Differential analysis of eradicated and resistant subclones following cancer treatment identifies that L1 activity associated with resistance is induced by current therapies and blocked by the antiretroviral drug azidothymidine.
Abstract
Treatment-eradicated cancer subclones have been reported in leukemia and have recently been detected in solid tumors. Here we introduce Differential Subclone Eradication and Resistance (DSER) analysis, a method developed to identify molecular targets for improved therapy by direct comparison of genomic features of eradicated and resistant subclones in pre- and posttreatment samples from a patient with BRCA2-deficient metastatic prostate cancer. FANCI and EYA4 were identified as candidate DNA repair–related targets for converting subclones from resistant to eradicable, and RNAi-mediated depletion of FANCI confirmed it as a potential target. The EYA4 alteration was associated with adjacent L1 transposon insertion during cancer evolution upon treatment, raising questions surrounding the role of therapy in L1 activation. Both carboplatin and enzalutamide turned on L1 transposon machinery in LNCaP and VCaP but not in PC3 and 22Rv1 prostate cancer cell lines. L1 activation in LNCaP and VCaP was inhibited by the antiretroviral drug azidothymidine. L1 activation was also detected postcastration in LuCaP 77 and LuCaP 105 xenograft models and postchemotherapy in previously published time-series transcriptomic data from SCC25 head and neck cancer cells. In conclusion, DSER provides an informative intermediate step toward effective precision cancer medicine and should be tested in future studies, especially those including dramatic but temporary metastatic tumor regression. L1 transposon activation may be a modifiable source of cancer genomic heterogeneity, suggesting the potential of leveraging newly discovered triggers and blockers of L1 activity to overcome therapy resistance.
Significance:
Differential analysis of eradicated and resistant subclones following cancer treatment identifies that L1 activity associated with resistance is induced by current therapies and blocked by the antiretroviral drug azidothymidine.
Introduction
Understanding the emergence of cancer-cell resistance to therapy is central to improving cancer outcomes. A resistant cancer-cell subclone is relatively easy to define—it is a population of cancer cells remaining after a patient has received therapy targeting the original cancer-cell population. By contrast, a cancer-cell subclone eradicated by therapy is relatively hard to define, because it requires reasonable proof that the eradicated subclone no longer exists in the patient after a specific therapy. It is also critical to recognize that key characteristics of an eradicated subclone cannot be imputed solely from characteristics in a resistant subclone, because the point at which characteristics defining resistance arose is not known a priori.
The first cancer subclones genomically proven to be eradicated by specific therapy were reported in 2015 in leukemia (1), where the focus was largely on the emergence of resistance rather than on characteristics of the eradicated subclone. We recently reported a clinically important subclone eradicated by carboplatin chemotherapy in a metastatic prostate cancer (mPC) patient “A34” (2). To our knowledge, this is the first reported genomic evidence of subclone eradication in a solid tumor, a metastatic prostate cancer where we previously reported somatic L1 retrotransposon activity as a source of traceable genomic heterogeneity (3).
Taken together, these findings led us to hypothesize that (i) Prospectively planned, side-by-side Differential Subclone Eradication and Resistance (DSER) genomic analysis in individual patients with partial responses could provide a uniquely powerful intermediate step for advancing precision cancer medicine, and (ii) L1 activation itself is a dynamic source of genomic heterogeneity leading to eradicability or resistance, and perhaps this response could be blocked by existing medications.
Materials and Methods
A34 samples and DNA data
Tissue and blood samples from patient A34 were collected as part of the Project to Eliminate lethal Cancer (PELICAN) integrated clinical-molecular autopsy study of lethal prostate cancer (Table 1). The patient gave informed written consent to participate in the John Hopkins Medicine Institutional Review Board (IRB)–approved study. Detailed specimen isolation and analysis methods are contained in Woodcock and colleagues (2) and in Supplementary Methods.
Table 1.
A34 samples and data studied.
| Available sequencing data | |||||||
|---|---|---|---|---|---|---|---|
| Sample from A34 | Collection time-point | Letter ID | Tumor cellularity | Whole genome | Deep targeted | Methylation | Substitutions from WGS |
| SacralBoneMet | 11 years prior to death | e | 0.86 | Yes | Yes | Yes | 5,181 |
| Serum | 11 years prior to death | — | 0.04 | No | Yes | No | — |
| Prostate | 9 years prior to death | X | 0.70 | No | Yes | No | — |
| Prostate | 9 years prior to death | Y | 0.67 | No | Yes | No | — |
| Prostate | 9 years prior to death | Z | 0.84 | No | Yes | No | — |
| LiverMet1 | Autopsy | c | 0.84 | Yes | Yes | Yes | 12,745 |
| LiverMet12 | Autopsy | d | 0.85 | Yes | Yes | Yes | 13,762 |
| LiverMet3 | Autopsy | a | 0.79 | Yes | Yes | Yes | 12,332 |
| Serum | Autopsy | — | 0.56 | No | Yes | Yes | — |
| Plasma | Autopsy | — | 0.63 | No | Yes | Yes | — |
Note: For each of the 10 samples used in the study, the identifiers and time of sample collection is indicated, along with sequencing data types available. Purity for samples with WGS data was inferred using the Battenberg algorithm (https://github.com/cancerit/cgpBattenberg) and purity for samples without WGS data was based on the variant allele frequency information and the DPClust algorithm as described in Woodcock and colleagues (2). Numbers of substitutions per sample are based on the WGS data as analyzed in Gundem and colleagues (8). The number of variants per subclone can be found in Supplementary Table S4. WGS reference normal spleen DNA is not included in the table.
Identification of L1 insertion sites
Whole-genome sequencing (WGS) reads aligned to hg19 from A34 metastatic samples were analyzed for somatic L1 insertions (solo-L1 insertions or L1-mediated transductions) using TraFiC-mem v1.1 (Supplementary Methods, Supplementary Table S1; ref. 3).
Generation and analysis of CpG methylation data
From A34 metastatic and autopsy blood DNA samples, paired-end reads generated from Illumina TruSeq Methyl Capture EPIC libraries were quality controlled, trimmed, and aligned to hg19 genome using Bismark v0.22.3 and Bowtie v2.3.4.1. MethylKit v1.11.0 was used for differential methylation analysis (Supplementary Methods).
Cell culture
LNCaP and 22Rv1 cells were cultured in a humidified CO2-incubator at 37°C in Gibco RPMI 1640 (1X) media (Thermo Fisher Scientific) supplemented with 10% FBS (Gibco standard FBS, Thermo Fisher Scientific), 2 mmol/L L-glutamine (Gibco, Thermo Fisher Scientific), and combination of 100 U/mL Penicillin and 100 μg/mL Streptomycin (Gibco Pen Strep, Thermo Fisher Scientific). For VCaP cells, DMEM (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS (GE Healthcare HyClone) and for PC3 F-12 K medium supplemented with 10% FBS, and antibiotics as for LNCaP growth medium was used. Cell lines were obtained from ATCC, authenticated using STR (short tandem repeat) markers and Mycoplasma tested (Supplementary Methods).
Exposure of cell lines to carboplatin/enzalutamide alone
Carboplatin or enzalutamide (ENZ) dissolved in DMSO and culture medium without FBS was added after a 24-hour initial incubation period. To control cells DMSO diluted in culture medium was added to a final concentration of 0.02%. LNCaP prostate cancer cells were transfected with L1 plasmid or positive and negative control plasmids and treated with carboplatin (5 μmol/L) or ENZ (10 μmol/L) the day after transfection, and cells were monitored for 5 days.
Exposure of cell lines to carboplatin/ENZ and azidothymidine
First, the potential cell toxicity of azidothymidine (AZT) alone was determined and no effects of AZT on cell viability were seen up to 50 μmol/L tested in LNCaP or VCaP cells (Supplementary Fig. S1). Next, AZT was introduced to both LNCaP and VCaP cells alone and in combination with carboplatin or ENZ.
L1-EGFP retrotransposition assay
The setup of the retrotransposition assay and the creation of the plasmids containing an L1RP (4) element tagged with GFP were as described by Ostertag and colleagues (5) and implemented as shown in Faulkner and colleagues (6) with modifications (Supplementary Methods).
siRNA silencing
LNCaP or PC3 cells were reverse transfected with 25 nmol/L siRNAs against FANCI (Dharmacon, ON-TARGETplus SMARTpool, L-022320–01–0005) or Scr control (Dharmacon, nontargeting pool), using OPTI-MEM and Lipofectamine 2000 transfection reagent (Invitrogen) in 3 biological replicates on 12-well plates. Samples were collected after 72 hours for mRNA isolation (Supplementary Table S2).
Cell proliferation assay
For cell proliferation assays, cells were reverse transfected with 25 nmol/L siRNAs against FANCI (Dharmacon, ON-TARGETplus SMARTpool, L-022320–01–0005) or Scr control (Dharmacon, nontargeting pool), using OPTI-MEM and Lipofectamine 2000 transfection reagent (Invitrogen) in 4 biological replicates in a 384-well plate. LNCaP cells were plated on the wells (1000 cells/well) in antibiotic-free regular growth medium (Supplementary Methods).
qRT-PCR
The isolation of RNA from cell lines was conducted using TriPure Isolation Reagent (Roche) following the manufacturer's protocol. The concentration of the RNA samples was measured using NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer, followed by dilution of the samples to 1 μg/μL. The conversion of 1 μg of RNA to cDNA was done using Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer's instructions (Supplementary Methods, Supplementary Table S3).
Western blot
Whole-cell lysates (WCL) were prepared using SDS sample buffer (66 mmol/L Tris-HCl pH 6.8, 13% glyserol, 2.1% SDS, and 0,01% Bromophenol Blue) with protease inhibitor added (cOmplete Protease Inhibitor Cocktail, Roche; Supplementary Methods).
Estimation of L1 mRNA levels from RNA sequencing
RNA sequencing (RNA-seq) reads were quality controlled (TrimGalore v0.6.5), trimmed (Cutadapt v1.18), and aligned to the hg38 genome (STAR v2.7.8 with Gencode Release 33 annotations). featureCounts v2.0.2 (7) was used to quantify reads within 146 putatively retrotransposition-active human L1 elements with intact ORF1 and ORF2 as annotated in the L1Base2 database. Counts were normalized in each sample to represent the number of reads mapping to the putatively active L1 elements per million aligned reads in the sample (Supplementary Methods).
Results
Distinct eradicated and resistant subclones
Patient A34 presented with 19 distinct subclones across his primary tumor and metastases that could be identified via analysis of DNA single-nucleotide variants and indels using the DPClust method as previously shown (Supplementary Table S4; ref. 2). Four cancer subclones specific to the sacral bone metastasis removed at surgery 11 years prior to death were also present in serum and transurethral resection of the prostate cancer samples prior to carboplatin chemotherapy, but absent from three liver metastases and serum and plasma sampled at autopsy, consistent with eradication by carboplatin chemotherapy (Fig. 1A; Supplementary Fig. S2; ref. 2). Neither androgen deprivation therapy nor radiation therapy were the cause of this subclone eradication because one of the eradicated subclones is still detected in samples Z, X, and Y that were obtained by transurethral resection from the prostate after androgen deprivation and after radiation therapy to the prostate, sacrum, lung, and penis (Fig. 1A; Supplementary Fig. S2). Resistant subclones (but not eradicated subclones) were detected in the liver metastases and autopsy blood samples, with subclone detection in autopsy serum and plasma consistent with separate shedding of tumor DNA into blood by distinct liver metastases. Based on these findings, if eradicated subclones were still present in A34′s body at the time of death, there is a reasonable expectation that they would have shed tumor DNA detectable by the deep targeted sequencing performed, and no such signals were detected (Fig. 1A).
Figure 1.
Genomic evolutionary analysis of primary and metastatic samples and subclones in patient A34 reveals distinct genomic features in resistant and eradicated subclones. A, Subclonal structure of the lethal metastatic prostate cancer of patient A34, modified from Woodcock and colleagues (2), presented as a phylogenetic tree and subclone and sample button plot. Of the 19 total subclones, 17 were used to construct the phylogenetic tree (two subclones were excluded as they contained >50% indels). The red box denotes subclones eradicated by treatment that were not detected in autopsy samples, and the light blue box outlines subclones found at autopsy that resisted chemotherapy. In the phylogenetic tree, dotted lines connect the final subclone of a lineage with a letter denoting the sample or samples in which it was observed. The color of each subclone reflects its location as shown in the key. Red numbers indicate the total number of substitutions present at each point in the phylogenetic tree based on WGS data. The button plot also illustrates the subclones detected in each sample, with the area of each circle corresponding to the cancer cell fraction (CCF) of the subclone [CCF of 1 is found in the top (truncal) nodes of the plot]. Circles outlined with dashes indicate subclones not detected in a sample. Below the button plot, the treatment exposure of samples from patient A34 is shown as a matrix. Eradicated subclones continued to exist after exposure to external beam radiation and androgen deprivation, and therefore subclone eradication is associated with carboplatin plus etoposide chemotherapy. Samples are labeled according to the time of collection (yrsptd, years prior to death) and location. The SacralBoneMet sample is a spinal cord compressing sacral nerve root metastasis removed 11 yrsptd of the patient. B, Circos plot tree of L1 insertion events in the WGS samples from A34. Curved lines are L1 transductions with a determined source element, with each line colored according to the chromosome of its source. Black triangles denote solo L1 integration sites (source element unknown). Somatic L1 integration in TARID specific to the eradicated subclone is marked. C, Barplots of methylation percentage in the CpG island in the promoter region of the EYA4 gene, which may be affected by the L1 insertion into the 3′ adjacent sense–antisense paired TARID gene in the SacralBoneMet sample. Solid colored bars indicate significantly hypermethylated sites in the SacralBoneMet (black) relative to the same genomic positions in the LiverMet samples and autopsy blood samples (cyan). Violin plots show methylation percentage at the significantly hypermethylated CpG sites in the EYA4 gene promoter, potentially as a consequence of L1 insertion into TARID. D, Knockdown of FANCI reduces the proliferation of LNCaP but not PC3 cells. Scrambled (Scr) control siRNA was used as reference. The cell confluence was determined after a 5-day knockdown using IncuCyte S3 Image analysis tools. Asterisks indicate significant difference between sample conditions based on t test. **, P < 0.01.
Genomic lesions potentially conferring eradicability
We analyzed A34 WGS and deep targeted sequence data previously reported (2, 8), and supplemented this with genome-wide CpG methylation data to investigate the potential causes of selective cancer subclone eradication and resistance. Despite the presence of 4825 total substitutions in the eradicated subclones, with identified truncal drivers including biallelic inactivation of BRCA2, PTEN LOH, mutations in CEBPA and ARID1A and subclonal FOXA1 amplification, subclones 15, 14, 13, and 12 were eradicated by carboplatin and etoposide chemotherapy (Fig. 1A; Supplementary Fig. S2; refs. 2, 8).
Since BRCA2-deficient tumors have recently been proven responsive to chemotherapy leveraging their decreased DNA-repair capacity (9, 10), we hypothesized that the eradicated subclones may have had additional genomic lesions conferring increased sensitivity to DNA damage. We searched for alterations in currently known members of DNA repair pathways. The chemotherapy-eradicated subclone harbored a 40 Mbp heterozygous deletion in the q-arm of chromosome 15 that contains 295 protein-coding genes, 4 of which have functions involving DNA repair (NEIL1, FANCI, POLG, and BLM; Supplementary Figs. S3 and S4A). Of these, FANCI is of particular interest as it belongs to the same Fanconi anemia gene family as BRCA2, regulates recruitment of the Fanconi anemia core complex at sites of DNA damage independent of FANCD2 (11), and has been previously shown to reduce BRCA1/2–deficient cell survival when depleted in an ovarian cancer model (12). Additionally, FANCI is the only DNA repair gene within the region deemed essential for prostate cancer cells in CRISPR knockout experiments from the PICKLES database (Supplementary Fig. S4B; ref. 13).
Extending the analysis to somatic L1 transposon transduction during the evolution of metastases in A34, we used updated L1 detection algorithms to increase the resolution of these findings, identifying a total of 50 L1 insertion events across A34 cancer DNA samples (Fig. 1B; Supplementary Tables S1–S5). Notably, we observed repeated unique transductions from the same source elements on chrXp22.2, chr22q12.1, chr13q21.2, and chr5q21.3 across the L1 somatic evolutionary tree (Fig. 1B; Supplementary Table S1). All of these elements have recently been shown to be recurrent sources of L1 transductions in multiple cancer types, including prostate, with the elements on chrXp22.2 and chr22q12.1 being particularly frequent sources of transductions (14).
The eradicated sacral bone metastasis subclone harbored a unique profile of 7 L1 insertions not found in the resistant subclones, one of which was a 595 bp L1 transduction from the chromosome Xp22.2 source element into the TARID gene intron 3 (Fig. 1B). TARID functions as a promoter demethylase and forms a sense–antisense gene pair with the gene EYA4, suggesting that TARID may have a role in EYA4 transcriptional regulation (15, 16). Accordingly, we found the EYA4 promoter significantly hypermethylated in the eradicated sacral bone metastasis compared with the resistant liver metastases and autopsy blood samples [66/122 (54%) CpG sites; Fig. 1C]. Transcriptome analysis of the sacral bone metastasis was not possible because no remaining tissue material is available, but the increased methylation of the EYA4 promoter region nonetheless points to possible lower levels of expression of EYA4. Previous studies have found cells with an absence of EYA4 expression to be more sensitive to DNA damage upon exposure to cisplatin (17, 18).
As a test of the hypothesis that decreased FANCI function could have contributed to selective subclone eradication, we exposed prostate cancer cell lines PC3 and LNCaP to FANCI siRNA with and without carboplatin treatment. The inhibition of FANCI resulted in a significant decrease in the proliferation of LNCaP cells while no effect was observed in PC3 cells (Fig. 1D; Supplementary Fig. S4C–S4D, Supplementary Table S2), confirming FANCI's role in maintaining LNCaP capacity to undergo cell division. Exposure of siFANCI LNCaP cells to 10 μmol/L carboplatin additionally significantly decreased proliferation (Supplementary Fig. S4D). Similar to BRCA2 somatic inactivation in A34, LNCaP (but not PC3) contains deficient RAD50 and CHEK2 functions upstream of the BRCA complex, again supporting increased dependency on FANCI in BRCA-deficient tumors (19).
Genomic lesions potentially conferring gradual increase in resistance to chemotherapy in resistant subclones
The resistant subclones identified in liver metastases, serum, and plasma contained 8915 total substitutions (Fig. 1A). Additional potential drivers in these subclones include RB1 S485F mutation, 17p LOH, PDE4B biallelic loss, and amplification of NCOA2 and FOXA1. RB1 S485F is not currently contained in COSMIC (20), but is predicted to be deleterious by MutationTaster2 (21), Provean (22), and the International Cancer Genome Consortium Data Portal.
We also analyzed L1 insertion events as possible contributors to resistance to chemotherapy, since previous studies have implicated L1 insertion events as the mediators of mutations, deletions, or rearrangements in the genome (14), but found none to be shared by all resistant subclones.
From the clinical timeline (Supplementary Fig. S2), the first round of carboplatin/etoposide was associated with the longest (approx. 4-year) response window and appears likely to have been the round when the subclones 15, 14, 13, and 12 were eradicated. Subsequent rounds of carboplatin/etoposide provided a decrementing response, consistent with Darwinian selection of subclones resistant to carboplatin/etoposide.
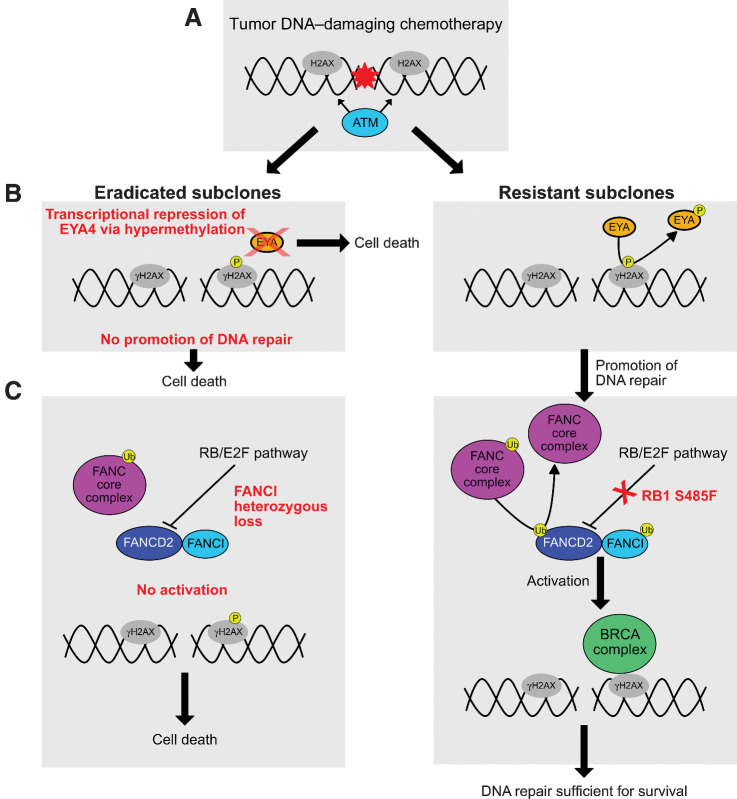
Taken together, the comparative genomic analyses of eradicated and resistant subclones in A34′s metastatic cancer support the concept of high sensitivity to DNA-damaging chemotherapy due to truncal BRCA2 biallelic inactivation in all subclones, with enhanced sensitivity to chemotherapy in the eradicated subclones due to relatively deficient FANCI and EYA4 function and incrementally increasing resistance to carboplatin/etoposide conferred to resistant subclones harboring RB1 S485 mutation and other detected alterations. A visual summary of the proposed DSER analysis for A34 is shown in Fig. 2A–C.
Figure 2.
DSER analysis in Case A34. A, Carboplatin and etoposide induce DNA double-strand breaks, causing histone H2AX to be targeted by ATM at its S139 phosphorylation site to form γH2AX. B, EYA proteins mediate the dephosphorylation of γH2AX at the Y142 residue to promote the repair response to DNA damage. A lack of EYA protein in the eradicated subclones and consequent lack of dephosphorylation enhances cell death relative to the resistant subclones. If EYA-mediated dephosphorylation occurs, the DNA repair response proceeds. C, Ubiquitination of the complex formed by FANCD2 and FANCI is required for activation of the BRCA DNA repair complex. If the ubiquitination is inhibited, this promotes cell death. Since the presence of FANCI is needed for FANCD2 ubiquitination, and compensatory increase in FANCD2 activity is known to occur in BRCA2-deficient tumors (12), absence (and/or reduced levels) of FANCI would likely lead to enhanced cell death in the face of DNA damaging chemotherapy. The RB/E2F pathway is implicated as a negative regulator of FANCD2 transcription. Deleterious RB1 S485F (present in all three resistant liver metastases) could lead to increased FANCD2 levels and enhanced DNA-repair response, leading to subclone survival during DNA-damaging chemotherapy.
Carboplatin and ENZ induce L1 in prostate cancer cells
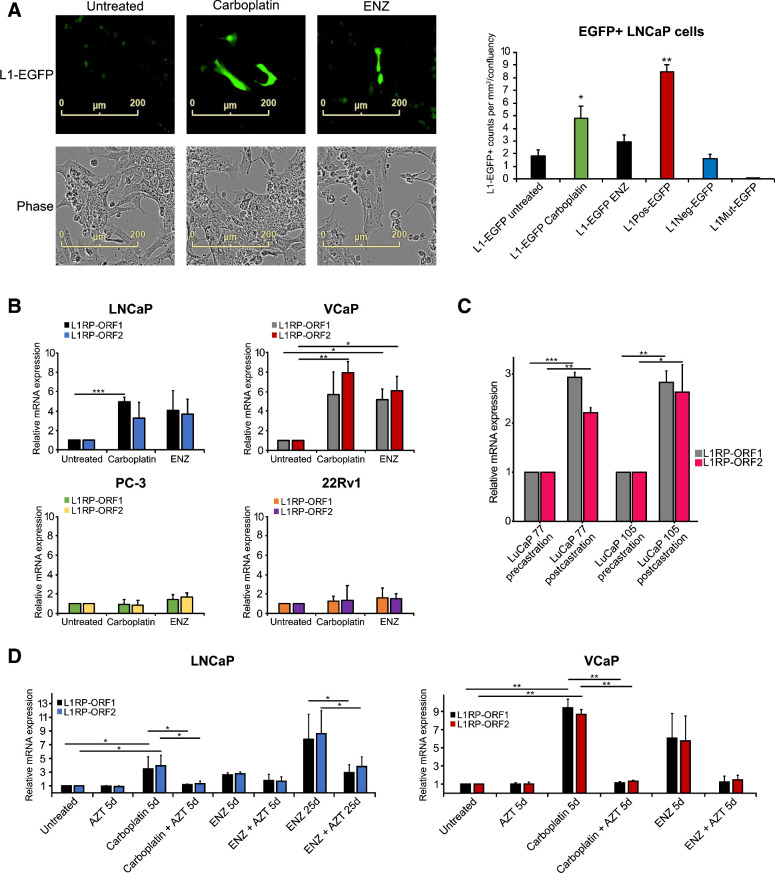
Since the pedigree of somatic L1 transposon integration in A34 metastatic cancer cells is substantially different in eradicated and resistant subclones and may be associated with differences in response to chemotherapy, we hypothesized that L1 activity is triggered by androgen deprivation or chemotherapy itself. To test this hypothesis, we exposed prostate cancer cell lines to carboplatin or ENZ and studied L1 retrotransposon activation using a retrotransposition L1-EGFP reporter assay (Fig. 3A, Supplementary Fig. S5A and S5B). L1 retrotransposition indicated by green fluorescence appeared in LNCaP prostate cancer cells 5 days after commencement of exposure to carboplatin (Fig. 3A). With the same assay, exposure of LNCaP to ENZ showed a relative increase in L1-EGFP reporter, below statistical significance relative to control cells (Fig. 3A). We then examined L1 ORF1 and ORF2 mRNA levels in untreated, 5-day carboplatin-treated, and 5-day ENZ-treated LNCaP, VCaP, PC3, and 22Rv1 cells measured by qPCR. Carboplatin induced an approximately 4-fold increase in ORF1 and ORF2 expression in LNCaP cells (P = 0.004 for ORF1 and P = 0.139 for ORF2, t test) and a 7-fold increase in expression in VCaP cells (P = 0.073 for ORF1 and P = 0.009 for ORF2, t test), but not in PC3 and 22Rv1 prostate cancer cells (Fig. 3B). Notably, ENZ induced ORF1 and ORF2 expression by 5.5-fold only in VCaP cells (P = 0.022 for ORF1 and P = 0.026 ORF2, t test; Fig. 3B). These findings taken together with the L1 insertion events observed in A34 suggest that induction of L1 transposon activity by chemotherapy and/or androgen-deprivation therapy may be a contributor to genomic heterogeneity in cancer cell populations harbored in individual patients' tumors.
Figure 3.
Carboplatin and enzalutamide induce L1 transposon activity in prostate cancer cells, and this activity can be blocked by AZT. A, The effects of carboplatin (5 μmol/L) or ENZ (10 μmol/L) on L1 activity in LNCaP prostate cancer cells analyzed using a retrotransposition L1-EGFP reporter assay and IncuCyte S3 imaging equipped with a green fluorescence channel. Representative IncuCyte cell images are shown for untreated, ENZ-, and carboplatin-exposed LNCaP cells with green channel only (L1-EGFP) and phase contrast. The control plasmids used included two negative controls (L1Neg-EGFP and L1Mut-EGFP) and a positive-control plasmid (L1Pos-EGFP). The barplot shows L1-positive EGFP+ counts per mm2/confluency quantified using IncuCyte. See also Supplementary Fig. S5. B, The effects of carboplatin or ENZ on ORF1 and ORF2 mRNA expression in LNCaP, VCaP, PC3, and 22Rv1 prostate cancer cells. C, L1 ORF1 and ORF2 mRNA levels from pre- and postcastration xenograft samples from LuCaP 77 and LuCaP 105 (23). D, The effects of carboplatin and AZT (50 μmol/L) or ENZ and AZT alone and in combination for 5 and 25 days on ORF1 and ORF2 expression in LNCaP cells and for 5 days in VCaP cells. Asterisks indicate significant difference between treatment conditions based on t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test if L1 induction is also occurring in tumor tissues as a result of antiandrogen treatment, we examined L1 ORF1 and ORF2 mRNA levels using qPCR in patient-derived LuCaP xenograft models, expanded in intact and castrate mice (LuCaP 77 and 105; ref. 23). Castration resulted in an approximately 3-fold increase in ORF1 and ORF2 expression in both xenografts (P < 0.001 for ORF1 and P = 0.002 for ORF2 in LuCaP 77, P = 0.005 for ORF1 and P = 0.037 for ORF2 in LUCaP 105; Fig. 3C), consistent with the cell line data. In a complementary approach, we additionally quantified L1 expression (see Methods) from previously published RNA-seq data from the same xenograft models (24) where castrated mice had higher levels of L1 mRNA (Supplementary Fig. S6A). We also analyzed previously published data from a head and neck cancer cell line SCC25 treated with PBS or with cetuximab for 11 weeks (25) and examined transcriptomically once per week. Cetuximab-treated cells showed higher L1 mRNA levels during the first 5 weeks of treatment (P = 0.023, paired t test; Supplementary Fig. S6B). Taken together, these results suggest that L1 activation under stress may be a common phenomenon in cancer.
Antiretroviral drug AZT reverses treatment-induced L1 activity in prostate cancer cells
As an antiretroviral compound, nucleoside analogue reverse transcriptase inhibitor (nRTI) AZT has previously been reported to suppress L1 retrotransposition in HeLa cells (26). We hypothesized that AZT may be able to reduce carboplatin- and ENZ-induced L1 activity. Thus, we explored the effect of AZT on both ORF1 and ORF2 expression alone and in combination with carboplatin or ENZ in LNCaP and VCaP prostate cancer cells in vitro. In addition, as ENZ-induced ORF1 and ORF2 expression was only seen in VCaP cells after a 5-day delay, we included a longer 25-day exposure period for ENZ and AZT alone and in combination in LNCaP cells to determine whether ORF1 and ORF2 expression is altered in response to acquired resistance to ENZ. Next, AZT was introduced to both LNCaP and VCaP cells alone and in combination with carboplatin or ENZ. The results revealed that while carboplatin alone induced ORF1 and ORF2 mRNA expression approximately 4-fold compared with control-treated LNCaP (P = 0.036 for ORF1 and P = 0.013 for ORF2, t test) and 9-fold compared with control-treated VCaP cells (P = 0.004 for ORF1 and P = 0.002 for ORF2, t test), AZT alone did not have any significant effect on basal ORF1 and ORF2 mRNA expression (Fig. 3D). Moreover, combined administration of carboplatin and AZT kept the ORF1 and ORF2 levels at the basal level and no induction of ORF1 or ORF2 expression was detected in the combination treated LNCaP (P = 0.045 for ORF1 and P = 0.017 for ORF2, t test) or VCaP cells (P = 0.004 for ORF1 and P = 0.001 for ORF2, t test; Fig. 3D). Interestingly, while no significant changes of ENZ on ORF1 and ORF2 mRNA expression were seen after a 5-day delay in LNCaP cells, a longer 25-day ENZ incubation significantly induced ORF1 and ORF2 mRNA expression by 8-fold in LNCaP cells (P = 0.014 for ORF1 and P = 0.007 for ORF2, t test). Further, addition of AZT reduced the ENZ-induced ORF1 and ORF2 mRNA expression by 60%, leading to only 3-fold ORF1 and ORF2 induction in ENZ and AZT combination-treated LNCaP cells (P = 0.038 for ORF1 and P = 0.029 for ORF2, t test; Fig. 3D). AZT was also able to block L1 transposon activity in the L1-EGFP reporter assay (P < 0.05, t test) (Supplementary Fig. S6C). Additionally, AZT blocked the ENZ induced ORF1 and ORF2 mRNA expression in VCaP cells (Fig. 3D). On the protein level, carboplatin treatment of LNCaP cells significantly increased the level of L1 ORF1p produced by the cells, while further addition of AZT returned ORF1p levels to normal (Supplementary Fig. S6D). These results suggest that AZT prevents carboplatin- and ENZ-induced ORF1 and ORF2 expression and could be a modulator of cancer genome heterogeneity induction by L1 activity in cancer cells in patients during therapy.
Discussion
By combining deep longitudinal analysis of clinical, liquid biopsy, and tumor genomic data in patient A34′s mPC, we identified what we believe to be the first genomically documented case of differential cancer subclone eradication and resistance in a solid tumor (2). In the current study, we sought to delve deeper, to identify potential underlying causes of differential susceptibility to therapy, and to test the idea that DSER analysis could provide a highly informative intermediate step toward effective precision cancer medicine. DSER can be defined as a direct comparison of molecular attributes of pre- and posttreatment cancer subclones eradicated by and resistant to therapy to identify molecular targets for therapeutic conversion of resistant subclones to an eradicable state.
Our results revealed that in the eradicated cancer subclones of patient A34, the already-reduced DNA damage repair capacity provided by BRCA2 biallelic loss may have been potentiated by reduced activity of EYA4 and/or FANCI. The resistant subclones may have survived chemotherapy because of a RB1 S485F mutation. As a precision-medicine hypothesis-generating system, we believe this type of analysis could form a new and much needed bridge to more rapid progress in precision medicine, both in individual patients and in advancing mechanistic understanding in general. In patient A34 or cells in vitro mimicking A34, could the addition of drugs blocking the FANCD2 or FANCI, and/or drugs blocking EYA4 activity force the resistant subclones to be eradicated by platinum/etoposide or DNA-repair–inhibiting drugs such as olaparib? Because the root observations are in vivo and naturally developed in human, the differences observed using DSER are arguably more likely to identify useful vulnerabilities than studies starting with cell line or animal studies. Moreover, when hypotheses generated by DSER are tested in vitro and in animal studies, the power to unravel important mechanisms is likely to be substantially amplified.
How can we test the hypothesis that broadly applied DSER could accelerate progress in precision cancer medicine? The most dramatic recent advances in metastatic cancer treatment have occurred in cancers harboring DNA-repair defects (10) and in those responsive to recently developed immunotherapy methods, but these treatments typically do not cure the patient. This makes them the best place to apply DSER. The tools required to do DSER on a broad scale are relatively modest. First, whole genome-sequenced samples of tumor and blood are required prior to treatment and at the time of relapse. What can be learned about any eradicated and resistant subclones identified in the samples will be directly proportional to the quality and sufficiency of the samples for integrated clinical and molecular analysis. Whether or not subclone eradication is common enough with current partially effective therapies to warrant broader expansion of a DSER approach should become evident with as few as perhaps 20 patients with each tumor-treatment–type combination. This method could be added to existing clinical trials and could be a specific additional focus of existing large studies of metastatic cancer such as those reported by Robinson and colleagues (27) and Swanton and colleagues (28).
Performing DSER in patient A34 caused us to ask whether L1 transposon activation itself is a targetable source of cancer genomic heterogeneity leading to differential sensitivity to therapy.
Our results are to our knowledge the first to show that either carboplatin or enzalutamide can turn on L1 transposon machinery in cancer cells, and that this induced activation can be blocked in vitro by the antiretroviral drug AZT. We were surprised to discover that L1 activation after carboplatin occurs only after a 5-day delay in 2 of the 4 prostate cancer cell lines tested. The delay may explain why L1 activation was not detected in a previous study where osteosarcoma cells were studied immediately after cisplatin exposure (29). These findings indicate that induced L1 activity in response to stresses such as carboplatin chemotherapy or androgen deprivation may contribute to genomic heterogeneity-driven resistance to therapy in some cancers.
What remains unknown about somatic L1 induction in cancer is what initiates the initial binding of RNA Polymerase II to begin the transposition process. Our results provide a foundation for mechanistic studies of L1 induction, and could help reveal why the prevalence of somatic L1 induction varies among cancer-cell types (14). In prostate cancer, we speculate that differences in driver genes and remaining androgen receptor (AR) response pathways could underlie the L1 inducibility shown in LNCaP/VCaP cells that is lacking in PC3/22Rv1 cell lines.
It appears possible that in A34 L1 activity induced by carboplatin and etoposide chemotherapy could have contributed to genomic instability leading both to the eradicable and resistant subclones. Testing whether or not chemotherapy-induced L1 activity is clinically important or not could be tested in vivo by comparing ctDNA response with chemotherapy in patients treated with prechemo suppression of L1 by AZT versus placebo. AZT is currently included in several clinical trials in leukemia, lymphoma, and other cancers. The impact of AZT on L1 activation could be studied in samples from patients enrolled in those trials (30).
In summary, future work testing of DSER-based identification of novel cancer therapeutic targets, and deeper mechanistic studies of the role and manipulability of L1 induction in cancer evolution and therapy could accelerate progress in development of effective precision oncology.
Authors' Disclosures
K. Ketola reports grants from Academy of Finland, Sigrid Jusélius Foundation, and grants from Cancer Society of Finland during the conduct of the study. S. Taavitsainen reports grants from Finnish Cultural Foundation during the conduct of the study. R. Kaarijärvi reports grants from Finnish Cultural Foundation during the conduct of the study. M. Nykter reports grants from Cancer Society of Finland, Sigrid Jusélius Foundation, Academy of Finland, Jane And Aatos Erkko Foundation, Finnish Cancer Institute, and grants from Tampere University Hospital during the conduct of the study; grants from EU Horizon 2020 and Business Finland outside the submitted work; in addition, M. Nykter has a patent for Differential Subclone Eradication and Resistance Analysis pending. G.S. Bova reports grants from Cancer Society of Finland, Sigrid Jusélius Foundation, Finnish Cultural Foundation, and grants from Academy of Finland during the conduct of the study; in addition, G.S. Bova has a patent for Differential Subclone Eradication and Resistance Analysis pending. No disclosures were reported by the other authors.
Supplementary Material
Ketola et al Supplementary Material
L1 retrotransposition evens identified by TraFiC analysis in A34 in the current study.
FANCI siRNA experimental data depicted in Supp Fig S3
qPCR primers used for L1-ORF1, L1-ORF2, FANCI, and GAPDH.
SNVs and indels in A34 subclones. Eradicated subclones are subclone numbers 12, 13, 14, and 15.
Concordance of L1 insertion calls from the current study with insertion calls reported in Tubio et al.
Acknowledgments
The authors thank A34 and his family for participating in the PELICAN (Project to ELIminate lethal CANcer) integrated clinical-molecular autopsy study of prostate cancer. They thank M. A. Eisenberger, M. A. Carducci, V. Sinibaldi, T. B. Smyth, and G. J. Mamo for oncologic and urologic clinical support; W.B. Isaacs, P. Martikainen, R. Kylatie, M. Pirinen, H. Kallio, A. Koskenalho, J. Silander, G. Hutchins, and B. Crain for study support; LuCaP 77 and 105 xenograft tissue samples analyzed were generously provided by Robert L. Vessella (University of Washington Department of Urology). Computation was supported by Tampere University, Tampere, Finland, CSC Finland, and the Finnish Institute for Molecular Medicine, Helsinki, Finland. This work was carried out with the support of UEF Cell and Tissue Imaging Unit, University of Eastern Finland, Finland and Biocenter Finland. The study was financially supported by Cancer Society of Finland (2013-present); Sigrid Jusélius Foundation (2016-present); Finnish Cultural Foundation (2020-present); The Academy of Finland (2011-present); Cancer Research UK (2011–2014), PELICAN Autopsy Study family members and friends (1998–2004); John and Kathe Dyson (2000); US National Cancer Institute CA92234 (2000–2005); American Cancer Society (1998–2000); Johns Hopkins University Department of Pathology (1997–2011); Women's Board of Johns Hopkins Hospital (1998); The Grove Foundation (1998); Association for the Cure of Cancer of the Prostate (1994–1998); Brady Urological Institute (1991–1998), American Foundation for Urologic Disease (1991–1994).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
K. Ketola: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. H. Kaljunen: Formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Taavitsainen: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. R. Kaarijärvi: Investigation, visualization, writing–review and editing. E. Järvelä: Investigation, visualization, writing–review and editing. B. Rodríguez-Martín Software, formal analysis, investigation, visualization, writing–review and editing. K. Haase: Formal analysis, investigation, visualization, writing–original draft, writing–review and editing. D.J. Woodcock: Software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. J. Tubio: Software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. D.C. Wedge: Software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. M. Nykter: Conceptualization, resources, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. G. Bova: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 2015;6:6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woodcock DJ, Riabchenko E, Taavitsainen S, Kankainen M, Gundem G, Brewer DS, et al. Prostate cancer evolution from multilineage primary to single lineage metastases with implications for liquid biopsy. Nat Commun 2020;11:5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, et al. Mobile DNA in cancer. extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014;345:1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet 1999;8:1557–60. [DOI] [PubMed] [Google Scholar]
- 5. Ostertag EM, Kazazian HH. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 2001;11:2059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faulkner GJ, Billon V. L1 retrotransposition in the soma: a field jumping ahead. Mobile DNA 2018;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 8. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pomerantz MM, Spisák S, Jia L, Cronin AM, Csabai I, Ledet E, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 2017;123:3532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 11. Castella M, Jacquemont C, Thompson EL, Yeo JE, Cheung RS, Huang J-W, et al. FANCI regulates recruitment of the FA core complex at sites of DNA damage independently of FANCD2. PLOS Genetics 2015;11:e1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kais Z, Rondinelli B, Holmes A, O'Leary C, Kozono D, D'Andrea AD, et al. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep 2016;15:2488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 14. Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet 2020;1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013;14:880–93. [DOI] [PubMed] [Google Scholar]
- 16. Wood EJ, Chin-Inmanu K, Jia H, Lipovich L. Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse. Front Genet 2013;4:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson IM, Vucic EA, Enfield KSS, Thu KL, Zhang YA, Chari R, et al. EYA4 is inactivated biallelically at a high frequency in sporadic lung cancer and is associated with familial lung cancer risk. Oncogene 2014;33:4464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo M, Li Y, Shi X, Yang W, Zhou F, Sun N, et al. Aberrant methylation of EYA4 promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Cancer Sci 2018;109:1811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jividen K, Kedzierska KZ, Yang C-S, Szlachta K, Ratan A, Paschal BM. Genomic analysis of DNA repair genes and androgen signaling in prostate cancer. BMC Cancer. 2018;18:960. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6180441/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature Methods 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 22. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLOS ONE 2012;7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease an–d serve as models for evaluating cancer therapeutics. Prostate. 2017;77:654–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Annala M, Kivinummi K, Tuominen J, Karakurt S, Granberg K, Latonen L, et al. Recurrent SKIL-activating rearrangements in ETS-negative prostate cancer. Oncotarget 2015;6:6235–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein-O' Brien G, Kagohara LT, Li S, Thakar M, Ranaweera R, Ozawa H, et al. Integrated time course omics analysis distinguishes immediate therapeutic response from acquired resistance. Genome Med 2018;10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, Ostrowski MA. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One 2008;3:e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson DR, Wu Y-M, Lonigro RJ, Vats P, Cobain E, Everett J, et al. Integrative clinical genomics of metastatic cancer. Nature 2017;548:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swanton C. Take lessons from cancer evolution to the clinic. Nature 2020;581:382–3. [DOI] [PubMed] [Google Scholar]
- 29. Farkash EA, Kao GD, Horman SR, Prak ETL. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res 2006;34:1196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Armando RG, Gómez DLM, Gomez DE. New drugs are not enough-drug repositioning in oncology: an update. Int J Oncol 2020;56:651–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ketola et al Supplementary Material
L1 retrotransposition evens identified by TraFiC analysis in A34 in the current study.
FANCI siRNA experimental data depicted in Supp Fig S3
qPCR primers used for L1-ORF1, L1-ORF2, FANCI, and GAPDH.
SNVs and indels in A34 subclones. Eradicated subclones are subclone numbers 12, 13, 14, and 15.
Concordance of L1 insertion calls from the current study with insertion calls reported in Tubio et al.



![Figure 1. Genomic evolutionary analysis of primary and metastatic samples and subclones in patient A34 reveals distinct genomic features in resistant and eradicated subclones. A, Subclonal structure of the lethal metastatic prostate cancer of patient A34, modified from Woodcock and colleagues (2), presented as a phylogenetic tree and subclone and sample button plot. Of the 19 total subclones, 17 were used to construct the phylogenetic tree (two subclones were excluded as they contained >50% indels). The red box denotes subclones eradicated by treatment that were not detected in autopsy samples, and the light blue box outlines subclones found at autopsy that resisted chemotherapy. In the phylogenetic tree, dotted lines connect the final subclone of a lineage with a letter denoting the sample or samples in which it was observed. The color of each subclone reflects its location as shown in the key. Red numbers indicate the total number of substitutions present at each point in the phylogenetic tree based on WGS data. The button plot also illustrates the subclones detected in each sample, with the area of each circle corresponding to the cancer cell fraction (CCF) of the subclone [CCF of 1 is found in the top (truncal) nodes of the plot]. Circles outlined with dashes indicate subclones not detected in a sample. Below the button plot, the treatment exposure of samples from patient A34 is shown as a matrix. Eradicated subclones continued to exist after exposure to external beam radiation and androgen deprivation, and therefore subclone eradication is associated with carboplatin plus etoposide chemotherapy. Samples are labeled according to the time of collection (yrsptd, years prior to death) and location. The SacralBoneMet sample is a spinal cord compressing sacral nerve root metastasis removed 11 yrsptd of the patient. B, Circos plot tree of L1 insertion events in the WGS samples from A34. Curved lines are L1 transductions with a determined source element, with each line colored according to the chromosome of its source. Black triangles denote solo L1 integration sites (source element unknown). Somatic L1 integration in TARID specific to the eradicated subclone is marked. C, Barplots of methylation percentage in the CpG island in the promoter region of the EYA4 gene, which may be affected by the L1 insertion into the 3′ adjacent sense–antisense paired TARID gene in the SacralBoneMet sample. Solid colored bars indicate significantly hypermethylated sites in the SacralBoneMet (black) relative to the same genomic positions in the LiverMet samples and autopsy blood samples (cyan). Violin plots show methylation percentage at the significantly hypermethylated CpG sites in the EYA4 gene promoter, potentially as a consequence of L1 insertion into TARID. D, Knockdown of FANCI reduces the proliferation of LNCaP but not PC3 cells. Scrambled (Scr) control siRNA was used as reference. The cell confluence was determined after a 5-day knockdown using IncuCyte S3 Image analysis tools. Asterisks indicate significant difference between sample conditions based on t test. **, P < 0.01.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4039/9397610/ee0a6df92262/4901fig1.jpg)