Abstract
We have previously described a Pseudomonas aeruginosa gene, ptxR, which enhances exotoxin A production at the transcriptional level. We have also described another gene, ptxS, which is transcribed divergently from ptxR and interferes with the enhancement of exotoxin A synthesis by ptxR. However, the mechanisms through which ptxR and/or ptxS are regulated is not known. In this study, we attempted (by using the DNA gel shift assay) to determine if P. aeruginosa contains a potential regulatory protein that binds specifically to the ptxR or ptxS upstream region. In the initial analysis, different-sized gel shift bands were detected when a probe containing the ptxR-ptxS intergenic region was incubated with the lysate of P. aeruginosa PAO1. The strongest binding activity was detected with a smaller fragment that represents the ptxS upstream region. Additional deletion analysis localized the binding to a 52-bp fragment immediately upstream of ptxS. The gel shift band was not detected when the 52-bp fragment was incubated with the lysate of the ptxS isogenic mutant PAO1::ptxS. However, the binding band was regenerated when a plasmid carrying ptxS intact was introduced into PAO1::ptxS. In addition, the gel shift band was detected when the 52-bp fragment was incubated with a lysate of Escherichia coli in which ptxS was overexpressed from the T7 promoter. The effect of PtxS on ptxS expression was examined by using a ptxS-lacZ fusion plasmid. The level of β-galactosidase activity produced by PAO1::ptxS carrying the fusion plasmid was four- to fivefold higher than that produced by PAO1 carrying the same plasmid. Using DNase I footprinting analysis, the binding region was specified to a 20-bp fragment. Within the fragment, a 14-bp palindromic sequence exists that may function as a PtxS binding site. These results suggest that PtxS autoregulates its synthesis by binding to a specific sequence within the ptxS upstream region.
Pseudomonas aeruginosa is a gram-negative pathogen that causes a variety of infections, including wound infections, nosocomial infections, and lung infections in cystic fibrosis patients (3, 34). The ability of P. aeruginosa to cause these infections is due to the production of several extracellular virulence factors (35). Among these different virulence factors, exotoxin A is considered the most toxic. Exotoxin A is an ADP-ribosyltransferase enzyme that catalyzes the transfer of the ADP-ribosyl moiety of NAD+ onto elongation factor 2 of eukaryotic cells, causing cessation of the protein synthesis process and subsequent cell death (12). In vitro production of exotoxin A by P. aeruginosa is controlled by different environmental factors. These factors include the level of iron in the growth medium, the growth temperature, and the presence of certain amino acids and nucleotides in the growth medium (2, 13).
Exotoxin A production by P. aeruginosa is a complicated process that involves several positive and negative regulatory genes (7, 19, 31, 33). One of the most extensively analyzed of these genes is regA, which is required for maximum production of exotoxin A by P. aeruginosa (33). A P. aeruginosa mutant defective in regA produces neither regA nor toxA mRNA (24, 33). The regA gene, which codes for a 28-kDa protein, enhances exotoxin A production at the transcriptional level (33). We have previously described another toxA regulatory gene, ptxR (9). The presence of a plasmid carrying ptxR in P. aeruginosa enhances exotoxin A production four- to fivefold (9). The ptxR gene encodes a 34-kDa protein that belongs to the LysR family of transcriptional activators and enhances toxA transcription through regA (9). However, the exact mechanism of this regulation is still unknown. In addition to exotoxin A, ptxR positively regulates the production of the P. aeruginosa siderophores pyochelin and pyoverdine (6). However, the mechanism of this regulation appears to be different from that of exotoxin A. Although ptxR enhances exotoxin A production in P. aeruginosa, it does not interfere with the negative regulation of exotoxin A synthesis by iron (9). In contrast, siderophore production in P. aeruginosa carrying a ptxR plasmid is deregulated with respect to iron (6).
We have recently described another toxA regulatory gene, ptxS, which interferes with the effect of ptxR on toxA transcription (6). The product of ptxS, PtxS, is a 37-kDa protein which belongs to the GalR family of transcriptional repressors (6). Although the exact mechanism of PtxS function is not known, available evidence suggests that ptxS negatively regulates the transcription of ptxR (6). The ptxS gene, which is located 5′ of ptxR, is transcribed in the opposite orientation of ptxR (6). Between the ptxR and ptxS translational start codons is a 562-bp intergenic region (6). Despite the analysis of ptxR and ptxS functions, the mechanisms through which these genes are regulated are unknown. In this study, we attempted to determine if P. aeruginosa contains a protein that specifically binds to either the ptxR or the ptxS upstream region (a potential DNA binding regulatory protein). Our results showed that PtxS negatively autoregulates its own synthesis by binding specifically to the ptxS upstream region.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium (1% Bacto Tryptone [Difco Laboratories, Detroit, Mich.], 0.5% yeast extract, 1% NaCl). For binding experiments, P. aeruginosa was grown in Chelex-treated Trypticase soy broth dialysate (BBL Microbiology Systems, Cockeysville, Md.) to which 1% glycerol and 0.05 M monosodium glutamate were added (TSB-DC) (17). Cultures were grown at 37°C with vigorous aeration. Antibiotics were used at the following concentrations: ampicillin, 75 μg/ml (E. coli); carbenicillin, 100 (E. coli) or 300 (P. aeruginosa) μg/ml; rifampin, 80 μg/ml (P. aeruginosa).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Prototroph | 11 |
| PAO1::ptxS | ΔptxS::Ω Smra | 6 |
| Escherichia coli | ||
| DH5α | supE44 thi-1 recA1 gyrA Nalra | 1 |
| K38 | HfrC; host for pT7 expression system | 26 |
| Plasmids | ||
| pUC18 | Cbr Apr ColE1; general cloning vectora | Stratagene |
| pAH56 | pUC18 carrying 2.2-kbp BamHI/HindIII fragment which contains ptxS | 9 |
| pSW205 | β-Galactosidase cloning vector carrying 1.8-kb P. aeruginosa stability fragment | 24 |
| pBS8-4 | Cbr; ptxS-lacZ fusion in pSW205 | This study |
| pT7-5 | Cloning vector for T7 expression system | 26 |
| pJAC17 | pT7-5 carrying HincII-HindIII fragment containing intact ptxS open reading frame | 6 |
Abbreviations: Δ, deletion; Ω, 2.0-kb SmaI omega fragment; Sm, streptomycin; Nal, nalidixic acid; Ap, ampicillin; Cb, carbenicillin; ColE1, ColE1 origin of replication; r, resistant.
Preparation of cell extracts.
P. aeruginosa PAO1 was grown in 100 ml of TSB-DC at 30°C for 14 h. The cells were harvested, resuspended in 10 ml of distilled water (10× concentration), and lysed by being passed twice through a French pressure cell at 1,000 lb/in2 (American Instrument Company, Silver Spring, Md.). Lysed cells were centrifuged at 240 × g, and the supernatant fraction (which represents the cell lysate) was isolated and divided into several aliquots. The amount of protein in each sample was determined by the method of Lowry et al. (14).
Gel shift assay.
DNA fragments containing different segments of the ptxS upstream region were obtained by either restriction digestions or PCR (32). The fragments were labeled with [γ-32P]ATP (Amersham, Arlington Heights, Ill.) by the end-labeling technique using T4 polynucleotide kinase (1). The binding experiments were performed as previously described (1). The binding reaction mixture (20-μl total volume) contained DNA binding buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 10 mM KCl, 0.1 mM dithiothreitol, 5% glycerol, 50 μg of bovine serum albumin per ml), 1 μg of poly(dI-dC) (Boehringer Mannheim, Indianapolis, Ind.) per ml (for nonspecific binding), and 25 to 50 μg of the 10× PAO1 lysate. Approximately 107 cpm of radiolabeled DNA probe was added, and the reaction mixture was incubated at 30°C for 30 min. Approximately 3 μl of the tracking dye (50% sucrose, 0.6% bromphenol blue) was added to the reaction mixture, and it was loaded onto an 8% polyacrylamide gel in 1× Tris-borate-EDTA buffer (1). The gel was electrophoresed at 180 V for 2.5 h. The gels were dried and exposed to X-ray film.
DNase I footprint analysis.
The 103-bp fragment which corresponds to the ptxS upstream region was synthesized by PCR. The fragment was end labeled by using T4 polynucleotide kinase (1). One label was then removed by using the KpnI restriction enzyme. The singly labeled DNA fragment was incubated with 5 to 25 μg of the partially purified protein by using a reaction mixture identical to that described above for the gel shift assay. After 30 min of incubation at 30°C, the reaction mixture was manipulated as listed in the manufacturer’s instructions (Core Footprinting System; Promega, Madison, Wis.). Fifty microliters of a Ca2+-Mg2+ solution was added, and the mixture was incubated for 1 min at room temperature; approximately 3.5 × 10−2 U of DNase I was added, and the mixture was incubated for 1 min at room temperature; and the reaction was stopped by the addition of 75 μl of the Stop Solution supplied in the footprinting kit. The reaction was phenol extracted and precipitated in ethanol. The DNA was resuspended in loading buffer and separated on a 10% sequencing gel.
Construction of pBS8-4.
The 740-bp BamHI/ScaI fragment containing 726 bp of the ptxS upstream region and 14 bp of the ptxS structural gene was isolated from pJAC13 (6). The 3′ and 5′ ends were converted to blunt ends, and the fragment was cloned into the SmaI site of pUC18. The ptxS upstream region was then isolated from the resulting recombinant plasmid as a BamHI/EcoRI fragment. The 5′ ends were converted to blunt ends, and the fragment was cloned in the SmaI site of the lacZ translational fusion vector pSW205 (23). Through these different manipulations, a 7-bp pUC18 sequence was added to the 14 bp of the ptxS structural gene. Nucleotide sequence analysis was performed to confirm the construction of pBS8-4.
Enzyme assay.
Levels of β-galactosidase activity were determined as previously described (16). A 1-ml pellet was washed and resuspended in 100 μl of distilled water, and the membranes were disrupted by sonication. Statistical analysis was done by the paired t test (22) using the computer program InStat 2.01 (GraphPad Software, San Diego, Calif.).
Expression experiments.
PtxS was overproduced in E. coli K38 by using the T7 expression system as previously described (6, 26).
RESULTS
Identification of the potential DNA binding protein.
Despite our previous analysis of the P. aeruginosa ptxR gene, its effect on the expression of the toxA and regA genes, and the interaction between the ptxR and ptxS genes, we still do not know the factors or the specific conditions that regulate the expression of either gene. One possible mechanism for such regulation is through transcriptional factors (positive or negative) that specifically bind to the upstream region of ptxR, ptxS, or both. Computer analysis revealed that the ptxR and ptxS upstream regions contain specific sequences that may represent binding sites for regulatory proteins (6). Therefore, we have utilized the DNA gel shift assay to determine if P. aeruginosa contains a protein(s) that specifically binds to the ptxS or ptxR upstream region. The source of the putative protein was the lysate of P. aeruginosa PAO1 (both ptxR and ptxS were originally isolated from PAO1) that was grown in an iron-deficient medium (TSB-DC). The probe for initial gel shift experiments was the 740-bp BamHI-KpnI fragment containing 553 bp of the ptxR-ptxS intergenic region (ptxR and ptxS are divergently transcribed) (6) and 167 bp of the ptxR structural gene (Fig. 1). Two gel shift bands were detected when the P. aeruginosa lysate was incubated with the 740-bp fragment (data not shown). Since this intergenic fragment contains both the ptxS and ptxR upstream regions, we attempted to assign the observed binding to either the ptxR or the ptxS upstream region by dividing the fragment. The 740-bp fragment was divided into three smaller fragments by using available restriction sites: a 257-bp BamHI-DpnI fragment containing the putative upstream region of ptxR and a portion of its open reading frame, a 157-bp DpnI-DpnI intergenic fragment, and a 304-bp DpnI-KpnI fragment containing the putative upstream region of ptxS (Fig. 1). Two gel shift bands with strong intensity were detected when the 304-bp probe was incubated with the PAO1 lysate (25). A fainter gel shift band was detected when the 157-bp probe was incubated with the PAO1 lysate (25). Since the binding to the 304-bp probe was consistently detected and remained very intense, further experiments were conducted focusing on the localization and purification of the potential DNA binding protein(s). Additional subcloning and gel shift experiments further found that the binding activity could be assigned to two separate DNA regions: a 201-bp fragment and a 103-bp fragment (25; Fig. 2A). Since the 103-bp fragment was located immediately upstream of ptxS and represented a putative ptxS regulatory region, we chose to focus on the purification of this potential DNA binding protein and define the specific sequence to which it binds. The 103-bp fragment was subsequently divided into two smaller fragments: a 52-bp fragment and a 63-bp fragment (Fig. 1). Since no suitable restriction sites were available to generate these two fragments, they were synthesized by PCR. A gel shift band was detected with the 52-bp fragment only (Fig. 2A). The binding specificity was confirmed by competition experiments using the 103-bp probe and the unlabeled 52- or 63-bp fragment. Only when an excess of the unlabeled 52-bp fragment was added to the binding reaction mixture (103-bp probe and PAO1 lysate) did the binding band disappear (Fig. 2B). These results suggest that the PAO1 lysate contains a potential DNA binding protein that specifically binds to the ptxS upstream region.
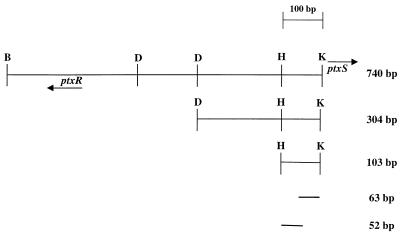
FIG. 1.
Diagram of the different probes (within the ptxR-ptxS intergenic region) that were used in the gel shift assays. The 304- and 103-bp fragments of the ptxS upstream region were generated by restriction digestion using available restriction sites (B, BamHI; D, DpnI; H, HincII; K, KpnI). The 52- and 63-bp fragments were synthesized by PCR. Arrows indicate the direction of transcription of ptxR and ptxS.
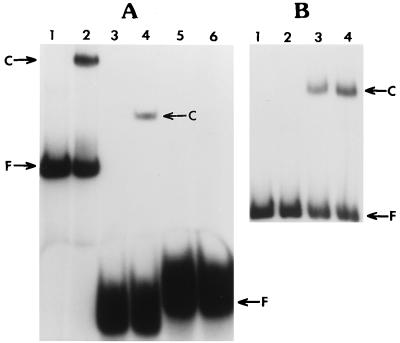
FIG. 2.
(A) Electrophoretic mobility shift assay of three different probes incubated with the lysate of P. aeruginosa PAO1. The binding reaction mixtures are described in Materials and Methods. Each DNA-protein reaction mixture contained 25 μg of the PAO1 lysate. F, free probe; C, DNA-protein complex. Lanes: 1, 103-bp probe alone; 2, 103-bp probe plus PAO1 lysate; 3, 52-bp probe alone; 4, 52-bp probe plus PAO1 lysate; 5, 63-bp probe alone; 6, 63-bp probe plus PAO1 lysate. (B) Competitive gel shift assay using the labeled 103-bp probe and the unlabeled 52- or 63-bp fragment. The DNA-protein binding reaction mixtures contained 25 μg of PAO1 lysate. Lanes: 1, 103-bp probe alone; 2, 103-bp probe plus PAO1 lysate plus 52-bp fragment; 3, 103-bp probe plus PAO1 lysate plus 63-bp fragment; 4, 103-bp probe plus PAO1 lysate (control).
Binding of PtxS to the ptxS upstream region.
Homology searches showed that PtxS belongs to the family of GalR-LacI repressors (6). Many proteins of this family are known to autoregulate their own synthesis (8, 15, 28). This autoregulation is accomplished through the specific binding of these proteins to the promoter region of their own genes (8, 15). If, similar to these proteins, PtxS autoregulates its own synthesis in P. aeruginosa, the specific gel shift band that the PAO1 lysate produces with the 52-bp fragment may represent binding of PtxS to its promoter region. To examine this possibility, we have utilized P. aeruginosa PAO1::ptxS, which is a ptxS isogenic mutant of PAO1 (6). This mutant was constructed by the gene replacement technique as previously described (6). The construction of the mutant was confirmed by Southern blot hybridization experiments using a specific ptxS probe (6; data not shown). Both PAO1 and PAO1::ptxS were grown in TSB-DC medium, and the lysates were examined for 52-bp fragment binding activity. As seen in Fig. 3A, the specific gel shift band was detected with the PAO1 lysate only. Complementation experiments were conducted to confirm that the absence of the binding activity in the PAO1::ptxS lysate was due to the loss of ptxS. For these experiments, plasmid pAH56, which carries an intact copy of ptxS (9), was introduced into PAO1::ptxS. A specific gel shift band was detected when the lysate of PAO1::ptxS/pAH56 was incubated with the 52-bp fragment (Fig. 3A). This band parallels the typical gel shift band that we usually detect when the PAO1 lysate is incubated with the 52-bp fragment (Fig. 3A). These results suggest that ptxS contributes to the observed binding to the ptxS upstream region.
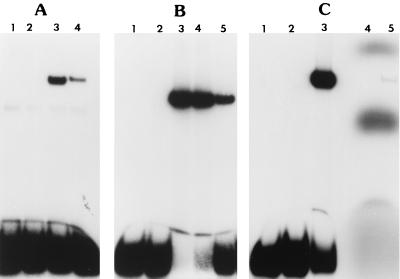
FIG. 3.
Gel shift assays to determine if the DNA-protein complex that was detected with the 52-bp probe represents binding of PtxS to the ptxS upstream region. (A) The lysate from a ptxS isogenic mutant (PAO1::ptxS) resulted in no specific gel shift band. The gel shift band was regenerated when a plasmid (pAH56) carrying an intact copy of ptxS was introduced into the mutant. The 52-bp probe was incubated with either no lysate (lane 1), PAO1::ptxS lysate (lane 2), PAO1 lysate (lane 3), or PAO1::ptxS/pAH56 lysate (lane 4). Each reaction mixture contained 25 μg of cell lysate. (B) PtxS synthesized in E. coli (by the T7 expression system) binds specifically to the 52-bp probe. The lysate obtained from E. coli K38 containing either pJAC17 (ptxS expression plasmid) or pT7-5 (vector control) was incubated with the 52-bp probe. Lanes: 1, 52-bp probe alone; 2, 52-bp probe plus K38/pT7-5 lysate (10 μg); 3, 52-bp probe plus K38/pJAC17 lysate (3.0 μg); 4, 52-bp probe plus K38/pJAC17 lysate (1.5 μg); 5, 52-bp probe plus PAO1 lysate (25 μg). (C) Incubation of radiolabeled PtxS (using the T7 expression system) with the unlabeled 52-bp fragment. In parallel with the typical gel shift assay using the labeled 52-bp probe and K38/pJAC17 lysate (lanes 1 to 3), radiolabeled PtxS was incubated with the unlabeled 52-bp probe and assayed for binding activity (lane 5). The position of the band exactly parallels that of the typical gel shift band. Lanes: 1, labeled 52-bp probe alone; 2, 52-bp probe plus K38/pT7-5 lysate (10 μg); 3, 52-bp probe plus K38/pJAC17 lysate (3 μg); 4, labeled PtxS alone; 5, labeled PtxS plus unlabeled 52-bp fragment.
Confirmation of PtxS binding to the ptxS upstream region.
The above-described results showed that a functional ptxS gene is required to produce the 52-bp fragment-specific binding activity. However, PtxS may either bind directly to the 52-bp fragment or induce another P. aeruginosa protein to bind. Thus, to confirm that the observed gel shift band is due to PtxS, we conducted additional gel shift experiments using the lysate of E. coli K38/pJAC17, in which ptxS is expressed from the T7 promoter (6). Plasmid pJAC17 was constructed by cloning of the HincII-HindIII fragment which carries the intact ptxS open reading frame in the SmaI-HindIII sites of pT7-5 (6). Expression experiments showed that K38/pJAC17 produced a 38-kDa protein (6). Thus, PtxS was synthesized in K38/pJAC17 using the previously described protocol (6) except that labeled cysteine and methionine were replaced with unlabeled cysteine and methionine. K38 carrying the cloning vector pT7-5 was used as a negative control. As shown in Fig. 3B, the lysate of K38/pT7-5 produced no gel shift band with the 52-bp fragment, whereas the lysate of K38/pJAC17 produced the typical gel shift band (Fig. 3B). To provide further evidence that the binding protein in the lysate of K38/pJAC17 is PtxS, PtxS was selectively labeled by using 35S-labeled cysteine and methionine as previously described. The lysates of the labeled cells were incubated with the unlabeled 52-bp fragment. In a parallel experiment, the lysate of K38/pJAC17 that was prepared with unlabeled amino acids was incubated with the 32P-labeled 52-bp fragment. The incubation of the 35S-labeled lysate with the unlabeled 52-bp fragment produced a specific gel shift band (Fig. 3C). This band migrated in exactly the same position as the typical gel shift band that was produced by incubation of the unlabeled lysate with the 32P-labeled 52-bp fragment (Fig. 3C). These results strongly suggest that the PtxS protein specifically binds to the ptxS upstream region.
Autoregulation of PtxS synthesis.
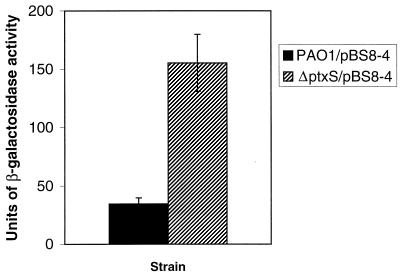
After proving that PtxS binds specifically to the ptxS upstream region, it was important to determine if the binding of PtxS causes autoregulation of PtxS synthesis. To examine this possibility, we utilized the ptxS-lacZ fusion plasmid pBS8-4 (see Materials and Methods for construction). Plasmid pBS8-4 was introduced into both PAO1 and PAO1::ptxS. PAO1/pBS8-4 and PAO1::ptxS/pBS8-4 were grown in TSB-DC medium for 14 h, and the level of β-galactosidase activity was determined as previously described (16). As depicted in Fig. 4, the level of β-galactosidase activity produced by PAO1::ptxS/pBS8-4 was four- to fivefold higher than that produced by PAO1/pBS8-4. These results suggest that PtxS negatively autoregulates its own synthesis in P. aeruginosa.
FIG. 4.
Autoregulation of ptxS expression in P. aeruginosa. Plasmid pBS8-4 (which carries a ptxS-lacZ fusion) was introduced into PAO1 and PAO1::ptxS. Cells were grown in TSB-DC medium for 14 h, and the level of β-galactosidase activity was determined as previously described (16). PAO1 carrying pSW205 (negative control) produced no β-galactosidase activity. The values are averages of three independent experiments ± the standard error of the mean.
Identification of the specific ptxS sequence to which PtxS binds.
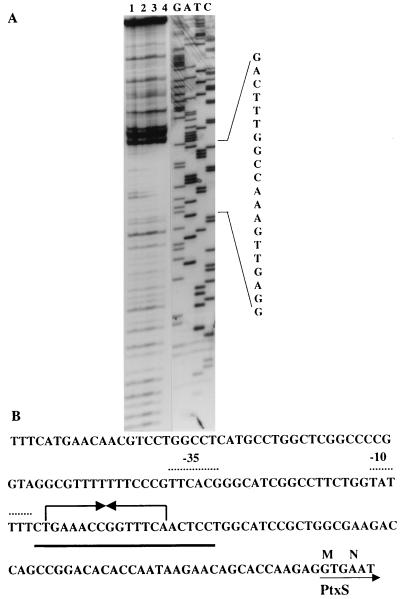
The precise ptxS sequence to which PtxS binds was identified by DNase I protection analysis as previously described using PtxS synthesized by E. coli K38/pJAC17 (6). PtxS protected a 20-bp region from DNase I digestion (Fig. 5A). Within the 20-bp protected region, there is a 14-bp sequence of dyad symmetry (Fig. 5B). We have confirmed that ptxS is expressed in E. coli by using a ptxS-lacZ fusion (data not shown). However, we have not been able to determine the ptxS transcriptional start site. Since ptxS is expressed in E. coli, we tried to determine if the ptxS promoter resembles the E. coli ς70 promoters (10). We have identified potential −10 and −35 sequences within the 20-bp protected region. The −10 sequence (which is located 97 bp from the GTG translation start codon) contains four of the six bases (TATAAT) that are found within the −10 sequence of ς70 promoters, whereas the −35 site contains five of the six bases (TTGACA) that are found within the −35 sequence of ς70 promoters (Fig. 5B). In addition, similar to that of ς70 promoters, the distance between the −10 and −35 sites is 17 bp (Fig. 5B).
FIG. 5.
Determination of the specific nucleotide sequence within the ptxS upstream region to which PtxS binds. (A) DNase I protection analysis of PtxS binding to the ptxS upstream region. The 103-bp fragment of the ptxS upstream region was singly end labeled with [γ-32P]ATP and incubated with increasing concentrations of E. coli K38/pJAC17 lysate in which ptxS was overexpressed from the T7 promoter. Lanes: 1, 103-bp probe; 2 to 4, 103-bp probe plus 10, 15, and 25 μg of lysate, respectively. All reaction mixtures were treated with DNase I. The complementary nucleotide sequence of the fragment is shown. In addition, specific nucleotides that constitute the DNase I-protected region are indicated on the right. (B) Nucleotide sequence of the ptxS upstream region. The solid line indicates the 20-bp DNase I-protected region. The 14-bp palindrome is shown by opposing arrows. The potential −10 and −35 sites (based on comparisons with ς70 promoters) are identified by dotted lines.
DISCUSSION
Results obtained in this study confirm that PtxS negatively autoregulates its synthesis in P. aeruginosa. Based on our previous analysis, we have suggested that PtxS belongs to the GalR-LacI family of transcriptional repressors (6). The most significant homology between PtxS and proteins of the GalR-LacI family is within the helix-turn-helix motif (DNA binding motif) (6). Among the different proteins within this family, three are known to negatively autoregulate their own synthesis: the galactose isorepressor GalS (28), the cytidine repressor CytR (8), and the purine regulon repressor PurR (15). Several previous transcriptional studies have shown that the negative autoregulation of GalS, CytR, and PurR is weak (about two- to threefold) (8, 15, 28). However, these studies utilized transcriptional fusions that were carried either in the chromosome or on low-copy-number plasmids. For example, the level of galS expression from a galS-lacZ fusion carried in the chromosome of an E. coli galS mutant was increased by twofold (28). Similarly, in a cytR mutant, the levels of cytR expression from a cytR-lacZ fusion carried on a low-copy-number plasmid was increased by twofold (8). In comparison, the increase in the level of ptxS expression in PAO1::ptxS is slightly higher (four- to fivefold) (Fig. 4). The copy number of the ptxS-lacZ fusion plasmid (pBS8-4), and other plasmids that carry the 1.8-kb PstI stability fragment (18, 30), is not known. However, if the copy number of pBS8-4 is assumed to be high, the autoregulation of PtxS is similar to that of CytR and GalS. Alternatively, if the copy number is low, the levels of ptxS autoregulation may be even more significant. Currently, we are constructing a ptxS-lacZ fusion by using the low-copy-number, broad-host-range lacZ transcriptional fusion vector pMP190 (21). This fusion should help us directly compare the levels of autoregulation between ptxS and the other three galR-lacI genes.
The ptxS upstream region shares several characteristic features with the upstream regions of galS, cytR, and purR. For example, all three genes autoregulate their own synthesis by binding to specific regions (operator sites) within their genes (8, 15, 28). Many proteins of the GalR-LacI family bind to multiple operator sites either within the upstream region or within the structural region of the genes that they regulate (27). The presence of multiple operator sites is thought to be important in augmenting the repression of the regulated genes (5). For example, galE, galS, galP, and galR are known to contain two operator sites, i.e., one within the promoter region and another within the structural gene (29). The purR gene contains two operator sites that are located downstream of the transcription initiation site (15, 20). The cytR gene contains only one operator site upstream of the transcription start site (8). Each operator site contains a consensus binding sequence that is specific for each protein (8, 15, 20, 28). The nucleotides within these consensus sequences are arranged in dyad symmetry, or a palindrome, to allow their respective proteins to bind as dimers (29). As we have shown in this study, PtxS binds to a single site within the 103-bp fragment upstream of the GTG translational start codon (Fig. 5B). This ptxS binding region contains a 14-bp palindromic sequence that may represent a potential PtxS operator site (Fig. 5B). The 5′ half of this palindromic sequence contains all three conserved nucleotides ( … AAC) that are usually detected within the DNA binding half sites for several proteins of the GalR-LacI family (27). The location of the PtxS operator site with respect to the ptxS transcriptional start site is not known. Despite several attempts, we were not able to determine the ptxS transcription start site. However, if ptxS has a ς70 promoter that utilizes the potential −10 and −35 sites that we have identified (Fig. 5B), the ptxS transcriptional start site may be within the DNase I-protected region. If this proves to be true, then unlike galS, purR, and cytR, the ptxS operator site would be within the ptxS transcriptional start site.
PtxS may autoregulate its own synthesis by a mechanism that resembles those utilized by some proteins of the GalR-LacI family. However, the specific environmental signal to which PtxS responds, as well as the exact mechanism through which PtxS regulates its target genes (other than ptxS), is not known. Most proteins of the GalR-LacI family function in response to certain environmental signals called effectors. These effectors include carbohydrates, nucleosides, and modified amino acids (4, 27). Within the structure of many of the GalR-LacI proteins, there are conserved regions to which different effectors may bind (27). As we have shown previously, the strongest homology between PtxS and the different GalR-LacI proteins is within the amino-terminal DNA binding motif (6). Although a second region of homology has been identified, this region does not involve the effector binding sequences (6). The lack of conserved effector sequences within PtxS suggests that the protein does not utilize the effector binding system for its function. PtxS was originally identified through its negative effect on exotoxin A synthesis (6). Exotoxin A synthesis in P. aeruginosa is regulated by different environmental signals (especially iron, which negatively regulates toxA expression) (2, 13, 33). However, PtxS binding to its promoter was not affected by the presence of iron in the growth medium (data not shown). Whether iron affects PtxS binding to other genes remains to be determined.
With respect to the PtxS target gene, we have previously shown that ptxS modifies the function of the toxA transcriptional activator ptxR (6). The ptxS gene is divergently transcribed from ptxR from the opposite strand (6). Based on recent transcriptional analysis of ptxR (using a ptxR-lacZ fusion plasmid), we have suggested that PtxS may interfere with ptxR expression in P. aeruginosa (6). However, this effect is not likely to be accomplished through direct binding of PtxS to the ptxR promoter. PtxS (that was produced in E. coli by the T7 expression system) did not bind to different segments of the ptxR upstream region (data not shown). In addition, computer analysis revealed that the ptxR upstream region lacks the 14-bp palindromic sequence (PtxS operator site) (data not shown). Therefore, it is possible that PtxS regulates ptxR expression through another regulatory gene.
ACKNOWLEDGMENTS
This study was supported by a Public Health Service grant (AI-33386) to Abdul Hamood. Britta Swanson was supported by a research fellowship from the Cystic Fibrosis Foundation.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moor D, Seidman J, Smith J, Strauhle K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1988. [Google Scholar]
- 2.Bjorn M J, Iglewski B H, Ives S K, Sadoff J C, Vasil M L. Effect of iron on yields of exotoxin A in cultures of Pseudomonas aeruginosa PA103. Infect Immun. 1977;19:785–791. doi: 10.1128/iai.19.3.785-791.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodey G, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:297–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 4.Bouchez D, Tourneur J. Organization of the agropine synthesis region of the T-DNA of the Ri plasmid from Agrobacterium rhizogenes. Plasmid. 1991;25:27–39. doi: 10.1016/0147-619x(91)90004-g. [DOI] [PubMed] [Google Scholar]
- 5.Choy H E, Adhya S. RNA polymerase idling and clearance in gal promoters: use of supercoiled mini-circle DNA template made in vivo. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colmer J A, Hamood A N. Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR. Mol Gen Genet. 1998;258:250–259. doi: 10.1007/s004380050729. [DOI] [PubMed] [Google Scholar]
- 7.Gambello M J, Kaye S, Iglewski B H. lasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and enhancer of exotoxin A synthesis. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlach P, Valentin-Hansen P, Bremer E. Transcriptional regulation of the cytR gene of Escherichia coli: autoregulation and positive control by the cAMP/CAP complex. Mol Microbiol. 1990;4:479–488. doi: 10.1111/j.1365-2958.1990.tb00614.x. [DOI] [PubMed] [Google Scholar]
- 9.Hamood A N, Colmer J A, Ochsner U A, Vasil M L. Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol. 1996;21:97–110. doi: 10.1046/j.1365-2958.1996.6251337.x. [DOI] [PubMed] [Google Scholar]
- 10.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglewski B H, Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc Natl Acad Sci USA. 1975;2:2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P V. Exotoxin A of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
- 14.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 15.Meng L M, Kilstrup M, Nygaard P. Autoregulation of PurR repressor synthesis and involvement of purR in the regulation of purB, purC, purL, purMN, and guaBA expression in Escherichia coli. Eur J Biochem. 1990;187:373–379. doi: 10.1111/j.1432-1033.1990.tb15314.x. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1973. [Google Scholar]
- 17.Ohman D E, Sadoff J C, Iglewski B H. Exotoxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince R W, Cox C D, Vasil M L. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol. 1993;175:2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolfes R J, Zalkin H. Autoregulation of Escherichia coli purR requires two control sites downstream of the promoter. J Bacteriol. 1990;172:5758–5766. doi: 10.1128/jb.172.10.5758-5766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 22.Steel R G, Torrie J H. Principles and procedures of statistics. A biometrical approach. 2nd ed. New York, N.Y: McGraw-Hill, Inc.; 1980. [Google Scholar]
- 23.Storey D G, Frank D W, Farinha M A, Kropinski A M, Iglewski B H. Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol. 1990;4:499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 24.Storey D G, Raivio T, Frank D W, Wick M J, Kaye S, Iglewski B H. Effect of regB on expression from P1 and P2 promoters of the Pseudomonas aeruginosa regAB operon. J Bacteriol. 1991;173:6088–6094. doi: 10.1128/jb.173.19.6088-6094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson, B. L., and A. N. Hamood. Unpublished data.
- 26.Tabor S, Richardson C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 28.Weickert M J, Adhya S. Control of transcription of the Gal repressor and isorepressor gene in Escherichia coli. J Bacteriol. 1993;175:251–258. doi: 10.1128/jb.175.1.251-258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weickert M J, Adhya S. The galactose regulon of Escherichia coli. Mol Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 30.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 31.West S E H, Sample A K, Runyen-Janecky L J. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White B A. PCR protocols: current methods and applications. Totowa, N.J: Humana Press; 1993. [Google Scholar]
- 33.Wick M J, Frank D W, Storey D G, Iglewski B H. Structure, function, and regulation of Pseudomonas aeruginosa exotoxin A. Annu Rev Microbiol. 1990;44:335–363. doi: 10.1146/annurev.mi.44.100190.002003. [DOI] [PubMed] [Google Scholar]
- 34.Woods R E. Pseudomonas: the compromised host. Hosp Pract. 1976;11:91–100. doi: 10.1080/21548331.1976.11706983. [DOI] [PubMed] [Google Scholar]
- 35.Woods R E, Iglewski B H. Toxins of Pseudomonas aeruginosa: new perspectives. Rev Infect Dis. 1983;5:S715–S722. doi: 10.1093/clinids/5.supplement_4.s715. [DOI] [PubMed] [Google Scholar]