Abstract
Tumor suppressors represent a critical line of defense against tumorigenesis. Their mechanisms of action and the pathways they are involved in provide important insights into cancer progression, vulnerabilities, and treatment options. Although nuclear and cytosolic tumor suppressors have been extensively investigated, relatively little is known about tumor suppressors localized within the mitochondria. However, recent research has begun to uncover the roles of these important proteins in suppressing tumorigenesis. Here, we review this newly developing field and summarize available information on mitochondrial tumor suppressors.
Introduction
Cancer has been affecting people since the beginning of our history. Despite considerable effort in the last decades to find a cure, cancer-related death remains one of the leading causes of death worldwide. Around 18 million people are diagnosed with cancer worldwide and 9.6 million people succumb to this disease annually. The most common cancer types affecting humans are breast, colorectal, lung, and prostate cancer (1). Apart from the enormous toll cancer takes on lives of people, it also has a major impact on the world's economy; the total cost of cancer in Europe alone (in 2018) is estimated at 199 billion EUR (2). All these crucial factors create a great incentive to devise novel cancer treatments and diagnostic methods. Carcinogenesis, simplistically, requires two fundamental processes to occur: activation of proto-oncogenes to drive the cells' excessive proliferation and deactivation of tumor suppressor genes. Tumor suppressors are early defense molecules, which function as cellular brakes, stopping cells from uncontrollably dividing, therefore halting their progression toward carcinogenesis. Tumor suppressors were not specifically evolved to fulfil the tumor suppressor role; instead, each of them has a distinct and important role in cellular biology, which helps them to protect the cell from cellular transformation. Knudson's pioneering studies of retinoblastomas led to the discovery of the first tumor suppressor, the retinoblastoma gene and protein (3–6), which functions as a regulator of cell-cycle progression. The discovery of another famous tumor suppressor—p53, often referred to as the “guardian of the genome”—followed shortly thereafter (7–11). P53 was evolved to protect the genome of the cell from DNA damage, thus preventing genomic instability that precedes cellular transformation. Since then, many tumor suppressor genes and proteins have been discovered, including VHL (Von Hippel–Lindau), APC (adenomatous polyposis coli), and PTEN (phosphatase and tensin homolog). Although the great majority of the known tumor suppressor proteins reside in cytosol or nucleus, there is an expanding list of new, mitochondria-localized proteins that exert tumor-suppressive effects. Even though mitochondria are not considered the ultimate drivers of carcinogenesis, it is widely accepted that changes in mitochondrial processes, such as metabolic reprogramming and reactive oxygen species (ROS) dysregulation, can greatly facilitate this process.
The roles of the mitochondrial tumor suppressors are varied and include alterations in metabolic pathways, cellular differentiation, immune response, redox status, lipid biosynthesis, and mitochondrial dynamics. As such, they could provide an important link to cancer-associated processes such as metabolic reprogramming, the Warburg effect, inflammation, and stemness. Here, we review several members of the mitochondrial tumor suppressor family: lactamase-like B protein (LACTB), proline oxidase (POX), fumarate hydratase (FH), sirtuins (SIRT), fusion protein 1/tumor suppressor candidate 2 (FUS1/TUSC2), succinate dehydrogenase (SDH), and microtubule-associated scaffold protein 1 (MTUS1; Fig. 1; Supplementary Table S1). It is well documented that the function of many tumor suppressors is often context dependent; while these proteins can suppress tumorigenicity in certain contexts, such as specific tissues, tumor types, and (epi) genetic backgrounds, they can promote tumorigenicity in others. Therefore, to provide a complete review of the aforementioned tumor suppressors, we also briefly discuss the specific physiologic contexts where some of these proteins display a tumor promoter function, thus illustrating the complexity of tumor biology.
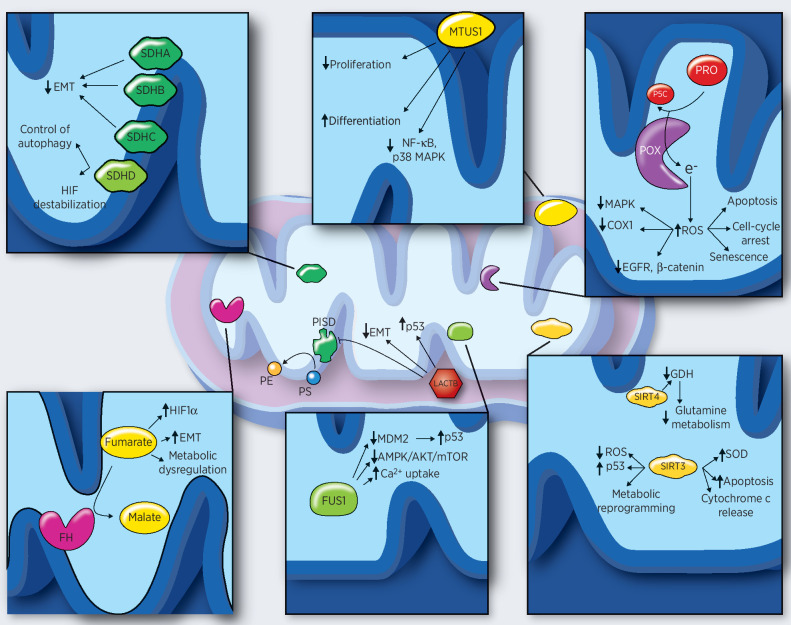
Figure 1.
Individual mitochondrial tumor suppressors and their main mechanisms of action. LACTB expression leads to stabilization of P53, downregulation of PISD and EMT, and induction of differentiation. SDHA, SDHB, and SDHC decrease EMT, whereas SDHD controls the autophagy and induces HIF1α destabilization. MTUS1 controls NF-κB and MAPK signaling pathways and is able to induce differentiation and repress cancer cell proliferation. Increased ROS levels, due to the enzymatic activity of POX, impair MAPK, EGFR, and Wnt/β-catenin signaling pathways; decrease COX1 levels; and induce apoptosis, cell-cycle arrest, and senescence. FH catalyzes the reaction from fumarate to malate. Accumulation of fumarate, due to FH inactivation, induces HIF1 stabilization, EMT, and metabolic dysregulation. FUS1 stabilizes p53 through MDM2, inhibits AMPK/AKT/mTOR pathways, and enhances Ca2+ uptake. SIRT3 activity leads to p53 stabilization decrease in ROS levels, metabolic reprogramming, apoptosis, and increase in SOD levels. SIRT4 downregulates the glutamine metabolism through GDH. GDH, glutamate dehydrogenase; MDM2, mouse double minute 2; PE, phosphatidylethanolamine; P5C, Δ1-pyroline-5-carboxylate; PISD, phosphatidylserine decarboxylase; PRO, proline; PS, phosphatidylserine; SOD, superoxide dismutase.
This review only describes those mitochondrial proteins whose tumor-suppressive function has been confirmed by four or more independent studies. Because the available information about these proteins is still sparse and fragmentary, it was our ambition to lay a foundation and create a resource upon which future research could be built.
LACTB
Evolutionary origin
The eukaryotic serine β-lactamase–like protein (LACTB) is a mitochondrial protein, evolutionary related to the bacterial penicillin-binding/β-lactamase proteins family (12). It is most likely derived from bacteria by horizontal or endosymbiotic gene transfer. Its conserved catalytic serine residue suggests that it still possesses peptidase or esterase activity. In bacteria, these classes of proteins are involved in bacterial cell wall biogenesis and peptidoglycan synthesis. However, because eukaryotic cells lack cell wall structures, the role of LACTB there remains unknown and is of great interest (12).
Localization and characterization
Mapped to the 15q22.1 chromosome, the human LACTB is a 547 amino acid long ubiquitously expressed protein, the highest expression being in skeletal muscles, heart, and kidneys (13, 14). LACTB is localized in the mitochondrial intermembrane space, and its localization depends on the N-terminal 62 AA-long mitochondrial targeting sequence (15).
In 2009, Polianskyte and colleagues demonstrated that LACTB can give rise to filaments formed by tetrameric subunits, suggesting its role in intramitochondrial membrane organization and micro-compartmentalization (15). LACTB belongs among the essential factors for proper activity of electron transport chain's (ETC) complex I, pointing to a regulatory role of LACTB in the respiratory chain (16). LACTB was also shown to be associated with mitochondrial ribosome (17). Transgenic mice with upregulated LACTB levels varied notably from the control mice, their fat-mass-to-lean-mass ratios being 20% higher than the wild type control. This suggests LACTB's role in obesity and fatty acid metabolism (18, 19). A study in 2016 showed that LACTB influences the progression of atherosclerotic plaques through modulating the levels of inflammatory chemokines, implying a role in regulation of immunity (20).
LACTB as a tumor suppressor
In 2017, while researching the expression profiles of “cancer-resistant” tissues, Keckesova and colleagues identified LACTB as a potential tumor suppressor (Fig. 2). Subsequent in vivo, in vitro, and clinical studies demonstrated LACTB's tumor-suppressive function in breast cancer (14). LACTB is downregulated in various breast cancer cell lines as well as in human breast cancer samples, where it showed 34% to 42% protein level decrease compared with normal breast tissue. LACTB overexpression, in a panel of tumorigenic and nontumorigenic cell lines, led to a decrease of proliferation rate in cancer cells, while nontumorigenic cells were minimally affected. This effect was also observed in vivo; the induction of LACTB in already formed tumors had a negative effect on their growth (14). LACTB knockdown in normal human epithelial cells in combination with overexpression of an oncogene (such as HRAS G12V or MYCT58A) led to malignant transformation of these cells, resulting in tumor formation, further confirming the tumor suppressive role of LACTB (14). Dysregulation of LACTB in breast cancer is associated with high expression of miR-374a, which promotes cancer cell survival and growth in vitro and in vivo. MiR-374a knockdown suppresses the cells' proliferative and colony formation activity, as well as migration and invasion capacity. LACTB silencing, however, reverses these changes (21). Further in vitro and in vivo studies demonstrated that LACTB has a negative impact on cancer growth, migration, and/or invasion also in gliomas, hepatocellular carcinomas, and colorectal cancer (22–25). In glioma A172 cells, the overexpression of LACTB significantly inhibited their proliferation. Its expression was shown to be involved in the invasion and angiogenesis process through the downregulation of proliferating cell nuclear antigen (PCNA), matrix metalloproteinase 2 and 9 (MMP2, MMP9), VEGF, proteins important in glioma cell proliferation, invasion, and angiogenesis (22). LACTB is also strongly downregulated in colorectal cancer, which is considered one of the most aggressive malignancies, and low levels of LACTB predict a poor prognosis in patients (23, 25). Interestingly, the downregulation of LACTB in colorectal cancer is not due to genetic mutations, but depends on epigenetic changes in its promoter: mainly promoter methylation and histone H3 hypoacetylation (25).
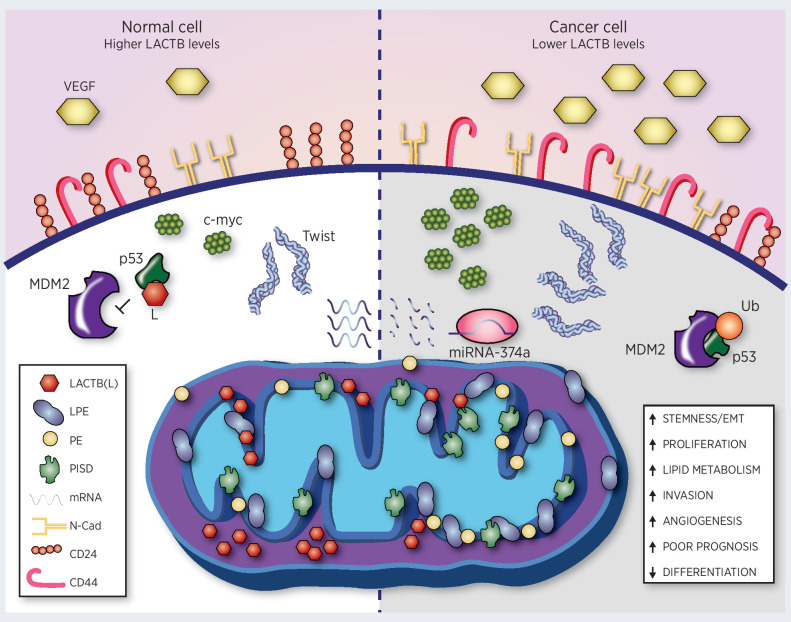
Figure 2.
LACTB effects in a normal cell and cancer cell. LACTB downregulation in cancer cells (through expression of miRNA-374a) leads to an increase in stemness/EMT (through increases of EMT factors such as Twist), angiogenesis (through increased levels of VEGF), proliferation (through increased levels of myc), invasion, lipid metabolism, and a decrease in differentiation. Lower LACTB levels in cancer cells also leads to degradation of p53 through mouse double minute 2 (MDM2). Increasing levels of LACTB to the physiologic levels in normal cells leads to decreases of mitochondrial lipid metabolism (through downregulation of PISD, PE, and LPE in mitochondrial membranes), stabilization of p53, and induction of differentiation (as manifested by increase in epithelial markers and decrease in mesenchymal markers). N-Cad, N-cadherin.
In vitro and in vivo experiments showed that in colorectal cancer cells, LACTB suppresses tumor growth and metastasis and can directly interact with p53 protein thus preventing its degradation by mouse double minute 2 (MDM2). This axis can potentiate the tumor suppressive effects promoted by both, p53 and LACTB, proteins. Moreover, p53 depletion reduces the tumor suppressive activity of LACTB when it is overexpressed in colorectal cancer (25). While this study provided interesting mechanistic insight into LACTB function, it did not clarify whether the interaction between p53 and LACTB occurs inside the mitochondria or in the cytoplasm. Thus far, two studies attempted to elucidate the function of tumor-suppressive function of LACTB outside the mitochondria with opposing conclusion. In breast cancer, deletion of mitochondrial localization sequence within LACTB abrogated its ability of tumor suppression, pointing to the importance of LACTB's mitochondrial localization for its tumor suppressor function (14). However, another study, performed in melanomas, showed cytoplasmic interaction of LACTB with serine/threonine–protein phosphatase (PP1A). This interaction was shown to prevent the binding between yes-associated protein (YAP) and PP1A in melanocytes thus repressing their malignant transformation (26). Future studies in this area are needed to clarify the importance of nonmitochondrial LACTB in tumor suppression and to define specific contexts when such nonmitochondrial tumor suppression occurs.
Cell differentiation
Breast cancer cells that survived LACTB induction displayed a more differentiated, epithelial-like morphology (14). This change in morphology was accompanied by an increase in the levels of epithelial differentiation markers, such as CD24 and epithelial cell adhesion molecule (EPCAM), and a decrease of certain mesenchymal markers [CD44, zinc finger E-box-binding homeobox 1 (ZEB1)]. This resulted in decreased proliferation and impaired tumorigenesis. A similar observation was noted in tumor tissues (14). The ability of LACTB to promote a differentiated phenotype was also observed in colorectal cancer (23, 25). Zeng and colleagues in 2018 demonstrated that in HCT116 and HCT8 cancer cells, stable LACTB overexpression promotes a morphologic change from a “spindle-like” into the tight “cell-to-cell” adhesion phenotype, while LACTB knockout reverses this process (25). These results showed that LACTB overexpression increased the expression of epithelial cell markers such as E-cadherin and β-catenin. In contrast, N-cadherin, vimentin, c-Myc, cyclin D1, and Twist1 mesenchymal markers were strongly downregulated, revealing a contribution of LACTB to the inhibition of epithelial–mesenchymal transition (EMT) in colorectal cancer (23, 25).
The role of LACTB in differentiation was also confirmed in normal cells, namely in skeletal muscle cells (14, 27). During the myoblast differentiation process in C2C12 cells, LACTB levels gradually increase. This process is negatively regulated by miR-351-5p expression. Overexpression of miR-351-5p downregulated LACTB and its inhibition had the opposite effect. When LACTB was silenced, the proliferation rate of C2C12 myoblasts increased (through upregulation of cyclin-regulated factors), which led to inhibition of the differentiation process and impaired myogenesis (27).
Autophagy
LACTB in colorectal cancer promotes autophagy through increasing the LC3-II/LC3-I ratio and the Unc51-like autophagy activating kinase-1 (ULK1) expression levels. The promotion of autophagy by LACTB and the consequent inhibition of EMT (in colorectal cancer) was shown to be the result of the inactivation of the phosphoinositide 3-kinase/protein kinase B/mTOR (PI3K/AKT/mTOR) signaling pathway via regulation of the PIK3R3 level and downregulation of PI3K (23).
Lipid metabolism
The ability of LACTB to promote loss of tumorigenicity and onset of differentiation in breast cancer cells is partly dependent on its ability to reprogram the lipid metabolism (14). This is achieved through LACTB-dependent downregulation of the lipid-synthesizing mitochondrial phosphatidylserine decarboxylase (PISD) enzyme, which leads, in turn, to subsequent changes in the levels of mitochondrial lyso-phosphatidylethanolamine (LPE) and phosphatidylethanolamine (PE; Fig. 2). In hepatocellular carcinoma, metabolic dysregulation is an essential part of cancer progression and hepatocellular carcinoma is typically characterized by metabolic disorders (28). It was demonstrated that LACTB mRNA and protein levels were both downregulated in hepatocellular carcinoma, and decreased LACTB expression was associated with poor prognosis. On the other hand, its overexpression inhibited cancer cell proliferation, migration, invasion, and tumor growth (24). The expression and activity of important enzymes involved in lipid metabolism pathways in hepatocellular carcinoma (such as carnitine palmitoyltransferase 1A, acyl-coenzyme A dehydrogenase, and others) were shown to be significantly related to LACTB expression, reinforcing LACTB's role in lipid metabolism (24).
LACTB as a tumor promoter
LACTB is involved in cancer progression in nasopharyngeal carcinoma and pancreatic adenocarcinoma where elevated LACTB expression is associated with poor survival rate (29). In the nasopharyngeal carcinoma context, LACTB expression promoted metastasis while suppression of LACTB reduced cellular mobility and metastatic ability. LACTB suppression was shown to enhance histone H3 stability and its acetylation, thus inhibiting the activation of receptor tyrosine–protein kinase/epidermal growth factor receptor–extracellular signal-regulated kinase (ERBB3/EGFR-ERK) pathway (30).
POX
Another tumor suppressor protein localized in mitochondria is POX. POX, also known as proline dehydrogenase (PRODH), is the first enzyme of the proline metabolic pathway. Localized in the inner mitochondrial membrane, POX catalyzes the oxidation of proline into Δ1-pyroline-5-carboxylate (P5C). Electrons produced by this reaction are transferred onto ubiquinone, explaining the dependence of POX on complex III and IV for its proper activity (31). The connection with the electron transport chain, however, seems to be more profound. POX's expression in DLD-1 (human colorectal cancer) cells negatively regulated the expression of components of the ETC and was inhibited by complex II inhibitors and succinate (31).
POX as a tumor suppressor
The interest in POX as a potential tumor suppressor was sparked in 1997 by a study from Polyak and colleagues, which identified POX as one of the several genes whose expression was significantly increased by the apoptosis-causing p53 tumor suppressor (32). Indeed, p53 response elements were identified in both promotor and intronic regions of the POX gene (33, 34). Decreased levels of POX have been detected in multiple tumor tissue samples, most notably in kidney and digestive tract tissues (35). In renal cell carcinoma (RCC), protein levels of POX are negatively regulated by miR-23b* (36). A follow up in vitro study suggested that the c-myc oncogene can indirectly downregulate POX levels by the miR-23b* both in P493 and PC3 cancer cells (37).
Apoptosis
Although POX was initially discovered as one of the genes induced during p53-dependent apoptosis, it is also able to mediate apoptosis independently of p53, and its proapoptotic, tumor-suppressive effects have been confirmed both in vitro and in vivo (35, 38–40). As it turns out, ROS play a pivotal role in POX's mechanism of action (Fig. 1). Thus, increased POX expression is accompanied by a surge in ROS generation (41, 42), the presence of which has been found essential for the apoptotic effects of POX in cancer cells (42–45). Mechanistically, at least in some instances, ROS generated by POX is able to mobilize Ca2+, which in turn activates calcineurin and nuclear factor of activated T cells (NFAT), both of which were crucial for POX-mediated apoptosis in lung, renal, colon, and ovarian carcinoma cells (44). Interestingly, POX has been shown to activate the extrinsic apoptotic pathways in human colon cancer cells as well. Its expression was accompanied by increased cleavage of caspase-8 and higher levels of death receptor 5 (DR5) and TNF-related apoptosis-inducing ligand (TRAIL) mRNA which seems to be, at least partially, mediated by NFAT (45). Another study showed that troglitazone, a PPARγ ligand, also induced the expression of POX and increased its catalytic activity (46). The induction of PPARγ led to an increased formation of ROS by POX and onset of apoptosis in multiple human cancer cell lines (47, 48).
Signaling pathways
In colorectal cancer, the phosphorylation status and hence the activity of mitogen-activated protein kinase kinase (MEK), ERK (45), EGFR and glycogen synthase kinase-3β (thus the stability of β-catenin; ref. 42) was negatively affected by POX expression. Cyclooxygenase-2, which is frequently upregulated in colorectal cancer and often contributes to cancer development (49), was also downregulated as was the production of prostaglandin E2 (PGE2; ref. 42). Constitutively active MEK partially blocked apoptosis induced by POX (45) as did the addition of PGE2 (42). These effects were, to some degree, reversed by the introduction of manganese superoxide dismutase (MnSOD; refs. 42, 45), further reinforcing the importance of ROS as the main mediator of POX's tumor suppressive function. Hypoxia-inducible factor 1α (HIF1α) levels were shown to be greatly reduced by POX in both normoxic and hypoxic conditions in colon cancer cells. This is achieved through POX-dependent upregulation of α-ketoglutarate, a critical substrate for prolyl hydroxylation and degradation of HIF1α (35). This effect has been reversed by the aforementioned miR-23b*, where miR-23b* did induce the expression of HIF1α by inhibiting POX (36).
Cell-cycle arrest and senescence
In addition to apoptosis, POX was able to induce G2 arrest in human colon cancer cells. The arrest was accompanied by an upregulation of genes from the growth arrest and DNA damage–inducible gene family (GADD) such as GADD45α, GADDβ, GADDγ, and GADD34 (35). POX activity has been shown to be vital in inducing senescence in both transformed (human osteosarcoma, U2OS) and untransformed (human fibroblast, Hs68) cell lines in response to DNA damage and POX's ability to generate ROS was essential for this effect. Furthermore, ROS produced by the overexpression of POX could cause DNA damage itself (50).
POX as a tumor promoter
In certain contexts, namely during hypoxia and/or nutrient stress, POX was shown to act as a promoter of tumor survival. Under hypoxia, the expression of POX rose considerably in various cancer cell lines in vitro and in the hypoxic tumor microenvironment of the breast cancer MDA-MB-231 cells in vivo. The increased levels of POX contributed to the survival of cancer cells under hypoxia and glucose deprivation. In the hypoxic conditions, this was explained mechanistically by the ability of POX to induce ROS-dependent protective autophagy instead of apoptosis in cancer cells (51). Under nutrient stress, glucose-starved or rapamycin treated colorectal cancer cells displayed significant increase in the catalytic activity of POX. This led to the degradation of proline, which can be obtained by cells from breakdown of the extracellular matrix. Subsequent increase in ATP levels allowed the survival of cancer cells. These results indicated that the induction of proline cycle initiated by POX under conditions of nutrient stress may be a mechanism by which cells switch to a catabolic mode for maintaining cellular energy levels (52). Similar results were also observed in pancreatic ductal adenocarcinoma cells, where under conditions of nutrient stress POX-dependent oxidation of collagen-derived proline promoted cancer cell survival and proliferation (53). Cancer progression was also shown to be mediated by POX-dependent proline catabolism in non–small cancer lung cells (NSCLC). In this cancer type, POX is upregulated by lymphoid-specific helicase (LSH), an epigenetic driver of NSCLC, in a p53-dependent manner. The activity of POX then promotes EMT as well as expression of inflammatory cytokines [most likely by producing ROS and affecting inhibitor of nuclear factor kappa-B subunit alpha (IKKα) phosphorylation], whereas its knockdown reduced cell growth, impaired colony formation, downregulated markers of mesenchymal state, and upregulated epithelial marker (E-cadherin; ref. 54).
FH
FH, also known as fumarase, is a member of the tricarboxylic acid (TCA) cycle that catalyzes reversible hydration of fumarate to malate. Although the TCA cycle takes place in mitochondrial matrix, a so called “echoform” of FH can also be found in cytosol and can localize to nucleus upon DNA damage, where it participates in DNA damage response (55, 56).
FH as a tumor suppressor
Mutations in FH predispose to malignant paragangliomas and pheochromocytomas (57, 58) as well as to hereditary leiomyomatosis and renal cell carcinoma (HLRCC; ref. 59, 60). FH loss has also been reported in glioma, ependymoma, adrenocortical carcinoma, neuroblastomas, osteosarcoma, and Ewing sarcoma (61).
The underlying mechanisms of FH's tumor suppressive role are yet to be completely understood; accumulation of fumarate (through inactivation of FH), however, seems to be beneficial to cancer cells and could be considered the common denominator of most negative (i.e., cancer-promoting) effects in FH-defective cancer cells (Fig. 1). These effects can be roughly divided into two categories: fumarate itself directly acting as an inhibitor of various enzymes and fumarate as a source of cysteine modifications that can hinder protein activity. The accumulated fumarate can spontaneously modify proteins via Michael addition reaction with the sulfhydryl group of cysteine, thus creating S-(2-succinyl) cysteine (2SC; ref. 62). This modification inactivates proteins such as glyceraldehyde-3-phosphate dehydrogenase (with a possible link to diabetes and mitochondrial stress; refs. 62, 63), mitochondrial aconitase 2 (64), iron regulatory protein 2 (65), and Kelch-like ECH-associated protein 1 [KEAP1; which is a negative regulator of nuclear factor erythroid 2-related factor 2 (NRF2); ref. 66]. 2SC modification can be considered a reliable biomarker for detection of FH-defective tissues (67).
HIF1α
HIF1α expression in hypoxic regions triggers cancer plasticity and heterogeneity, promoting an aggressive and metastatic phenotype. Together with NF-B, they regulate over 1,000 genes, enhancing cell survival through the expression of growth factors, such as vascular VEGF, which contribute to a new vascularization within the tumor (68). In renal cancer, higher levels of stabilized HIF1α were detected in HLRCC-derived cell lines with inactive FH (69) as well as in FH-defective fibroblasts (70). HLRCC exhibits higher angiogenesis in association with increased transcript expression from the VEGF and BNIP3 hypoxia-responsive genes (71). In addition, HIF stabilization, in FH knockdown mouse embryonic fibroblasts (MEF), led to increased expression of MET oncogene, which in turn, helped further stabilize HIF, creating a feed-forward loop. Expression of MET positively affected motility and could contribute to tumor progression (72). Thus far, there are two proposed mechanisms of HIF stabilization in FH-defective cells: it has been shown that fumarate acts as an inhibitor of HIF prolyl hydroxylase thus blocking the path to HIF degradation (69, 73). A different study suggests that increased ROS (formed, at least partially, by NADPH oxidase and protein kinase C-δ activity in FH-deficient cells) is responsible for HIF prolyl hydroxylase inhibition, possibly though oxidizing cellular pool of Fe2+ to Fe3+, thus depriving HIF prolyl hydroxylase of its cofactor (74). FH-deficient cancer cells were, in fact, found to be iron deficient and had notably higher levels of HIF1α (75), pointing to a role of FH in iron metabolism.
Iron metabolism
Fumarate was found to inhibit the activity of ETC's complex II directly, via product inhibition, and complex I indirectly, thus reducing cellular respiration. As complex I contains iron–sulphur clusters, and some targets of fumarate-caused succinylation are proteins important for iron–sulphur clusters biogenesis, this could provide an explanation of complex I inactivity in FH-defective cancer cells (76). In HLRCC, accumulated fumarate dysregulates iron metabolism in two major ways, by covalently modifying iron regulatory protein 2, which is consequently unable to repress ferritin translation (65), and by enhancing ferritin translation via the activation of NRF2; ferritin then promotes the expression of forehead box protein M1 (FOXM1), which in turn promotes the proliferation (65). Moreover, the upregulated ferritin can lower cytosolic iron levels; this seems to correlate with the observation that FH-deficient cancer cells had lower cytosolic iron levels (75). This was further explained by reduced AMP-activated protein kinase (AMPK) levels leading to lower expression of iron divalent metal transporter 1 (DMT1; ref. 75), which seems to suggest that the dysregulation of iron metabolism in FH-defective cells happen via multiple ways.
EMT
FH deficiency was shown to induce the process of EMT, which promotes migration, invasion, and cancer progression. Fumarate acts as an inhibitor of α-ketoglutarate–dependent dioxygenases [such as ten-eleven translocation (Tet) enzymes responsible for DNA demethylation], thus affecting genome-wide histone methylation (77). Vimentin and other EMT-related genes were upregulated in Fh1−/− normal and cancer renal cells. The reintroduction of Fh1 into these cells was sufficient to diminish the expression of EMT-related genes and restore the expression of E-cadherin, leading to more epithelial-like morphology of the cells. Further experiments showed that the fumarate-induced inhibition of Tet enzymes and subsequent DNA hypermethylation, which led to downregulation of antimetastatic miRNAs from miR-200 family, was responsible for the expression of EMT-related transcription factors (78). The importance of FH inhibition in EMT is further reinforced in the example of nasopharyngeal carcinoma. LSH, which acts as a chromatin modifier, is upregulated in nasopharyngeal carcinoma and is important for its progression. LSH mediates silencing of FH transcription, which then promotes EMT and cancer progression through modulation of IKKα activity (79).
DNA repair
FH was shown to be able to promote DNA repair. Upon ionization-induced DNA damage, DNA-dependent protein kinase (DNA-PK) that participates in the nonhomologous end joining (NHEJ) DNA repair phosphorylates nuclear FH, which then associates with chromatin at the double-strand break (DSB) region. Locally produced fumarate enhances the recruitment of DNA–PK complex at the DSB region which promotes NHEJ-mediated DNA repair and survival (80). The involvement of FH in DNA damage response seems to be highly conserved as it has been primarily discovered in yeast, where it was shown to participate in homologous recombination (HR; refs. 81, 82). Subsequent studies confirmed and extended the importance of FH and fumarate in DNA repair and genomic stability (83, 84).
Oxidative stress
Accumulated fumarate can upregulate antioxidant response element-controlled genes (85). This might be accomplished by covalently modifying cysteine residues of KEAP1 (succinylation), which is then unable to repress NRF2-mediated antioxidant response pathway (66). An antioxidant phenotype could prevent ROS-induced DNA damage, thus providing cells with growth advantage. On the other hand, fumarate was also found to succinylate gluthathione (GSH), leading to persistent state of oxidative stress and causing senescence in vitro and in vivo (86). Fumarate can also indirectly activate Abelson tyrosine-protein kinase 1 (ABL1) proto-oncogene possibly via oxidative stress–dependent mechanism (87).
Modulation of aerobic glycolysis and AMPK signaling
FH is an important metabolic enzyme and its dysregulation in cancer cells is accompanied by considerable metabolic reprogramming of cancer cell metabolism. FH-defective HLRCC cells (UOK262) had significant changes in protein phosphorylation, leading to inhibition of pyruvate dehydrogenase's enzymatic activity, thus preventing carbon from entering the TCA cycle (88). This led to enhanced aerobic glycolysis and higher lactate production to compensate for the defects in energy production (75, 89). It is worth mentioning, however, that the rate of oxidative phosphorylation was not insignificant. Pentose phosphate pathway, specifically its oxidative branch, was also enhanced, leading to increased amounts of ribose and NADPH, the latter of which can be used in fatty acid synthesis (89). Cells with defective FH also used other compensatory mechanisms, such as glutamine-dependent reductive carboxylation (to produce Ac-CoA and citrate) and oxidative metabolism of glutamine to fumarate to restore the energy homeostasis (90).
AMPK is generally regarded as a master sensor of metabolism (91). The accumulated fumarate protected both FH-defective fibroblasts and renal cells from apoptosis by activating AMPK (70). In support of these findings comes a screen of synthetic lethality, where it was revealed that the inhibition of adenylate cyclase in FH-deficient cell lines (human embryonic kidney cells and HLRCC-derived UOK262) negatively affects the cells' survival (92). In contrast, a different study found that AMPK levels in FH-deficient kidney cancer cells were reduced and the cells exhibited dysregulation in iron metabolism (deficiency in cytosolic iron) as well as the activation of anabolic proteins (acetyl-CoA carboxylase and ribosomal protein S6) and iron regulatory proteins, all of which was accompanied by increased expression of HIF1α. The activation of AMPK and silencing of HIF1α decreased the tumorigenicity of these cells (75).
FH as a tumor promoter
Paradoxically, while cancer arises from errors in the DNA repair mechanisms, to divide correctly, cancer cells also need to have functional DNA repair. As such, the abovementioned DNA repair function of FH is sometimes beneficial to cancer cells. For example, in gastric cancer tissue, FH mRNA and protein levels are significantly increased. Its knockdown impaired DNA damage repair and resulted in better response to cisplatin-mediated chemotherapy both in vitro and in vivo (93). FH was also able to promote cell survival upon exposure to ionizing radiation in other cancer cell lines, such as U2OS from human osteosarcoma and GSC11 (human primary glioblastoma cells; ref. 80).
Thus, we have a paradoxical behavior of FH, where in cancer cells which do express FH, mitochondrially localized FH acts as a tumor suppressor (through HIF1 stabilization and other mechanisms) and nuclear-localized FH acts as a tumor promoter (through DNA repair mechanism). As to which of these two branches of FH action manifests more in cancer cells is currently thought to be context dependent and influenced by localization of FH, concentration of fumarate and malate and the stage of tumor development (61, 94, 95).
Sirtuins
The highly conserved sirtuin protein family has several roles spanning from metabolism to oxidative stress (96–98). Seven human sirtuins have been described, three of which, SIRT3, SIRT4, and SIRT5, were identified also in mitochondria (99–102). The biological function of these proteins is NAD+-dependent deacetylase and mono-ADP-ribosyltransferase (103–105). There is growing evidence, however, that they may possess other enzymatic activities such as desuccinylation and demalonylation (106). Despite a relatively large amount of information known about these proteins, there is still a lively debate about their complex role in cancer. As our review is primarily concerned with mitochondrial tumor suppressor, we will only focus on the function and properties of sirtuins 3 and 4 (Fig. 1).
Sirtuin 3 as a tumor suppressor
Studies have shown SIRT3 to be downregulated in various cancers, such as mantle cell lymphoma, leukemia, hepatocellular carcinoma, pancreatic cancer, lung cancer, or gastric cancer (107–111). Sirt3−/− MEFs presented gene instability, and a single oncogene expression by viral transduction induced cell transformation in vitro. In addition, Sirt3−/− mice generated estrogen receptor/progesterone receptor (ER/PR)–positive tumors in the mammary gland over 24 months (112). A dataset study performed with genomic RNA from 38 normal and 36 human breast cancer samples showed that SIRT3 expression is decreased in cancer compared with the normal tissue (112).
Glycolysis regulation
In mantle cell lymphoma, SIRT3 protein levels are reduced and relate to a poor clinical outcome (107). Similar results were found in chronic lymphocytic leukemia samples and malignant B cells (107). The absence of SIRT3 was accompanied by an increase in superoxide dismutase 2 (SOD2) and isocitrate dehydrogenase 2 (IDH2) acetylation as well as an increase in ROS level. Conversely, when SIRT3 is overexpressed, these cells exhibit a decrease of the Warburg effect (decreased lactate production and downregulation of several glycolytic genes), which is further diminished by a drop in glucose uptake (107), supporting the idea that SIRT3 negatively regulates glycolysis. SIRT3 mediates the metabolic reprogramming through the destabilization of HIF1α (113, 114). Expression of SIRT3 is downregulated in human breast cancer and its loss correlates with upregulation of HIF1α target genes, such as GLUT1, HK2, PGK1, PDK1, LDHA, and VEGFA. On the other hand, overexpression of SIRT3 represses glycolysis and proliferation in breast cancer cells, confirming the metabolic mechanism for tumor suppression (113). Expression of SIRT3 can be controlled by the ZEB1 (109), a key regulator in metastasis and EMT (115). It has been demonstrated that ZEB1 can interact with the methyl-CpG binding domain protein 1 (MBD1) and suppress, through an interaction with the promoter in the E-box and Z-box, SIRT3 expression in a pancreatic cancer cell line, enhancing the aerobic glycolysis (109).
Activation of apoptosis
When compared with normal adjacent tissue, SIRT3 expression is downregulated in human lung adenocarcinoma and its reexpression significantly inhibits the cancer cells growth through Annexin V and caspase 3–mediated apoptosis. In this cell model, SIRT3 overexpression not only increased the BAX/BCL-2 and BAD/BCL-X/L ratios but also increased the levels of p53 and p21 and decreased ROS levels (110). In chronic myelogenous leukemia and promyelocytic human leukemia, SIRT3 activates apoptosis through caspase-3, Bcl-2, and cytochrome c pathway (116).
Modulation of cellular signaling pathways
In gastric cancer, SIRT3 protein and mRNA levels are significantly reduced both in cancer tissues and cell lines and overexpression of SIRT3 decreases cell proliferation and colony numbers (111). Conversely, its knockdown promotes cell growth and colony formation ability. Mechanistically, it was shown that SIRT3 inhibits the expression of Notch-1 both at mRNA and protein levels. Moreover, Notch-1 overexpression drastically reduces the inhibitory effects of SIRT3 on tumor cell proliferation (111). In addition, in cells from hepatocellular carcinoma, overexpressing SIRT3 leads to higher levels of p53 most likely through MDM2 downregulation (108). SIRT3 seems to play an important role in a plethora of pathways as its overexpression reduces the NAD+ level, activates the AKT and c-Jun N-terminal kinase (JNK) signaling pathways, and represses the ERK 1/2 pathway (108). JNK signaling pathway enhances apoptosis (117), whereas AKT and ERK stimulate cell growth (118). Imbalance in these pathways would affect cell proliferation and cell growth (in hepatocellular carcinoma), indicating the tumor-suppressive role of SIRT3.
Sirtuin 3 as a tumor promoter
The oncogenic role of SIRT3 was demonstrated in oral squamous cell carcinoma (OSCC), in which SIRT3 protein levels are elevated; its downregulation enhanced apoptosis and induced colony formation. Furthermore, cells with low levels of SIRT3 are more sensitive to cisplatin and ionizing radiation, indicating that presence of SIRT3 can induce resistance to chemotherapy and apoptosis in these cancer cells (119). The tumor promoting properties of SIRT3 were also reported in melanomas and RCC, where mRNA and protein levels of SIRT3 were increased compared with their nontumorigenic counterparts (119–121). Knockdown of SIRT3 in multiple melanoma cell lines resulted in decreased proliferation, inhibited colony formation, and reduced migration capacity and these cells exhibited senescence-like phenotype and dysregulation of the cell cycle. In a xenograft mouse model, SIRT3 knockdown caused a decrease in tumor growth (122). In RCC cell lines, SIRT3 was shown to have a role in glutamine oxidation, as SIRT3 knockdown had impaired proliferation, glutamine oxidation, and significantly reduced activity of glutamate dehydrogenase (GDH; ref. 123). SIRT3 has also been identified as rescuing EJ bladder carcinoma cells from p53-induced senescence and growth arrest by interacting with p53′s MASD (mitochondria-associated senescence domain) region between amino acid 64 and 209. It was shown to exert deacetylation activity in vitro on p53 peptide sequences (124).
Sirtuin 4
Sirtuin 4 is widely expressed in multiple tissues such as pancreas or human muscle (125). As is the case with SIRT3, its main function is also related to metabolism; it decreases the activity of GDH (126) and inhibits insulin secretion in pancreatic β cells (127). Under nutrient-depleted conditions, SIRT4 is activated and deacetylates malonyl-CoA decarboxylase, thus inhibiting the conversion of malonyl-CoA to acetyl-CoA, a crucial reaction in lipid metabolism. Indeed, mice lacking SIRT4 show an increase in malonyl-CoA decarboxylase activity and display an imbalance in lipid metabolism, protecting them against diet-induced obesity (128). It has been found that SIRT4 also has a lysine deacylase enzymatic activity; it removes acyl moieties from lysine residues (methylglutaryl-, hydroxymethylglutaryl-, and 3-methylglutaconyl-lysine), thus affecting leucine metabolism and insulin secretion (129).
Sirtuin 4 as a tumor suppressor
Glutamine metabolism
SIRT4 was found to be downregulated in hepatocellular carcinoma, colorectal, gastric, and lung cancer and esophageal squamous cell carcinoma (130–133). In gastric cancer, its downregulation was related with depth invasion and lymph node–positive cases (134). SIRT4 was discovered as a tumor suppressor involved in cellular metabolic response to DNA damage in lung cancer. The mechanism of action involves regulation of glutamine metabolism. This occurs via inhibition of GDH activity, which converts α-ketoglutarate (α-KG) into l-glutamine (135). Moreover, SIRT4 reconstitution decreased glutamine uptake, but not glucose, and repressed proliferation (135). Since this initial study, the involvement of SIRT4 in glutamine metabolism was observed and confirmed in other cellular models. A microarray study of 89 colorectal cancer cases showed that SIRT4 protein levels were decreased compared with the normal tissue and this decrease correlated with a worse prognosis. Its overexpression inhibited the proliferation of colorectal cancer cells in vitro and in vivo (131). Modulation of glutamine metabolism was once again found to be important for these effects as SIRT4 weakened the ability of colorectal cancer cells to utilize glutamine and enhanced cell death caused by glucose metabolism inhibitor 2-deoxyglucose. SIRT4′s role in metabolic regulation was further confirmed by its ability to inhibit pyruvate dehydrogenase (PDH) complex through the hydrolysis of the lipoamide cofactor (136), which binds to the E2 catalytic subunit and it is essential for PDH function (137). In colorectal cancer, SIRT4 influences EMT, leading to upregulation of E-cadherin expression and inhibition of migration, proliferation, and invasion by negatively affecting glutamine metabolism (138). This occurs through SIRT4′s repression of the activity of GDH. Supplementation of cells with α-KG abrogates the expression of E-cadherin by SIRT4 (138). The EMT-suppressive role of SIRT4 was further confirmed in gastric cancer, where overexpression of SIRT4 increased the expression of E-cadherin, and downregulated N-cadherin and vimentin (132). SIRT4 is also downregulated in lung cancer (133) and lung cancer cells transfected with SIRT4 exhibited inhibition of cell proliferation, cell cycle, cell invasion, and migration through the inhibition of the mitochondrial fission protein dynamin-1-like protein (Drp1) phosphorylation and downregulation of MEK/ERK signaling pathway (133).
Sirtuin 4 as a tumor promoter
Recent studies in esophageal squamous cell carcinoma (ESCC) demonstrated, that SIRT4 is upregulated in tumor tissues when compared with the adjacent normal tissue and SIRT4 levels inversely correlated with the mean survival time of ESCC patients (139). A similar phenotype was found in breast cancer, where SIRT4 proteins levels were increased compared with the nonneoplastic tissue counterparts and promoted migration, invasion, and proliferation of breast cancer cells (140). In a cell line derived from liver carcinoma, HepG2, SIRT4 overexpression under cellular stress conditions increased the cell survival and protected cancer cells against stress-induced cell death. Furthermore, SIRT4 promoted growth after DNA-damaging conditions such as radiation, UV irradiation, and cisplatin exposure (141).
FUS1/TUSC2
FUS1 (fusion 1 protein), also known as TUSC2 (tumor suppressor candidate 2) is a small, 110 AA-long, ubiquitously and highly conserved mitochondrial protein. It has been described as a regulator of Ca2+ ion uptake in the mitochondria, having a protective role in autoimmunity, cancer, and inflammation (142).
FUS1/TUSC2 as a tumor suppressor
FUS1, localized on the chromosome 3p21.3 (143), is frequently involved in cancer. Its expression is absent or reduced in majority of lung cancers (144). In nasopharyngeal carcinoma, its downregulation is controlled by miR-663b (145). miR-93, miR98, and miR-197 were also shown to bind to the 3′UTR region of the FUS1 transcript and decrease its expression (146). The expression of FUS1 is negatively regulated by sequence elements in both, 5′UTR and 3′UTR, regions of FUS1 mRNA (147). The tumor-suppressive properties of FUS1 were demonstrated through overexpression studies in lung cancer cells, which resulted in induction of apoptosis and inhibition of cell growth due to a cell-cycle arrest (Fig. 1; refs. 148, 149).
Stabilization of p53
Expression of both FUS1 and p53 synergistically decreased tumor growth and induced apoptosis in NSCLC. Levels of MDM2 were decreased when cells were cotransfected with p53 and FUS1, suggesting that the tumor suppressive role may be linked to the accumulation of p53 through FUS1-mediated downregulation of MDM2 (150).
Modulation of cellular signaling pathways
FUS1 was characterized as an inhibitor of several tyrosine kinases such as c-Abl, EGFR, platelet-derived growth factor receptor (PDGFR), and c-Kit (149, 151). Moreover, it was found that FUS1 is linked to the AMPK/AKT/mTOR signaling pathway in liver kinase B1 (LKB1)–defective NSCLC cells (152). LKB1 is a tumor suppressor, which is mutated or inactivated in 50% of NSCLC and was shown to activate the apoptotic regulator AMPK (153, 154) what leads to inhibition of AKT. FUS1 increases the effects of the AKT inhibitor, MK2206, by enhancing cell's sensitivity to this inhibitor. Combination of both FUS1 reexpression and MK2206 decreased tumor growth in a human LKB1-defective H322 xenograft mouse model. In vitro experiments showed that increased AMPK phosphorylation and the resulting increased kinase activity are responsible for this effect. AMPK knockout disrupted the FUS1–MK2206 cooperation. Moreover, combination of MK2206 treatment and FUS1 reexpression also decreased the phosphorylation and the enzymatic activity of mTOR and AKT. These results establish a connection between FUS1 expression and the AMPK/AKT/mTOR signaling pathway axis in the LKB1-deficient cells (152).
Calcium homeostasis and immunity
In mitochondria, FUS1 regulates calcium homeostasis. Epithelial cells, splenocytes, and activated CD4+ T cells with FUS1 knockout exhibited reduced calcium uptake into mitochondria what led to increased mitochondrial membrane potential and ROS generation. Increased ROS generation and NF-κB activation can lead to downregulation of programmed cell death protein 1 (PD-1) and programmed-death ligand 1 (PD-L1) levels in T lymphocytes (142), thus enhancing the autoimmune response and suppressing tumor immune evasion. FUS1 protein levels are increased during immune activation and are repressed by asbestos, tobacco exposure, or other environmental assaults in vivo (155). Fus1−/− mice showed an increase in chronic inflammation, suppression of antitumor defense, dysregulation of mitochondrial membrane potential, increased oxidative stress and genotoxicity, and dysregulation of the mitochondrial uncoupling protein 2 (UCP2) expression (155). These experiments support the role of FUS1 as a link between the mitochondrial homeostasis and the inflammatory response.
SDH
SDH is an enzymatic complex located within the inner mitochondrial membrane, responsible for the oxidation of succinate to fumarate accompanied by the reduction of ubiquinone to ubiquinol in electron transport chain. SDH is a heteromeric complex formed by four nuclear-encoded subunits: SDHA, SDHB, SDHC, and SDHD (156, 157). All subunits are essential for its proper function; for this reason, deleterious mutations in any of the SDH genes invariably result in a decreased SDH activity. Defects of SDH are comparatively rare in humans (156). Mutations or its inhibition leads to accumulation of succinate in mitochondria, causing disruption in ATP generation and mitochondrial impairment (156, 157). SDH is known to be involved in neuroprotection and its mutations can correlate with neurodegenerative disorders such as Parkinson disease (158). Moreover, inherited defects of SDH in humans are associated with encephalomyelopathy namely Leigh syndrome (157).
SDH as a tumor suppressor
Heterozygous germline mutations in SDHA, SDHB, SDHC, and SDHD have been identified to cause hereditary paragangliomas, pheochromocytomas, gastrointestinal stromal tumor (GIST), RCC, and breast cancer (159–161). Because of SDH's role in the electron transport chain, it has been suggested that ROS production can be induced by SDH-inactivating mutations showing the potentially big impact of an enzymatic complex on the genomic instability, apoptosis, and neoplastic transformation due to ROS accumulation (Fig. 1; refs. 162, 163). In cells with inhibited SDH, succinate accumulated in mitochondria is transported to the cytosol. Elevated cytosolic levels of succinate inhibit the activity of HIF prolyl-hydroxylase (PHD). This process stabilizes HIF1α what leads to expression of genes involved in the promotion of angiogenesis, metastasis, and metabolism, contributing to more aggressive tumor phenotypes (164).
SDHA and SDHB
The subunit A and B of the SDH complex are affected by germline mutations such as missense or nonsense mutations resulting in a loss of expression of both subunits (159, 165). It was also demonstrated that the SDHB promoter could be subjected to epigenetic modifications in primary neuroblastomas and phaeochromocytomas (166). The mutations of SDHB subunit are mostly present in paraganglioma, phaeochromocytoma, and in neuroblastoma cancer development, but SDHB and SDHA are also involved in breast cancer albeit with low frequencies; SDHA is mutated in 3.19% and SDHB in 0.1% of patients with breast cancer (161). SDHB is known to be involved in GIST. Twenty-seven percent of patients with GIST lack SDHA expression, which can be caused by heterozygous and somatic loss of both alleles (165).
EMT regulation
Subunit B is involved in the EMT process in various cancer types. A study published in 2016, demonstrated that SDHB downregulation facilitates the EMT process through hyperactivation of TGFβ signaling pathway and an increase in SNAIL1-SMAD3/4 mRNA and protein levels in colorectal cancer (167). SDHB-deficient metastatic pheochromocytomas/paragangliomas displayed a dramatic change in SNAI1/2 localization, which was translocated to the nucleus in all metastatic tumors. This translocation reflects the activation of SNAI1/2 as a transcription factor, which promotes the initial steps of the EMT (168). A connection between low SDH expression and EMT has also been described in serous ovarian cancer (162). In mice, Sdhb knockdown promotes histones hypermethylation (such as hypermethylation of H3K27), which, through promotion of EMT, contributes to the induction of a tumorigenic and metastatic phenotype. SDHB knockdown led to upregulation of various transcription factors involved in EMT, in particular to a dramatic increase of TWIST2 mRNA levels with a consequent downregulation of E-cadherin. These global epigenetic changes contributed to the reprogrammed carbon source utilization and tumor progression of ovarian cells in mice (162).
SDHC
SDHC also seems to be involved in gastrointestinal and breast cancer (169, 170). In breast cancer, its low expression promotes the expression of EMT genes such as vimentin (VIM), SNAI2, TWIST, and AXL (170).
SDHD
Mutations of subunit SDHD cause hereditary paraganglioma (171). Furthermore, SDHD mutations are associated with sporadic thyroid cancer (172). The mechanism, which promotes cancer progression in thyroid cells, is connected to the aberrant autophagy process in cells expressing mutated variant of SDHD. When functioning properly, SDHD prevents the degradation of forkhead box protein O3 (FOXO3a), a positive autophagy regulator (172). In cutaneous melanomas, SDHD expression is downregulated due to promoter mutations. The lower SDHD expression is correlated with a worse prognostic feature (173, 174).
MTUS1
MTUS1, also known as mitochondrial tumor suppressor 1, was discovered in 2003 and is located on chromosome 8p21.3–22 (175). The MTUS1 gene encodes six isoforms also known as Angiotensin-II type 2 receptor-interacting proteins (ATIP), signaling transducers that interact with AT2-receptor: ATIP1, ATIP2, ATIP3a, ATIP3b, ATIP4, and MTUS1 transcript variant 7 (176, 177). MTUS1 was named “mitochondrial tumor suppressor gene 1” according to its function, but its role is not strictly of a tumor suppressor, as it is involved in vascular remodeling, adipocyte differentiation, lymphocytosis, and cardiac hypertrophy (178). In endothelial cells, MTUS1 knockdown inhibits endothelial tube formation and migration. Moreover, MTUS1 is important for mitochondrial dynamics, motility and ROS production (178). MTUS1 is ubiquitously expressed in all tissues (175). Its major transcripts ATIP1, ATIP3, and ATIP4 have different tissue distribution. ATIP1 and 4 are abundantly present in the brain (cerebellum and fetal brain). ATIP3 is expressed in prostate, bladder, ovary, colon, and breast (179). In 2016, Wang and colleagues, described MTUS1 as a mitochondrial protein able to regulate the cytokine production, thus attributing it with an anti-inflammatory role. Silencing MTUS1 in endothelial cells activates NF-κB and p38MAPK, which are the main signaling pathways responsible for the inflammatory response in cells (180).
MTUS1 as a tumor suppressor
MTUS1 deficiency is associated with multiple types of cancer. A significant (50%) downregulation of MTUS1 was found in patients suffering from colon cancer (181). This downregulation was not a consequence of mutations in the gene but rather occurred on a post-transcriptional level. The downregulation resulted in increased cellular proliferation, whereas MTUS1 overexpression reduced the proliferation (181). MTUS1 expression is also reduced in gastric cancer and is associated with lymph node metastasis and poor overall survival rate (182). Furthermore, MTUS1 mRNA expression is downregulated in patients with RCC and correlates with poor prognosis, whereas its expression resulted in significantly longer survival time (183).
In human prostate cancer, the downregulation of ATIP/MTUS1 is influenced by EGF (184). Treatment with EGF decreased the mRNA levels of ATIP1 with a subsequent stimulation of cell growth. This effect was reversed by the activation of the AT2 receptor, which, via an unknown mechanism, promotes the formation of the ATIP/SHP-1 (Src homology 2 domain-containing protein–tyrosine phosphatase 1) complex. This complex, upon translocation to nucleus, promotes the inhibition of the proliferation-signaling cascade. The formation of this complex prevents the ATIP downregulation by EGF, leading to a decrease of the proliferation rate in prostate cancer cells (184). MTUS1 expression levels are also regulated by DNA methylation; in NSCLC, its promoter is methylated causing protein downregulation, whereas inhibition of DNA methylation restored MTUS1 expression levels (185). Another mode of MTUS1 regulation comes from miRNAs control, specifically in osteosarcoma, breast cancer, colorectal carcinoma, and lung cancer (186–189). In osteosarcoma, MTUS1 is a direct target of miR-765. The overexpression of the miR-765 promotes cell proliferation, migration, and invasion and is accompanied by activation of the ERK/EMT pathway (188). MiR-19a and miR-19b coregulate MTUS1 to promote cell proliferation and migration in lung cancer (186).
Differentiation
Failure to undergo cellular differentiation is one of the key events during cancer formation and in 2003, a study from Seibold and colleagues demonstrated a potential role of MTUS1 in this fundamental process (175). In human umbilical vein endothelial cells (HUVEC), under 3D culture conditions, MTUS1 is upregulated during the initial stages of the differentiation process. Similarly, the mRNA levels of MTUS1 in colon tumor tissue and different pancreatic tumor cell lines showed low expression in undifferentiated proliferating cancer cells and higher expression in differentiated and slowly proliferating cancer cells (175, 181). Further studies confirmed that downregulation of MTUS1/ATIP is a frequent event during the progression of oral tongue squamous cell carcinoma where this process is correlated with poor differentiation and enhanced proliferation (190).
Conclusion
Even though mitochondrial tumor suppressors are a diverse group of proteins, they share some underlying characteristics (Supplementary Table S1). First, several of them either directly (POX, SDH, FH) or indirectly (LACTB, sirtuins) participate in metabolic pathways, showing the importance of metabolic dysregulation in cancer. Second, many of them are connected to ROS generation (POX, SIRT3) as well as HIF signaling (FH, SDH) confirming that the cellular reprogramming triggered by (pseudo)hypoxia is an important element of tumor development. Third, several of them are connected to the notorious p53 tumor suppressor, either being induced by it (POX) or modulating its activity (LACTB, SIRT3, FUS1). Because p53 is mutated in a large number of cancers, these interactions could prove to be crucial in improving our concept of tumor suppression in general. Because the discovery and characterization of mitochondrial tumor suppressors is a new and emerging field, its translation into a therapeutic setting is still in its beginnings. Concerning LACTB tumor suppressor pathway, one possible therapeutic approach involves the ability of LACTB to downregulate/inhibit the mitochondrial PISD enzyme, thus promoting cancer cell death and/or differentiation. Indeed, it was shown that doxorubicin, a drug widely used in chemotherapy, is able to modify the mitochondrial membrane composition via PISD pathway inducing cell death in HeLa cells (191). In colorectal cancer, LACTB inhibits EMT and proliferation through PI3K inactivation and the use of PI3K inhibitors was already suggested as a possible therapeutic intervention in colorectal cancer (23). Untangling the dual, physiologic, and cancer type–specific role of POX (and proline metabolism as a whole) could lead to novel ways of treatment. The inhibition of collagen biosynthesis together with upregulation of POX might represent one such example (192). Furthermore, several downstream effectors of POX, such as COX-2, β-catenin, and EGFR, are already being used and researched as promising therapeutic targets in various types of cancer (193–195). Alterations in DNA damage response that accompany FH deficiency (and subsequent fumarate accumulation) could be exploited in therapy since these cells were shown to be vulnerable to synthetic-lethal targeting with PARP inhibitors (83). Because FH is an enzyme of a fundamental metabolic pathway, looking for potential therapy targets in the “rewired” metabolic pathways has likewise proven successful. Synthetic lethality has been discovered between FH and adenylate cyclases as well as genes of heme metabolism (92). As reviewed in Kancherla and colleagues, several other therapies have been proposed to target the FH tumor-suppressive pathway (196). FH deficiency leads to HIF accumulation and increased hypermethylation. Bevacizumab and erlotinib are used in clinic to treat patients with increased HIF levels. Regarding the hypermethylation, a current phase II clinical trial aims to evaluate the efficacy of SGI-110 (guadecitabine), a DNA methyltransferase inhibitor (clinical trial identifier: NCT03165721), in patients with HLRCC-associated RCC. Increased MET oncogene expression has also been detected in FH-deficient cells. An ongoing phase II clinical trial is aiming to compare survival between RCC patients treated with c-Met inhibitors versus sunitinib, the standard of care for RCC (clinical trial identifier: NCT02761057). Given the physiologic importance of sirtuins in the mitochondria and their key role in cancer, they have become attractive targets for drug design. The first described sirtuin activator, called resveratrol, is a plant metabolite present in grapes. Although it was originally described to activate human sirtuin 1 (197), later it also reported to affect the activity of sirtuin 3 and sirtuin 5 (198). However, several serious adverse reactions were reported that prevented its use in clinic and new analogues are being developed that could activate sirtuins at much lower doses (199). There is a growing field of nature-derived sirtuin 3 activators, such as honokiol (200, 201) and oroxylin A (202) that could prove useful in therapy development. Downstream effectors of sirtuins might also be taken into consideration. For instance, SIRT3 induces apoptosis through BCL, caspase-3 and p53 stabilization might increase the SIRT3 downstream effects. SIRT4 inhibited cells invasiveness through ERK–Drp1 signaling pathway, impairing mitochondrial dynamics. Inhibition of Drp1 by Mdivi-1 blocked cell cycle progression through G2–M arrest (203). Regarding FUS1 protein, a phase I clinical trial of FUS1-nanoparticles mediating functional gene transfer in humans was already completed (204), clinical trial identifier: NCT00059605. Combination of FUS1-nanoparticles with cisplatin or erlotinib showed an enhancement of the therapeutic effects of these drugs, pointing out the important role of FUS1 in modulating chemosensitivity of lung cancer cells (205, 206). Taking into account that c-Abl is suppressed by FUS1 in lung cancer, inhibitors targeting c-Abl, such as GNF5, might be used in future for treatment of lung cancer (207). Concerning SDH tumor-suppressive pathway, the most promising approaches are based on the prevention of the succinate accumulation, which is thought to be the major actor of SDH-related oncogenic processes. Some of the therapeutic approaches in SDH-mutated metastatic cancers involve targeting metabolic reprogramming, redox imbalance, pseudohypoxia, or epigenetic reprograming. Among these drugs are inhibitors of oxidative phosphorylation, HIF2a and PARP, or demethylating agents that currently are or will soon be tested in clinical trials (208).
Although the studies of mitochondrial tumor suppressors are still at a relatively early stage, they already provided many valuable insights into biology and dependencies of cancer cells and deepened our appreciation on the important role of mitochondria in tumor progression. Numerous in vitro and in vivo studies highlighted the importance of these tumor suppressors in various biological processes, ranging from regulation of metabolic pathways, ROS production, and hypoxia to promotion of differentiation, apoptosis, and tumor-suppressive metabolic states. These studies also led to emergence of many, as yet unanswered question regarding their role and mechanism of action in cancer cells. How are these tumor suppressors inactivated in cancer cells, and what are the genetic and epigenetic variations of these genes in human cancers? What are the identities of their physiologic upstream regulators and how do changes in mitochondrial processes lead to cancer cell differentiation? Which one of the oncogenic cascades activated in the absence of these tumor suppressors should be targeted to give us the best therapeutic benefits? Does the inactivation of these genes occur in the early stages of transformation or do they emerge at a later stage of tumor progression? Do these mitochondrial tumors suppressor interact with each other? Given the fact that many of them share similar mechanisms of action, it is probable that they might interact and modulate each other's activity, thus creating a general tumor-suppressive landscape within mitochondria. Answers to these questions are important for fully defining the role of these tumor suppressors in tumorigenesis and for applying this knowledge toward more efficient cancer treatments.
Authors' Disclosures
No disclosures were reported.
Acknowledgments
The authors thank Tom DiCesare for graphical assistance with presented images. This work was supported by the Czech Science Foundation (18–24473Y), IOCB Postdoctoral Fellowship (RVO 61388963), EMBO Installation Grant and Ministry of Education, Youth and Sports, and the European Social Fund [OP RDE; Project: “IOCB MSCA Mobility III” (No. CZ.02.2.69/0.0/0.0/19_074/0016322)].
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Hofmarcher T, Lindgren P, Wilking N, Jönsson B. The cost of cancer in Europe 2018. Eur J Cancer 2020;129:41–9. [DOI] [PubMed] [Google Scholar]
- 3. Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986;323:643–6. [DOI] [PubMed] [Google Scholar]
- 5. Dryja TP, Friend S, Weinberg RA. Genetic sequences that predispose to retinoblastoma and osteosarcoma. Symp Fundam Cancer Res 1986;39:115–9. [PubMed] [Google Scholar]
- 6. Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995;81:323–30. [DOI] [PubMed] [Google Scholar]
- 7. Linzer DI, Levine AJ. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979;17:43–52. [DOI] [PubMed] [Google Scholar]
- 8. Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979;278:261–3. [DOI] [PubMed] [Google Scholar]
- 9. Kress M, May E, Cassingena R, May P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J Virol 1979;31:472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell 1989;57:1083–93. [DOI] [PubMed] [Google Scholar]
- 11. Levine AJ, Finlay CA, Hinds PW. P53 is a tumor suppressor gene. Cell 2004;116:S67–9. [DOI] [PubMed] [Google Scholar]
- 12. Peitsaro N, Polianskyte Z, Tuimala J, Porn-Ares I, Liobikas J, Speer O, et al. Evolution of a family of metazoan active-site-serine enzymes from penicillin-binding proteins: a novel facet of the bacterial legacy. BMC Evol Biol 2008;8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith TS, Southan C, Ellington K, Campbell D, Tew DG, Debouck C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine beta-lactamase-like protein with an amino-terminal transmembrane domain. Genomics 2001;78:12–4. [DOI] [PubMed] [Google Scholar]
- 14. Keckesova Z, Donaher JL, De Cock J, Freinkman E, Lingrell S, Bachovchin DA, et al. LACTB is a tumor suppressor that modulates lipid metabolism and cell state. Nature 2017;543:681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polianskyte Z, Peitsaro N, Dapkunas A, Liobikas J, Soliymani R, Lalowski M, et al. LACTB is a filament-forming protein localized in mitochondria. Proc Natl Acad Sci U S A 2009;106:18960–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008;134:112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 2001;276:43958–69. [DOI] [PubMed] [Google Scholar]
- 18. Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, et al. Variations in DNA elucidate molecular networks that cause disease. Nature 2008;452:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet 2009;41:415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu JB, Yao X, Xiu J, Y H. MicroRNA-125b-5p attenuates lipopolysaccharide-induced monocyte chemoattractant protein-1 production by targeting inhibiting LACTB in THP-1 macrophages. Arch Biochem Biophys 2016;590:64–71. [DOI] [PubMed] [Google Scholar]
- 21. Zhang J, He Y, Yu Y, Chen X, Cui G, Wang W, et al. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med 2018;7:3351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li HT, Dong DY, Liu Q, Xu YQ, Chen L. Overexpression of LACTB, a mitochondrial protein that inhibits proliferation and invasion in glioma cells. Oncol Res 2019;27:423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu W, Yu M, Qin J, Luo Y, Zhong M. LACTB regulates PIK3R3 to promote autophagy and inhibit EMT and proliferation through the PI3K/AKT/mTOR signaling pathway in colorectal cancer. Cancer Manag Res 2020;12:5181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue C, He Y, Zhu W, Chen X, Yu Y, Hu Q, et al. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am J Transl Res 2018;10:4152–62. [PMC free article] [PubMed] [Google Scholar]
- 25. Zeng K, Chen X, Hu X, Liu X, Xu T, Sun H, et al. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and degradation. Oncogene 2018;37:5534–51. [DOI] [PubMed] [Google Scholar]
- 26. Ma Y, Wang L, He F, Yang J, Ding Y, Ge S, et al. LACTB suppresses melanoma progression by attenuating PP1A and YAP interaction. Cancer Lett 2021;506:67–82. [DOI] [PubMed] [Google Scholar]
- 27. Du J, Zhang P, Zhao X, He J, Xu Y, Zou Q, et al. MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-beta and is regulated by lnc-mg. FASEB J 2019;33:1911–26. [DOI] [PubMed] [Google Scholar]
- 28. Yang X, Zhang D, Liu S, Li X, Hu W, Han C. KLF4 suppresses the migration of hepatocellular carcinoma by transcriptionally upregulating monoglyceride lipase. Am J Cancer Res 2018;8:1019–29. [PMC free article] [PubMed] [Google Scholar]
- 29. Xie J, Peng Y, Chen X, Li Q, Jian B, Wen Z, et al. LACTB mRNA expression is increased in pancreatic adenocarcinoma and high expression indicates a poor prognosis. PLoS One 2021;16:e0245908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng LX, Wang MD, Xie P, Yang JP, Sun R, Zheng LS, et al. LACTB promotes metastasis of nasopharyngeal carcinoma via activation of ERBB3/EGFR-ERK signaling resulting in unfavorable patient survival. Cancer Lett 2021;498:165–77. [DOI] [PubMed] [Google Scholar]
- 31. Hancock CN, Liu W, Alvord WG, Phang JM. Co-regulation of mitochondrial respiration by proline dehydrogenase/oxidase and succinate. Amino Acids 2016;48:859–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature 1997;389:300–5. [DOI] [PubMed] [Google Scholar]
- 33. Raimondi I, Ciribilli Y, Monti P, Bisio A, Pollegioni L, Fronza G, et al. P53 family members modulate the expression of PRODH, but not PRODH2, via intronic p53 response elements. PLoS One 2013;8:e69152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maxwell SA, Kochevar GJ. Identification of a p53-response element in the promoter of the proline oxidase gene. Biochem Biophys Res Commun 2008;369:308–13. [DOI] [PubMed] [Google Scholar]
- 35. Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, Phang JM. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res 2009;69:6414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu W, Zabirnyk O, Wang H, Shiao YH, Nickerson ML, Khalil S, et al. miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene 2010;29:4914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A 2012;109:8983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci U S A 2000;97:13009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maxwell SA, Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. J Biol Chem 2003;278:9784–9. [DOI] [PubMed] [Google Scholar]
- 40. Hu CA, Donald SP, Yu J, Lin WW, Liu Z, Steel G, et al. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Mol Cell Biochem 2007;295:85–92. [DOI] [PubMed] [Google Scholar]
- 41. Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, et al. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res 2001;61:1810–5. [PubMed] [Google Scholar]
- 42. Liu Y, Borchert GL, Surazynski A, Phang JM. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene 2008;27:6729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, et al. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis 2005;26:1335–42. [DOI] [PubMed] [Google Scholar]
- 44. Rivera A, Maxwell SA. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J Biol Chem 2005;280:29346–54. [DOI] [PubMed] [Google Scholar]
- 45. Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene 2006;25:5640–7. [DOI] [PubMed] [Google Scholar]
- 46. Pandhare J, Cooper SK, Phang JM. Proline oxidase, a proapoptotic gene, is induced by troglitazone: evidence for both peroxisome proliferator-activated receptor gamma-dependent and -independent mechanisms. J Biol Chem 2006;281:2044–52. [DOI] [PubMed] [Google Scholar]
- 47. Kim KY, Ahn JH, Cheon HG. Apoptotic action of peroxisome proliferator-activated receptor-gamma activation in human non small-cell lung cancer is mediated via proline oxidase-induced reactive oxygen species formation. Mol Pharmacol 2007;72:674–85. [DOI] [PubMed] [Google Scholar]
- 48. Wang J, Lv X, Shi J, Hu X, Du Y. Troglitazone induced apoptosis via PPARγ activated POX-induced ROS formation in HT29 cells. Biomed Environ Sci 2011;24:391–9. [DOI] [PubMed] [Google Scholar]
- 49. Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 1994;107:1183–8. [DOI] [PubMed] [Google Scholar]
- 50. Nagano T, Nakashima A, Onishi K, Kawai K, Awai Y, Kinugasa M, et al. Proline dehydrogenase promotes senescence through the generation of reactive oxygen species. J Cell Sci 2017;130:1413–20. [DOI] [PubMed] [Google Scholar]
- 51. Liu W, Glunde K, Bhujwalla ZM, Raman V, Sharma A, Phang JM. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Res 2012;72:3677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pandhare J, Donald SP, Cooper SK, Phang JM. Regulation and function of proline oxidase under nutrient stress. J Cell Biochem 2009;107:759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Olivares O, Mayers JR, Gouirand V, Torrence ME, Gicquel T, Borge L, et al. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat Commun 2017;8:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y, Mao C, Wang M, Liu N, Ouyang L, Liu S, et al. Cancer progression is mediated by proline catabolism in non-small cell lung cancer. Oncogene 2020;39:2358–76. [DOI] [PubMed] [Google Scholar]
- 55. Dik E, Naamati A, Asraf H, Lehming N, Pines O. Human fumarate hydratase is dual localized by an alternative transcription initiation mechanism. Traffic 2016;17:720–32. [DOI] [PubMed] [Google Scholar]
- 56. Yogev O, Naamati A, Pines O. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J 2011;278:4230–42. [DOI] [PubMed] [Google Scholar]
- 57. Castro-Vega LJ, Buffet A, De Cubas AA, Cascón A, Menara M, Khalifa E, et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet 2014;23:2440–6. [DOI] [PubMed] [Google Scholar]
- 58. Clark GR, Sciacovelli M, Gaude E, Walsh DM, Kirby G, Simpson MA, et al. Germline FH mutations presenting with pheochromocytoma. J Clin Endocrinol Metab 2014;99:E2046–50. [DOI] [PubMed] [Google Scholar]
- 59. Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406–10. [DOI] [PubMed] [Google Scholar]
- 60. Alam NA, Olpin S, Rowan A, Kelsell D, Leigh IM, Tomlinson IP, et al. Missense mutations in fumarate hydratase in multiple cutaneous and uterine leiomyomatosis and renal cell cancer. J Mol Diagn 2005;7:437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: a multifaceted tumor suppressor. Semin Cell Dev Biol 2020;98:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alderson NL, Wang Y, Blatnik M, Frizzell N, Walla MD, Lyons TJ, et al. S-(2 -Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch Biochem Biophys 2006;450:1–8. [DOI] [PubMed] [Google Scholar]
- 63. Blatnik M, Frizzell N, Thorpe SR, Baynes JW. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by fumarate in diabetes. Diabetes 2008;57:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ternette N, Yang M, Laroyia M, Kitagawa M, O'Flaherty L, Wolhulter K, et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep 2013;3:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kerins MA-O, Vashisht AA, Liang BX, Duckworth SJ, Praslicka BJ, Wohlschlegel JA, et al. Fumarate mediates a chronic proliferative signal in fumarate hydratase-inactivated cancer cells by increasing transcription and translation of ferritin genes. Mol Cell Biol 2017;37:e00079–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Adam J, Hatipoglu E, O'Flaherty L, Ternette N, Sahgal N, Lockstone H, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 2011;20:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bardella C, El-Bahrawy M, Frizzell N, Adam J, Ternette N, Hatipoglu E, et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J Pathol 2011;225:4–11. [DOI] [PubMed] [Google Scholar]
- 68. Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015;3:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 2005;8:143–53. [DOI] [PubMed] [Google Scholar]
- 70. Bardella C, Olivero M, Lorenzato A, Geuna M, Adam J, O'Flaherty L, et al. Cells lacking the fumarase tumor suppressor are protected from apoptosis through a hypoxia-inducible factor-independent, AMPK-dependent mechanism. Mol Cell Biol 2012;32:3081–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pollard P, Wortham N, Barclay E, Alam A, Elia G, Manek S, et al. Evidence of increased microvessel density and activation of the hypoxia pathway in tumors from the hereditary leiomyomatosis and renal cell cancer syndrome. J Pathol 2005;205:41–9. [DOI] [PubMed] [Google Scholar]
- 72. Costa B, Dettori D, Lorenzato A, Bardella C, Coltella N, Martino C, et al. Fumarase tumor suppressor gene and MET oncogene cooperate in upholding transformation and tumorigenesis. FASEB J 2010;24:2680–8. [DOI] [PubMed] [Google Scholar]
- 73. Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumors which result from germline FH and SDH mutations. Hum Mol Genet 2005;14:2231–9. [DOI] [PubMed] [Google Scholar]
- 74. Sudarshan S, Sourbier C, Kong HS, Block K, V Romero VA, Yang Y, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol 2009;29:4080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, et al. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 2011;20:315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tyrakis PA, Yurkovich ME, Sciacovelli M, Papachristou EK, Bridges HR, Gaude E, et al. Fumarate hydratase loss causes combined respiratory chain defects. Cell Rep 2017;21:1036–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012;26:1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016;537:544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. He X, Yan B, Liu S, Jia J, Lai W, Xin X, et al. Chromatin remodeling factor LSH drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res 2016;76:5743–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jiang Y, Qian X, Shen J, Wang Y, Li X, Liu R, et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat Cell Biol 2015;17:1158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yogev O, Yogev O, Singer E, Shaulian E, Goldberg M, Fox TD, et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol 2010;8:e1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Leshets M, Ramamurthy D, Lisby M, Lehming N, Pines O. Fumarase is involved in DNA double-strand break resection through a functional interaction with Sae2. Curr Genet 2018;64:697–712. [DOI] [PubMed] [Google Scholar]
- 83. Sulkowski PL, Sundaram RK, Oeck S, Corso CD, Liu Y, Noorbakhsh S, et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat Genet 2018;50:1086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Johnson TI, Costa ASH, Ferguson AN, Frezza C. Fumarate hydratase loss promotes mitotic entry in the presence of DNA damage after ionising radiation. Cell Death Dis 2018;9:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011;20:511–23. [DOI] [PubMed] [Google Scholar]
- 86. Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun 2015;6:6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sourbier C, Ricketts CJ, Matsumoto S, Crooks DR, Liao PJ, Mannes PZ, et al. Targeting ABL1-mediated oxidative stress adaptation in fumarate hydratase-deficient cancer. Cancer Cell 2014;26:840–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gonçalves E, Sciacovelli M, Costa ASH, Tran MGB, Johnson TI, Machado D, et al. Post-translational regulation of metabolism in fumarate hydratase deficient cancer cells. Metab Eng 2018;45:149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang Y, Lane AN, Ricketts CJ, Sourbier C, Wei MH, Shuch B, et al. Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS One 2013;8:e72179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumor cells with defective mitochondria. Nature 2011;481:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 2017;66:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Boettcher M, Lawson A, Ladenburger V, Fredebohm J, Wolf J, Hoheisel JD, et al. High throughput synthetic lethality screen reveals a tumorigenic role of adenylate cyclase in fumarate hydratase-deficient cancer cells. BMC Genomics 2014;15:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yu HE, Wang F, Yu F, Zeng ZL, Wang Y, Lu YX, et al. Suppression of fumarate hydratase activity increases the efficacy of cisplatin-mediated chemotherapy in gastric cancer. Cell Death Dis 2019;10:413. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94. Leshets M, Silas YBH, Lehming N, Pines O. Fumarase: From the TCA Cycle to DNA damage response and tumor suppression. Front Mol Biosci 2018;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang M, Soga T, Pollard PJ, Adam J. The emerging role of fumarate as an oncometabolite. Front Oncol 2012;2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ye X, Li M, Hou T, Gao T, Zhu WG, Yang Y. Sirtuins in glucose and lipid metabolism. Oncotarget 2017;8:1845–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. George J, Ahmad N. Mitochondrial sirtuins in cancer: emerging roles and therapeutic potential. Cancer Res 2016;76:2500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, Mack NJ, Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal 2018;28:643–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Du Y, Hu H, Hua C, Du K, Wei T. Tissue distribution, subcellular localization, and enzymatic activity analysis of human SIRT5 isoforms. Biochem Biophys Res Commun 2018;503:763–9. [DOI] [PubMed] [Google Scholar]
- 100. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005;16:4623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 2002;158:647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 2007;21:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun 1999;260:273–9. [DOI] [PubMed] [Google Scholar]
- 104. Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A 2000;97:14178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000;403:795–800. [DOI] [PubMed] [Google Scholar]
- 106. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011;334:806–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yu W, Denu RA, Krautkramer KA, Grindle KM, Yang DT, Asimakopoulos F, et al. Loss of SIRT3 provides growth advantage for B cell malignancies. J Biol Chem 2016;291:3268–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang YY, Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun 2012;423:26–31. [DOI] [PubMed] [Google Scholar]
- 109. Xu WY, Hu QS, Qin Y, Zhang B, Liu WS, Ni QX, et al. Zinc finger E-box-binding homeobox 1 mediates aerobic glycolysis via suppression of sirtuin 3 in pancreatic cancer. World J Gastroenterol 2018;24:4893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, et al. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep 2013;30:1323–8. [DOI] [PubMed] [Google Scholar]
- 111. Wang L, Wang WY, Cao LP. SIRT3 inhibits cell proliferation in human gastric cancer through down-regulation of Notch-1. Int J Clin Exp Med 2015;8:5263–71. [PMC free article] [PubMed] [Google Scholar]
- 112. Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010;17:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 2011;19:416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011;30:2986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Krebs AM, Mitschke J, Lasierra LM, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 2017;19:518–29. [DOI] [PubMed] [Google Scholar]
- 116. Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, et al. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem 2009;106:643–50. [DOI] [PubMed] [Google Scholar]
- 117. Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene 2008;27:6245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Winter JN, Jefferson LS, Kimball SR. ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am J Physiol Cell Physiol 2011;300:C1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 2011;117:1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 2008;382:790–801. [DOI] [PubMed] [Google Scholar]
- 121. Kamarajan P, Alhazzazi TY, Danciu T, D'Silva NJ, Verdin E, Kapila YL. Receptor-interacting protein (RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and tumorigenesis. Cancer 2012;118:5800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. George J, Nihal M, Singh CK, Zhong W, Liu X, Ahmad N. Pro-proliferative function of mitochondrial sirtuin deacetylase SIRT3 in human melanoma. J Invest Dermatol 2016;136:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Choi J, Koh E, Lee YS, Lee HW, Kang HG, Yoon YE, et al. Mitochondrial Sirt3 supports cell proliferation by regulating glutamine-dependent oxidation in renal cell carcinoma. Biochem Biophys Res Commun 2016;474:547–53. [DOI] [PubMed] [Google Scholar]
- 124. Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One 2010;5:e10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem 2007;282:33583–92. [DOI] [PubMed] [Google Scholar]
- 126. Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 2006;126:941–54. [DOI] [PubMed] [Google Scholar]
- 127. Argmann C, Auwerx J. Insulin secretion: SIRT4 gets in on the act. Cell 2006;126:837–9. [DOI] [PubMed] [Google Scholar]
- 128. Laurent G, German NJ, Saha AK, de Boer VCJ, Davies M, Koves TR, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 2013;50:686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab 2017;25:838–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Huang G, Lai X, Chen Z, Yu Z, Zhou D, Wang P, et al. Sirtuin-4 (SIRT4) is downregulated in hepatocellular carcinoma and associated with clinical stage. Int J Clin Exp Pathol 2016;9:6511–7. [Google Scholar]
- 131. Huang G, Cheng J, Yu F, Liu X, Yuan C, Liu C, et al. Clinical and therapeutic significance of sirtuin-4 expression in colorectal cancer. Oncol Rep 2016;35:2801–10. [DOI] [PubMed] [Google Scholar]
- 132. Sun H, Huang D, Liu G, Jian F, Zhu J, Zhang L. SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion. Onco Targets Ther 2018;11:3959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Fu L, Dong Q, He J, Wang X, Xing J, Wang E, et al. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene 2017;36:2724–36. [DOI] [PubMed] [Google Scholar]
- 134. Huang G, Cui F, Yu F, Lu H, Zhang M, Tang H, et al. Sirtuin-4 (SIRT4) is downregulated and associated with some clinicopathological features in gastric adenocarcinoma. Biomed Pharmacother 2015;72:135–9. [DOI] [PubMed] [Google Scholar]
- 135. Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 2013;23:450–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 2014;159:1615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry 1991;30:8501–12. [DOI] [PubMed] [Google Scholar]
- 138. Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, et al. Tumor-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer 2015;113:492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lai X, Yu Z, Chen X, Huang G. SIRT4 is upregulated in Chinese patients with esophageal cancer. Int J Clin Exp Pathol 2016;9:10543–9. [Google Scholar]
- 140. Huang G, Lin Y, Zhu G. SIRT4 is upregulated in breast cancer and promotes the proliferation, migration and invasion of breast cancer cells. Int J Clin Exp Pathol 2017;10:11849–56. [PMC free article] [PubMed] [Google Scholar]
- 141. Jeong SM, Hwang S, Seong RH. SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun 2016;470:251–6. [DOI] [PubMed] [Google Scholar]
- 142. Uzhachenko R, Ivanov SV, Yarbrough WG, Shanker A, Medzhitov R, Ivanova AV. Fus1/Tusc2 is a novel regulator of mitochondrial calcium handling, Ca2+-coupled mitochondrial processes, and Ca2+-dependent NFAT and NF-κB pathways in CD4+ T cells. Antioxid Redox Signal 2014;20:1533–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res 2000;60:6116–33. [PubMed] [Google Scholar]
- 144. Prudkin L, Behrens C, Liu DD, Zhou X, Ozburn NC, Bekele BN, et al. Loss and reduction of FUS1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin Cancer Res 2008;14:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liang S, Zhang N, Deng Y, Chen L, Zhang Y, Zheng Z, et al. miR-663b promotes tumor cell proliferation, migration and invasion in nasopharyngeal carcinoma through targeting TUSC2. Exp Ther Med 2017;14:1095–103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146. Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 2009;7:1234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Lin J, Xu K, Gitanjali J, Roth JA, Ji L. Regulation of tumor suppressor gene FUS1 expression by the untranslated regions of mRNA in human lung cancer cells. Biochem Biophys Res Commun 2011;410:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kondo M, Ji L, Kamibayashi C, Tomizawa Y, Randle D, Sekido Y, et al. Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene 2001;20:6258–62. [DOI] [PubMed] [Google Scholar]
- 149. Ji L, Roth JA. Tumor suppressor FUS1 signaling pathway. J Thorac Oncol 2008;3:327–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Deng WG, Kawashima H, Wu G, Jayachandran G, Xu K, Minna JD, et al. Synergistic tumor suppression by coexpression of FUS1 and p53 is associated with down-regulation of murine double minute-2 and activation of the apoptotic protease-activating factor 1-dependent apoptotic pathway in human non-small cell lung cancer cells. Cancer Res 2007;67:709–17. [DOI] [PubMed] [Google Scholar]
- 151. Lin J, Sun T, Ji L, Deng W, Roth J, Minna J, et al. Oncogenic activation of c-Abl in non-small cell lung cancer cells lacking FUS1 expression: inhibition of c-Abl by the tumor suppressor gene product Fus1. Oncogene 2007;26:6989–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Meng J, Majidi M, Fang B, Ji L, Bekele BN, Minna JD, et al. The tumor suppressor gene TUSC2 (FUS1) sensitizes NSCLC to the AKT inhibitor MK2206 in LKB1-dependent manner. PLoS One 2013;8:e77067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Shah U, Sharpless NE, Hayes DN. LKB1 and lung cancer: more than the usual suspects. Cancer Res 2008;68:3562–5. [DOI] [PubMed] [Google Scholar]
- 154. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumor suppression. Nat Rev Cancer 2009;9:563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Uzhachenko R, Issaeva N, Boyd K, Ivanov SV, Carbone DP, Ivanova AV. Tumor suppressor Fus1 provides a molecular link between inflammatory response and mitochondrial homeostasis. J Pathol 2012;227:456–69. [DOI] [PubMed] [Google Scholar]
- 156. Rustin P, Munnich A, Rötig A. Succinate dehydrogenase and human diseases: new insights into a well-known enzyme. Eur J Hum Genet 2002;10:289–91. [DOI] [PubMed] [Google Scholar]
- 157. Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion 2010;10:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Farshbaf MJ. Succinate dehydrogenase in Parkinson's disease. Front Biol 2017;12:175–82. [Google Scholar]
- 159. Bayley JP, Devilee P, Taschner PE. The SDH mutation database: an online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Med Genet 2005;6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gimenez-Roqueplo A, Favier J, Rustin P, Rieubland C, Crespin M, Nau V, et al. Mutations in the SDHB gene are associated with extra-adrenal and/or malignant phaeochromocytomas. Cancer Res 2003;63:5615–21. [PubMed] [Google Scholar]
- 161. Kim S, Kim DH, Jung WH, Koo JS. Succinate dehydrogenase expression in breast cancer. Springerplus 2013;2:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Apuria P, Lunt S, Väremo L, Vergnes L, Gozo M, Beach J, et al. Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab 2014;2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, Ishii N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res 2005;65:203–9. [PubMed] [Google Scholar]
- 164. Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005;7:77–85. [DOI] [PubMed] [Google Scholar]
- 165. Wagner AJ, Remillard SP, Zhang YX, Doyle LA, George S, Hornick JL. Loss of expression of SDHA predicts SDHA mutations in gastrointestinal stromal tumors. Mod Pathol 2013;26:289–94. [DOI] [PubMed] [Google Scholar]
- 166. Astuti D, Morris M, Krona C, Abel F, Gentle D, Martinsson T, et al. Investigation of the role of SDHB inactivation in sporadic phaeochromocytoma and neuroblastoma. Br J Cancer 2004;91:1835–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang H, Chen Y, Wu G. SDHB deficiency promotes TGFbeta-mediated invasion and metastasis of colorectal cancer through transcriptional repression complex SNAIL1-SMAD3/4. Transl Oncol 2016;9:512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libe R, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab 2012;97:E954–62. [DOI] [PubMed] [Google Scholar]
- 169. Killian JK, Miettinen M, Walker R, Wang Y, Zhu YJ, Waterfall1 JJ, et al. Recurrent epimutation of SDHC in gastrointestinal stromal tumors. Sci Transl Med 2014;24:268ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Rosland GV, Dyrstad SE, Tusubira D, Helwa R, Tan TZ, Lotsberg ML, et al. Epithelial to mesenchymal transition (EMT) is associated with attenuation of succinate dehydrogenase (SDH) in breast cancer through reduced expression of SDHC. Cancer Metab 2019;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 2000;287:848–51. [DOI] [PubMed] [Google Scholar]
- 172. Yu W, Ni Y, Saji M, Ringel MD, Jaini R, Eng C. Cowden syndrome-associated germline succinate dehydrogenase complex subunit D (SDHD) variants cause PTEN-mediated down-regulation of autophagy in thyroid cancer cells. Hum Mol Genet 2017;26:1365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Populo H, Batista R, Sampaio C, Pardal J, Lopes JM, Soares P. SDHD promoter mutations are rare events in cutaneous melanomas but SDHD protein expression is downregulated in advanced cutaneous melanoma. PLoS One 2017;12:e0180392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhang T, Xu M, Makowski MM, Lee C, Kovacs M, Fang J, et al. SDHD promoter mutations ablate GABP transcription factor binding in melanoma. Cancer Res 2017;77:1649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M. Identification of a new tumor suppressor gene located at chromosome 8p21.3–22. FASEB J 2003;17:1180–2. [DOI] [PubMed] [Google Scholar]
- 176. Bozgeyik I, Yumrutas O, Bozgeyik E. MTUS1, a gene encoding angiotensin-II type 2 (AT2) receptor-interacting proteins, in health and disease, with special emphasis on its role in carcinogenesis. Gene 2017;626:54–63. [DOI] [PubMed] [Google Scholar]
- 177. Di Benedetto M, Pineau P, Nouet S, Berhouet S, Seitz I, Louis S, et al. Mutation analysis of the 8p22 candidate tumor suppressor gene ATIP/MTUS1 in hepatocellular carcinoma. Mol Cell Endocrinol 2006;252:207–15. [DOI] [PubMed] [Google Scholar]
- 178. Wang Y, Huang Y, Liu Y, Li J, Hao Y, Yin P, et al. Microtubule associated tumor suppressor 1 interacts with mitofusins to regulate mitochondrial morphology in endothelial cells. FASEB J 2018;32:4504–18. [DOI] [PubMed] [Google Scholar]
- 179. Di Benedetto M, Bièche I, Deshayes F, Vacher S, Nouet S, Collura V, et al. Structural organization and expression of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene 2006;380:127–36. [DOI] [PubMed] [Google Scholar]
- 180. Wang Y, Dai X, Liu Y, Li J, Liu Z, Yin P, et al. MTUS1 silencing promotes E-selectin production through p38 MAPK-dependent CREB ubiquitination in endothelial cells. J Mol Cell Cardiol 2016;101:1–10. [DOI] [PubMed] [Google Scholar]
- 181. Zuern C, Heimrich J, Kaufmann R, Richter K, Settmacher U, Wanner C, et al. Down-regulation of MTUS1 in human colon tumors. Oncol Rep 2009;23:183–9. [PubMed] [Google Scholar]
- 182. Li X, Liu H, Yu T, Dong Z, Tang L, Sun X. Loss of MTUS1 in gastric cancer promotes tumor growth and metastasis. Neoplasma 2014;61:128–35. [DOI] [PubMed] [Google Scholar]
- 183. Sim J, Wi YC, Park HY, Park SY, Yoon YE, Bang S, et al. Clinicopathological significance of MTUS1 expression in patients with renal cell carcinoma. Anticancer Res 2020;40:2961–7. [DOI] [PubMed] [Google Scholar]
- 184. Louis SN, Chow L, Rezmann L, Krezel MA, Catt KJ, Tikellis C, et al. Expression and function of ATIP/MTUS1 in human prostate cancer cell lines. Prostate 2010;70:1563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Parbin S, Pradhan N, Das L, Saha P, Deb M, Sengupta D, et al. DNA methylation regulates microtubule-associated tumor suppressor 1 in human non-small cell lung carcinoma. Exp Cell Res 2019;374:323–32. [DOI] [PubMed] [Google Scholar]
- 186. Gu Y, Liu S, Zhang X, Chen G, Liang H, Yu M, et al. Oncogenic miR-19a and miR-19b co-regulate tumor suppressor MTUS1 to promote cell proliferation and migration in lung cancer. Protein Cell 2017;8:455–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kara M, Kaplan M, Bozgeyik I, Ozcan O, Celik OI, Bozgeyik E, et al. MTUS1 tumor suppressor and its miRNA regulators in fibroadenoma and breast cancer. Gene 2016;587:173–7. [DOI] [PubMed] [Google Scholar]
- 188. Lv D-B, Zhang J-Y, Gao K, Yu Z-H, Sheng W-C, Yang G, et al. MicroRNA-765 targets MTUS1 to promote the progression of osteosarcoma via mediating ERK/EMT pathway. Eur Rev Med Pharmacol Sci 2019;23:4618–28. [DOI] [PubMed] [Google Scholar]
- 189. Ozcan O, Kara M, Yumrutas O, Bozgeyik E, Bozgeyik I, Celik OI. MTUS1 and its targeting miRNAs in colorectal carcinoma: significant associations. Tumor Biol 2016;37:6637–45. [DOI] [PubMed] [Google Scholar]
- 190. Ding X, Zhang N, Cai Y, Li S, Zheng C, Jin Y, et al. Down-regulation of tumor suppressor MTUS1/ATIP is associated with enhanced proliferation, poor differentiation and poor prognosis in oral tongue squamous cell carcinoma. Mol Oncol 2012;6:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Bellance N, Furt F, Melser S, Lalou C, Thoraval D, Maneta-Peyret L, et al. Doxorubicin inhibits phosphatidylserine decarboxylase and modifies mitochondrial membrane composition in HeLa cells. Int J Mol Sci 2020;21:1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Palka J, Oscilowska I, Szoka L. Collagen metabolism as a regulator of proline dehydrogenase/proline oxidase-dependent apoptosis/autophagy. Amino Acids 2021;4:1438–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Xu YQ, Long X, Han M, Huang MQ, Lu JF, Sun XD, et al. Clinical benefit of COX-2 inhibitors in the adjuvant chemotherapy of advanced non-small cell lung cancer: a systematic review and meta-analysis. World J Clin Cases 2021;9:581–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Jung YS, Park JI. Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp Mol Med 2020;52:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wheeler D, Dunn E, Harari P. Understanding resistance to EGFR inhibitors—impact on future treatment strategies. Nat Rev Clin Oncol 2010;7:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kancherla P, Daneshvar M, Sager RA, Mollapour M, Bratslavsky G. Fumarate hydratase as a therapeutic target in renal cancer. Expert Opin Ther Targets 2020;24:923–36. [DOI] [PubMed] [Google Scholar]
- 197. Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov 2006;5:493–506. [DOI] [PubMed] [Google Scholar]
- 198. Gertz M, Nguyen GTT, Fischer F, Suenkel B, Schlicker C, Fränzel B, et al. A molecular mechanism for direct sirtuin activation by resveratrol. PLoS One 2012;7:e49761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Shaito A, Posadino AM, Younes N, Hasan H, Halabi S, Alhababi D, et al. Potential adverse effects of resveratrol: a literature review. Int J Mol Sci 2020;21:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, et al. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun 2015;6:6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Pillai VB, Kanwal A, Fang YH, Sharp WW, Samant S, Arbiser J, et al. Honokiol, an activator of Sirtuin-3 (SIRT3) preserves mitochondria and protects the heart from doxorubicin-induced cardiomyopathy in mice. Oncotarget 2017;8:34082–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Wei L, Zhou Y, Dai Q, Qiao C, Zhao L, Hui H, et al. Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis 2013;4:e601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Marsboom G, Toth PT, Ryan JJ, Hong Z, Wu X, Fang YH, et al. Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ Res 2012;110:1484–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Lu C, Stewart DJ, Lee JJ, Ji L, Ramesh R, Jayachandran G, et al. Phase I clinical trial of systemically administered TUSC2(FUS1)-nanoparticles mediating functional gene transfer in humans. PLoS One 2012;7:e34833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Deng WG, Wu G, Ueda K, Xu K, Roth JA, Ji L. Enhancement of antitumor activity of cisplatin in human lung cancer cells by tumor suppressor FUS1. Cancer Gene Ther 2008;15:29–39. [DOI] [PubMed] [Google Scholar]
- 206. Xiaobo C, Majidi M, Feng M, Shao R, Wang J, Zhao Y, et al. TUSC2(FUS1)-erlotinib induced vulnerabilities in epidermal growth factor receptor(EGFR) wildtype non-small cell lung cancer(NSCLC) targeted by the repurposed drug auranofin. Sci Rep 2016;6:35741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Khatri A, Gu JJ, McKernan CM, Xu X, Pendergast AM. ABL kinase inhibition sensitizes primary lung adenocarcinomas to chemotherapy by promoting tumor cell differentiation. Oncotarget 2019;10:1874–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Moog S, Lussey-Lepoutre C, Favier J. Epigenetic and metabolic reprogramming of SDH-deficient paragangliomas. Endocr Relat Cancer 2020;27:R451–R63. [DOI] [PubMed] [Google Scholar]