Abstract
The eukaryotic endoplasmic reticulum (ER) maintains protein homeostasis by eliminating unwanted proteins through the evolutionarily conserved ER-associated degradation (ERAD) pathway. During ERAD, maturation-defective and surplus polypeptides are evicted from the ER lumen and/or lipid bilayer through the process of retrotranslocation and ultimately degraded by the proteasome. An integral facet of the ERAD mechanism is the ubiquitin system, composed of the ubiquitin modifier and the factors for assembling, processing and binding ubiquitin chains on conjugated substrates. Beyond simply marking polypeptides for degradation, the ubiquitin system is functionally intertwined with retrotranslocation machinery to transport polypeptides across the ER membrane.
To ensure that proteins progressing through the ER are properly folded and functional, eukaryotic cells have evolved a sophisticated quality-control mechanism that can eliminate terminally misfolded or unassembled polypeptides whose propensity for aggregation may cause cytotoxicity. This mechanism, known as ERAD, also has active roles in regulating levels of enzymes or signaling molecules in response to perturbed metabolic states1,2. As an adaptable and responsive mechanism of remediation, ERAD not only enables the necessary flux of functional proteins through the secretory pathway but also supports the survival of individual cells and development of multicellular organisms during environmental changes3.
The discovery of the ERAD process dates back to the late 1980s, when several groups reported that incorrectly folded polypeptides or unassembled subunits of protein complexes often failed to progress beyond the ER, with many being targeted for rapid degradation4–6. These studies hinted at the existence of a protein quality-control mechanism at the level of the ER, but the identity of the responsible protease remained elusive until the mid-1990s, when a series of seminal studies implicated components of the ubiquitin-proteasome system (UPS)7–12. Since then, a panoply of tools, including small-molecule inhibitors, viral proteins, genetic screens in yeast and, more recently, complex genetic interaction maps, proteomics and advanced biochemical strategies have been used to identify many if not most components (Tables 1 and 2) and to delineate important interactions required for recognition, targeting and delivery of misfolded proteins from the ER to the UPS.
Table 1.
Ubiquitin conjugation, binding and processing proteins in ERAD
| Protein (human) | Other names | Protein (yeast) | Function | Ub-related domains | Other domains | L | M | C | TM |
|---|---|---|---|---|---|---|---|---|---|
| Uba1 | Uba1p | Ubiquitin-activating enzyme | X | – | |||||
| Ube2j1, Ube2j2 | Ubc6e | Ubc6p | Ubiquitin-conjugating enzyme | UBCc | X | X | TA | ||
| Ube2g2 | Ubc7p | Ubiquitin-conjugating enzyme | UBCc | X | – | ||||
| Hrd1 | Synoviolin | Hrd1p | Ubiquitin ligase | RING | VBMa | X | X | X | 6 |
| TEB4 | March IV | Doa10p | Ubiquitin ligase | RING | X | X | X | 12 | |
| gp78 | AMFR, RNF45 | Ubiquitin ligase | RING, CUEb, G2BR | VIMa | X | X | X | 6 | |
| RNF5 | RMA1 | Ubiquitin ligase | RING | X | X | X | 2 | ||
| RNF170 | Ubiquitin ligase | RING | X | X | X | 3 | |||
| Trc8 | RNF138 | Ubiquitin ligase | RING | X | X | X | 8 | ||
| RNF103 | Kf-1 | Ubiquitin ligase | RING | X | X | X | 4 | ||
| RFP2 | Ubiquitin ligase | RING | B-box | X | X | X | T1 | ||
| Fbx2 | Ubiquitin ligase | F-Box | X | – | |||||
| Fbx6 | Ubiquitin ligase | F-Box | X | – | |||||
| Parkin | PARK2 | Ubiquitin ligase | RING, UBL | IBR | X | – | |||
| CHIP | Ubiquitin ligase | U-Box | TPR | X | – | ||||
| UBE4a | Ufd2p | Ubiquitin ligase | U-Box | X | – | ||||
| USP13 | Deubiquitinase | UBAb (2) | X | – | |||||
| USP25 | Deubiquitinase | UIMb (2), UBAb | SIM | X | – | ||||
| YOD1 | OTUD2 | Deubiquitinase | OTU, UBX | X | – | ||||
| Ataxin-3 | Deubiquitinase | UIMb | Josephin, VBMa | X | – | ||||
| OTUB1 | Deubiquitinase | OTU | X | – | |||||
| VCIP135 | VCPIP | Deubiquitinase | OTU | X | – | ||||
| HERP | Usa1p | Scaffolding | UBL | X | X | MA | |||
| TMUB1 | Scaffolding | UBL | NES | X | X | X | 2 | ||
| FAM8A1 | Ubiquitin-ligase cofactor | X | X | X | 3 | ||||
| AUP1 | Cue1p | E2 recruitment, ubiquitin binding | CUEb, G2BR | X | X | MA |
Common and alternate mammalian ERAD-component names are given, along with yeast orthologs. Putative function within ERAD is shown, in addition to ubiquitin-related and other domains (numbers of domains are shown in parentheses). Cellular exposure to ER lumen (L), ER membrane (M) or cytoplasm (C) is indicated by ‘X’. Links with the ER membranes are listed under TM: numbers, polytopic TM domains; T1, type I; TA, tail-anchored; MA, membrane associated.
p97/VCP-binding domain.
Ubiquitin-binding domain.
Table 2.
ERAD components involved in targeting to the UPS
| Protein (human) | Other names | Protein (yeast) | Function | Ub-related domains | Other domains | L | M | C | TM |
|---|---|---|---|---|---|---|---|---|---|
| Substrate recognition and targeting | |||||||||
| SEL1 | Hrd3p | Substrate binding and recruitment | Sel1-like (12) | X | X | X | T1 | ||
| INSIG1, INSIG2 | Substrate binding and recruitment | X | X | X | 2 | ||||
| BAP31 | Substrate binding and recruitment | X | X | X | 3 | ||||
| ERLIN1, ERLIN2 | Substrate binding and recruitment | X | X | X | 4 | ||||
| OS-9 | Yos9p | Glycan binding | MRH | X | – | ||||
| XTP3-B | Glycan binding | MRH(2) | X | – | |||||
| ERMan1 | Mns1p | Glycan trimming | X | – | |||||
| EDEM1, EDEM2, EDEM3 | Htm1p | Glycan trimming | X | – | |||||
| PDI | P4HB | Disulfide-bond rearrangement | X | – | |||||
| BiP | GRP78 | Kar2p | Chaperone Hsp70 | X | – | ||||
| Hsp70 | Ssa1p | Chaperone Hsp70 | X | – | |||||
| GRP94 | gp96 | Chaperone Hsp90 | X | – | |||||
| ERdj4 | Chaperone Hsp40 | J | X | – | |||||
| ERdj5 | DNAJC10 | Scj1p | Chaperone Hsp40 | J, Trx | X | – | |||
| DNAJB12 | Chaperone Hsp40 | J | X | X | T2 | ||||
| Retrotranslocation and dislocation | |||||||||
| Derlin1, Derlin2, Derlin3 | Der1p, Dfm1p | Rhomboid pseudoprotease | SHPa | X | X | X | 6 | ||
| UBAC2 | Dsc2p | Rhomboid pseudoprotease | UBAb | X | X | X | 6 | ||
| RHBDL4 | Rhomboid protease | UIMb | VBMa | X | X | X | 6 | ||
| p97/VCP | Cdc48p | AAA+ ATPase | AAA | X | – | ||||
| TorsinA | DYT1 | AAA+ ATPase | AAA | X | – | ||||
| UbxD2 | Erasin | p97/VCP adaptor | UBX | X | X | MA | |||
| UbxD8 | ETEA | Ubx2p | p97/VCP adaptor | UBAb, UBX | X | X | MA | ||
| VIMP | SelS | p97/VCP adaptor | VIMa | X | X | T2 | |||
| Ufd1 | Ufd1p | p97/VCP adaptor | UT3b | BS1a | X | – | |||
| Npl4 | Npl4p | p97/VCP adaptor | NZFb, UBX | X | – | ||||
| NGly1 | PNGase1 | Png1p | Deglycosylation | PUBa, PAW | X | – | |||
| Substrate delivery to the proteasome | |||||||||
| Ubiquilin-1 | Dsk2p | Shuttling factor | UBL, UBAb | X | – | ||||
| hHR23A, hHR23B | Rad23p | Shuttling factor | UBL, UBAb | X | – | ||||
| DNAJB2 | HSJ1 | Shuttling factor | UIMb (2) | J | X | X | GG | ||
| Ubl4A | GET5 | Shuttling factor | UBL | X | – | ||||
| Trc35 | GET4 | Shuttling factor | X | – | |||||
| Bag6 | BAT3, Scythe | Shuttling factor, chaperone | UBL | X | – | ||||
| SGTA | Shuttling factor, chaperone | UBL binding domain | X | – | |||||
Common and alternate mammalian ERAD-component names are given, along with yeast orthologs. Putative function within ERAD is shown, in addition to ubiquitin-related and other domains (numbers of domains are shown in parentheses). Cellular exposure to ER lumen (L), ER membrane (M) or cytoplasm (C) is indicated by ‘X’. Links with the ER membranes are listed under TM: numbers, polytopic TM domains; T1, type I; T2, type II; MA, membrane associated; GG, geranylgeranylated.
p97/VCP-binding domain.
Ubiquitin-binding domain.
Intriguingly, no evidence indicates that the ER lumen contains any UPS components such as ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2s) or the proteasome, probably because these proteins lack appropriate ER-targeting signals. Accordingly, ERAD substrates are either partially (for integral membrane substrates) or completely (for luminal substrates) segregated from the UPS machinery by the lipid bilayer. Thus, a transport mechanism able to move polypeptides across the ER membrane is implicitly required for degradation, at least for soluble luminal ERAD substrates. Because protein translocation at the ER traditionally refers to vectorial insertion of ribosome-bound nascent chains through the Sec61 translocon into the ER lumen or lipid bilayer, the terms ‘retrotranslocation’ and ‘dislocation’ are used to describe the reverse movement of polypeptides during ERAD. The former refers to the movement of a luminal entity across the lipid bilayer, whereas the latter describes the dislodging of membrane proteins into the cytosol in ERAD13. Recent studies underscore an elaborate molecular design that integrates many UPS components with a complex retrotranslocation apparatus to channel misfolded proteins of the secretory pathway for degradation. Our discussions below reflect the growing understanding of the integrated roles of UPS components in ERAD.
The ubiquitin system in ERAD
Ubiquitin and ubiquitin-like proteins in ERAD.
The 76-residue polypeptide modifier ubiquitin can be conjugated to lysine or, in some cases, to serine, threonine or cysteine residues of a substrate14–16. Ubiquitin is added either as a monomer (monoubiquitination) or extended to form chains (polyubiquitination) through one of its seven lysine residues or its N terminus. Both the length (monomer versus chain) and the linkages of ubiquitin chains can affect the fate, stability, subcellular localization and activity of the modified substrates17,18. With a few exceptions, inhibiting ubiquitination prevents not only degradation but also retrotranslocation of most substrates from the ER13, thus indicating that ubiquitin conjugation is vital for continuous processing of ERAD substrates. Ubiquitin can be conjugated to both ERAD substrates13 and the machinery19, but the linkages, frequency and overall impact of ubiquitination on ERAD machinery are not well defined. Eukaryotic cells can also modify substrates with small ubiquitin-like (UBL) proteins that have similarities in both sequence and structure to ubiquitin20. A role for the small ubiquitin-like modifier (SUMO) in promoting ubiquitination of an ERAD substrate by a SUMO-dependent ubiquitin ligase (RNF4) was suggested recently21, but a general requirement of SUMO or RNF4 in ERAD has not been demonstrated.
The ubiquitination machinery in ERAD.
Ubiquitin conjugation requires the actions of an enzyme cascade. Once the E1 enzyme activates ubiquitin, a ubiquitin-conjugating enzyme (E2) can act in conjunction with a ubiquitin ligase (E3) to transfer ubiquitin to a selected substrate22. In budding yeast, two ER-anchored E3s (Hrd1p and Doa10p) are responsible for most (if not all) substrate ubiquitination in ERAD23,24 (Table 1), but additional E3 ligases may also contribute to ERAD under special conditions25. Both genetic and biochemical studies support a spatially deterministic mode of substrate selection for ERAD in yeast, whereby Hrd1p ubiquitinates proteins misfolded in either their luminal (ERAD-L) or transmembrane domains (ERAD-M), and Doa10p targets those with domains misfolded in the cytosol (ERAD-C)26,27. Hrd1p and Doa10p are both conserved evolutionarily (Table 1), yet mammalian cells use an expanded collection of ERAD-related E3s to deal with folding crisis at the ER28. Interestingly, there has not been a commensurate diversification of E2s used, with only Ube2j1, Ube2j2 (refs. 29,30) and Ube2g2 (ref. 31) (orthologs of Ubc6p and Ubc7p, respectively) implicated in mammalian ERAD so far (Table 1). Presumably, the E3 capacity has expanded in response to the larger genomes, multicellular nature and longer lifespans of mammals, which collectively confer a higher risk of protein misfolding. With more E3s available, the compartment-based targeting model valid for yeast is not entirely adequate to explain substrate-E3 relationships in mammals. A misfolded luminal substrate whose degradation requires Hrd1 can be switched to depend on another E3, gp78, by simply tethering the substrate to the ER membrane32. Moreover, multiple E3s frequently contribute to the turnover of the same substrate (either in parallel or in tandem), thus reflecting either an opportunistic mode of E3 action afforded by substrate proximity or a complex hierarchical organization of prioritized substrate-E3 relationships33–35.
To build extended ubiquitin chains competent for recognition by the proteasome, substrates need a minimal ‘dwell time’ with their E3s so that multiple rounds of interactions with cognate E2s can be engaged. However, the structural diversity and sheer quantity of potential substrates that may require remediation (>7,000 unique proteins) means that many of the E3s involved in mammalian ERAD will need to be able to degrade a range of substrates in bulk without forming specific substrate-enzyme relationships. This implies that ERAD E3s use low-stringency selection criteria and may form only transient interactions with substrates. Several strategies have been evolved to maximize polyubiquitination efficiency in ERAD (Box 1).
Box 1. Different strategies of building ubiquitin chains for ER quality control.
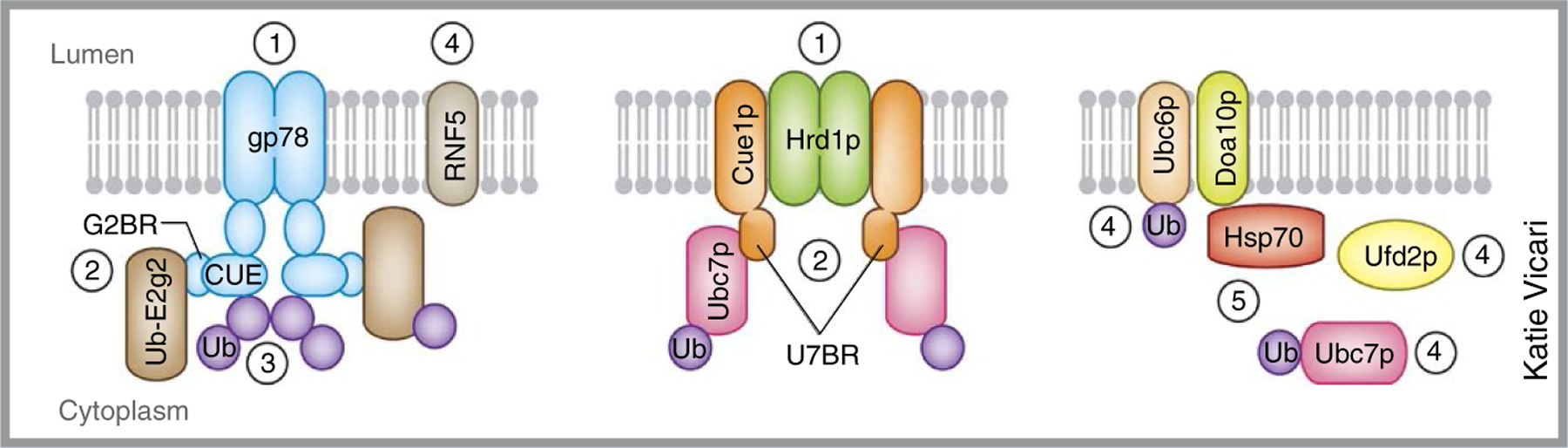
(1) Polytopic ERAD E3s can form functional homo-oligomers to enhance ubiquitination activity71,82. Oligomerization of gp78 increases polyubiquitination efficiency, probably by promoting chain preassembly on the active site of the E2 Ube2g2, which can be transferred en bloc to a substrate90. (2) Unlike many other E3s, some ERAD E3s recruit cognate E2s through high-affinity interactions. The yeast Hrd1p interactor Cue1p contains a high-affinity binding region for the E2 Ubc7p (U7BR) that is required for efficient ubiquitination and degradation of Hrd1 substrates121,122. Although there is no bona fide Cue1p homolog, two mammalian proteins, gp78 and AUP1, contain a similar Ube2g2-binding region (G2BR). Both U7BR and G2BR domains bind the ‘back side’ of the corresponding E2s, inducing allosteric changes in E2 that increase ubiquitin loading and transfer121,123,124. Stable E3-E2 interactions, when acting in the context of an E3 homo-oligomer, may enhance ubiquitination efficiency either by forming preassembled ubiquitin chains or by simultaneously positioning multiple E2s in proximity to substrates. (3) The G2BR also acts in conjunction with ubiquitin-binding domains (UBDs) to further improve chain-building efficiency. Both Cue1p and gp78 contain ubiquitin-binding CUE domains that promote chain elongation125–127. (4) Functional cooperation among ubiquitinating enzymes (E2s or E3s) or between E3s and molecular chaperones may be another mechanism that promotes ubiquitination. Whereas one E2 (for example, Ubc6p) may be dedicated solely to chain initiation, another (for example, Ubc7p) may serve to elongate those chains. Likewise, one E3 (for example, RNF5) can initiate ubiquitin-chain formation, whereas another E3 (for example, gp78) elongates the chains, functioning similarly to an E4 (refs. 128–130). (5) Doa10p also associates with the cytosolic chaperone Hsp70, which promotes polyubiquitination, probably by increasing the substrate dwell time on Doa10p130. Similar functional interplays between an E3 and a chaperone have also been demonstrated in mammalian ERAD131.
Deubiquitinases in ERAD.
Ubiquitin chain length, linkage and branching are highly dynamic and are governed by the opposing enzymatic actions of the ubiquitin-conjugating cascade and the families of deubiquitinases (DUBs). DUBs are isopeptidases that remove ubiquitin conjugates from substrates or disassemble ubiquitin chains36. Many DUBs implicated in ERAD, including YOD1 (also known as OTUD2)37, USP13 (ref. 38) and Ataxin-3 (ref. 39) (Table 1), bind the retrotranslocation-driving ATPase p97/VCP (Cdc48p in yeast) either directly or indirectly. A general role for DUBs within the ERAD mechanism is still uncertain, because they do not appear to be required for substrate processing in yeast. The lack of conserved involvement of DUBs within eukaryotic retrotranslocation mechanisms would argue against any fundamental roles for DUBs in retrotranslocation per se40. Instead, these enzymes are more likely to act indirectly to regulate the retrotranslocation process. In agreement with this notion, YOD1 (OTUD2) can negatively regulate retrotranslocation of a nonubiquitinated ERAD substrate41, thus suggesting that an ERAD-machinery component rather than an ERAD substrate may be regulated by DUBs, which in turn influences the retrotranslocation activity. More recently, another mammalian DUB, USP13, was found to interact directly with the E3 ligase gp78. This interaction is required to prevent gp78-mediated ubiquitination of a cytosolic chaperone in the Bag6 complex (discussed below), which, if not inhibited, could inactivate this ERAD-facilitating chaperone42. Thus, by suppressing spurious and counterproductive ubiquitin conjugation, the cooperation between USP13 and gp78 restricts gp78’s activity toward its intended targets to promote ERAD.
A DUB-E3 partnership could also improve the substrate selectivity for E3s in ER-protein quality control via another means. A major challenge of any protein quality-control system is to distinguish misfolded from folded polypeptides. Although gross changes can occur, quite often the structural differences between the two forms can be subtle. In an effort to reconstitute substrate discrimination by an SCF (Skp, cullin, F-box–containing) ubiquitin ligase complex in vitro, Hegde and colleagues used two closely related membrane-bound substrates, one targeted for degradation and the other exempted. They provide evidence that deubiquitination by cytosolic DUBs may amplify the difference in ubiquitination efficiency, thus allowing the misfolded form with slightly higher E3 affinity to be rapidly ubiquitinated and the other to be rescued by the DUBs43. In terms of energy consumption, this protein-turnover strategy appears costly, because many rounds of ubiquitination are wasted with repetitive addition and removal. However, because the end point of evolution is not to develop the most ‘economical’ biological process, what appears to be an energy-inefficient process can sometimes be justified if the trade-off yields an improvement to the overall fitness of an organism. One such example is the bulk degradation of many newly synthesized nascent polypeptides by the proteasome, a seemingly ‘wasteful’ process that actually provides an essential source of antigenic peptides required for host defense against invading pathogens44.
The ubiquitin-selective ATPase p97/VCP.
Once a portion of a luminal ERAD substrate that reaches the cytosol has been ubiquitinated, the polypeptide is dislocated from the lipid bilayer to complete retrotranslocation. The energy source driving the dislocation reaction appears to be provided by the highly conserved ATPase p97/VCP45–48 (Table 2). p97/VCP is a member of the large ATPase associated with diverse cellular activities (AAA+) family by virtue of the two AAA+ ATPase cassettes that it bears49. p97/VCP monomers oligomerize to form a homohexamer in which the ATPase domains are oriented as two stacked rings (D1 and D2) that form a centrally aligned pore, and the N-terminal domains flank the barrel formed by the D1 ring50,51. As one of the most abundant proteins in cells, p97/VCP is able to participate in a diverse range of cellular processes, including ERAD, along with a collection of cofactors recruited through p97/VCP-binding domains (for example, VIM, VBR and SHP)52. Many p97/VCP cofactors also contain ubiquitin-binding domains (UBDs) that bind directly to ubiquitinated substrates. For ERAD, ubiquitin binding through the dimeric Ufd1–Npl4 complex is responsible for recognition of substrates53,54. p97/VCP also associates with many E3s (for example, Hrd1 and gp78), ERAD accessory factors (for example, UbxD2, UbxD8 and VIMP) and DUBs (for example, Ataxin-3)49,55, thus establishing a platform for these factors to constitutively modulate ubiquitination at sites of retrotranslocation.
Links between p97/VCP and the ubiquitin system extend far beyond the ERAD pathway. In almost every process involving p97/VCP, the substrates engaged are either mono- or polyubiquitinated. p97/VCP appears to serve as a ubiquitin-selective chaperone that uses the energy from ATP hydrolysis to segregate its substrate proteins from large immobile structures or complexes once the substrates are properly marked by ubiquitin. This idea is supported by several in vitro studies demonstrating that purified p97/VCP together with appropriate cofactors can extract ubiquitinated proteins from either the ER membrane or chromatin DNA56–58. The mechanism underlying the ‘segregase’ activity of p97/VCP is unclear. Structural analogy between p97/VCP and other AAA+ ATPase members that function as protein ‘translocases’ encourages a model in which p97/VCP threads substrates through its central pore, but compelling evidence has yet to be presented to support such a mechanism. Although several conserved hydrophobic residues at the center of the D2 ring are crucial for substrate binding as well as for the function of p97/VCP in ERAD59, structural analyses determined that a zinc atom occludes the pore in p97/VCP at the D1 level50. This obstruction within the pore, together with the fact that ERAD substrates often contain partially folded elements and bulky N-linked oligosaccharides, makes it unlikely that substrates are able to pass entirely through a p97/VCP central cavity that narrows at the D1 ring. How p97/VCP efficiently engages and processes its substrates remains an outstanding question.
Ubiquitin receptors and UBL-bearing proteins.
To facilitate the coupling of retrotranslocation to degradation, ubiquitin chains conjugated to misfolded ER proteins must be recognized by cytosolic ‘ubiquitin receptors’—molecules that bind these adducts through an assortment of UBDs (for example, UBA, UIM, NZF and CUE)60. UBD-containing proteins appear to play a major part in shuttling substrates from sites of retrotranslocation at the ER membrane to the proteasome while protecting substrate ubiquitin chains from promiscuous DUB activity and/or promoting chain elongation. UBDs also facilitate the formation of protein networks by recognizing the UBL domains found in selected ERAD factors. For instance, the proteasome-associated, UBD-UBL–containing proteins hHR23A, hHR23B and ubiquilins have been proposed to function as a ‘substrate collector’ for the proteasome in both ERAD and other degradation pathways61–65 (Table 2). The UBL domains in these factors interact with the proteasome through a ubiquitin-interacting motif (UIM) in the S5a subunit of the proteasome66. Other UBL-containing proteins involved in ERAD include Herp67,68, TMUB1 (ref. 69) and two components of the recently identified cytosolic chaperone holdase complex, Bag6 and Ubl4A70 (Table 2). Herp and its yeast ortholog Usa1p appear to serve a crucial scaffolding function that assembles a functional retrotranslocation complex71,72. The UBL domain of Bag6 binds both the CUE domain of gp78 and the UBA domain of UbxD8, and these interactions position Bag6 at the sites of retrotranslocation marked by gp78, where it prevents protein aggregation to promote ERAD73. The UBL domain in the Bag6 cofactor Ubl4A also recruits small glutamine-rich tetratricopeptide repeat-containing protein-α (SGTA), which may serve as a cochaperone to assist Bag6 in ERAD74. Although UBLs share a similar protein fold with ubiquitin, the amino acid composition of the UBD binding surface on UBLs can vary substantially, thus ensuring specific recognition by selected UBDs74.
Roles of ubiquitin in retrotranslocation
Because polyubiquitination occurs when substrates reach the ER’s cytosolic face and before release into the cytosol, it is generally believed that ubiquitin conjugation to substrates is what drives the retrotranslocation process75. Thus, substrate topology and relative cytosolic exposure become important determinants of how and when ubiquitination occurs and, consequently, how retrotranslocation proceeds.
Modes of retrotranslocation.
The current view posits that ER luminal proteins are retrotranslocated first by movement of a segment (‘handle’) across the ER membrane via a protein-conducting channel. This reaction converts untethered substrates to a virtual membrane-bound state, allowing them to be accessed by the cytosolic retrotranslocation machinery that will pull them through the membrane (Fig. 1). For substrates integrated in the ER membrane, retrotranslocation may be initiated by application of force to one or more domains already exposed to the cytosol, as proposed for the ERAD-M substrates, or to a luminal domain that had been moved across the membrane, as suggested for a type I membrane protein76,77. The latter mechanism is conceptually identical to retrotranslocation of an ERAD-L substrate. Thus, integral membrane ERAD substrates can be processed via either ERAD-M or ERAD-L mechanisms, at least in mammals (Fig. 1).
Figure 1.
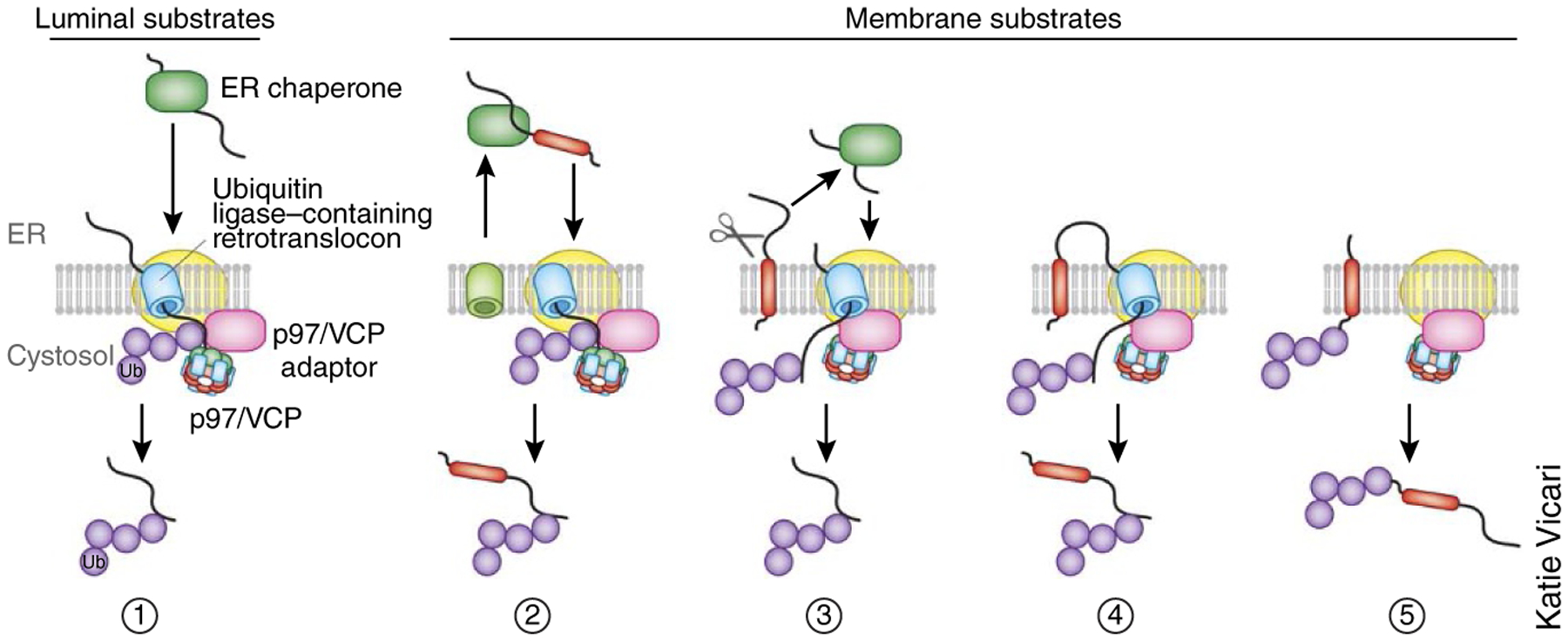
Distinct modes of retrotranslocation. Retrotranslocation can occur in principle via the following modes. (1) A luminal substrate must first be inserted into the ER membrane in a process that is probably facilitated by a membrane-bound retrotranslocon component. Once a portion of the substrate has been exposed to the cytosol, the ubiquitination machinery can access it to facilitate its ubiquitination and subsequent dislocation from the membrane. Retrotranslocation of membrane proteins may proceed via several routes. Ub, ubiquitin. (2) Type I membrane proteins containing charged residues in their TM domain might be completely translocated into the ER lumen and then retrotranslocated similarly to a luminal substrate. (3) Some membrane proteins may first be processed by an intramembranous protease to release its luminal domain, which is then retrotranslocated. (4) Some membrane proteins may translocate their ER-luminal domain out of the ER first via a proteinaceous channel, which is then ubiquitinated and pulled out of the membrane. The TM domain may enter the channel via a lateral gate before being dislocated from the membrane. (5) Some membrane proteins may be ubiquitinated in their cytosolic domain and then pulled into a proteinaceous channel before release from the ER membrane.
Direct extraction of hydrophobic transmembrane (TM) segments from lipid bilayers presents a substantial energetic barrier in any ERAD mechanism, yet ubiquitinated integral membrane substrates can be fully dislocated into the cytosol by the p97/VCP complex before their degradation57. Some type I membrane proteins appear to have solved this conundrum by first being processed by the ubiquitin-selective intramembranous rhomboid protease RHBDL4, whose cleavage effectively converts the residual luminal domain into an ERAD-L substrate78. The membrane substrates favored by RHBDL4 all contain one or more charged residues in their TM domains that are thought to be the ‘signal’ for engaging RHBDL4 (ref. 78). However, charged residues may also cause a type I membrane protein to fully translocate into the ER lumen before retrotranslocation is initiated, as demonstrated recently for TCRα79. Conceptually, a more general way to overcome the energetic barrier is to use a laterally gated protein channel from which TM domains are released into the cytosol one by one, as proposed previously80–82. In an entirely different strategy, ERAD machinery and substrates may preferentially localize to certain ER domains with lipid compositions that render membrane structures unstable, as hinted at by the observation that some ERAD factors (for example, AUP1 and UbxD8) are stably present in both ER and in lipid droplets83–85. The latter are lipid monolayer–encircled organelles that may originate from specialized lipid domains in the ER. Retrotranslocation may be favorable in these domains, dubbed ‘escape hatches’, because the unstable lipid structures can lower the energy barrier for dislodging TM domains of substrates from the membrane86. That said, no evidence suggests that lipid-droplet formation is required for ERAD of either luminal or membrane substrates, at least in yeast87. Although each of the aforementioned models offers attractive mechanisms, it is important to recognize that they are not necessarily mutually exclusive.
Ubiquitination site selection and chain linkage in retrotranslocation.
Given that the substrate-E3 relationships may be quite promiscuous in ERAD, would E3s still preferentially ubiquitinate specific residues within ERAD substrates? Retrotranslocation substrates are likely to emerge from the ER membrane in an unfolded or partially unfolded state. This lack of structure, together with the diversity of substrates, argues implicitly for ERAD E3s to have less stringent selection criteria than other E3s. With the sites of retrotranslocation effectively ‘guarded’ by ubiquitin ligases, it is conceivable that stochastic encounters with available acceptor residues in an ERAD substrate may decorate it with heterogeneous ubiquitination patterns on multiple residues. This ‘bulk ubiquitination’ mechanism stands in sharp contrast to the specific enzyme-client relationship maintained by some E3s that act on folded proteins, and it may be more akin to cotranslational ubiquitination of defective nascent polypeptides emerging from ribosomes88.
Regardless of which residues are modified, it is Lys48-linked chains that appear to be essential for retrotranslocation and degradation of most ERAD substrates53,89. This is consistent with in vitro studies demonstrating that a key ubiquitin conjugating enzyme Ube2g2 (Ubc7p in yeast) assembles Lys48-linked ubiquitin chains that are preferentially recognized by the UBDs in the p97/VCP complex90. Lys11-linked ubiquitin chains can also be assembled by Ubc6p and Doa10p in yeast91. Notably, the Ubc6p ortholog Ube2j1 is essential for degradation of multiple Hrd1-dependent substrates, but the chain linkages on these substrates have yet to be determined33.
Roles of ubiquitin ligases in retrotranslocation.
The identity of the putative protein channel responsible for retrotranslocating ERAD-L and ERAD-M substrates has yet to be unequivocally established. Several candidates for this proteinaceous channel or pore have been proposed, including the Sec61 complex, members of the Derlin family (or Der1p in yeast) and polytopic E3s such as Hrd1 and gp78 (ref. 92).
A role for the Sec61 complex in retrotranslocation was initially proposed on the basis of reported interactions of the translocon with some ERAD substrates93,94 and of genetic evidence that certain Sec61α-mutant yeast strains were defective in retrotranslocation95,96. However, structural studies of archaeal SecYEG, the Sec61 homolog, revealed a narrow tunnel97 that is unlikely to accommodate most ERAD substrates carrying bulky N-linked oligosaccharides or partially folded domains.
The Derlins (Der1p family) contain several polytopic membrane proteins (with either four or six TM segments) that share both structural homology and conserved domains with the family of Rhomboid pseudoproteases98. Functionally, Derlin-1 or Derlin-2 is required for retrotranslocation of several luminal ERAD substrates in mammals98,99, and in yeast, Der1p is essential for degradation of ERAD-L but not ERAD-M substrates27. Importantly, retrotranslocation of a nonubiquitinated ERAD substrate in an in vitro system requires Derlin-1 but not Sec61α100. Derlin-1 and Derlin-2, as well as Der1p, associate closely with ubiquitin ligases such as Hrd1 (refs. 101–104), gp78 (ref. 33) and RNF5 (ref. 34) as well as other ERAD factors, to form large complexes spanning the ER membrane33,102,105. These complexes may be the basis of retrotranslocation channels, or they may serve to couple the two spatially segregated ERAD events—namely the insertion of substrates into the membrane from the ER lumen and their subsequent ubiquitination upon emerging into the cytosol102,106.
The Hrd1 ubiquitin ligase has also surfaced as an attractive candidate for a retrotranslocation channel, at least for a subset of ERAD substrates. Initially identified in a genetic screen for yeast mutants defective in regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoAR)10,23,80, this multispanning ER-localized E3 is a key ERAD component in all eukaryotes, in which it is required for degradation of both ERAD-M and ERAD-L substrates26,27,33. For degradation of ERAD-M substrates, however, the Hrd1p cofactors Der1p and Usa1p are dispensable, and the role of another Hrd1p adaptor, Hrd3p, appears to be limited to stabilizing Hrd1p107. These observations led to the speculation that Hrd1p may function as the core component of a retrotranslocon, with its accessory factors scaffolding the channel complex, recruiting substrates and gating the entry of substrates into the Hrd1p-containing retrotranslocon82. Several lines of evidence support this model. First, degradation of ERAD-M and ERAD-L substrates can be further segregated in yeast by mutation to selected Hrd1p TM domains, which impair recognition and ubiquitination of only the ERAD-M substrate Hmg2p81. Next, the ERAD-L substrate misfolded carboxypeptidase Y (denoted CPY*) undergoing retrotranslocation could be cross-linked to Hrd1p TM domains in a process dependent on its ubiquitin-ligase activity and the ATPase activity of Cdc48p82. This suggests that ubiquitination-dependent Cdc48p activity serves as a cytosolic ‘ratchet’ stabilizing otherwise-transient interactions between the substrate and Hrd1p. However, the result does not distinguish whether the interaction occurs during substrate recognition or retrotranslocation. It is also noteworthy that an engineered ERAD substrate carrying the TM region of Hmg1p fused to the RING domain of Hrd1p is dislocated from the membrane and degraded even in the absence of Hrd1p, thus hinting at the existence of at least one Hrd1-independent route for retrotranslocation57.
To supplement Hrd1p activity, Saccharomyces cerevisiae also uses Doa10p, a second multispanning E3 that promotes degradation of ERAD-C substrates24,108. Like Hrd1p, Doa10p forms complexes within the lipid bilayer but shares only a few components with the Hrd1p complex, including Ubx2p (a ubiquitin regulatory X-domain-containing factor that recruits Cdc48p) and the E2 Ubc7p27. The mechanism underlying retrotranslocation initiation for ERAD-C substrates may be distinct from that of the Hrd1-dependent pathways because ERAD-C substrate recognition occurs in the cytosol. In this regard, the Doa10p-dependent pathway may be primarily responsible for degrading integral membrane proteins that are damaged in cytosol-exposed segments after these substrates have left the biosynthetic pathway. Indeed, manipulation of a Doa10p substrate to allow aberrant engagement of the Sec61 translocon may transiently displace a Doa10p ‘degron’ from the cytosol to the ER lumen, thus permitting it to engage Hrd1p for degradation109. The ERAD-M and ERAD-C pathways do not require substrate-recruiting factors in the ER lumen, thus suggesting that the ancient membrane quality-control pathways may implicate only multispanning E3 ubiquitin ligases, the Cdc48p complex and the proteasome, as exemplified in a recently discovered mitochondrial quality-control pathway110. Other elements (or modules) involved in degrading ERAD-L substrates may have been added to the ERAD-M machinery during evolution, to result in the expansion of substrate repertoire. The development of the ERAD-L mechanism also creates a new strategy for protein translocation across the lipid bilayer; this strategy appears to have been adopted by certain cells containing red alga–derived endosymbionts and by peroxisomes to move polypeptides between subcellular compartments111 (Box 2).
Box 2. A comparison of the ERAD-L mechanism with the peroxisome import pathway and the SELMA system.
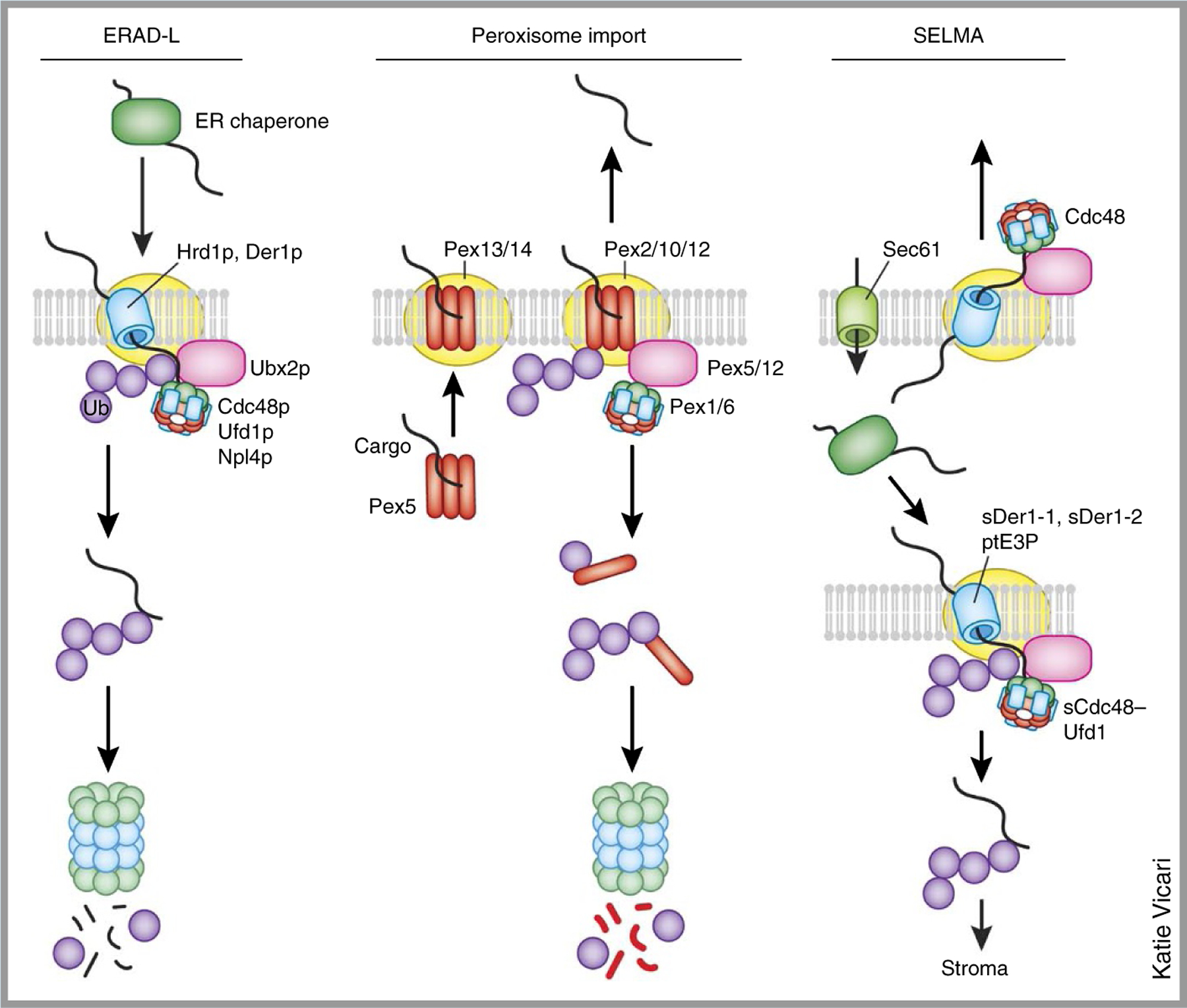
The ERAD-L mechanism shares a striking resemblance to a peroxisome protein–import pathway and the symbiont-specific ERAD-like machinery (SELMA). In yeast, ERAD-L requires a membrane complex comprising the substrate adaptor protein Hrd3p, Der1p and the ubiquitin ligase Hrd1p. The substrates are moved across the ER membrane via this large membrane complex. On the cytosolic face, an adaptor, Ubx2p, recruits the Cdc48p–Ufd1p–Npl4p complex to the ER membrane, which extracts the substrate from the membrane. The import of peroxisome proteins bearing a C-terminal peroxisomal targeting signal 1 (PTS1) begins with the recognition of cargo in the cytosol by a receptor protein, Pex5. Like Hrd3p, Pex5 uses several tetratricopeptide repeats (TPRs) to bind substrates. The Pex5–cargo complex is recruited to the membrane by a docking complex (Pex13/14) that also contains several RING-finger E3 ligases, Pex2, Pex10 and Pex12 (Pex2/10/12). Pex5 is believed to form a pore after integration into the membrane, and the release of substrate cargos into the peroxisome lumen is coupled to the ubiquitination and subsequent extraction of Pex5 from the membrane. Cytosolic AAA+ ATPase Pex1 and Pex6 (Pex1/6) mediate the dislocation of Pex5 from the membrane via a process analogous to the extraction of ubiquitinated ERAD substrates. The SELMA is a protein-translocation system located in the second-outermost membrane of certain plastids found in some alga and human parasites. Proteins in the stroma of these plastids need to traverse four layers of membrane. After Sec61-dependent transport into the ER lumen, they use the SELMA to reach the periplastidal compartment (PPC) for further translocation into the stroma. The system appears to be adapted from the ERAD-L machinery, because it uses two Der1-like proteins (sDer1–1 and sDer1–2), a membrane-bound E3 ligase (ptE3P) and a Cdc48-like complex (sCdc48–Ufd1) to move polypeptides across this membrane.
A modular organization of the ERAD machinery.
Breaking ERAD down into functionally distinct modules that sequentially process substrates has been a convenient strategy for understanding how individual factors contribute to the overall mechanism. These modules, spanning three cellular environments, execute the following functions in ERAD (Fig. 2): (i) recognition, (ii) retrotranslocation initiation, (iii) ubiquitination, (iv) extraction, dislocation and retrotranslocation, (v) delivery and (vi) degradation. Despite the dramatic expansion in ERAD-machinery components, the modular organization of the ERAD machinery is conserved from yeast to humans. The components engaged in ERAD fall into to one or more of these categories, with some components acting at the interfaces to bring these functional modules together either transiently or stably in order to orchestrate the concerted ERAD function33. The notion of modularity conjures images of a segmented series of events, with a substrate passed from one module to the next as it moves toward the proteasome. However, the degrading polypeptides may also engage different ERAD modules as though they were beads on a string.
Figure 2.
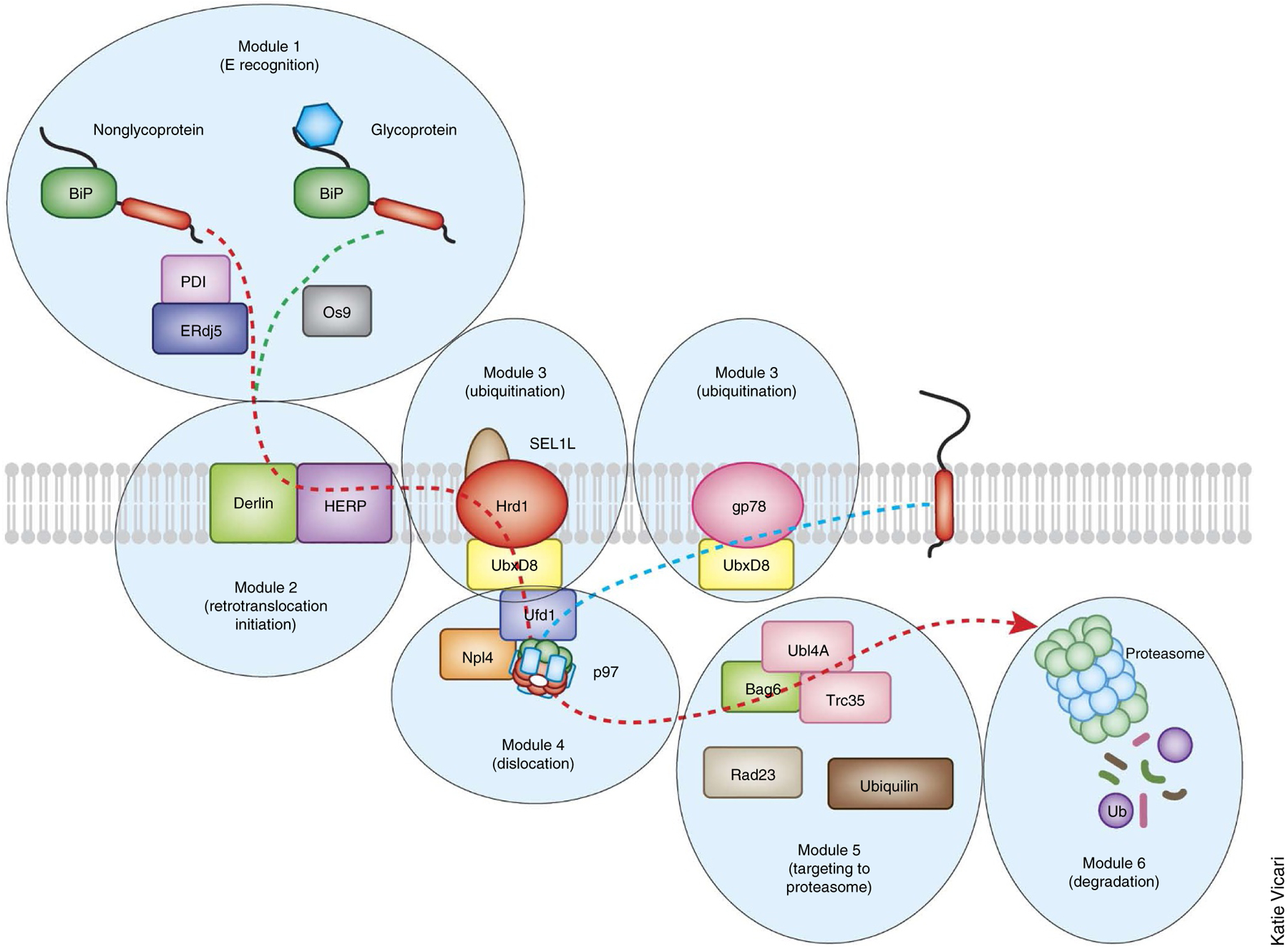
Modular organization of the mammalian ERAD system. A large number of ERAD factors are organized into several modules to mediate retrotranslocation. In general, proteins in the same module tend to form stable interactions, whereas factors from different modules bind each other in a dynamic manner, although there are exceptions. For instance, a large, stable membrane complex consisting of an E3 ligase and other factors is likely to mediate the retrotranslocation initiation and the ubiquitination modules. Within a given module, factors can also form parallel pathways to transport distinct classes of the substrates. For an ERAD-L mechanism, the module for substrate recognition and recruitment distinguishes misfolded proteins from folded ones in the lumen and then selectively targets them for insertion into the ER membranes. Subsequent ubiquitin conjugation is mediated by ubiquitination modules. This is followed by extraction from the ER membrane via the AAA+ ATPase p97/VCP complex. Dislocated products are shuttled to the proteasome, which may have been brought near to the retrotranslocation site through interaction with the ER membranes. The yeast ERAD system has a similar modular organization with fewer participating components. The retrotranslocation and degradation of ERAD-M and ERAD-C substrates do not seem to require the recognition and initiation modules in the ER. The dashed lines indicate the flow of the substrates.
Roles of ubiquitin in targeting substrates to the proteasome
Once retrotranslocated, substrates need to be rapidly degraded to prevent any exposed hydrophobic patches and/or mislocalized TM domains from aggregating in the cytosol. Because inhibition of proteasome activity leads to stabilization of most ERAD substrates in the ER lumen, it is presumed that retrotranslocation and degradation are tightly coupled. A recent study on the archaeal Cdc48p homolog and the 20S proteasome suggested that direct interactions between Cdc48p and the 20S proteasome can align the central axis of these two ring-like complexes, to form a continuous conduit through which substrates can be directly transferred from Cdc48p to the 20S proteasome112. Such interactions, if they exist in eukaryotes, would provide a simple coupling mechanism that links retrotranslocation to degradation. However, this model is hard to reconcile with biochemical and genetic data showing that p97/VCP does not form stable interactions with 20S proteasomes in eukaryotic cells113,114 and that ERAD requires both the 20S and the 19S proteasome9,33,47. Moreover, as mentioned above, structural studies of p97/VCP also fail to support the processing model based on substrate threading.
A more plausible mechanism is the ‘handoff’ model, in which a handful of p97/VCP and proteasome-associated factors shuttle substrates from p97/VCP to the proteasome. In addition to the aforementioned UBA-UBL proteins and the E4 Ufd2p, a special chaperone system comprising the Bag6–Ubl4A–Trc35 complex and the cochaperone SGTA was recently implicated in this process115. Bag6 is an abundant cytosolic chaperone that forms a large homo-oligomer via a large proline-rich segment. Although predicted to be unstructured, the proline-rich domain is sufficient to bind substrates bearing long hydrophobic segments and to maintain them in an unfolded yet soluble state73. In cells, this chaperone holdase activity is required to keep some retrotranslocated substrates in a soluble state competent for proteasome degradation70. The Bag6 adaptor Ubl4A uses a noncanonical UBL domain to bind a new UBD in SGTA in ERAD74. Bag6 also interacts with the proteasome and the proteasome adaptor proteins such as ubiquilin74,116. These interactions may be coupled to the transfer of substrates to the proteasome for degradation. The budding yeast does not contain a Bag6 homolog. A homologous system consisting of Sgt2p, Get5p and Get4p (homologs of SGTA, Ubl4A and Trc35, respectively) exists in yeast, but it is involved mainly in targeting tail-anchored proteins to the ER membrane115. However, it has been reported that a group of small cytosolic heat-shock proteins may serve an analogous chaperone function to channel retrotranslocated ERAD substrates for proteasome degradation117.
Conclusion and perspective
Efforts over the past two decades have led to the identification of most (if not all) components involved in ERAD of both yeast and mammals, and recent work has shed light on their organization into functional modules underlying the molecular mechanisms responsible for each molecular event of ERAD. Despite these advances, many fundamental questions about ERAD remain elusive. The mechanism for moving misfolded proteins across the ER membrane is still poorly characterized, thus leaving the identity of the retrotranslocon(s) as a point of conjecture and the potential modulation of the retrotranslocation machinery by ubiquitination and deubiquitination an open question. How the AAA+ ATPase p97/VCP physically extracts and transfers ubiquitinated polypeptides to the proteasome is also unclear. The answers to these questions will probably require establishment of in vitro biochemical systems capable of recapitulating the individual steps of ERAD. The in vitro ERAD assays available currently have relatively low efficiency, which has limited their ability to provide mechanistic understanding. Several recent studies have reported cell-based ERAD assays with higher sensitivity118–120, and these may be adaptable to improve the efficiency of in vitro assays. Also missing from the toolbox is a robust set of ERAD substrates suitable for developing sensitive in vitro assays. Identification of new substrates, particularly endogenous ones, is crucial to understanding E3 substrate recognition, and it may not only reveal further biological relevance of ERAD but also arm researchers with more powerful ‘weaponry’ to tackle the roles of ubiquitin in this important—but still rather mysterious—biological process.
ACKNOWLEDGMENTS
We thank J. Schulz, E. Fenech, K. Bryon-Dodd (University of Oxford) and J. Olzmann (University of California, Berkeley) for critical reading of the manuscript. Research in the laboratory of Y.Y. is supported by the intramural research program of the NIDDK, US National Institutes of Health. J.C.C. is supported by funding from the Ludwig Institute for Cancer Research and the UK Medical Research Council.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Brodsky JL & Wojcikiewicz RJ Substrate-specific mediators of ER associated degradation (ERAD). Curr. Opin. Cell Biol 21, 516–521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampton RY ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol 14, 476–482 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Ron D & Walter P Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol 8, 519–529 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Needham PG & Brodsky JL How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim. Biophys. Acta 1833, 2447–2457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippincott-Schwartz J, Bonifacino JS, Yuan LC & Klausner RD Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell 54, 209–220 (1988). [DOI] [PubMed] [Google Scholar]; Refs. 5 and 6 were the first to demonstrate that unassembled endogenous membrane proteins are rapidly degraded after import into the ER, thus defining a new pathway of protein degradation at the ER.
- 6.Bonifacino JS, Cosson P & Klausner RD Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63, 503–513 (1990). [DOI] [PubMed] [Google Scholar]
- 7.Ward CL, Omura S & Kopito RR Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 (1995). [DOI] [PubMed] [Google Scholar]; Refs. 7–12 demonstrated that ERAD requires the ubiquitin-proteasome system, thus suggesting that the substrates need to be retrotranslocated into the cytosol before degradation.
- 8.Sommer T & Jentsch S A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365, 176–179 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Hiller MM, Finger A, Schweiger M & Wolf DH ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Hampton RY, Gardner RG & Rine J Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7, 2029–2044 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen TJ et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129–135 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Wiertz EJ et al. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Tsai B, Ye Y & Rapoport TA Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol 3, 246–255 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Shimizu Y, Okuda-Shimizu Y & Hendershot LM Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40, 917–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol 177, 613–624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikura S, Weissman AM & Bonifacino JS Serine residues in the cytosolic tail of the T-cell antigen receptor α-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J. Biol. Chem 285, 23916–23924 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickart CM & Fushman D Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol 8, 610–616 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Li W & Ye Y Polyubiquitin chains: functions, structures, and mechanisms. Cell. Mol. Life Sci 65, 2397–2406 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Veen AG & Ploegh HL Ubiquitin-like proteins. Annu. Rev. Biochem 81, 323–357 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Ahner A et al. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier–dependent pathway. Mol. Biol. Cell 24, 74–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickart CM Mechanisms underlying ubiquitination. Annu. Rev. Biochem 70, 503–533 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Bays NW, Gardner RG, Seelig LP, Joazeiro CA & Hampton RY Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol 3, 24–29 (2001). [DOI] [PubMed] [Google Scholar]; Refs. 23 and 24 identified the two major ubiquitin ligases involved in ERAD in budding yeast.
- 24.Swanson R, Locher M & Hochstrasser M A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15, 2660–2674 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolz A, Besser S, Hottmann H & Wolf DH Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum–associated protein degradation. Proc. Natl. Acad. Sci. USA 110, 15271–15276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vashist S & Ng DT Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol 165, 41–52 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. 26 and 27 elucidated different routes by which distinct classes of misfolded ER proteins are retrotranslocated for degradation.
- 27.Carvalho P, Goder V & Rapoport TA Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Mehnert M, Sommer T & Jarosch E ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays 32, 905–913 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Mueller B, Klemm EJ, Spooner E, Claessen JH & Ploegh HL SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA 105, 12325–12330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X et al. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol 187, 655–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Du S & Fang S gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum. Curr. Protein Pept. Sci 13, 414–424 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Bernasconi R, Galli C, Calanca V, Nakajima T & Molinari M Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol 188, 223–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christianson JC et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol 14, 93–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper used a systems-biology approach to construct a complex functional-interaction map of the proteins involved in mammalian ERAD.
- 34.Younger JM et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126, 571–582 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Morito D et al. Gp78 cooperates with RMA1 in endoplasmic reticulum–associated degradation of CFTRDeltaF508. Mol. Biol. Cell 19, 1328–1336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komander D, Clague MJ & Urbe S Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol 10, 550–563 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Ernst R, Mueller B, Ploegh HL & Schlieker C The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 36, 28–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sowa ME, Bennett EJ, Gygi SP & Harper JW Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Li L & Ye Y Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol 174, 963–971 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ernst R et al. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 8, e1000605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernardi KM, Williams JM, Inoue T, Schultz A & Tsai B A deubiquitinase negatively regulates retro-translocation of non-ubiquitinated substrates. Mol. Biol. Cell 24, 3545–3556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y et al. USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. Elife 3, e01369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported that a machinery protein in ERAD could be subject to regulation by ubiquitination in a proteasome-independent manner.
- 43.Zhang ZR, Bonifacino JS & Hegde RS Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell 154, 609–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert U et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Ye Y, Meyer HH & Rapoport TA The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 (2001). [DOI] [PubMed] [Google Scholar]; Refs. 45, 46–48 and 54 demonstrated the involvement of the AAA+ ATPase p97/VCP in ERAD. Ref. 45 also used an in vitro retrotranslocation assay to show that p97/VCP is required to move ubiquitinated ERAD substrates from the membranes to the cytosol for degradation by the proteasome.
- 46.Rabinovich E, Kerem A, Frohlich KU, Diamant N & Bar-Nun S AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum–associated protein degradation. Mol. Cell. Biol 22, 626–634 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarosch E et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol 4, 134–139 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K & Hampton RY HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114–4128 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye Y Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J. Struct. Biol 156, 29–40 (2006). [DOI] [PubMed] [Google Scholar]
- 50.DeLaBarre B & Brunger AT Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol 10, 856–863 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Zhang X et al. Structure of the AAA ATPase p97. Mol. Cell 6, 1473–1484 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Meyer H, Bug M & Bremer S Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Flierman D, Ye Y, Dai M, Chau V & Rapoport TA Polyubiquitin serves as a recognition signal, rather than a ratcheting molecule, during retrotranslocation of proteins across the endoplasmic reticulum membrane. J. Biol. Chem 278, 34774–34782 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Ye Y, Meyer HH & Rapoport TA Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol 162, 71–84 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y & Ye Y Roles of p97-associated deubiquitinases in protein quality control at the endoplasmic reticulum. Curr. Protein Pept. Sci 13, 436–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rape M et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Garza RM, Sato BK & Hampton RY In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem 284, 14710–14722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raman M, Havens CG, Walter JC & Harper JW A genome-wide screen identifies p97 as an essential regulator of DNA damage–dependent CDT1 destruction. Mol. Cell 44, 72–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLaBarre B, Christianson JC, Kopito RR & Brunger AT Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell 22, 451–462 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Husnjak K & Dikic I Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem 81, 291–322 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Richly H et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 (2005). [DOI] [PubMed] [Google Scholar]; Refs. 61, 62 and 64 defined the role of a class of UBA- and UBL-containing proteins in targeting retrotranslocated products to the proteasome for degradation.
- 62.Verma R, Oania R, Graumann J & Deshaies RJ Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Kim I, Mi K & Rao H Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell 15, 3357–3365 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim I et al. The Png1-Rad23 complex regulates glycoprotein turnover. J. Cell Biol 172, 211–219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim PJ et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol 187, 201–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hiyama H et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem 274, 28019–28025 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Okuda-Shimizu Y & Hendershot LM Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell 28, 544–554 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulze A et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol 354, 1021–1027 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Jo Y, Sguigna PV & DeBose-Boyd RA Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem 286, 15022–15031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble States for proteasome degradation. Mol. Cell 42, 758–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated the involvement of a multifunctional cytosolic chaperone in maintaining the solubility of retrotranslocated polypeptides, which promotes their turnover in mammalian cells.
- 71.Horn SC et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 36, 782–793 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Huang CH, Chu YR, Ye Y & Chen X Role of HERP and a HERP-related protein in HRD1-dependent protein degradation at the endoplasmic reticulum. J. Biol. Chem 289, 4444–4454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Liu Y, Lee JG & Ye Y A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation. J. Biol. Chem 288, 18068–18076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Y, Cai M, Yang Y, Huang L & Ye Y SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2, 1633–1644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vembar SS & Brodsky JL One step at a time: endoplasmic reticulum–associated degradation. Nat. Rev. Mol. Cell Biol 9, 944–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shamu CE, Story CM, Rapoport TA & Ploegh HL The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol 147, 45–58 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burr ML et al. MHC class I molecules are preferentially ubiquitinated on endoplasmic reticulum luminal residues during HRD1 ubiquitin E3 ligase-mediated dislocation. Proc. Natl. Acad. Sci. USA 110, 14290–14295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fleig L et al. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol. Cell 47, 558–569 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Feige MJ & Hendershot LM Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol. Cell 51, 297–309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bordallo J, Plemper RK, Finger A & Wolf DH Der3p/Hrd1p is required for endoplasmic reticulum–associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9, 209–222 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sato BK, Schulz D, Do PH & Hampton RY Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol. Cell 34, 212–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carvalho P, Stanley AM & Rapoport TA Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579–591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. 82 and 106 used an elegant in vivo cross-linking approach to characterize the interactions between a retrotranslocation substrate and the Hrd1p ubiquitin-ligase complex. The evidence suggests that Hrd1p and Der1p play essential parts in substrate recognition and/or retrotranslocation.
- 83.Klemm EJ, Spooner E & Ploegh HL Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J. Biol. Chem 286, 37602–37614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jo Y, Hartman IZ & DeBose-Boyd RA Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 24, 169–183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olzmann JA, Richter CM & Kopito RR Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. USA 110, 1345–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ploegh HL A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448, 435–438 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Olzmann JA & Kopito RR Lipid droplet formation is dispensable for endoplasmic reticulum–associated degradation. J. Biol. Chem 286, 27872–27874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duttler S, Pechmann S & Frydman J Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sliter DA, Aguiar M, Gygi SP & Wojcikiewicz RJ Activated inositol 1,4,5-trisphosphate receptors are modified by homogeneous Lys-48- and Lys-63-linked ubiquitin chains, but only Lys-48-linked chains are required for degradation. J. Biol. Chem 286, 1074–1082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li W, Tu D, Brunger AT & Ye Y A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333–337 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Xu P et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hampton RY & Sommer T Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol 24, 460–466 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Wiertz EJ et al. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438 (1996). [DOI] [PubMed] [Google Scholar]
- 94.Scott DC & Schekman R Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J. Cell Biol 181, 1095–1105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pilon M, Schekman R & Romisch K Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16, 4540–4548 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou M & Schekman R The engagement of Sec61p in the ER dislocation process. Mol. Cell 4, 925–934 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Van den Berg B et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 98.Greenblatt EJ, Olzmann JA & Kopito RR Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α−1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol 18, 1147–1152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang CH, Hsiao HT, Chu YR, Ye Y & Chen X Derlin2 protein facilitates HRD1-mediated retro-translocation of sonic hedgehog at the endoplasmic reticulum. J. Biol. Chem 288, 25330–25339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wahlman J et al. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129, 943–955 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Knop M, Finger A, Braun T, Hellmuth K & Wolf DH Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15, 753–763 (1996). [PMC free article] [PubMed] [Google Scholar]; Refs. 101, 102 and 105 reported a family of conserved multispanning membrane proteins essential for retrotranslocation of a subset of ERAD substrates.
- 102.Ye Y, Shibata Y, Yun C, Ron D & Rapoport TA A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 (2004). [DOI] [PubMed] [Google Scholar]
- 103.Kothe M et al. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J. Biol. Chem 280, 28127–28132 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Gauss R, Sommer T & Jarosch E The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25, 1827–1835 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lilley BN & Ploegh HL A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 (2004). [DOI] [PubMed] [Google Scholar]
- 106.Mehnert M, Sommer T & Jarosch E Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat. Cell Biol 16, 77–86 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Gardner RG et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol 151, 69–82 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS & Carvalho P Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife 2, e00953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rubenstein EM, Kreft SG, Greenblatt W, Swanson R & Hochstrasser M Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol 197, 761–773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu S, Peng G, Wang Y, Fang S & Karbowski M The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell 22, 291–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bolte K et al. Making new out of old: recycling and modification of an ancient protein translocation system during eukaryotic evolution. Mechanistic comparison and phylogenetic analysis of ERAD, SELMA and the peroxisomal importomer. Bioessays 33, 368–376 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Barthelme D & Sauer RT Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine. Science 337, 843–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Isakov E & Stanhill A Stalled proteasomes are directly relieved by P97 recruitment. J. Biol. Chem 286, 30274–30283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Verma R et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee JG & Ye Y Bag6/Bat3/Scythe: a novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. Bioessays 35, 377–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Minami R et al. BAG-6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol 190, 637–650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ahner A, Nakatsukasa K, Zhang H, Frizzell RA & Brodsky JL Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation. Mol. Biol. Cell 18, 806–814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grotzke JE, Lu Q & Cresswell P Deglycosylation-dependent fluorescent proteins provide unique tools for the study of ER-associated degradation. Proc. Natl. Acad. Sci. USA 110, 3393–3398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X, Yu YY, Myers N & Hansen TH Decoupling the role of ubiquitination for the dislocation versus degradation of major histocompatibility complex (MHC) class I proteins during endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem 288, 23295–23306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhong Y & Fang S Live cell imaging of protein dislocation from the endoplasmic reticulum. J. Biol. Chem 287, 28057–28066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Metzger MB et al. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol. Cell 50, 516–527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kostova Z, Mariano J, Scholz S, Koenig C & Weissman AMA Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J. Cell Sci 122, 1374–1381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Das R et al. Allosteric activation of E2-RING finger–mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol. Cell 34, 674–685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li W et al. Mechanistic insights into active site–associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl. Acad. Sci. USA 106, 3722–3727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bagola K et al. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol. Cell 50, 528–539 (2013). [DOI] [PubMed] [Google Scholar]
- 126.Liu S et al. Promiscuous interactions of gp78 E3 ligase CUE domain with polyubiquitin chains. Structure 20, 2138–2150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen B et al. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA 103, 341–346 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koegl M et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644 (1999). [DOI] [PubMed] [Google Scholar]
- 129.Hatakeyama S, Yada M, Matsumoto M, Ishida N & Nakayama KI U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem 276, 33111–33120 (2001). [DOI] [PubMed] [Google Scholar]
- 130.Nakatsukasa K, Huyer G, Michaelis S & Brodsky JL Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meacham GC, Patterson C, Zhang W, Younger JM & Cyr DM The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol 3, 100–105 (2001). [DOI] [PubMed] [Google Scholar]


