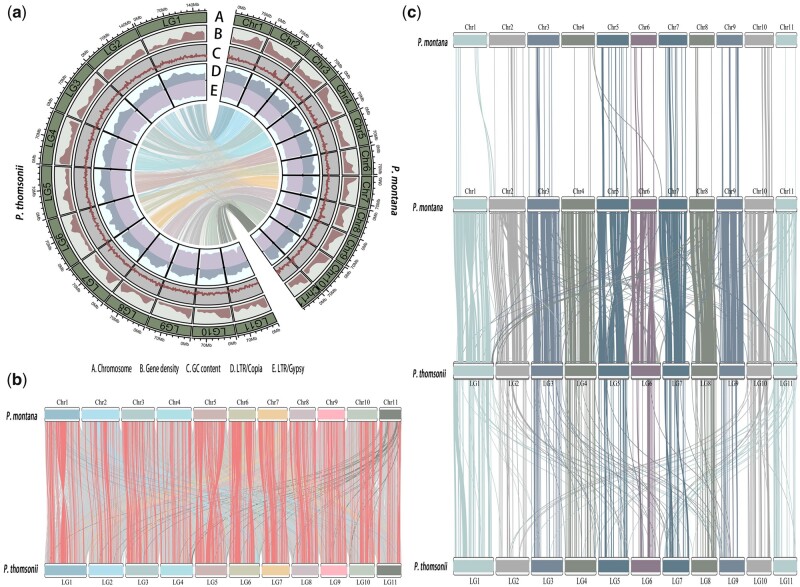
Figure 1.
The genome characteristics of P. montana and synteny between P. montana and P. thomsonii. (a) Distribution of genomic features within the P. montana and P. thomsonii genomes. A. Length of chromosome; B. Gene density with 1 Mb window size; C. GC content with 1 Mb window size; D. LTR/Copia density with 1 Mb window size; E. LTR/Gypsy density with 1 Mb window size; the lines in the innermost layer of the circle show the synteny between P. montana and P. thomsonii. (b) SVs including inversion, translocation (>10 kb) between P. montana and P. thomsonii; the gray lines represent normal collinearity and red lines show intrachromosomal inversion, while the other colors show interchromosomal SVs for each chromosome. (c) Collinear gene blocks within genome and between the genomes of P. montana and P. thomsonii (A color version of this figure appears in the online version of this article).
Furthermore, no-coding RNAs (ncRNA) account for 0.027% of the genome sequence, including 302 miRNA, 1,134 tRNA, 167 rRNA and 461 snRNA. Repeat sequences were annotated by homology (RepBase as database) and de novo annotations. Tandem repeats and transposable elements (TE) account for 77.75% (760.89 Mb) of the genome. In addition, long-terminal repeats (LTR) are the dominating component of TEs, taking account of 51.75% of the genome. Copia and Gypsy LTR retrotransposons are the major numbers of LTR, accounting for 16.49% and 15.85% of the genome, respectively. Whereas the proportions of DNA transposons and long-interspersed nuclear elements are 19.16% and 7.78%, respectively (Supplementary Table S7).
3.2. Comparative genomics analysis between P. montana and P. thomsonii
Compared with the P. thomsonii genome (1,381.61 Mb),11 the P. montana genome has smaller genome size and more compact gene distribution (Fig. 1a). A total of 318 Mb syntenic regions were observed between P. montana and P. thomsonii. Moreover, large number of SVs were identified between the two genomes (Fig. 1b), which is possibly caused by the more fragmented genome of P. thomsonii (contig N50 of 593.70 kb). Genome collinearity analysis identified a total of 19,629 genes (49.20% ortholog frequencies) are common between the two genomes (Fig. 1c). However, more gene duplication events were observed in P. thomsonii genome (9,568 collinear genes, 23.35% paralog frequencies) than that in P. montana genome (1,685 collinear genes, 4.34% paralog frequencies). There is no whole-genome duplication (WGD) occurring after their divergence, while the peaks of Ks at near zero seems to be caused by tandem or segment duplication47 (Supplementary Fig. S2). Taken together, compared with the P. montana genome, the larger genome size of P. thomsonii is approximately due to more repeat sequences and gene duplication events.
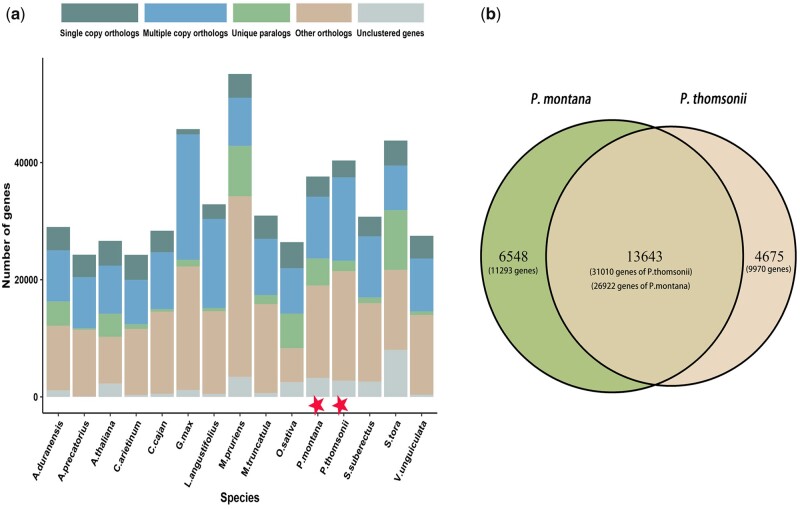
To investigate the gene family diversity of P. montana and P. thomsonii, identification and clustering of gene families were performed. Two model species (A. thaliana and O. sativa) and 13 legume species (A. precatorius, A. duranensis, C. cajan, C. arietinum, G. max, L. angustifolius, M. pruriens, M. truncatula, P. thomsonii, P. montana, S. tora, S. suberectus and V. unguiculata) were selected to perform comparative genomics analysis. A total of 515,730 genes were clustered into 27,243 orthogroups and several types of features including single-copy orthologs, multiple-copy orthologs, unique paralogs, other orthologs and unclustered genes were identified (Fig. 2a). More unique paralogs and less multiple-copy orthologs were identified in P. montana compared with that in P. thomsonii. A total of 6,548 and 4,675 variety-specific orthogroups were found for P. montana and P. thomsonii, containing 11,293 and 9,970 genes, respectively (Fig. 2b).
Figure 2.
Clusters of gene families. (a) Distribution of genes contained in different features within species. Asterisk highlights two important species (P. montana and P. thomsonii) in the present study. (b) Number of shared and non-shared gene families between P. montana and P. thomsonii and the number of genes contained in these families.
GO enrichment analysis for the members of variety-specific gene families indicated that the major members of variety-specific gene families for P. montana are involved in microtubule and cell wall organization, which play important roles in response to abiotic stresses.48 Whereas the major members of variety-specific gene families for P. thomsonii are related to the responses to auxin, defense, and oxidative stress, as well as recognition of pollen (Supplementary Fig. S3). These results suggested that the two varieties evolved differently specific functions to adapt to environments. Besides, KEGG enrichment analysis for the members of variety-specific gene families showed that several pathways related to the biosynthesis of bioactive metabolites were significantly enriched in P. thomsonii, including flavone, flavonoid and phenylpropanoid. However, these pathways were not significantly enriched by the variety-specific gene families in P. montana (Supplementary Fig. S3). These variety-specific genes possibly lead to the significantly higher nutritional and medicinal values of P. thomsonii compared with P. montana.
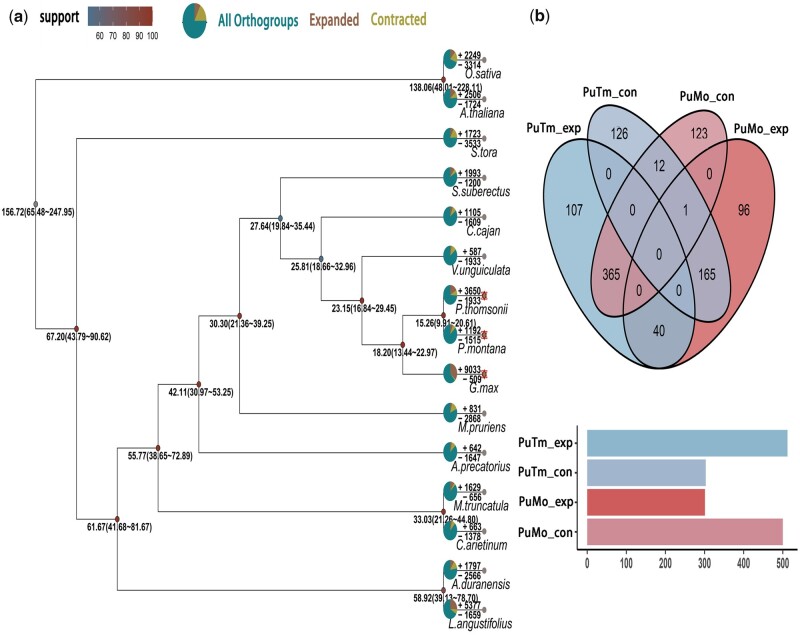
To investigate the expansion/contraction of gene families in P. montana, 194 single-copy orthologs shared by P. montana and the other 14 species were used for construction of phylogenetic tree and estimation of species divergence time. The results indicated that P. montana and P. thomsonii evolved as sister groups and diverged ∼15.26 million years ago (Mya). Both of the two varieties were most related to G. max and separated from G. max about 18.20 Mya (Fig. 3a). The total number of expanded and contracted gene families in P. thomsonii is three more times than that in P. montana. Nonetheless, the numbers of significantly expanded and contracted gene families in P. thomsonii are closed to the numbers of significantly contracted and expanded gene families in P. montana, respectively (Fig. 3b). More than three-quarter significantly expanded gene families in P. thomsonii experienced significant contraction in P. montana.
Figure 3.
Phylogenetic analysis, divergence time, and gene family expansions and contractions. (a) The phylogenetic tree was constructed on the basis of 194 single-copy orthologs using A. thaliana and O. sativa as the outgroups. Divergence times (Mya) are indicated by the numbers below the nodes. Gene family expansions and contractions are indicated by positive or negative values and pies, respectively. (b) Venn diagram shows the overlap gene families significantly (<0.05) expanded/contracted in P. montana (PuMo_exp and PuMo_con, respectively) and P. thomsonii (PuTm_exp and PuTm_con, respectively) and bar graph shows total number of significantly (<0.05) expanded/contracted gene families.
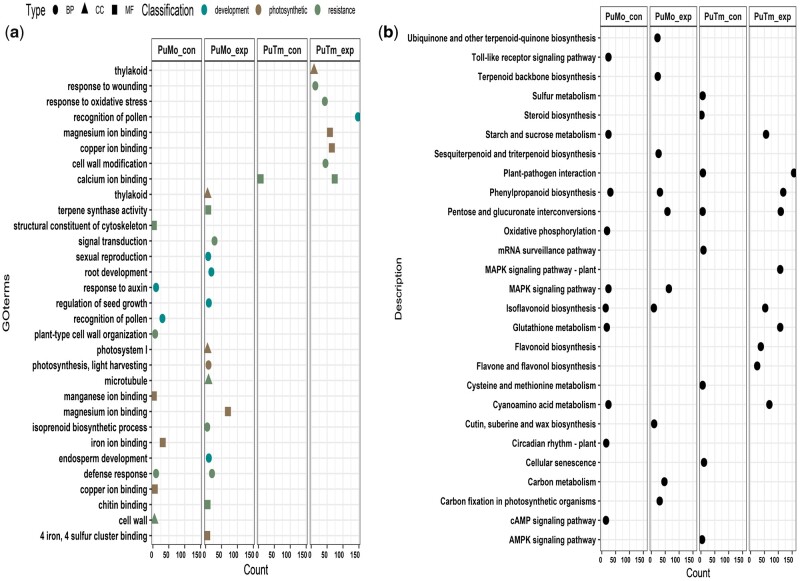
A total of 59 and 46 GO terms were significantly enriched by the members of the expanded (PuMo_exp) and contracted (PuMo_con) gene families in P. montana, respectively. While 61 and 8 GO terms were enriched for the members of the expanded (PuTm_exp) and contracted (PuTm_con) gene families in P. thomsonii, respectively (Supplementary file1). To better characterize the expanded/contracted gene families in both P. thomsonii and P. montana, the GO terms related to development, resistance and photosynthesis are showed in Fig. 4. More GO terms involving in the adaption and resistance were enriched by the members of expanded gene families than that of the contracted, which may lead to the strong vigour of P. thomsonii and P. montana compared with the other Leguminous plants. Especially, several genes belonging to P. montana expanded gene families were significantly enriched in ‘root development’ and ‘microtubule’, which possibly lead to the better cold-adaption of P. montana. KEGG enrichment analysis showed gene families related to starch and sucrose metabolism, phenylpropanoid and isoflavonoid biosynthesis were expanded in P. thomsonii while contracted in P. montana. These results further explain the phenomenon that there are more contents of valuable bioactive metabolites and starch in P. thomsonii than that in P. montana.
Figure 4.
GO (a) and KEGG (b) enrichment analyses of the genes in significantly expanded and contracted gene families in P. montana and P. thomsonii. For each gene set, only GO terms significantly enriched and possibly associated with development, photosynthetic and resistance, as well as the top 10 pathways significantly enriched in KEGG analysis are shown. Count indicates the number of genes associated with this GO term or KEGG pathway in the genes contained in expanded/contracted gene families.
3.3. Graphic genome of P. montana
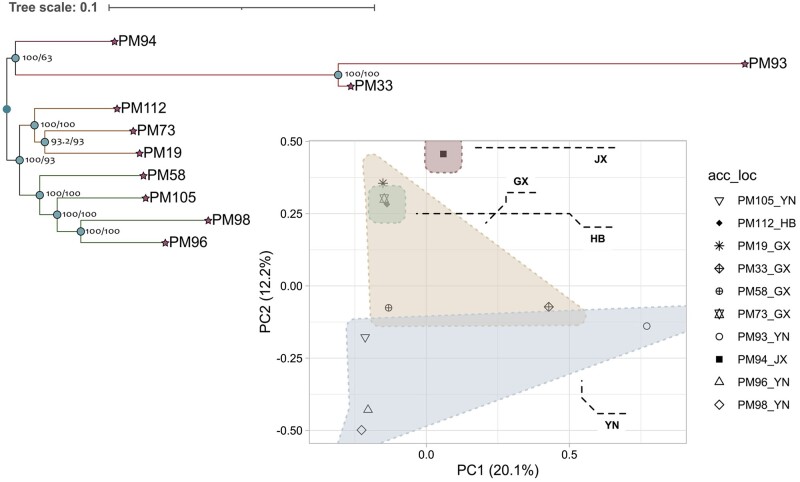
To estimate the genetic diversity of P. montana, the other 10 accessions from different regions were selected for next-generation sequencing. Using the genome of PM12 as the reference, variants among the accessions were identified by GATK. A total of 29.01 million SNPs and 6.45 million InDels were identified. Phylogenetic tree was constructed using the high-quality SNPs. The results showed that the 10 accessions were separated into 3 groups, demonstrating genetic differences among the P. montana accessions (Fig. 5). Moreover, PCA further showed the genetic differences of the 10 accessions are not totally agreement with geographical distribution, which is possibly due to the diverse altitudes or local environments in the same province.
Figure 5.
Phylogenetic tree and PCA constructed with SNPs. Numbers above the nodes are bootstrap values and Clusters are based on the province (the source of samples) including Guangxi (GX), Jiangxi (JX), Hubei (HB) and Yunnan (YN).
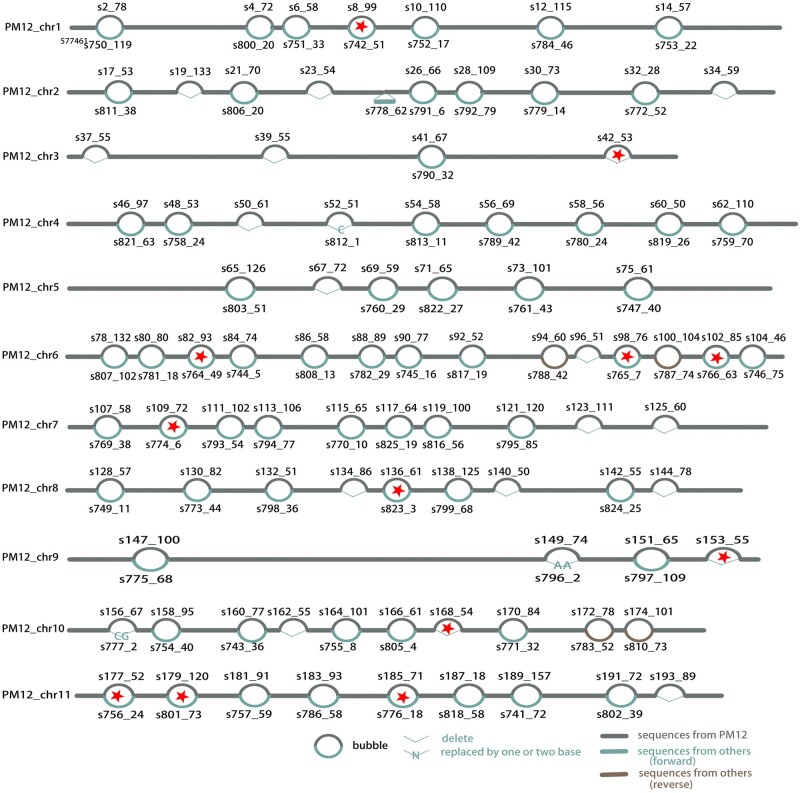
To represent the genetic diversity of P. montana, a graphic genome was constructed. The clean reads were firstly assembled into contigs and then scaffolded into chromosome-scale genomes by Ragtag using the de novo assembled genome of PM12 as the guided reference. The 10 reference-guide assembled genomes showed similar genome size and contiguity as the reference genome (Supplementary Table S3). Good collinearity was observed between the genomes of 10 accessions and the reference genome, further demonstrating high quality of the reference genome (Supplementary Fig. S4). Graphic genome of P. montana was constructed by minigraph program based on the chromosome-level genomes of 11 accessions. Then the graphic genome was used to call SVs (>50 bp). A total of 92 SVs were identified, including four inversions, 22 InDels and 66 substitutions. No shared SVs were found among the accessions (Supplementary file2). A total of 12 SVs occurred in gene body (Fig. 6). GO annotations of the 12 genes harboring SVs showed five of the eight genes functionally annotated were related to responses to stress and cytoskeleton and 4 of the 12 genes harboring SV were not functionally annotated (Supplementary File 2 and Table S8). These results demonstrated that the genetic background of P. montana accessions is relatively narrow, which is approximately due to the genetic property of clonal reproduction.
Figure 6.
Graphic genome of P. montana showing SVs. The number above or below the bubble shows the two segment names composing the bubble and the length of segment. Asterisk highlights the SV occurring in gene body.
4. Discussion
Pueraria montana is widely distributed in Asia. Although its value in medicine and nutrition is low, its genomic resources are very valuable and important for investigating bioactive metabolites and adaption of Pueraria, owing to the different metabolism and growth habits of P. montana compared with the other Pueraria varieties.8 To mine the genetic resources of P. montana, a chromosome-level genome of P. montana was de novo assembled in the present study. Then, comparative genomics analysis was performed between P. montana and P. thomsonii. At last, a graphic genome was constructed based on the 11 P. montana accessions. The results not only provided a high-quality genome and graphic genome for P. montana, but also clarified the genomic differences between P. montana and P. thomsonii, which are valuable for the related evolutionary studies and mining the excellent gene resources from P. montana.
A chromosome-scale genome with size of 978.59 Mb, contig N50 of 1.61 Mb, and scaffold N50 of 80.18 Mb was de novo assembled for P. montana in this study. The genome contiguity of P. montana was significantly improved compared with that of P. thomsonii (contig N50 of 593.70 kb).11 The BUSCO and re-mapping analysis further confirmed the completeness and accuracy of the assembled genome. Pueraria montana and P. thomsonii are the two varieties of P. lobata. However, significantly different genome sizes were observed between them. The previously assembled genome size of P. thomsonii was 1,381.61 Mb, which is significantly larger than that of P. montana. It is well known that genome size is a pivotal feature for biodiversity and meaningful for species evolution.49,50 Comparative genomics analysis in this study demonstrated that the expansion of P. thomsonii genome is due to the more repetitive sequences and duplicated genes. Similar results were also reported in Tung Tree,51 field pennycress52 and Chinese pine.53 There are several mechanisms for gene duplication, including WGD, tandem duplication, segment duplication and transposon- or retrotransposon-mediated duplication.54 According to the Ks analysis, there is no WGD event occurred in the two varieties after their divergence, whereas each has a peak at Ks near zero, which is possibly caused by tandem or segment duplication.
Significant genetic variations were observed between P. montana and P. thomsonii in this study. Compared with P. thomsonii, 6,548 gene families were specifically found in P. montana genome, most of which are related to basic functions and signaling pathways. Whereas, there are 4,675 variety-specific gene families in the P. thomsonii genome, which are significantly enriched in several metabolic pathways related to glutathione, phenylpropanoid and flavonoid. Furthermore, expansion of gene orthogroups, which are related to the biosynthesis of flavonoid, phenylpropanoid, flavone and isoflavonoid, as well as metabolism of starch and sucrose, was observed in P. thomsonii. However, contraction of the corresponding gene orthogroups was observed in P. montana. Previous metabolome analysis showed that the contents of amino acids, sugars, flavonoids and flavonoids in the root of P. lobata and P. thomsonii were remarkably higher than that of P. montana.8 Variation of secondary metabolism among different varieties are mainly owing to genetic diversity.55–57 The differentially expanded gene families and the variety-specific genes in P. montana genome possibly cause the metabolic differences between P. montana and P. thomsonii, resulting in their distinct nutritional and medicinal values.
It was reported that microtubules serving as sensors play an important role in tolerance to cold, salt and drought stress.58,59 GO enrichment analysis in the present study showed that several variety-specific and expanded gene families in P. montana genome are functionally related to microtubules and cytoskeleton. Previous studies demonstrated that partial and transient depolymerization of microtubules at the early stage of cold signaling may promote the opening of calcium channels, which act as a second messenger to transmit cold signaling into cells and promote the expression of cold-related genes.48,60 Therefore, the microtubule-related variety-specific genes in P. montana are likely associated with stronger cold-adaption in P. montana compared with P. thomsonii. These microtubule-related variety-specific genes can be used to improve the cold-adaption of the other species or varieties.
A graphic genome of P. montana was constructed using the assembled genome and the other 10 reference-guided genomes in this study. It was reported that using graphic genome as reference genome could improve the read-mapping sensitivity as well as the number and accuracy of the identified variants.61 Several graphic genomes have been constructed to represent comprehensive genetic variants and gene contents of the species with high genetic diversity, such as soybean, cattle and human.62–64Pueraria montana is a variety with low genetic variants possibly owing to its genetic property of clonal reproduction.65 Thus, the graphic genome of P. montana only contains 92 SVs (>50 bp), with far lower genetic diversity than that of the species mentioned above. Additionally, SVs identified in the graphic genome are mainly related to agronomic traits, domestication and adaptation.63,66 Among the 12 genes harboring SV in P. montana graphic genome, six genes were functionally annotated as relation to response to stimulus. These results will facilitate the utilization of excellent resistance genes in P. montana.
Supplementary data
Supplementary data are available at DNARES online.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31960420), Guangxi Natural Science Foundation Project (2021GXNSFBA220026), Science and Technology Development Fund of Guangxi Academy of Agricultural Sciences (Guinongke 2021JM11) and Special project for basic scientific research of Guangxi Academy of Agricultural Sciences (Guinongke 2021YT057).
Accession number
JANIXD000000000.
Authors’ contributions
Qiusheng Kong and Huabing Yan conceived and designed the study and revised the manuscript. Changjuan Mo participated in data analysis, writing and revising the manuscript. Zhengdan Wu, Xiaohong Shang, Minghua Wei, Haiyan Wang and Liang Xiao participated in data analysis. Pingli Shi, Sheng Cao, Liuying Lu and Wendan Zeng contributed to the sample preparation. All authors read and approved the final manuscript.
Conflict of interest
None declared.
Contributor Information
Changjuan Mo, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, China.
Zhengdan Wu, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Xiaohong Shang, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Pingli Shi, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Minghua Wei, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, China.
Haiyan Wang, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, China.
Liang Xiao, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Sheng Cao, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Liuying Lu, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Wendan Zeng, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China.
Huabing Yan, Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning 530007, China; State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi University, Nanning 530007, China.
Qiusheng Kong, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, China.
Data Availability
All clean PacBio Sequel, BGI and Hi-C sequence reads of genome, RNA-seq paired-end reads and Iso-Seq of transcriptome for the P. montana have been deposited at DDBJ/ENA/GenBank under the accession from SRR20067666 to SRR20067682 in BioProject PRJCA009679 as well as the Genome Warehouse in National Genomics Data Center under accession number from SRX16105738 to SRX16105754 in BioProject PRJNA855556 in National Center for Biotechnology Information. The whole genome sequence data reported in this paper has been deposited at DDBJ/ENA/GenBank under the accession JANIXD000000000 as well as the Genome Warehouse in National Genomics Data Center under accession number GWHBJCY00000000 which is publicly accessible at https://ngdc.cncb.ac.cn/gwh.
References
- 1. Zhao Z., Guo P., Brand E.. 2018, A concise classification of bencao (materia medica), Chin. Med., 13, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang S., Zhang S., Wang S., Gao P., Dai L.. 2020, A comprehensive review on Pueraria: insights on its chemistry and medicinal value, Biomed. Pharmacother., 131, 110734. [DOI] [PubMed] [Google Scholar]
- 3. Wong K.H., Li G.Q., Li K.M., Razmovski-Naumovski V., Chan K.. 2011, Kudzu root: traditional uses and potential medicinal benefits in diabetes and cardiovascular diseases, J. Ethnopharmacol., 134, 584–607. [DOI] [PubMed] [Google Scholar]
- 4. van der Maesen L. J. G. 1985, A Revision of the Genus Pueraria DC. with Some Notes on Teyleria Backer (Agricultural University Wageningen Papers), vol.35, p.62. Pudoc Scientific Publishers: The Netherlands. [Google Scholar]
- 5. Chen S.B., Liu H.P., Tian R.T., et al. 2006, High-performance thin-layer chromatographic fingerprints of isoflavonoids for distinguishing between Radix Puerariae lobate and Radix Puerariae thomsonii, J. Chromatogr. A, 1121, 114–9. [DOI] [PubMed] [Google Scholar]
- 6. Liu D., Ma L., Zhou Z., et al. 2021, Starch and mineral element accumulation during root tuber expansion period of Pueraria thomsonii Benth, Food Chem., 343, 128445. [DOI] [PubMed] [Google Scholar]
- 7. Adolfo L.M., Rao X., Dixon R.A.. 2022, Identification of Pueraria spp. through DNA barcoding and comparative transcriptomics, BMC Plant Biol., 22, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shang X., Huang D., Wang Y., et al. 2021, Identification of nutritional ingredients and medicinal components of Pueraria lobata and its varieties using UPLC-MS/MS-based metabolomics, Molecules, 26, 6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coiner H.A., Hayhoe K., Ziska L.H., Van Dorn J., Sage R.F.. 2018, Tolerance of subzero winter cold in kudzu (Pueraria montana var. lobata), Oecologia, 187, 839–49. [DOI] [PubMed] [Google Scholar]
- 10. Tungmunnithum D., Intharuksa A., Sasaki Y.. 2020, A promising view of Kudzu Plant, Pueraria montana var. lobata (Willd.) Sanjappa & Pradeep: flavonoid phytochemical compounds, taxonomic data, traditional uses and potential biological activities for future cosmetic application, Cosmetics, 7, 12. [Google Scholar]
- 11. Shang X., Yi X., Xiao L., et al. 2022, Chromosomal-level genome and multi-omics dataset of Pueraria lobata var. thomsonii provide new insights into legume family and the isoflavone and Puerarin biosynthesis pathways, Hortic Res., 9, uhab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker B.J., Abeel T., Shea T., et al. 2014, Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement, PLoS One, 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belaghzal H., Dekker J., Gibcus J.H.. 2017, Hi-C 2.0: an optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation, Methods, 123, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vasimuddin M., Misra S., Li H., Aluru S.. 2019, Efficient architecture-aware acceleration of BWA-MEM for multicore systems, Int. Parall. Distrib. P., 34, 314–24. [Google Scholar]
- 15. Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J.. 2013, Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions, Nat. Biotechnol., 31, 1119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Durand N.C., Shamim M.S., Machol I., et al. 2016, Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments, Cell Syst., 3, 95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 18. Gurevich A., Saveliev V., Vyahhi N., Tesler G.. 2013, QUAST: quality assessment tool for genome assemblies, Bioinformatics, 29, 1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S., Zhou Y., Chen Y., Gu J.. 2018, fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, 34, i884–i90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kallenborn F., Hildebrandt A., Schmidt B.. 2021, CARE: context-aware sequencing read error correction, Bioinformatics, 37, 889–95. [DOI] [PubMed] [Google Scholar]
- 21. Swat S., Laskowski A., Badura J., et al. 2021, Genome-scale de novo assembly using ALGA, Bioinformatics, 37, 1644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alonge M., Soyk S., Ramakrishnan S., et al. 2019, RaGOO: fast and accurate reference-guided scaffolding of draft genomes, Genome Biol., 20, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H. 2018, Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics, 34, 3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Z., Wang H.. 2007, LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons, Nucleic Acids Res., 35, W265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benson G. 1999, Tandem repeats finder: a program to analyze DNA sequences, Nucleic Acids Res., 27, 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowe T.M., Eddy S.R.. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence, Nucleic Acids Res., 25, 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A.. 2005, Rfam: annotating non-coding RNAs in complete genomes, Nucleic Acids Res., 33, D121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B.. 2006, AUGUSTUS: ab initio prediction of alternative transcripts, Nucleic Acids Res., 34, W435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Besemer J., Borodovsky M.. 2005, GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses, Nucleic Acids Res., 33, W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu T.D., Watanabe C.K.. 2005, GMAP: a genomic mapping and alignment program for mRNA and EST sequences, Bioinformatics, 21, 1859–75. [DOI] [PubMed] [Google Scholar]
- 31. Avram O., Kigel A., Vaisman-Mentesh A., et al. 2021, PASA: proteomic analysis of serum antibodies web server, PLoS Comput. Biol., 17, e1008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haas B.J., Salzberg S.L., Zhu W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments, Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Darling A.E., Marçais G., Delcher A.L., et al. 2018, MUMmer4: a fast and versatile genome alignment system, PLoS Comput. Biol., 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buchfink B., Xie C., Huson D.H.. 2015, Fast and sensitive protein alignment using DIAMOND, Nat. Methods, 12, 59–60. [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Tang H., Debarry J.D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen C., Chen H., Zhang Y., et al. 2020, TBtools: an integrative toolkit developed for interactive analyses of big biological data, Mol. Plant., 13, 1194–202. [DOI] [PubMed] [Google Scholar]
- 37. Emms D.M., Kelly S.. 2019, OrthoFinder: phylogenetic orthology inference for comparative genomics, Genome Biol., 20, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar R.C. 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 32, 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Darriba D., Taboada G.L., Doallo R., Posada D.. 2011, ProtTest 3: fast selection of best-fit models of protein evolution, Bioinformatics, 27, 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stamatakis A. 2014, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics, 30, 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cali D.S., Kim J.S., Ghose S., Alkan C., Mutlu O.. 2019, Nanopore sequencing technology and tools for genome assembly: computational analysis of the current state, bottlenecks and future directions, Brief Bioinform., 20, 1542–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z. 2007, PAML 4: phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 24, 1586–91. [DOI] [PubMed] [Google Scholar]
- 43. Mendes F.K., Vanderpool D., Fulton B., Hahn M.W.. 2021, CAFE 5 models variation in evolutionary rates among gene families, Bioinformatics, 36, 5516–8. [DOI] [PubMed] [Google Scholar]
- 44. Wu T., Hu E., Xu S., et al. 2021, clusterProfiler 4.0: a universal enrichment tool for interpreting omics data, Innovation (Camb)., 2, 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H., Feng X., Chu C.. 2020, The design and construction of reference pangenome graphs with minigraph, Genome Biol., 21, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minh B.Q., Schmidt H.A., Chernomor O., et al. 2020, IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era, Mol. Biol. Evol., 37, 1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakano M., Hirakawa H., Fukai E., et al. 2021, A chromosome-level genome sequence of Chrysanthemum seticuspe, a model species for hexaploid cultivated chrysanthemum, Commun. Biol., 4, 1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nick P. 2013, Microtubules, signalling and abiotic stress, Plant J., 75, 309–23. [DOI] [PubMed] [Google Scholar]
- 49. Vitales D., Fernandez P., Garnatje T., Garcia S.. 2019, Progress in the study of genome size evolution in Asteraceae: analysis of the last update, Database (Oxford)., 2019, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ibarra-Laclette E., Lyons E., Hernandez-Guzman G., et al. 2013, Architecture and evolution of a minute plant genome, Nature, 498, 94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L., Liu M., Long H., et al. 2019, Tung Tree (Vernicia fordii) genome provides a resource for understanding genome evolution and improved oil production, Genomics Proteomics Bioinformatics, 17, 558–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geng Y., Guan Y., Qiong L., et al. 2021, Genomic analysis of field pennycress (Thlaspi arvense) provides insights into mechanisms of adaptation to high elevation, BMC Biol., 19, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Niu S., Li J., Bo W., et al. 2022, The Chinese pine genome and methylome unveil key features of conifer evolution, Cell, 185, 204–17.e14. [DOI] [PubMed] [Google Scholar]
- 54. Panchy N., Lehti-Shiu M., Shiu S.H.. 2016, Evolution of gene duplication in plants, Plant Physiol., 171, 2294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W., Wen L., Chen Z., et al. 2021, Study on metabolic variation in whole grains of four proso millet varieties reveals metabolites important for antioxidant properties and quality traits, Food Chem., 357, 129791. [DOI] [PubMed] [Google Scholar]
- 56. Tang Y.C., Liu Y.J., He G.R., et al. 2021, Comprehensive analysis of secondary metabolites in the extracts from different lily bulbs and their antioxidant ability, Antioxidants (Basel)., 10, 1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Veremeichik G.N., Grigorchuk V.P., Butovets E.S., et al. 2021, Isoflavonoid biosynthesis in cultivated and wild soybeans grown in the field under adverse climate conditions, Food Chem., 342, 128292. [DOI] [PubMed] [Google Scholar]
- 58. Chun H.J., Baek D., Jin B.J., et al. 2021, Microtubule dynamics plays a vital role in plant adaptation and tolerance to salt stress, Int. J. Mol. Sci., 22, 5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma H., Liu M.. 2019, The microtubule cytoskeleton acts as a sensor for stress response signaling in plants, Mol. Biol. Rep., 46, 5603–8. [DOI] [PubMed] [Google Scholar]
- 60. Wang L., Sadeghnezhad E., Riemann M., Nick P.. 2019, Microtubule dynamics modulate sensing during cold acclimation in grapevine suspension cells, Plant Sci., 280, 18–30. [DOI] [PubMed] [Google Scholar]
- 61. Rakocevic G., Semenyuk V., Lee W.-P., et al. 2019, Fast and accurate genomic analyses using genome graphs, Nat. Genet., 51, 354–62. [DOI] [PubMed] [Google Scholar]
- 62. Paten B., Novak A.M., Eizenga J.M., Garrison E.. 2017, Genome graphs and the evolution of genome inference, Genome Res., 27, 665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu Y.C., Du H.L., Li P.C., et al. 2020, Pan-genome of wild and cultivated soybeans, Cell, 182, 162–76.e13. [DOI] [PubMed] [Google Scholar]
- 64. Talenti A., Powell J., Hemmink J.D., et al. 2022, A cattle graph genome incorporating global breed diversity, Nat. Commun., 13, 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bentley K.E., Mauricio R.. 2016, High degree of clonal reproduction and lack of large-scale geographic patterning mark the introduced range of the invasive vine, Kudzu (Pueraria montana var. lobata), in North America, Am. J. Bot., 103, 1499–507. [DOI] [PubMed] [Google Scholar]
- 66. Li H., Wang S., Chai S., et al. 2022, Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber, Nat. Commun., 13, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All clean PacBio Sequel, BGI and Hi-C sequence reads of genome, RNA-seq paired-end reads and Iso-Seq of transcriptome for the P. montana have been deposited at DDBJ/ENA/GenBank under the accession from SRR20067666 to SRR20067682 in BioProject PRJCA009679 as well as the Genome Warehouse in National Genomics Data Center under accession number from SRX16105738 to SRX16105754 in BioProject PRJNA855556 in National Center for Biotechnology Information. The whole genome sequence data reported in this paper has been deposited at DDBJ/ENA/GenBank under the accession JANIXD000000000 as well as the Genome Warehouse in National Genomics Data Center under accession number GWHBJCY00000000 which is publicly accessible at https://ngdc.cncb.ac.cn/gwh.