Abstract
Several genetic polymorphisms of the innate immune system have been described to increase the risk of cytomegalovirus (CMV) infection in transplant patients. The aim of this study was to assess the impact of a polygenic score to predict CMV infection and disease in high risk CMV transplant recipients (heart, liver, kidney or pancreas). On hundred and sixteen CMV-seronegative recipients of grafts from CMV-seropositive donors undergoing heart, liver, and kidney or pancreas transplantation from 7 centres were prospectively included for this purpose during a 2-year period. All recipients received 100-day prophylaxis with valganciclovir. CMV infection occurred in 61 patients (53%) at 163 median days from transplant, 33 asymptomatic replication (28%) and 28 CMV disease (24%). Eleven patients (9%) had recurrent CMV infection. Clinically and/or functionally relevant single nucleotide polymorphisms (SNPs) from TLR2, TLR3, TLR4, TLR7, TLR9, AIM2, MBL2, IL28, IFI16, MYD88, IRAK2 and IRAK4 were assessed by real time polymerase chain reaction (RT-PCR) or sequence-based typing (PCR-SBT). A polygenic score including the TLR4 (rs4986790/rs4986791), TLR9 (rs3775291), TLR3 (rs3775296), AIM2 (rs855873), TLR7 (rs179008), MBL (OO/OA/XAO), IFNL3/IL28B (rs12979860) and IFI16 (rs6940) SNPs was built based on the risk of CMV infection and disease. The CMV score predicted the risk of CMV disease with an AUC of the model of 0.68, with sensitivity and specificity of 64.3 and 71.6%, respectively. Even though further studies are needed to validate this score, its use would represent an effective model to develop more robust scores predicting the risk of CMV disease in donor/recipient mismatch (D+/R-) transplant recipients.
Keywords: cytomegalovirus, solid organ transplantation, infectious disease, innate immunity, multicenter study
Introduction
Cytomegalovirus (CMV) is the most common opportunistic pathogen in solid organ transplant (SOT) recipients. While CMV infection can occur through reactivation of latent virus in seropositive recipients, CMV transmission from the donor is the riskiest clinical scenario in CMV seronegative transplant recipients because of the lack of a pre-existing CMV-specific immunity. CMV infection develops in 36% to 100% of SOT recipients, with rates of symptomatic disease between 11%-72% without prophylaxis (1, 2). In the case of donor/recipient CMV mismatch (D+/R-), more than 50% would develop CMV disease if no antiviral prophylaxis is administered (2). Some strategies have been developed in recent years in order to prevent CMV infection and disease such as extending antiviral prophylaxis from 3 to 6 months (3), using regimens containing mTOR inhibitors (4) or measuring CMV-specific T-cells immunity (5). However, CMV infection is still worrisome in SOT recipients and new biomarkers are necessary (6).
Immunosuppression predominantly impairs the adaptive immune response by blocking lymphocyte activation signalling pathways, depleting lymphocytes, or diverting lymphocyte trafficking (7). With the use of pharmacologic immunosuppression, the innate immune responses are critical in the defence against infection (8). Innate immune system components of either secreted (e.g., type I interferons, IFNs), membrane-bound (e.g., toll-like receptors; TLRs, mannose-binding lectin; MBL) or cytoplasmic (e.g., DNA cytoplasmic sensors) nature, are responsible for sensing and containment of the viral infection during the lag phase needed for adaptive immunity to become operative (9, 10). In the case of CMV, a large number of studies have reported the effect of single nucleotide polymorphisms (SNPs) in a variety of innate immune component genes (with especial relevance of TLR4 and MBL2) on the risk of CMV infection (11–13). In the setting of organ transplant, donor and/or recipients genes encoding pattern recognition molecules and receptors such as TLR (TLR2, 4 and 9) or MLB (MBL2), interferon (IFNL3/IL28B), cytokines (IL12, IL10), cytoplasmic sensors (AIM2, IFI16, IRAK2, IRAK4) and immune regulatory membrane-associated proteins (human programmed death-1, PD1) have been linked with an increased risk of CMV infection and disease among different transplant populations either solid organ or allogenic stem cell transplant patients (14–25).
TLR2, TLR3, TLR4 and TLR9 are pattern recognition receptors with a key role in innate immunity against viral infections. Accordingly, TLR genetic polymorphisms have been reported to impact the course of CMV infection. The heterozygous TLR3 rs3775296 genotype is overrepresented in Polish children with CMV infection compared with uninfected cases (26). Likewise, TLR9 variants (T/C, rs187084; C/T, rs352140) are associated with CMV disease in children (27) and TLR2 variants (A>G, rs5743708) are associated with increased risk of congenital HCMV infection in a Polish cohort (28).
Although TLR4 is not directly involved in the recognition of CMV, TLR4 stimulation increases the number of CMV-specific CD4+ and CD8+ T cells by enhancing antigen presentation by dendritic cells (29). TLR4 polymorphisms (rs4986790 and rs4986791) were associated with higher risk of CMV disease in kidney transplant recipient cohorts (30, 31).
TLR7 is also involved in viral recognition including CMV (29, 32), and it induces different IFN types such as IFN-λ3/IL-28B (33, 34). In fact, the rs12979860T allele of IFNL3IL28B has a protective effect against CMV infection in allogeneic stem cell transplant patients (35). However, studies evaluating the impact of TLR7 polymorphism on CMV infection risk are still pending.
MyD88, IRAK2 and IRAK4 are key downstream mediators of TLR signalling and are involved in fast CMV infection responses (36, 37). The IRAK2 hypofunctional Leu392Val (rs3844283) variant has been associated with reduced spontaneous viral clearance (38).
Other receptors are involved in CMV recognition, including MBL, which recognizes glycoproteins present in the envelope of the virion. Hence, MBL2 genetic variants that lead to low-MBL levels were associated with an increased risk of CMV infection and disease (31, 39).
Recognition of cytoplasmic DNA is also a remarkable function in the initiation of innate immune responses. Absent in melanoma 2 (AIM2) and IFI16 are cytoplasmic sensors serving this role, and are involved in the control of CMV infection (40–43).
On this basis, we aimed to explore the impact of innate immunity on the risk of CMV infection and disease in patients at high risk to develop CMV infection such as CMV-seronegative recipients that received a CMV-seropositive donor in a prospective cohort of solid organ transplant recipients. To that end, we investigated SNPs from innate immune genes critically involved in eliciting effective anti-viral immune responses (IFNL3/IL28B, MBL2, TLR2, TLR3, TLR4, TLR7, TLR9, MYD88, IRAK2, IRAK4, AIM2, IFI16) to further explore their individual or combined potential impact in the risk of CMV infection.
Material and Methods
Setting and Study Population
We conducted a multicenter prospective observational study in seven Spanish hospitals (Hospital Clínic de Barcelona, Hospital de Bellvitge, Hospital Marqués de Valdecilla, Hospital 12 de Octubre, Hospital de Cruces, Hospital Virgen del Rocio and Hospital Reina Sofía) with an active transplant program. All adult CMV mismatch transplant recipients (CMV-seronegative recipients of grafts from CMV-seropositive donors) that signed informed consent document and agreed to participate in the study were prospectively included. Exclusion criteria were not to meet the inclusion criteria previously described and not to sign informed consent document. All patients were followed for 12 months post-transplant. Demographic data, type of transplant, immunosuppressive regimens, occurrence of biopsy-proven acute allograft rejection and infection episodes were prospectively recorded. The study was approved by each participating hospital research ethic committee. All patients signed informed consent at inclusion.
Definitions
CMV DNAemia was defined as a positive CMV PCR in plasma without clinical symptoms. Symptomatic CMV infection was categorized as CMV disease and defined as CMV “viral syndrome” or tissue invasive disease according to published guidelines (44). CMV “viral syndrome” required the following criteria: (1) positive DNAemia for CMV; (2) a temperature > 38°C with no other accountable source; and (3) a leukocyte count < 4000/mm (3), an atypical lymphocyte concentration >3%, an elevation of transaminases or a platelet count <100,000/mm (3). A diagnosis of tissue-invasive disease required histopathological evidence of CMV (identification of inclusion bodies or viral antigens in biopsy material or bronchoalveolar lavage specimen cells by immunocytochemistry) with or without a positive PCR of CMV in the tissue. Late-onset CMV disease was defined as CMV disease occurring after prophylaxis completion.
Any CMV viral load by PCR without any previous positive CMV viral load post transplantation was considered primary CMV infection. A positive CMV PCR of 1000 IU/mL or higher after a primary infection with confirmed clearance (negative CMV viral load) was considered significant. Cytomegalovirus viral load was measured by quantitative real time polymerase chain reaction (qPCR) Cobas® CMV (Roche®, Switzerland) according to the manufacturer’s instructions. The test can quantify CMV DNA over the range of 34.5 - 1E+07 IU/mL.
Transplant Infection Prophylaxis Protocol
SOT patients received perioperative antibacterial prophylaxis for up to 48h after transplantation depending on each center’s protocol. During the study period, Pneumocystis jirovecii prophylaxis with trimethoprim-sulfametoxazole (one double strength tablet once three times a week) was given in the first six months after transplantation. All patients received prophylaxis with oral valganciclovir (900 mg once daily or dose adjusted by kidney function if there was renal impairment) or intravenous ganciclovir (5 mg/kg daily or dose adjusted by kidney function) for 100 days according to international guidelines (44). After antiviral prophylaxis completion, a preemptive strategy was applied (surveillance after prophylaxis) (44). A CMV PCR was performed every 15 days in the first month and then monthly for 12 months. Anti-fungal prophylaxis was given in high-risk recipients according to the American Society of Transplantation Infectious diseases Community of practice recommendations (45).
Immunogenetic Analyses
Genomic DNA was extracted from a 1.5 mL whole blood sample using the QIAmp DNA blood mini nucleic acid extraction kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer’s instructions, and stored at -80°C until use.
Genotyping of MBL2 was done by a polymerase chain reaction and sequence‐based typing (PCR160 SBT) technique as previously reported (46). Briefly, six single nucleotide polymorphisms (SNPs) in 161 the promoter region (-550 G>C, [H>L]; -221 C>G, [X>L]; +4 C>T, [P>Q]), and exon 1 (codon 52 CGT [Leu]>TGT [Arg], [A>D]; codon 54 GGC [Gly]>GAC [Asp], [A>B]; codon 57 GGA [Gly]>GAA [Glu], [A>C]) of the MBL2 gene were analyzed. Variants at codons 52 (Arg; D), 54 (Asp; B) and 57 (Glu; C), are major determinants of low serum MBL levels (47) and are collectively named O, while A indicates the wild-type variants. SNPs at positions –550, −221 and +4 also influence serum MBL levels in individuals with the A variant. However, the functional effects of [H>L] and [P>Q] SNPs appear to be minor compared to [X>L], with X being the allele associated with lower MBL2 expression. Accordingly, individuals were genotypically classified as high- (A/A, A/XA), intermediate- (XA/XA, A/O) or low- (XA/O, O/O) MBL producers.
Genotyping of SNPs in the TLR2 (rs5743708, CGG[Arg753]>CAG[Gln]), TLR3 (rs3775296, intron 1 + 95 C>A; rs3775291, CTC[Leu412]>TTC[Phe]), TLR4 (rs4986790, GAT[Asp299]>GGT[Gly]; rs4986791, ACC[Thr399]>ATC[Ile]), TLR7 (rs179008, CAA[Gln11]>CTA[Leu]), TLR9 (rs5743836, 5’UTR -1486 T>C; rs187084, 5’UTR -1237 T>C; rs352140, CCG[Pro545]>CCA[Pro]), MyD88 (rs6853, 3’UTR A>G), IRAK2 (rs3844283, CTG[Leu392]>GTG[Val]), IRAK4 (rs4251513, intron G>C), AIM2 (rs855873, intron G>A) IFI16 (rs6940, ACT[Thr723]>TCT[Ser]) and IFNL3/IL28B (rs12979860, intron C>T) genes were performed by allelic discrimination using TaqMan SNP-genotyping Assays (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA, USA) on a LightCycler 480 Instrument II (Roche) according to manufacturer’s instructions. Allelic frequencies for the autosomal SNPs genotyped are shown in Supplementary Table S3 . All of them were in Hardy-Weinberg equilibrium except for the IRAK4 rs4251513 SNP.
Statistical Methods
Statistical tests were performed using SPSS Version 19 (SPSS, Chicago, IL). For comparisons of study groups, two-sided Mann–Whitney U-Test for nonparametric independent samples was used. Clinical and infection-specific characteristics were compared across groups using Fisher’s exact test or χ (2) test for categorical variables, and Student’s t-test for continuous variables. Two-sided p-values <0.05 were considered statistical significant.
The CMV genetic risk score was constructed using a logistic regression model with all independent variables. The coefficients of the independent variables with confidence intervals of 80% not including zero were extracted and a score was obtained including these selected variables depending on the coefficient weight. The discriminatory power of the score was evaluated by the area under the receiver operating characteristics (ROC) curve and the 95% confidence interval (CI). Then, a cut-off value to estimate the diagnostic sensitivity and specificity in the validation set was selected.
The sample size of the study was calculated accounting an estimate 40% incidence of late CMV infection in patients with any genetic polymorphism compared to 15% of patients with a wild type genotype. From published data, 10% of transplant subjects will have a genetic polymorphism. To demonstrate the aforementioned risk difference with an alpha risk of 5% and a power of 80%, we needed to include 129 patients with a solid organ transplant and a D +/R- serological pattern.
Results
During the 2-year recruitment period (2013-2015), 116 CMV mismatch (D+/R-) transplant recipients from 7 Spanish hospitals were included. Table 1 shows the baseline, demographic and clinical characteristics of enrolled patients at inclusion. Sixty-one patients (53%) had at least one episode of CMV infection, 33 of them (28%) clinically asymptomatic and 28 categorized as CMV disease (24%), none of them occurring while on antiviral prophylaxis. Eleven patients (9%) had recurrent CMV infection. Median days from transplantation to CMV infection was 163 (SD 73). Forty-nine patients (42%) presented at least one episode of bacterial infection during the study period, 8 (7%) non-CMV viral infections and 7 (6%) invasive fungal infections.
Table 1.
Baseline, demographic and clinical characteristics of patients.
| Variable | n=116 |
|---|---|
| Age (median, SD) | 49 ±14.1 |
| Gender, male, n(%) | 93 (80) |
| Days of follow-up (median, SD) | 684 (497) |
| Type of transplantation, n(%) | |
| ◾ Kidney | 66 (57) |
| ◾ Liver | 34 (29) |
| ◾ Heart | 10 (9) |
| ◾ Multivisceral transplantation | 6 (5) |
| Induction immunosuppressive treatment, n(%) | |
| ◾ None | 39 (34) |
| ◾ Lymphocyte-depleting antibody | 45 (39) |
| ◾ Anti-thymocyte globulin | 31 (28) |
| Maintenance immunosupressive treatment, n(%) | |
| ◾ Calcineurin inhibitors + mycophenolate mofetil +prednisone | 96 (83) |
| ◾ Calcineurin inhibitors + mTOR inhibitors+prednisone | 18 (15) |
| ◾ mTOR inhibitors + mycophenolate mofetil + prednisone | 1 (1) |
| Postransplantation non- infectious complications, n(%) | |
| ◾ Acute rejection (only biopsy proven) | 16 (14) |
| ◾ Hemodialysis | 22 (19) |
| ◾ Surgical reintervention related to transplantation | 22 (19) |
| ◾ Graft loss | 3 (2) |
| ◾ Death | 1 (1) |
| Cytomegalovirus infection, n(%) | 61 (53) |
| First episode | |
| Asymptomatic replication | 33 (28) |
| CMV disease | 28 (24) |
| CMV syndrome | 11 (10) |
| End-organ CMV disease | 17 (15) |
| Median days post transplantation (SD) | 163 (73) |
| Viral load (median, SD) | 66351(188736) |
| Second episode | 11 (10) |
| Asymptomatic replication | 9 (8) |
| CMV disease | 2 (2) |
| CMV syndrome | 1 (1) |
| End-organ CMV disease | 1 (1) |
| Median days post transplantation (SD) | 239 (72) |
| Viral load (median, SD) | 71647 (175673) |
| Third episode | |
| Asymptomatic replication | 4 (3) |
| Median days post transplantation (SD) | 289 (42) |
| Viral load (median, SD) | 52354 (61217) |
| Bacterial infection, n(%) | 49 (42) |
| Other viral infections , n(%) | 8 (7) |
| Invasive fungal infection, n(%) | 7 (6) |
Table 2 shows the distribution of TLR2, TLR3, TLR4, TLR7, TLR9, AIM2, MBL2, IFI16, IFNL3/IL28B, MYD88, IRAK2 and IRAK4 genotypes by group. No differences in CMV infection, either asymptomatic CMV infection or disease, was found between groups. Nevertheless, we performed a subanalysis according to the type of organ transplanted ( Supplementary Tables S1 , S2 ) and found that kidney and liver recipients presenting with TLR4 rs4986790/rs4986791 polymorphism presented more frequently CMV infection comparing with those with wild type (70% vs 41% and 100% vs 51% respectively, p=0.05). Additionally, IFNL3/IL28B rs12979860 polymorphism was considered a protector factor against CMV infection in liver recipients (27% vs 74%, p=0.03).
Table 2.
Univariate analysis of CMV infection according to TLR 2, 3, 4, 7, 9, AIM2, MBL2, IFI16, IL28B, MYD88, IRAK2 and 4 genotypes.
| SNP genotype | Asymptomatic CMV infection | p | CMV disease | p | CMV infection | p | |
|---|---|---|---|---|---|---|---|
| Viral syndrome | Tissue-invasivedisease | ||||||
| TLR2 rs5743708 | |||||||
| Wild type GG (n=111) | 31 (28%) | 0.6 | 10 (9%) | 17 (15%) | 0.5 | 58 (52%) | 0.5 |
| Variant GA (n=5) | 2 (40%) | 1 (20%) | 0 | 3 (60%) | |||
| TLR3 rs3775296 | |||||||
| Wild type CC (n=74) | 22 (30%) | 0.4 | 7 (9%) | 9 (12%) | 0.3 | 38 (38%) | 0.4 |
| Variant AA or AC (n=42) | 11(33%) | 4 (9%) | 8 (19%) | 23 (55%) | |||
| Homozygous AA (n=2) | 0 | 0.2 | 0 | 0 | 0 | 0.9 | |
| Heterozygous AC (n=40) | 11(27%) | 4 (10%) | 8(20%) | 23 (57%) | |||
| TLR3 rs3775291 | |||||||
| Wild type CC (n=51) | 15 (29%) | 0.5 | 4 (8%) | 7 (13%) | 0.8 | 26 (51%) | 0.4 |
| Variant TT or CT (n=65) | 18 (28%) | 7 (11%) | 10 (15%) | 35 (54%) | |||
| Homozygous TT (n=18) | 6 (33%) | 0.9 | 1 (6%) | 2 (11%) | 9 (50%) | 0.9 | |
| Heterozygous CT (n=67) | 12 (18%) | 6 (9%) | 8 (12%) | 26 (39%) | |||
| TLR4 rs4986790 / rs4986791 | |||||||
| Wild type AA / CC (n=97) | 28 (29%) | 0.3 | 8 (8%) | 13 (13%) | 0.6 | 49 (50%) | 0.3 |
| Variant CC or AC / TT or CT (n=19) | 5 (26%) | 3 (16%) | 4 (21%) | 12 (63%) | |||
| Homozygous CC / TT (n=2) | 0 | 0.4 | 1 (50%) | 0 | 0.3 | 1 (50%) | 0.5 |
| Heterozygous AC / CT (n=17) | 5 (29%) | 2 (12%) | 4 (24%) | 11 (65%) | |||
| TLR7 ex3 rs179008 | |||||||
| Wild type AA (n=87) | 25 (29%) | 0.3 | 7 (8%) | 12 (14%) | 0.7 | 44 (51%) | 0.4 |
| Variant AT or TT (n=29) | 8 (28%) | 4 (14%) | 5 (17%) | 17 (59%) | |||
| Homozygous TT (n=26) | 7 (27%) | 0.7 | 4 (15%) | 3 (12%) | 0.4 | 14 (54%) | 0.2 |
| Heterozygous AT (n=3) | 1 (33%) | 0 | 2 (67%) | 3 (100%) | |||
| TLR9 ex4 rs3775291 | |||||||
| Wild type AA (n=33) | 9 (27%) | 0.3 | 6 (18%) | 4 (12%) | 0.2 | 19 (57%) | 0.3 |
| Variant AG or GG (n=83) | 24 (29%) | 5 (6%) | 13 (16%) | 42 (51%) | |||
| Homozygous GG (n=22) | 6 (27%) | 0.6 | 3 (14%) | 3 (14%) | 0.2 | 12 (55%) | 0.7 |
| Heterozygous AG (n=61) | 18 (30%) | 2 (3%) | 10 (16%) | 30 (49%) | |||
| AIM2 rs855873 | |||||||
| Wild type GG (n=98) | 30 (31%) | 0.3 | 9 (9%) | 14 (14%) | 0.6 | 53 (54%) | 0.3 |
| Variant AG-AA (n=18) | 3 (17%) | 2 (11%) | 3 (17%) | 8 (44%) | |||
| MBL2 ex1 | |||||||
| High A/A or XA/A (n=65) | 16 (25%) | 0.9 | 5 (8%) | 12 (25%) | 0.2 | 33 (51%) | 0.9 |
| Intermediate A/0 or XA/XA (n=36) | 13 (36%) | 3 (8%) | 2 (6%) | 18 (50%) | |||
| Low 0/0 or XA/0 (n=15) | 4 (27%) | 3 (20%) | 3 (20%) | 10 (66%) | |||
| IFI16 rs6940 | |||||||
| Wild type AA (n=98) | 26 (27%) | 0.2 | 10 (10%) | 15 (15%) | 0.5 | 51 (52%) | 0.5 |
| Variant AT or TT (n=18) | 7 (39%) | 1 (6%) | 2 (11%) | 10 (55%) | |||
| IL28 rs12979860 | |||||||
| Wild type CC (n=64) | 17 (27%) | 0.5 | 7 (11%) | 10 (16%) | 0.4 | 34 (53%) | 0.5 |
| Variant CT or TT (n=52) | 16 (31%) | 4 (8%) | 7 (13%) | 27 (52%) | |||
| Homocygous TT (n=8) | 4 (9%) | 0.7 | – | – | 0.4 | 4 (50%) | 1 |
| Heterozygous CT (n=44) | 12 (27%) | 4 (9%) | 7 (16%) | 23 (52%) | |||
| MYD88 rs6853 | |||||||
| Wild type AA (n=82) | 25 (30%) | 0.5 | 9 (11%) | 11 (13%) | 0.6 | 45 (55%) | 0.3 |
| Variant GG or AG (n=34) | 8 (24%) | 2 (6%) | 6 (18%) | 16 (47%) | |||
| Homozygous GG (n=2) | – | 0.5 | – | 1 (50%) | 0.5 | 1 (50%) | 0.7 |
| Heterozygous AG (n=32) | 8 (25%) | 2 (6%) | 5 (16%) | 15 (47%) | |||
| IRAK2 rs3844283 | |||||||
| Wild type CC (n=50) | 16 (32%) | 0.3 | 6 (12%) | 4 (8%) | 0.2 | 26 (52%) | 0.9 |
| Variant GG or CG (n=66) | 17 (26%) | 5 (8%) | 13 (20%) | 35 (53%) | |||
| Homozygous GG (n=9) | 2 (22%) | 0.6 | – | 2 (22%) | 0.4 | 4 (44%) | 0.7 |
| Heterozygous CG (n=57) | 15 (26%) | 5 (9%) | 11 (19%) | 31 (54%) | |||
| IRAK4 rs4251513 | |||||||
| Wild type CC (n=38) | 13 (34%) | 0.8 | 3 (8%) | 7 (18%) | 0.7 | 23 (60%) | 0.2 |
| Variant GG or CG (n=78) | 20 (26%) | 8 (10%) | 10 (13%) | 48 (61%) | |||
| Homozygous GG (n=34) | 11 (32%) | 0.4 | 4 (12%) | 5 (15%) | 0.7 | 20 (59%) | 0.7 |
| Heterozygous CG (n=44) | 9 (20%) | 4 (9%) | 5 (11%) | 18 (41%) | |||
The CMV polygenic score was built through a stepwise elimination process ( Table 3 ). The final calculated score is described by the following formula: Genetic risk score for CMV disease = 0.68 x TLR4 rs4986790/rs4986791 – 0.56 x TLR9 rs3775291 + 0.34 x TLR3 rs3775296+ 0.47 x AIM2 rs855873+ 0.71 x TLR7 rs179008-0.43 x MBL OO/OA/XAO – 0.49 x IFNL3/IL28B rs12979860 – 0.55 x IFI16 rs6940.
Table 3.
Calculation of the CMV genetic risk score: variables selected in the logistic regression model.
| Variable | Coefficient (β) | Standard error | Wald χ2 | p value |
|---|---|---|---|---|
| TLR3 rs3775296 | 0.34 | 0.22 | ||
| TLR4 rs4986790/rs4986791 | 0.68 | 0.32 | ||
| TLR7 rs179008 | 0.71 | 0.39 | ||
| TLR9 rs3775291 | 0.56 | 0.30 | ||
| AIM2 rs855873 | 0.47 | 0.25 | ||
| MBL2 (O/O, O/A, XA/O) | 0.43 | 0.24 | ||
| IFI16 rs6940 | -0.55 | 0.28 | ||
| IFNL3/IL28 rs12979860 | -0.49 | 0.25 | ||
| 8.2 | 0.004 |
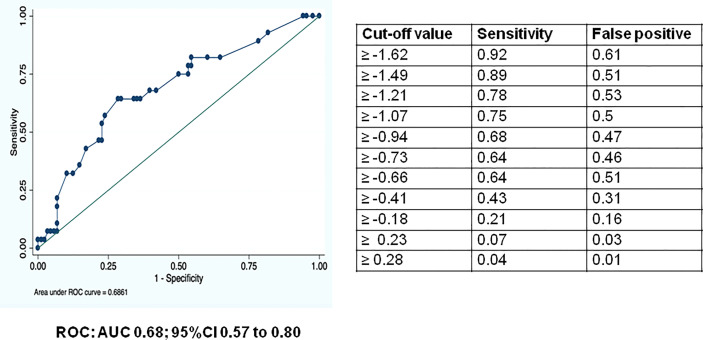
The discriminatory power of the score was assessed by the area under the receiver operation characteristics (ROC) curve and area under the ROC curve (AUCs). The discriminatory power of the CMV polygenic risk score and cut-off values for the ROC curve are shown in Figure 1 . The optimal estimated cut-off value of the score could be established in -0.36, conferring a sensitivity of the score of 62.3% (CI 53.5-71.1), a specificity of 52.73% (43.6-61.8), a positive predictive value of 0.59 (CI 0.5-0.7) and a negative predictive value of 0.56 (CI 0.4-0.6). Nevertheless, in order to improve specificity, the cut-off of 0 confers a sensitivity of the score of 43% (CI 33.6-51.6), a specificity of 71% (CI 62.6-79.2), a positive predictive value of 0.62 (CI 0.5-0.7) and a negative predictive value of 0.53 (0.4-0.6).
Figure 1.
Receiver operating characteristics (ROC) curve and area under the ROC curve (AUC) for assessing the discriminatory power of the risk score model for the prediction of CMV disease in SOT recipients, cut-off values for the ROC curve. ROC: AUC 0.68; 95%CI 0.57 to 0.80.
The values of sensitivity, specificity, positive and negative predictive values of the score are shown in Table 4 . TLR4 SNPs rs4986790/rs4986791 had the highest predictive value among these parameters, especially in terms of specificity (87.3%).
Table 4.
Area under the ROC curve (AUCs) of innate immune receptor gene SNPs for the prediction of CMV disease patients in SOT recipients with sensitivity and specificity.
| Gene SNP | AUCs (95%CI) | SEN % | SPE % | PPV % | NPV % | LR+ | LR- |
|---|---|---|---|---|---|---|---|
| TLR3 3775296 | 0.51 (0.42, 0.61) | 37.7 | 65.4 | 54.8 | 48.6 | 1.09 | 0.95 |
| TLR4 rs4986790/rs4986791 | 0.53 (0.44, 0.63) | 19.7 | 87.3 | 63.2 | 49.5 | 1.54 | 0.92 |
| TLR7 rs179008 | 0.53 (0.44, 0.63) | 27.9 | 78.2 | 58.6 | 49.4 | 1.27 | 0.92 |
| TLR9 rs3775291 | 0.47 (0.38, 0.57) | 68.8 | 25.4 | 50.6 | 42.4 | 0.92 | 1.22 |
| AIM2 rs855873 | 0.47 (0.38, 0.56) | 13.1 | 81.8 | 44.4 | 45.9 | 0.72 | 1.06 |
| MBL2 (O/O, O/A, XA/O) | 0.51 (0.44, 0.57) | 16.4 | 85.4 | 56.6 | 48.0 | 1.13 | 0.98 |
| IFI16 rs6940 | 0.51 (0.41, 0.60) | 16.4 | 85.4 | 55.6 | 48.0 | 1.13 | 0.98 |
| IFNL3/IL28 rs12979860 | 0.49 (0.40, 0.58) | 44.2 | 54.5 | 51.9 | 46.9 | 0.97 | 1.02 |
*ROC receiver operation characteristics, SEN sensitivity, SPE specificity, PPV positive predictive value, NPV negative predictive value, LR+ positive likelihood ratio, LR- negative likelihood ratio.
Discussion
In this prospective study of a large multicenter cohort of SOT recipients at high risk to develop CMV infection and disease, we built a polygenic score based on innate immune receptors gene SNPs to predict the probability of developing post-transplant CMV infection and disease. Many previous studies have addressed the risk of CMV infection in transplant patients according to the innate immune single gene SNPs. Our score was based on these previous associations and SNPs in innate immune receptor genes with theoretical influence in the immune control of CMV replication.
Several individual TLRs are involved in sensing viral nucleic acids and structural components. TLR3 recognizes viral double-stranded RNA (dsRNA) during human CMV replication and activates antiviral immune responses through production of type I IFN and inflammatory cytokines. Interestingly, increased frequency of the heterozygous TLR3 rs3775296 genotype has been reported in Polish children with CMV infection compared with uninfected cases (26). In line with this, the same group also found association of TLR9 variants (T/C, rs187084; C/T, rs352140) with CMV disease in children (27) and of TLR2 (A>G, rs5743708) with increased risk of congenital HCMV infection in Polish fetuses and neonates (28). TLR4 is not directly involved in the recognition of CMV, but it has been demonstrated that TLR4-ligands enhance the ability of dendritic cells to present CMV antigens resulting in an increased number of antigen-specific activated CD4+ and CD8+ T cells (29). In our study TLR4 polymorphism had a specificity of 87% to detect CMV disease and what is more, in the subgroup of kidney and liver recipients TLR4 polymorphism was associated with CMV infection, in line with other studies performed in kidney transplant recipients’cohorts including non-only high risk patients (30, 31). In the same way, we found that TLR3, TLR7 and TLR9 polymorphisms increased the risk of CMV disease.
Recognition of cytoplasmic DNA is an important immunological signature that marks the initiation of innate immune response. Absent in melanoma 2 (AIM2) is a cytoplasmic sensor that recognizes DNA of microbial or host origin (40) and give rise to a multiprotein complex – the so called AIM2 inflammasome - which plays a pivotal role in the host immune response to multiple pathogens. Compared to wild-type cells, AIM2-deficient macrophages showed a limited ability to activate caspase-1, process IL-1β, and induce cell death. In addition, AIM2-deficient cells were unable to efficiently control CMV infection, as the transcription of virus DNA polymerase gene UL54 and major tegument protein gene UL83 were higher compared to wild-type cells (42). IFI16 is another cytoplasmic DNA sensor that acts as restriction factor for CMV replication (41). When IFI16 undergoes phosphorylation relocalizes to the cytoplasm of CMV-infected cells and restricts CMV replication by downregulating viral mRNAs and their protein expression (43). Importantly, at the present study we found that both AIM2 and IFI16 polymorphisms were related with CMV disease.
MBL is a serum C-type lectin produced in the liver that binds carbohydrates present on a wide variety of bacterial, fungal, viral and parasitic surfaces (48). The CMV envelope is extremely complex and shares capsid glycoproteins with similar structural properties to other herpes virus. The gM and gN glycoproteins are the most important and abundant in the envelope of the virion, and the gM/gN complex formation is essential for maintaining virion infectivity. Furthermore, other glycoproteins such as gB and gH mediate viral entry into cells and are potential targets for MBL binding. So, genotypes responsible for low-MBL levels were associated with an increased risk of CMV infection and disease (31, 39), accordingly with our results.
IFN-λ3/IL-28B is a type III interferon that plays a role in the TLR-induced anti-viral activity (49). A protective effect of the rs12979860T allele of the IFNL3IL28B against CMV infection in allogeneic stem cell transplant patients has been described (35), in agreement with the results of the present study.
Strategies for preventing CMV infection and disease in CMV mismatch patient are frequently debated in SOT recipients. Since the introduction of antiviral prophylaxis with i.v. ganciclovir or oral valganciclovir, a 100 to 200 day course of these drugs has been proposed for patients at high risk to develop CMV disease. However, this strategy is associated with high rates of toxicity (leucopoenia) leading to early discontinuation of prophylaxis. In addition, antiviral prophylaxis is associated with late-onset CMV disease and with an independently associated greater risk of mortality (50, 51). On the other hand, pre-emptive strategies have been historically associated with suboptimal results (52), although more recent data seems promising in liver transplant recipients (53). Furthermore, T cell immune functional assays such as ELISpot or QuantiFERON assays have shown association with higher incidence of CMV infection (5). Measurement of such parameters could potentially be used to inform the risk of infection as recommended by some international guidelines (44) although no interventional studies using these tests have been performed. Accordingly, there are some potential clinical scenarios where the CMV score could be used. Extending antiviral prophylaxis or using a longer pre-emptive strategy after antiviral prophylaxis could be considered in CMV-seronegative recipients of grafts from CMV-seropositive donors undergoing transplantation with high CMV score. Additionally, using mTOR inhibitors as maintenance immunosuppressive therapy due to protective anti CMV effect or using less intense immunosuppression are potential strategies for preventing CMV complication in patients at higher risk (4). Finally, this score can be a starting point for the creation of new scores to evaluate the risk of CMV infection and disease that include other parameters such as T cell immune functional assays and immunosuppressive regimen. Other proposal could be that this score can be used as a screening test and secondly, MBL levels and TLRs function could be measured by stimulation of the peripheral mononuclear cells with specific TLR-ligands in patients presenting with a CMV high-risk score to better assess the CMV risk.
The main strength of our study is the prospective inclusion of a large cohort of CMV mismatch transplant recipients from different centers with equivalent strategies of prophylaxis, which allowed obtaining granular data on immunosuppression and CMV events. Moreover, the genetic score predicts the risk of CMV disease using innate immune gene receptors polymorphisms. Nevertheless, an external validation of our results is mandatory in order to reaffirm our results.
Some caveats should be highlighted. Polygenic scores should be based on relevant genomic-wide associated studies (54). Our score was built based on previous published information analyzing single-gene SNPs in a variety of cohorts. Whole-genome sequencing studies to characterize the genetic profile associated to higher risk of CMV in transplant patients would require the coordination of a large international cohort. We propose a new line of research to apply precision medicine in predicting the management strategy of transplant patients at risk of CMV. The performance of our proposed polygenic score should be validated by other prospective cohorts. However, this pilot polygenic score could be improved by adding other candidate SNPs to the model. Furthermore, the heterogeneity of the cohort and the varying degree of CMV infection risk according to the type of allograft and immunosuppression represents another limitation to interpret the data. Certainly, building a model capable to control for all these variables would require a much larger cohort. Finally, other limitation of our study is that we performed the MBL genotyping only in the recipient. Taking into account that this protein is synthesized by the liver, MBL genotyping should have been performed in the donor in liver recipients.
To conclude, we propose a CMV polygenic score to predict the risk of CMV disease in CMV D+/R-, based on TLR4 (rs4986790/rs4986791), TLR9 (rs3775291), TLR3 (rs3775296), AIM2 (rs855873), TLR7 (rs179008), MBL (OO/OA/XAO), IFNL3/IL28B (rs12979860) and IFI16 (rs6940) genetic variants. Our score may help to identify patients at low risk of developing CMV disease that may benefit of shortening antiviral prophylaxis. On the other hand, those patients at high genetic risk of CMV may benefit of prolonged prophylaxis or even lifelong prophylaxis depending on the type of transplant. Further studies to validate the use of polygenic scores to predict the risk of CMV infection in SOT recipients are needed.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by Comitè d'Ètica de la Investigació amb medicaments (CEIm), Hospital Clinic de Barcelona (2012/7530). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MB, CC, FL and AM participated in research design. MB, CC and FL participated in the writing of the paper. MB, CC, LL, BS, JL, GS, MF-R, MF, SC, MM, EC, IO, MAM, FL SCL and AM participated in the performance of the research. MB, CC and JL contributed in analytic tools and data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Spanish Ministerio de Sanidad y Consumo (FIS PI12/01743), Instituto de Salud Carlos III, Fondo Europeo de Desarrollo Regional (FEDER). Unión Europea. “Una manera de hacer Europa”; Network for the Research in Infectious Diseases (REIPI) from the Instituto de Salud Carlos III, Madrid, Spain; and Ministerio de Ciencia e Innovación (PID2019-106658RB-I00, funded by MCIN/AEI/10.13039/501100011033). MF-R holds a research contract “Miguel Servet” (CP18/00073) from the Instituto de Salud Carlos III, Spanish Ministerio de Ciencia e Innovación.
Conflict of Interest
FL is founder and ad honorem scientific advisor or Sepsia Therapeutics S.L.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Appendix I: Members of the GESITRA-IC/SEIMC/REIPI investigators related to the study:
JM Aguado, Hospital Universitario “12 de Octubre”, Instituto de Investigación Hospital “12 de Octubre” (imas12) Madrid, Spain
J.Torre-Cisneros, Reina Sofía University Hospital/University of Cordoba, Cordoba, Spain.
J. Carratala, University Hospital, Bellvitge Biomedical Research Institute (IDIBELL), L’Hospitalet de Llobregat, Barcelona, Spain
C. Lumbreras, Hospital Universitario “12 de Octubre”, Instituto de Investigación Hospital “12 de Octubre” (imas12) Madrid, Spain
M. Montejo. Hospital Universitario de Cruces. Bilbao, Spain
C. Roca, Department of Medicine, University of Seville, Virgen del Rocío University Hospital, Institute of Biomedicine of Seville (IBiS),/CSIC/University of Seville,
M. Gutierrez University Hospital “Marqués de Valdecilla”, Instituto de Investigación “Marqués de Valdecilla” (IDIVAL), University of Cantabria, Santander, Spain
C. Rico. University Hospital “Marqués de Valdecilla”, Instituto de Investigación “Marqués de Valdecilla” (IDIVAL), University of Cantabria, Santander, Spain
L. Muñoz. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
A. Páez-Vega. Reina Sofía University Hospital/University of Cordoba, Cordoba, Spain.
E. Vidal Verdú. Reina Sofía University Hospital/University of Cordoba, Cordoba, Spain.
A.Rodríguez-Benot. Reina Sofía University Hospital/University of Cordoba, Cordoba, Spain.
J. Colmenero. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
F. Cofan. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
N. Armiger. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
M. Cardona. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
Y. Zboromyrska. Hospital Clinic - IDIBAPS, University of Barcelona, Spain
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.897912/full#supplementary-material
References
- 1. Fishman JA. Infection in Solid-Organ Transplant Recipients. N Engl J Med (2007) 357:2601–14. doi: 10.1056/NEJMra064928 [DOI] [PubMed] [Google Scholar]
- 2. Hodson EM, Jones CA, Webster AC, Strippoli GF, Barclay PG, Kable K, et al. Antiviral Medications to Prevent Cytomegalovirus Disease and Early Death in Recipients of Solid-Organ Transplants: A Systematic Review of Randomised Controlled Trials. Lancet (2005) 365:2105–15. doi: 10.1016/S0140-6736(05)66553-1 [DOI] [PubMed] [Google Scholar]
- 3. Humar A, Lebranchu Y, Vincenti F, Blumberg EA, Punch JD, Limaye AP, et al. The Efficacy and Safety of 200 Days Valganciclovir Cytomegalovirus Prophylaxis in High-Risk Kidney Transplant Recipients. Am J Transplant (2010) 10:1228–37. doi: 10.1111/j.1600-6143.2010.03074.x [DOI] [PubMed] [Google Scholar]
- 4. Cervera C, Cofan F, Hernandez C, Soy D, Marcos MA, Sanclemente G, et al. Effect of Mammalian Target of Rapamycin Inhibitors on Cytomegalovirus Infection in Kidney Transplant Recipients Receiving Polyclonal Antilymphocyte Globulins: A Propensity Score-Matching Analysis. Transpl Int (2016) 29:1216–25. doi: 10.1111/tri.12848 [DOI] [PubMed] [Google Scholar]
- 5. Manuel O, Husain S, Kumar D, Zayas C, Mawhorter S, Levi ME, et al. Assessment of Cytomegalovirus-Specific Cell-Mediated Immunity for the Prediction of Cytomegalovirus Disease in High-Risk Solid-Organ Transplant Recipients: A Multicenter Cohort Study. Clin Infect Dis (2013) 56:817–24. doi: 10.1093/cid/cis993 [DOI] [PubMed] [Google Scholar]
- 6. Manuel O, Avery RK. Update on Cytomegalovirus in Transplant Recipients: New Agents, Prophylaxis, and Cell-Mediated Immunity. Curr Opin Infect Dis (2021) 34:307–13. doi: 10.1097/QCO.0000000000000746 [DOI] [PubMed] [Google Scholar]
- 7. Béland S, Désy O, Vallin P, Basoni C, De Serres SA. Innate Immunity in Solid Organ Transplantation: An Update and Therapeutic Opportunities. Expert Rev Clin Immunol (2015) 11:377–89. doi: 10.1586/1744666X.2015.1008453 [DOI] [PubMed] [Google Scholar]
- 8. Medzhitov R, Janeway C. Innate Immunity. N Engl J Med (2000) 343:338–44. doi: 10.1056/NEJM200008033430506 [DOI] [PubMed] [Google Scholar]
- 9. Pichlmair A, Reis e Sousa C. Innate Recognition of Viruses. Immunity (2007) 27:370–83. doi: 10.1016/j.immuni.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 10. Takeuchi O, Akira S. Innate Immunity to Virus Infection. Immunol Rev (2009) 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sezgin E, An P, Winkler CA. Host Genetics of Cytomegalovirus (CMV) Pathogenesis. Front Genet (2019) 10:616. doi: 10.3389/fgene.2019.00616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernández-Ruiz M, Giménez E, Lora D, Aguado JM, Pascual M, Manuel O. Impact of MBL2 Gene Polymorphisms on the Risk of Infection in Solid Organ Transplant Recipients: A Systematic Review and Meta-Analysis. Am J Transplant (2019) 19:1072–85. doi: 10.1111/ajt.15160 [DOI] [PubMed] [Google Scholar]
- 13. Fernández-Ruiz M, Corrales I, Arias M, Campistol JM, Giménez E, Crespo J, et al. Association Between Individual and Combined SNPs in Genes Related to Innate Immunity and Incidence of CMV Infection in Seropositive Kidney Transplant Recipients. Am J Transplant (2015) 15:1323–35. doi: 10.1111/ajt.13107 [DOI] [PubMed] [Google Scholar]
- 14. Kijpittayarit S, Eid AJ, Brown RA, Paya CV, Razonable RR. Relationship Between Toll-Like Receptor 2 Polymorphism and Cytomegalovirus Disease After Liver Transplantation. Clin Infect Dis (2007) 44:1315–20. doi: 10.1086/514339 [DOI] [PubMed] [Google Scholar]
- 15. Hoffmann TW, Halimi JM, Büchler M, Velge-Roussel F, Goudeau A, Al Najjar A, et al. Association Between a Polymorphism in the IL-12p40 Gene and Cytomegalovirus Reactivation After Kidney Transplantation. Transplantation (2008) 85:1406–11. doi: 10.1097/TP.0b013e31816c7dc7 [DOI] [PubMed] [Google Scholar]
- 16. Mezger M, Steffens M, Semmler C, Arlt EM, Zimmer M, Kristjanson GI, et al. Investigation of Promoter Variations in Dendritic Cell-Specific ICAM3-Grabbing Non-Integrin (DC-SIGN) (CD209) and Their Relevance for Human Cytomegalovirus Reactivation and Disease After Allogeneic Stem-Cell Transplantation. Clin Microbiol Infect (2008) 14:228–34. doi: 10.1111/j.1469-0691.2007.01902.x [DOI] [PubMed] [Google Scholar]
- 17. Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, et al. Polymorphisms in Toll-Like Receptor Genes and Susceptibility to Infections in Allogeneic Stem Cell Transplantation. Exp Hematol (2009) 37:1022–9. doi: 10.1016/j.exphem.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 18. Brown RA, Gralewski JH, Razonable RR. The R753Q Polymorphism Abrogates Toll-Like Receptor 2 Signaling in Response to Human Cytomegalovirus. Clin Infect Dis (2009) 49:e96–9. doi: 10.1086/644501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cervera C, Lozano F, Linares L, Antón A, Balderramo D, Suárez B, et al. Influence of Mannose-Binding Lectin Gene Polymorphisms on the Invasiveness of Cytomegalovirus Disease After Solid Organ Transplantation. Transplant Proc (2009) 41:2259–61. doi: 10.1016/j.transproceed.2009.06.056 [DOI] [PubMed] [Google Scholar]
- 20. Hoffmann TW, Halimi JM, Büchler M, Velge-Roussel F, Goudeau A, Al-Najjar A, et al. Association Between a Polymorphism in the Human Programmed Death-1 (PD-1) Gene and Cytomegalovirus Infection After Kidney Transplantation. J Med Genet (2010) 47:54–8. doi: 10.1136/jmg.2009.068841 [DOI] [PubMed] [Google Scholar]
- 21. De Rooij BJF, van der Beek MT, Van Hoek B, Vossen ACTM, Rogier TenHove W, Roos A, et al. Mannose-Binding Lectin and Ficolin-2 Gene Polymorphisms Predispose to Cytomegalovirus (Re)Infection After Orthotopic Liver Transplantation. J Hepatol (2011) 55:800–7. doi: 10.1016/j.jhep.2011.01.039 [DOI] [PubMed] [Google Scholar]
- 22. Mitsani D, Nguyen MH, Girnita DM, Spichty K, Kwak EJ, Silveira FP, et al. A Polymorphism Linked to Elevated Levels of Interferon-γ Is Associated With an Increased Risk of Cytomegalovirus Disease Among Caucasian Lung Transplant Recipients at a Single Center. J Heart Lung Transplant (2011) 30:523–9. doi: 10.1016/j.healun.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 23. Kang SH, Abdel-Massih RC, Brown RA, Dierkhising RA, Kremers WK, Razonable RR. Homozygosity for the Toll-Like Receptor 2 R753Q Single-Nucleotide Polymorphism Is a Risk Factor for Cytomegalovirus Disease After Liver Transplantation. J Infect Dis (2012) 205:639–46. doi: 10.1093/infdis/jir819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egli A, Levin A, Santer DM, Joyce M, O’Shea D, Thomas BS, et al. Immunomodulatory Function of Interleukin 28B During Primary Infection With Cytomegalovirus. J Infect Dis (2014) 210:717–27. doi: 10.1093/infdis/jiu144 [DOI] [PubMed] [Google Scholar]
- 25. Loeffler J, Steffens M, Arlt EM, Toliat MR, Mezger M, Suk A, et al. Polymorphisms in the Genes Encoding Chemokine Receptor 5, Interleukin-10, and Monocyte Chemoattractant Protein 1 Contribute to Cytomegalovirus Reactivation and Disease After Allogeneic Stem Cell Transplantation. J Clin Microbiol (2006) 44:1847–50. doi: 10.1128/JCM.44.5.1847-1850.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Studziñska M, Jabłoñska A, Wiśniewska-Ligier M, Nowakowska D, Gaj Z, Leśnikowski ZJ, et al. Association of TLR3 L412F Polymorphism With Cytomegalovirus Infection in Children. PLos One (2017) 12:e0169420. doi: 10.1371/journal.pone.0169420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paradowska E, Jabłońska A, Studzińska M, Skowrońska K, Suski PS, Wiśniewska-Ligier M, et al. TLR9 -1486t/C and 2848C/T SNPs Are Associated With Human Cytomegalovirus Infection in Infants. PLos One (2016) 11:e0154100. doi: 10.1371/journal.pone.0154100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wujcicka W, Paradowska E, Studzińska M, Wilczyński J, Nowakowska D. TLR9 2848 GA Heterozygotic Status Possibly Predisposes Fetuses and Newborns to Congenital Infection with Human Cytomegalovirus. PLoS One (2015) doi: 10.1371/journal.pone.0122831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loré K, Betts MR, Brenchley JM, Kuruppu J, Khojasteh S, Perfetto S, et al. Toll-Like Receptor Ligands Modulate Dendritic Cells to Augment Cytomegalovirus- and HIV-1-Specific T Cell Responses. J Immunol (2003) 171:4320–8. doi: 10.4049/jimmunol.171.8.4320 [DOI] [PubMed] [Google Scholar]
- 30. Ducloux D, Deschamps M, Yannaraki M, Ferrand C, Bamoulid J, Saas P, et al. Relevance of Toll-Like Receptor-4 Polymorphisms in Renal Transplantation. Kidney Int (2005) 67:2454–61. doi: 10.1111/j.1523-1755.2005.00354.x [DOI] [PubMed] [Google Scholar]
- 31. Cervera C, Lozano F, Saval N, Gimferrer I, Ibañez A, Suárez B, et al. The Influence of Innate Immunity Gene Receptors Polymorphisms in Renal Transplant Infections. Transplantation (2007) 83:1493–500. doi: 10.1097/01.tp.0000264999.71318.2b [DOI] [PubMed] [Google Scholar]
- 32. Zucchini N, Bessou G, Traub S, Robbins SH, Uematsu S, Akira S, et al. Cutting Edge: Overlapping Functions of TLR7 and TLR9 for Innate Defense Against a Herpesvirus Infection. J Immunol (2008) 180:5799–803. doi: 10.4049/jimmunol.180.9.5799 [DOI] [PubMed] [Google Scholar]
- 33. Dominguez-Molina B, Machmach K, Perales C, Tarancon-Diez L, Gallego I, Sheldon JL, et al. Toll-Like Receptor 7 (TLR-7) and TLR-9 Agonists Improve Hepatitis C Virus Replication and Infectivity Inhibition by Plasmacytoid Dendritic Cells. J Virol (2018) 92:e01219-18. doi: 10.1128/JVI.01219-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneider M, Matiqi T, Kundi M, Rieder FJJ, Andreas M, Strassl R, et al. Clinical Significance of the Single Nucleotide Polymorphism TLR2 R753Q in Heart Transplant Recipients at Risk for Cytomegalovirus Disease. J Clin Virol (2016) 84:64–9. doi: 10.1016/j.jcv.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bravo D, Solano C, Giménez E, Remigia MJ, Corrales I, Amat P, et al. Effect of the IL28B Rs12979860 C/T Polymorphism on the Incidence and Features of Active Cytomegalovirus Infection in Allogeneic Stem Cell Transplant Patients. J Med Virol (2014) 86:838–44. doi: 10.1002/jmv.23865 [DOI] [PubMed] [Google Scholar]
- 36. Hokeness-Antonelli KL, Crane MJ, Dragoi AM, Chu W-M, Salazar-Mather TP. IFN-αβ-Mediated Inflammatory Responses and Antiviral Defense in Liver Is TLR9-Independent But MyD88-Dependent During Murine Cytomegalovirus Infection. J Immunol (2007) 179:6176–83. doi: 10.4049/jimmunol.179.9.6176 [DOI] [PubMed] [Google Scholar]
- 37. Gay NJ, Gangloff M, O’Neill LAJ. What the Myddosome Structure Tells Us About the Initiation of Innate Immunity. Trends Immunol (2011) 32:104–9. doi: 10.1016/j.it.2010.12.005 [DOI] [PubMed] [Google Scholar]
- 38. Wang H, El Maadidi S, Fischer J, Grabski E, Dickhöfer S, Klimosch S, et al. A Frequent Hypofunctional IRAK2 Variant Is Associated With Reduced Spontaneous Hepatitis C Virus Clearance. Hepatology (2015) 62:1375–87. doi: 10.1002/hep.28105 [DOI] [PubMed] [Google Scholar]
- 39. Manuel O, Pascual M, Trendelenburg M, Meylan PR. Association Between Mannose-Binding Lectin Deficiency and Cytomegalovirus Infection After Kidney Transplantation. Transplantation (2007) 83:359–62. doi: 10.1097/01.tp.0000251721.90688.c2 [DOI] [PubMed] [Google Scholar]
- 40. Man SM, Karki R, Kanneganti TD. AIM2 Inflammasome in Infection, Cancer, and Autoimmunity: Role in DNA Sensing, Inflammation, and Innate Immunity. Eur J Immunol (2016) 46:269–80. doi: 10.1002/eji.201545839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gariano GR, Dell’Oste V, Bronzini M, Gatti D, Luganini A, de Andrea M, et al. The Intracellular DNA Sensor IFI16 Gene Acts as Restriction Factor for Human Cytomegalovirus Replication. PLos Pathog (2012) 88(12):6970–82. doi: 10.1371/journal.ppat.1002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang Y, Liu L, Ma D, Liao Y, Lu Y, Huang H, et al. Human Cytomegalovirus Triggers the Assembly of AIM2 Inflammasome in THP-1-Derived Macrophages. J Med Virol (2017) 89:2188–95. doi: 10.1002/jmv.24846 [DOI] [PubMed] [Google Scholar]
- 43. Dell’Oste V, Gatti D, Gugliesi F, De Andrea M, Bawadekar M, Lo Cigno I, et al. Innate Nuclear Sensor IFI16 Translocates Into the Cytoplasm During the Early Stage of In Vitro Human Cytomegalovirus Infection and Is Entrapped in the Egressing Virions During the Late Stage. J Virol (2014) 88:6970–82. doi: 10.1128/JVI.00384-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kotton CN, Kumar D, Caliendo AM, Huprikar S, Chou S, Danziger-Isakov L, et al. The Third International Consensus Guidelines on the Management of Cytomegalovirus in Solid-Organ Transplantation. Transplantation (2018) 102:900–31. doi: 10.1097/TP.0000000000002191 [DOI] [PubMed] [Google Scholar]
- 45. Husain S, Camargo JF. Invasive Aspergillosis in Solid-Organ Transplant Recipients: Guidelines From the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant (2019) 33:e13544. doi: 10.1111/ctr.13544 [DOI] [PubMed] [Google Scholar]
- 46. Sanclemente G, Moreno A, Navasa M, Lozano F, Cervera C. Genetic Variants of Innate Immune Receptors and Infections After Liver Transplantation. World J Gastroenterol (2014) 20:11116–30. doi: 10.3748/wjg.v20.i32.11116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Madsen HO, Garred P, Thiel S, Kurtzhals JA, Lamm LU, Ryder LP, et al. Interplay Between Promoter and Structural Gene Variants Control Basal Serum Level of Mannan-Binding Protein. J Immunol (1995) 155:3013–20. [PubMed] [Google Scholar]
- 48. Jack DL, Klein NJ, Turner MW. Mannose-Binding Lectin: Targeting the Microbial World for Complement Attack and Opsonophagocytosis. Immunol Rev (2001) 180:86–99. doi: 10.1034/j.1600-065X.2001.1800108.x [DOI] [PubMed] [Google Scholar]
- 49. Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An Important Role for Type III Interferon (IFN-λ/IL-28) in TLR-Induced Antiviral Activity. J Immunol (2008) 180:2474–85. doi: 10.4049/jimmunol.180.4.2474 [DOI] [PubMed] [Google Scholar]
- 50. Limaye AP, Bakthavatsalam R, Kim HW, Kuhr CS, Halldorson JB, Healey PJ, et al. Late-Onset Cytomegalovirus Disease in Liver Transplant Recipients Despite Antiviral Prophylaxis. Transplantation (2004) 78:1390–6. doi: 10.1097/01.TP.0000145989.22373.03 [DOI] [PubMed] [Google Scholar]
- 51. Singh N. Late-Onset Cytomegalovirus Disease as a Significant Complication in Solid Organ Transplant Recipients Receiving Antiviral Prophylaxis: A Call to Heed the Mounting Evidence. Clin Infect Dis (2005) 40:704–8. doi: 10.1086/427506 [DOI] [PubMed] [Google Scholar]
- 52. Bodro M, Sabé N, Lladõ L, Baliellas C, Niubó J, Castellote J, et al. Prophylaxis Versus Preemptive Therapy for Cytomegalovirus Disease in High-Risk Liver Transplant Recipients. Liver Transplant (2012) 18:1093–99. doi: 10.1002/lt.23460 [DOI] [PubMed] [Google Scholar]
- 53. Singh N, Winston DJ, Razonable RR, Lyon GM, Silveira FP, Wagener MM. Effect of Preemptive Therapy vs Antiviral Prophylaxis on Cytomegalovirus Disease in Seronegative Liver Transplant Recipients With Seropositive Donors: A Randomized Clinical Trial. JAMA - J Am Med Assoc (2020) 323:1378–87. doi: 10.1001/jama.2020.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choi SW, Mak TSH, O’Reilly PF. Tutorial: A Guide to Performing Polygenic Risk Score Analyses. Nat Protoc (2020) 15:2759–72. doi: 10.1038/s41596-020-0353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.