CAFs secrete TGFβ to induce a solid-to-acinar transition in lung cancer cells, demonstrating how the tumor microenvironment influences histological patterns and tumor heterogeneity in lung adenocarcinoma.
Abstract
Invasive lung adenocarcinoma (LADC) can be classified histologically as lepidic, acinar, papillary, micropapillary, or solid. Most LADC tumors manifest several of these histological subtypes, with heterogeneity being related to therapeutic resistance. We report here that in immunodeficient mice, human LADC cells form tumors with distinct histological features, MUC5AC-expressing solid-type or cytokeratin 7 (CK7)-expressing acinar-type tumors, depending on the site of development, and that a solid-to-acinar transition (SAT) could be induced by the tumor microenvironment. The TGFβ-Smad signaling pathway was activated in both tumor and stromal cells of acinar-type tumors. Immortalized cancer-associated fibroblasts (CAF) derived from acinar-type tumors induced SAT in 3D cocultures with LADC cells. Exogenous TGFβ1 or overexpression of an active form of TGFβ1 increased CK7 expression and reduced MUC5AC expression in LADC cells, and knockdown of Tgfb1 mRNA in CAFs attenuated SAT induction. RNA-sequencing analysis suggested that angiogenesis and neutrophil recruitment are associated with SAT in vivo. Our data indicate that CAF-mediated paracrine TGFβ signaling induces remodeling of tumor tissue and determines the histological pattern of LADC, thereby contributing to tumor heterogeneity.
Significance:
CAFs secrete TGFβ to induce a solid-to-acinar transition in lung cancer cells, demonstrating how the tumor microenvironment influences histological patterns and tumor heterogeneity in lung adenocarcinoma.
Introduction
Intratumoral heterogeneity is a hallmark of lung adenocarcinoma (LADC), which accounts for approximately 50% of all lung cancers and continues to have a poor prognosis despite progress in multimodal treatments (1). LADC is classified into five histological subtypes—lepidic, papillary, acinar, micropapillary, and solid—according to the latest WHO classification. Although the relation between histological subtypes and prognosis for individuals with LADC has been investigated (2), histopathologic analysis has revealed spatial heterogeneity of LADC tissue (3), with >80% to 90% of such tumors containing components corresponding to several of the five histological subtypes (4). Such tumor heterogeneity might be a major contributor to therapeutic resistance. In addition to the genomic diversity of lung cancer cells (5), functional and phenotypic heterogeneity among tumor cells can arise from nongenetic factors (6–8). However, the molecular mechanisms that determine the individual histological subtypes of LADC have remained unknown.
Tumor tissue consists of cancer cells, stromal cells, and various extracellular components such as extracellular matrix (ECM). The tumor microenvironment—including fibroblasts, immune cells, and endothelial cells—has been found to influence tumor progression (9). In particular, cancer-associated fibroblasts (CAF) are thought to play a key role in lung tumor development (10, 11). CAFs modulate the tumor microenvironment by synthesizing and remodeling ECM as well as by producing growth factors, and they thereby influence tumor cell proliferation, migration, metastasis, metabolism, and stemness (12, 13). In colorectal cancer, CAFs have been shown to stimulate glucose uptake by tumor cells as well as to promote tumor progression and to increase intratumoral heterogeneity (14). Furthermore, single-cell RNA-sequencing (RNA-seq) analysis revealed that CAFs and CAF-derived TGFβ contribute to the heterogeneity of pancreatic ductal adenocarcinoma (15).
TGFβ is a multifunctional cytokine that binds to a serine-threonine kinase receptor (TGFBR) and thereby triggers the phosphorylation of intracellular Smad protein effectors. Phosphorylated Smads translocate to the nucleus, where they induce the transcription of various genes that regulate cellular responses such as growth arrest, apoptosis, differentiation, motility, invasion, epithelial–mesenchymal transition, ECM production, fibrosis, angiogenesis, and modulation of immune function (16, 17). Given that TGFβ signaling is disordered in cancer, it is an attractive therapeutic target (17, 18). However, the biological actions of TGFβ are complex, and they include both tumor promotion and suppression (16, 18, 19), with the result that inhibition of TGFβ signaling is not always therapeutic. It is therefore important to determine which cells in tumor tissue release and which cells respond to TGFβ, as well as to examine which tumor subtypes depend on TGFβ signaling. A better understanding of the role of TGFβ signaling in tumor and stromal cells of LADC will facilitate the development of therapies that target the TGFβ signaling pathway in patients with cancer.
We here investigated the role of environmental factors in LADC tumor heterogeneity with the use of A549 human LADC cells transplanted into immunodeficient mice as well as of three-dimensional (3D) cocultures of CAFs and A549, Calu-3 human LADC cells, or NCI-H292 human lung mucoepidermoid carcinoma cells. We found that human lung cancer cells form tumors with distinct histological features in mice depending on the site of implantation, and that TGFβ signaling dependent on CAFs promotes a transition in the histological features of the injected tumor cells from solid type to acinar type. This solid-to-acinar transition (SAT) includes changes in cell morphology and gene expression as well as in TGFβ signaling. The TGFβ signaling pathway was thus highly activated in acinar-type tumors, and inhibition of this pathway suppressed acinar tumor formation by LADC cells. Our data suggest that TGFβ signaling dependent on CAFs induces SAT in LADC cells, and that this transition may contribute to the histological heterogeneity of LADC tissue.
Materials and Methods
Cell culture
The human LADC cell lines A549 and Calu-3, human lung mucoepidermoid carcinoma cell line NCI-H292, and mouse embryonic fibroblast cell line NIH3T3 were obtained from the ATCC. A549 and A549-GFP cells were cultured in DMEM–Ham's F12 nutrient mixture (1:1, v/v; Fujifilm Wako) supplemented with 10% FCS. Calu-3 and Calu-3–GFP cells were cultured in Eagle's MEM (ATCC) supplemented with 10% FCS. NCI-H292 and NCI-H292–GFP cells were cultured in RPMI-1640 medium (Fujifilm Wako) supplemented with 10% FCS. NIH3T3 cells were cultured in DMEM (Fujifilm Wako) supplemented with 10% FCS. All cells were maintained at 37°C under a humidified atmosphere of 5% CO2, were authenticated by examination of in vitro growth characteristics and morphological properties, and were found to be Mycoplasma negative. Early-passage cells were used for experiments.
Mice
Four-week-old female Balb/c nu/nu immune-deficient mice were obtained from Charles River Japan. They were bred and maintained in the animal facility at Keio University or Kumamoto University according to institutional guidelines. All animal experiments were performed in accordance with protocols approved by the ethics committee of Keio University or Kumamoto University.
Human LADC tumor samples
Tumor specimens from patients with LADC treated surgically at Kumamoto University Hospital were fixed in 10% formalin and subjected to hematoxylin–eosin staining or fluorescent multiplex IHC. This aspect of the study was approved by the institutional review board of Kumamoto University Hospital (Institutional Review Board approval number, 402).
Cell transplantation
Lung cancer cells were injected into the pleural cavity (1 × 106 cells in 50 μL of PBS), below the renal capsule (1 × 106 cells in 10 μL of PBS), below the skin (1 × 106 cells in 100 μL of PBS), or into the left ventricle (1 × 105 cells in 100 μL of PBS) of 4-week-old female Balb/c nu/nu immune-deficient mice that had been anesthetized by intraperitoneal administration of a combination of medetomidine (Nippon Zenyaku Kogyo), midazolam (Sandoz), and butorphanol (Meiji Seika Pharma). The resulting tumors were removed at 6 weeks after cell injection.
For the establishment of solid- or acinar-type tumor cells, cranial bone metastases or kidney subcapsular tumors were isolated from mice that had been injected with A549 cells expressing green fluorescent protein (GFP) into the left ventricle or below the renal capsule, respectively. The isolated tissue was cut into small pieces and incubated for 60 minutes at 37°C in a dissociation medium consisting of a 1:9 (v/v) mixture of 10× collagenase/hyaluronidase (Stemcell Technologies) and of DMEM–Ham's F12 nutrient mixture (1:1, v/v; Fujifilm Wako) containing DNase I (200 μg/mL, Millipore Sigma). The dissociated cells were isolated by centrifugation at 300 × g for 3 minutes at room temperature, suspended in a red blood cell lysis buffer (154 mmol/L NH4Cl, 14 mmol/L NaHCO3, and 0.1 mmol/L EDTA), and incubated for 1 minutes at room temperature. The suspension was again centrifuged at 300 × g for 3 minutes at room temperature, and the resulting pellet was suspended in DMEM–Ham's F12 medium supplemented with 10% FCS and seeded in 100-mm culture dishes.
Mucinous differentiation
On the basis of previous studies (20, 21), cells were seeded in 6-well ultralow attachment plates (#3471, Corning) at a density of 50,000 cells/mL and cultured for 2 days in serum-free DMEM–Ham's F12 medium supplemented with EGF (20 ng/mL, PeproTech), basic fibroblast growth factor (bFGF; 20 ng/mL, PeproTech), heparan sulfate (200 ng/mL, Seikagaku), and B27 supplement (Thermo Fisher Scientific). The resulting spheres were collected by centrifugation (300 × g for 3 minutes at room temperature) and dissociated by exposure to trypsin-EDTA (Thermo Fisher Scientific), and the released cells were then subjected to the indicated analyses.
Establishment of immortalized CAFs
A549-GFP pleural tumors, which contained abundant CAFs, were cut into small pieces and subjected to dissociation and lysis of red blood cells as described above. The dissociated cells were suspended in DMEM–Ham's F12 medium supplemented with 10% FCS and seeded in a 100-mm culture dish coated with collagen type I (Iwaki). After culture for 3 days, GFP-negative stromal cells were collected with a MoFlo cell sorter (Beckman Coulter), and human TERT cDNA was introduced into the sorted cells by infection with a retrovirus vector (pMSCV-CLXPpuro-ACC-hTERT). We used an ecotropic retrovirus to infect the mouse-derived stromal cells selectively and to allow complete exclusion of human-derived A549 cells through selection of the infected cells with puromycin (5 μg/mL). CAFs were cultured in DMEM–Ham's F12 medium supplemented with 10% FCS and in 35- or 100-mm culture dishes coated with collagen type I.
Establishment of CAF-containing tumor colonies
Lung cancer cells and CAFs were incubated separately overnight in corresponding culture medium containing NanoShuttle-PL (1 μL/10,000 cells, Nano3D Biosciences). The cells were then washed with PBS and dissociated into single cells with the use of TrypLE Express (Thermo Fisher Scientific). Dissociated lung cancer cells and CAFs were mixed at a 1:1 ratio (1,500 cells each) and suspended in serum-free DMEM–Ham's F12 medium supplemented with epidermal growth factor (20 ng/mL), bFGF (20 ng/mL), heparan sulfate (200 ng/mL), B27 supplement, and 5% Matrigel (#354234, Corning). The cell suspension (100 μL/well) was dispensed into the wells of a 96-well round-bottom ultralow attachment plate (#7007, Corning), which was then placed on top of a 96-well spheroid drive (Nano3D Biosciences) for 15 minutes on ice before transfer to an incubator at 37°C. After incubation for 15 minutes, the spheroid drive was removed from the plate. For single-culture of lung cancer cells, the same number of cells (1,500) was similarly processed. After 3D culture for 4 days, tumor colonies were subjected to live imaging with a fluorescence microscope (BZ-X700, Keyence), and 3D reconstruction of the images was performed with a BZ-X analyzer (Keyence). They were then subjected to immunofluorescence staining with a clearing technique or to cell sorting.
Immunofluorescence staining of tumor colonies with a clearing technique
Tumor colonies were immersed in a cell recovery solution (Corning) for 60 minutes on ice before fixation with 4% paraformaldehyde in PBS also for 60 minutes on ice. The fixed samples were washed twice with PBS and then stained with the use of a Visikol HISTO tissue-clearing technique (Visikol). The samples were permeabilized with a graded series of ethanol solutions (50% ethanol in PBS, 80% ethanol in deionized water, and 100% ethanol), washed consecutively with 20% dimethyl sufoxide in ethanol, 80% ethanol in deionized water, 50% ethanol in PBS, PBS, and PBS containing 0.2% Triton X-100, and then incubated with gentle shaking for 30 minutes at room temperature in Visikol HISTO penetration buffer. For the prevention of nonspecific binding of antibodies, the samples were incubated with gentle shaking for 30 minutes at 37°C in Visikol HISTO blocking buffer. They were then exposed with gentle shaking overnight at room temperature to primary antibodies in Visikol HISTO antibody buffer. The primary antibodies included mouse anti-MUC5AC (ab3649, Abcam), rabbit anti-CK7 (ab181598, Abcam), chicken anti-GFP (ab13970, Abcam), rabbit anti-DsRed (600–401–379, Rockland Immunochemicals), and rabbit anti–cleaved caspase-3 (#9661, Cell Signaling Technology). The samples were washed five times with 10% Visikol HISTO washing buffer in PBS, exposed with gentle shaking overnight at room temperature to Alexa Fluor 488–conjugated anti-mouse (Thermo Fisher Scientific), Alexa Fluor 555–conjugated anti-rabbit (Thermo Fisher Scientific), and Alexa Fluor 647–conjugated anti-chicken (Abcam) secondary antibodies in Visikol HISTO antibody buffer, and washed 10 times with 10% Visikol HISTO washing buffer in PBS. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) in 10% Visikol HISTO washing buffer in PBS. The samples were then transferred to the wells of a 96-well glass-bottom plate (Iwaki) and incubated with Visikol HISTO-M. The stained tumor colonies were observed with a confocal fluorescence microscope (FV3000, Olympus).
Data access
Raw and processed RNA-seq data generated in this study are available at Gene Expression Omnibus under accession GSE169029.
Other methods
All other methods are included in Supplementary Information.
Results
A549 human LADC cells form tumors with distinct histological features in mice
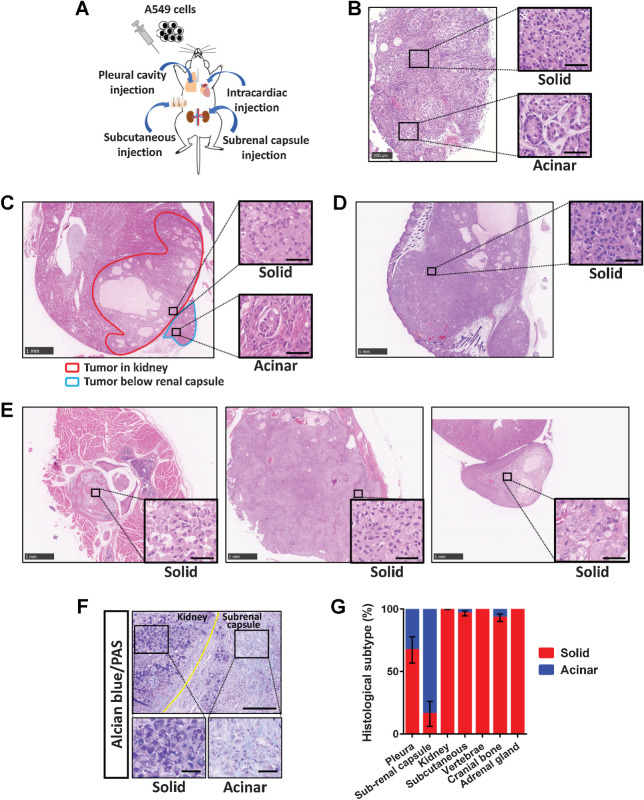
Human invasive LADC tissue often contains components corresponding to several of the five histological subtypes of such tumors (Supplementary Fig. S1A–S1E). To clarify the influence of environmental factors on LADC histology, we injected A549 human LADC cells into immunodeficient mice via four routes—pleural cavity, subcutaneous, left ventricle, or subrenal capsule injection (Fig. 1A)—and then examined the histology of the developed tumors. We found that the A549 cells formed tumors with distinct histological features, solid-type or acinar-type, depending on the site of tumor development (Fig. 1B–E). The solid-type tumors were composed of aggregates of uniform tumor cells positive for acidic mucin production (Fig. 1F), whereas the acinar-type tumors manifested glandlike structures encircled by stromal cells. Acinar-type cancer cells facing the stroma were positive for integrin α6, a basement membrane component (Supplementary Fig. S2A). In the pleural cavity, the developed tumors manifested both solid- and acinar-type histological components (Fig. 1B). Tumors that formed immediately below the renal capsule were predominantly acinar type, whereas those invading the kidney parenchyma were solid type (Fig. 1C). Subcutaneous tumors as well as those that formed in the vertebrae, cranial bone, or adrenal gland manifested solid-type histology (Fig. 1D and E). The proportions of histological subtypes for tumors formed at the various sites are shown in Fig. 1G.
Figure 1.
A549 cells form tumors of distinct histological subtypes depending on their site of development. A, Experimental scheme for establishment of A549 xenograft tumor models. A549 cells were injected into the pleural cavity, subcutaneously, into the left ventricle, or below the renal capsule of immunodeficient mice. B–E, Representative hematoxylin–eosin staining of tumors formed at 6 weeks after A549 cell injection via the four routes. B, A tumor that developed in the pleural cavity. The boxed regions on the left are shown at higher magnification on the right and correspond to solid- or acinar-type histology. Scale bars, 200 μm (left) and 50 μm (right). C, Tumors that developed in the kidney parenchyma (red outline) or below the renal capsule (blue outline) showed solid- and acinar-type histology, respectively. D, A subcutaneous tumor showing solid-type histology. E, Vertebral (left), cranial bone (middle), and adrenal gland (right) metastases formed after intracardiac injection of A549 cells all exhibited solid-type histology. Scale bars (C–E), 1,000 μm (low-power fields) and 50 μm (high-power fields). F, Alcian blue and periodic acid–Schiff staining of tumors formed after subrenal capsule injection of A549 cells. The tumor in the kidney (solid) showed a higher level of acidic mucin stained with Alcian blue than did that below the renal capsule (acinar). Scale bars, 200 μm (top) and 50 μm (bottom). G, The percentage area of each histological subtype within individual A549 xenograft tumors. Data are means ± SEM for three or four tumors at each site derived from at least three independent experiments.
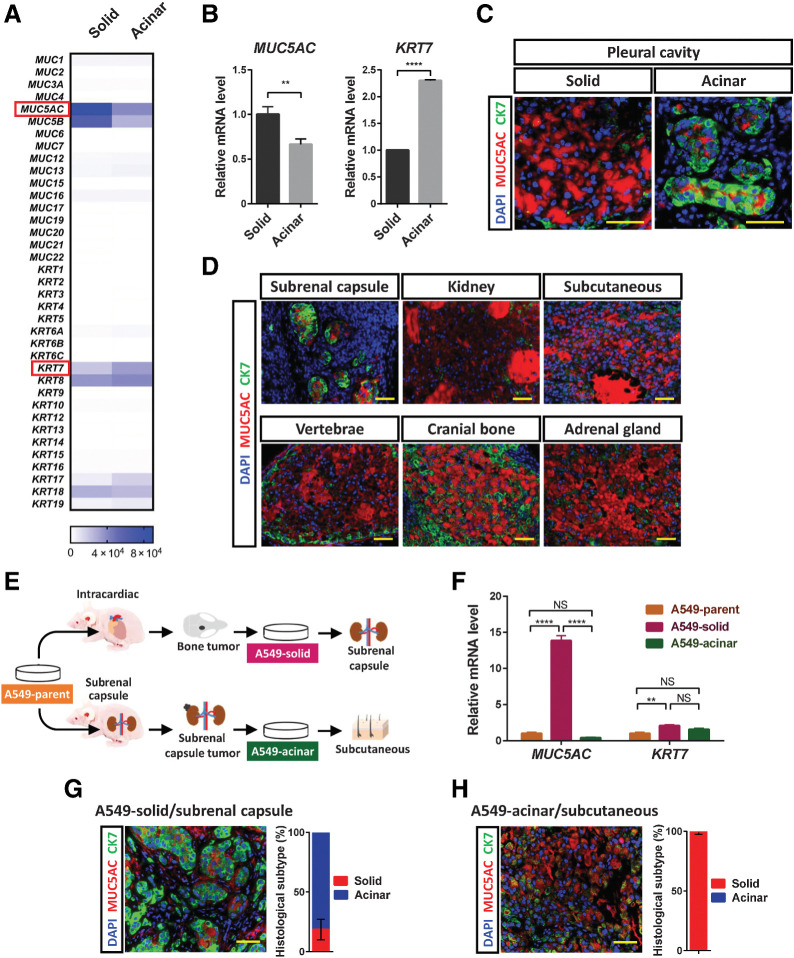
To characterize the biological features of the two histological subtypes of the A549 tumors, we isolated each type of tumor cell by laser microdissection from frozen sections of pleural tumors and performed RNA-seq analysis. Given that the expression of mucin and cytokeratin genes has been found to be related to the histological subtype of lung cancer, we compared gene expression patterns—in particular, the expression of mucin and cytokeratin genes—between solid-type and acinar-type tumor cells. We found that MUC5AC, which encodes a secretory mucin found predominantly in lung cancer (22), was expressed at a higher level in the solid-type tumor cells than in the acinar-type tumor cells. In contrast, KRT7, which encodes cytokeratin 7 (CK7), one of the most common cytokeratins expressed in LADC (23), was expressed at a higher level in the acinar-type tumor cells (Fig. 2A). We confirmed this differential expression of MUC5AC and KRT7 in the laser microdissection samples by reverse transcription (RT) and real-time PCR analysis (Fig. 2B). In addition to KRT7 mRNA, higher levels of KRT8 and KRT18 mRNA were expressed in the acinar-type tumor cells (Supplementary Fig. S2B). Immunohistofluorescence analysis also confirmed that MUC5AC and CK7 were preferentially expressed in solid-type and acinar-type tumor xenografts, respectively (Fig. 2C and D). Whereas peripheral acinar-type tumor cells in contact with stromal cells expressed CK7, however, central acinar-type tumor cells expressed MUC5AC, indicating that the expression of these proteins is spatially regulated.
Figure 2.
Environmental factors trigger SAT of A549 xenograft tumors. A, Heat map of the expression levels (counts) of mucin and cytokeratin genes determined by RNA-seq analysis of solid- or acinar-type tumor cells isolated by laser microdissection from pleural tumors formed by A549 cells in immunodeficient mice. Each sample is pooled from three mice in one experiment. B, RT and real-time PCR analysis of MUC5AC and KRT7 expression in solid- or acinar-type tumor cells isolated from pleural tumors by laser microdissection. The data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for solid-type tumor cells, and are means + SD for triplicate determinations. Each sample is pooled from three mice in one experiment. **, P < 0.01; ****, P < 0.0001 (unpaired two-tailed Student t test). C, Immunofluorescence staining for MUC5AC (red) and CK7 (green) in solid and acinar components of the same pleural tumor formed by A549 cells. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. D, Immunofluorescence staining for MUC5AC (red) and CK7 (green) in A549 tumors formed at the indicated sites. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. E, Experimental scheme for establishment of solid-type (A549-solid) and acinar-type (A549-acinar) tumor cells. For establishment of A549-solid cells, A549 cells (1 × 105) were injected into the left ventricle, and tumor cells were subsequently isolated from cranial bone metastases and were maintained in culture. For establishment of A549-acinar cells, A549 cells (1 × 106) were injected below the renal capsule, and cells were subsequently isolated from tumors that developed at the injection site and were maintained in culture. F, RT and real-time PCR analysis of MUC5AC and KRT7 expression in A549 parental cells, A549-solid cells, and A549-acinar cells. Data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for A549 parental cells, and are means + SD for triplicates of one of two independent experiments that yielded similar results. **, P < 0.01; ****, P < 0.0001; NS, not significant; by two-way ANOVA with Tukey multiple-comparison test. G and H, A549-solid cells were injected below the renal capsule (G), and A549-acinar cells were injected subcutaneously (H). The tumors that formed at each injection site were subjected to immunofluorescence staining for MUC5AC (red) and CK7 (green), and the percentage area of each histological subtype within individual tumors was determined. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The quantitative data are means ± SEM for three tumors at each site derived from three independent experiments.
To investigate further the role of the environment in determination of LADC histology, we first established two types of tumor-derived cells—solid-type cells isolated from cranial bone metastases (A549-solid) and acinar-type cells from subrenal capsule tumors (A549-acinar)—and then injected the two types of tumor cells into mice (Fig. 2E). Although the established A549-solid cells expressed MUC5AC at a high level (Fig. 2F), these cells were able to form CK7-expressing acinar-type tumors when injected below the kidney capsule (Fig. 2G), and we therefore termed this phenomenon SAT. In contrast, the A549-acinar cells formed solid-type tumors when injected subcutaneously (Fig. 2H). These results indicated that A549 LADC cells can develop into solid or acinar tumors, and that they can subsequently convert their histological subtype in a manner dependent on the environment that they colonize. Of note, GFP-labeled A549 (A549-GFP) cells in which MUC5AC had been disrupted with the use of the CRISPR-Cas9 system gave rise to histologically solid-type but not acinar-type tumors when injected below the renal capsule of mice (Supplementary Fig. S2C and S2D), indicating that downregulation of MUC5AC expression was not a trigger for SAT.
Lung cancer cell lines have the potential to undergo acinar-to-solid transition and SAT
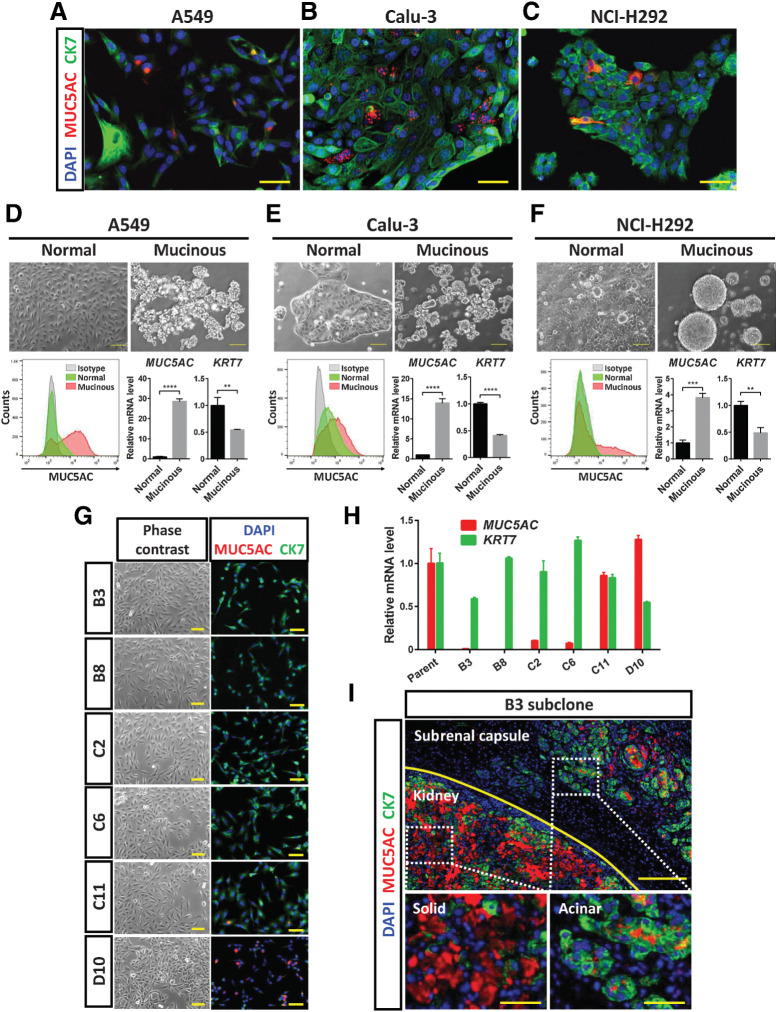
We found that three lung cancer cell lines (A549, Calu-3, and NCI-H292) were heterogeneous under adherent culture conditions. These cell lines comprised cells with different patterns of MUC5AC/CK7 expression as determined by immunocytofluorescence analysis, including MUC5AChigh/CK7low and MUC5AClow/CK7high cells (Fig. 3A–C). Cells expressing both MUC5AC and CK7 (stained yellow in Fig. 3A) might have a phenotype intermediate between solid and acinar, or manifest a hybrid SAT state. We then induced mucinous differentiation in these cell lines by culturing them under serum-free adhesion-restricted conditions. Such culture resulted in an increase in the expression of MUC5AC as well as in that of SPDEF, which encodes a master transcription factor for mucinous differentiation (24), indicating that such differentiation had indeed occurred (Supplementary Fig. S3A). In addition, flow cytometric analysis revealed that the number of MUC5AC+ cells increased in association with mucinous differentiation of A549, Calu-3, and NCI-H292 cells (Fig. 3D–F, Supplementary Fig. S3B). Changes in the relative abundance of MUC5AC and KRT7 mRNAs also showed that the cells underwent an acinar-to-solid transition (AST) in association with the induction of mucinous differentiation (Fig. 3D–F).
Figure 3.
Lung cancer cell lines have the ability to undergo AST and SAT. A–C, Immunofluorescence staining for MUC5AC (red) and CK7 (green) in A549, Calu-3, and NCI-H292 lung cancer cells, respectively. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. D–F, A549, Calu-3, and NCI-H292 cells, respectively, were cultured for 48 hours under normal or serum-free adhesion-restricted (mucinous differentiation) conditions. They were then examined by phase-contrast microscopy (top; scale bars, 100 μm) and subjected both to flow cytometric analysis of MUC5AC expression (bottom left) and to RT and real-time PCR analysis of MUC5AC and KRT7 expression (bottom right). The microscopic and flow cytometric data are representative of three independent experiments. The mRNA data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for cells cultured under the normal condition, and are means + SD from three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (unpaired two-tailed Student t test). G, Phase-contrast microscopy and immunofluorescence staining for MUC5AC (red) and CK7 (green) in six A549 subclones established by single-cell cloning. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. H, RT and real-time PCR analysis of MUC5AC and KRT7 expression in A549 parental cells and the six subclones. The data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for parental cells, and are means + SD of triplicates from one experiment. I, Immunofluorescence staining for MUC5AC (red) and CK7 (green) in tumors formed after injection of B3 subclone cells below the renal capsule of an immunodeficient mouse. Nuclei were stained with DAPI (blue). The top image shows the boundary (yellow line) between the kidney parenchyma and subrenal capsule regions. The kidney and subrenal capsule tumors are shown at higher magnification in the bottom left and bottom right images, respectively. The data are representative of two independent experiments. Scale bars, 200 μm (top) and 50 μm (bottom).
We performed single-cell cloning of A549 cells to determine whether AST/SAT occurs via cell selection or induction. We isolated six subclones that showed different expression patterns of MUC5AC and CK7 (Fig. 3G and H). All subclones formed solid-type and acinar-type tumors in the kidney parenchyma and below the renal capsule, respectively, after subrenal capsule injection in immunodeficient mice (Supplementary Fig. S3C). Although the B3 and B8 subclones expressed CK7 but not MUC5AC, they formed both CK7-expressing acinar-type and MUC5AC-expressing solid-type tumors in mice (Fig. 3I). In addition, the B3 subclone (MUC5AC negative) had the ability to undergo mucinous differentiation in vitro (Supplementary Fig. S3D). These data suggested that lung cancer cells possess phenotypic plasticity, including the ability to adopt solid- or acinar-type histology, depending on their environment.
CAFs induce histological transition of lung cancer cells
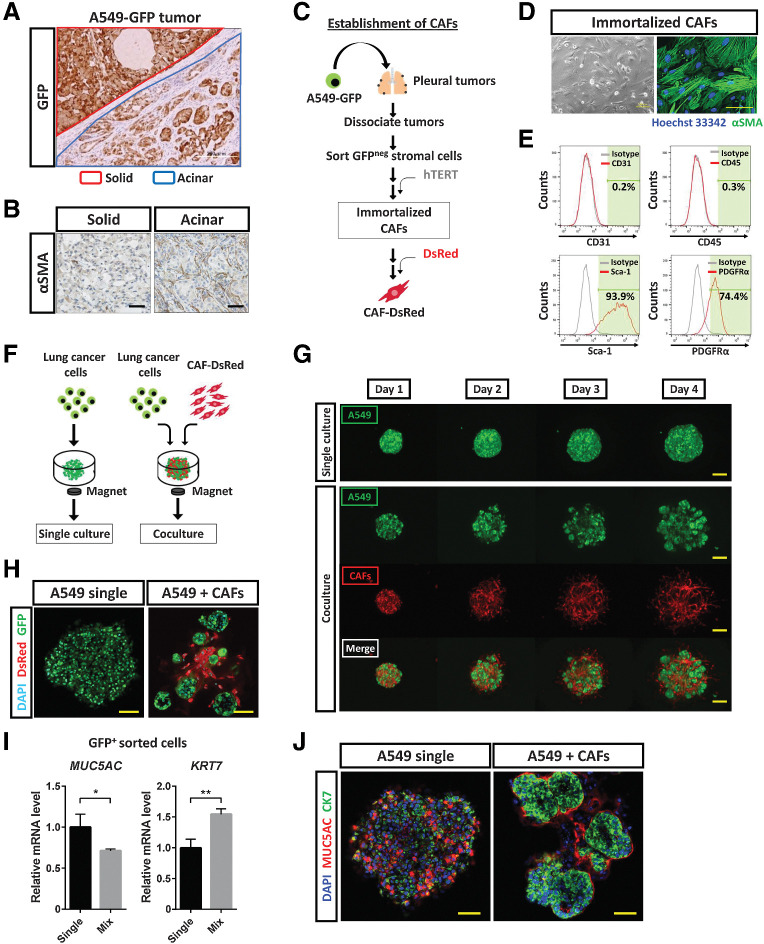
To understand the cellular mechanisms underlying AST or SAT, we focused on the stromal components of acinar-type tumor tissue, given that CK7-expressing acinar-type tumors manifested glandlike structures surrounded by stromal cells. Subrenal capsule injection of A549-GFP cells into immunodeficient mice resulted in the development of solid-type tumors consisting of almost all A549-GFP cells as well as of acinar-type tumors consisting of a substantial number of GFP-negative stromal cells in addition to A549-GFP cells (Fig. 4A). The acinar-type tumors also contained many fibroblasts positive for α–smooth muscle actin (αSMA; Fig. 4B) as well as abundant ECM—in particular, type I collagen (Supplementary Fig. S4A)—suggesting that CAFs might contribute to the tumor histological subtype.
Figure 4.
CAFs induce histological transition of lung cancer cells. A, Immunohistochemical staining for GFP in tumors developed after subrenal capsular injection of A549-GFP cells in immunodeficient mice. The red line demarcates a solid-type tumor in the kidney parenchyma, and the blue line an acinar-type tumor below the renal capsule. Scale bar, 200 μm. The image is representative of three independent experiments. B, Immunohistochemical staining for αSMA in solid and acinar tumors formed after subrenal capsular injection of A549-GFP cells. Scale bars, 50 μm. The images are representative of three independent experiments. C, Experimental scheme for establishment of immortalized CAFs. A549-GFP cells were injected into the pleural cavity of immunodeficient mice. After 6 weeks, pleural tumors were isolated and dissociated into single cells, GFP-negative stromal cells were collected by fluorescence-activated cell sorting, and human TERT cDNA was introduced into the stromal cells. The resulting immortalized CAFs were then labeled with DsRed. D, Phase-contrast microscopy and immunofluorescence staining for αSMA (green) in immortalized CAFs. Nuclei were stained with Hoechst 33342 (blue) in the immunofluorescence image. Scale bars, 100 μm (left) and 50 μm (right). The images are representative of three independent experiments. E, Flow cytometric analysis of CD31, CD45, Sca-1, and PDGFRα in immortalized CAFs. Data are representative of two independent experiments. F, Experimental scheme for 3D culture of GFP-labeled lung cancer cells with or without DsRed-labeled immortalized CAFs in medium containing 5% Matrigel for 4 days. G, Live imaging of GFP and DsRed fluorescence for A549-GFP colonies formed in the absence or presence of DsRed-labeled CAFs during 3D culture. Scale bars, 200 μm. The images are representative of three independent experiments. H, Confocal microscopy of GFP (green) and DsRed (red) immunofluorescence for tumor colonies formed by A549-GFP cells cultured alone or together with DsRed-labeled CAFs for 4 days. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The images are representative of three independent experiments. I, RT and real-time PCR analysis of MUC5AC and KRT7 expression in GFP+ cells sorted from A549-GFP tumor colonies formed as in H. Data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for the cells derived from CAF-free colonies, and are means + SD from two independent experiments. *, P < 0.05; **, P < 0.01 (unpaired two-tailed Student t test). J, Confocal microscopy of MUC5AC (red) and CK7 (green) immunofluorescence for tumor colonies formed as in H. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The images are representative of two independent experiments.
We isolated CAFs from acinar-type pleural tumor tissue formed by A549 cells and then immortalized them by introducing human TERT cDNA and labeled them with DsRed (Fig. 4C). The established CAFs expressed αSMA (Fig. 4D), and flow cytometric analysis (Fig. 4E) revealed that they corresponded to a fibroblast-enriched cell fraction (CD31−CD45−Sca-1+; ref. 25) and were positive for platelet-derived growth factor receptor α (PDGFRα), a representative marker for fibroblasts, including CAFs (26). We then established a 3D coculture system in which LADC cells were mixed with CAFs in vitro (Fig. 4F), and we found that the αSMA-positive CAFs induced the formation of acini by A549 cells in this system (Fig. 4G), showing that CAFs and cancer cells interact to direct tumor histology toward the acinar type. Confocal microscopy of the colonies formed in cocultures of A549-GFP cells and CAFs indeed revealed glandlike structures (Fig. 4H). Moreover, coculture of Calu-3 cells or NCI-H292 cells with CAFs also gave rise to colonies that manifested glandlike structures (Supplementary Fig. S4B–S4E). We isolated A549-GFP cells from colonies formed in cocultures and found that the interaction of these cells with CAFs resulted in downregulation of MUC5AC expression and upregulation of KRT7 expression (Fig. 4I). Confocal immunofluorescence microscopy confirmed similar effects at the protein level (Fig. 4J). These data thus indicated that interaction between cancer cells and CAFs contributes to determination of the histological subtype of LADC by promoting SAT.
TGFβ signaling is activated in acinar-type tumor cells
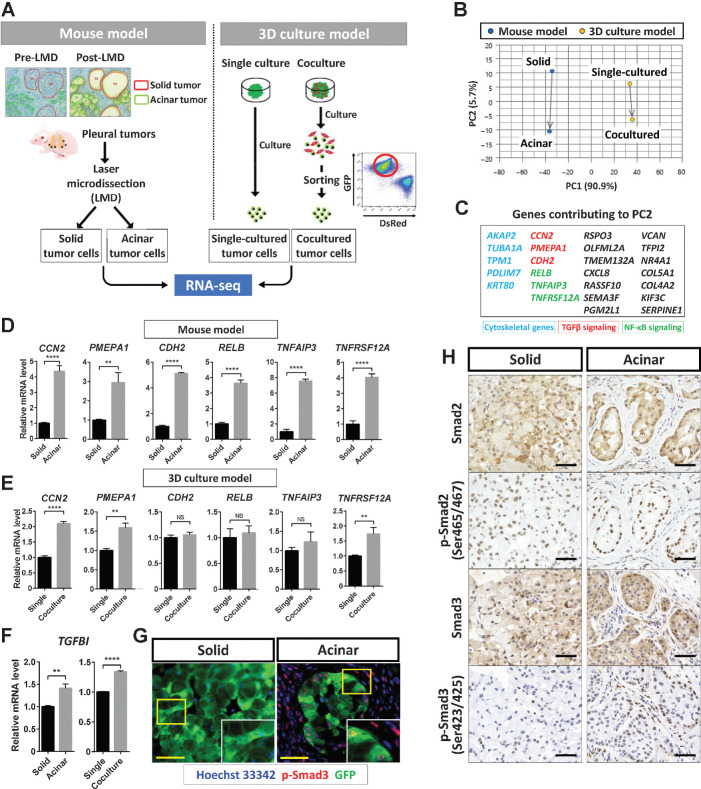
To clarify the molecular mechanisms contributing to SAT induction, we performed RNA-seq analysis of four A549 tumor cell samples derived from solid- or acinar-type tumor tissue formed in vivo or from 3D tumor colonies formed with or without CAFs in vitro (Fig. 5A), and we then subjected the expression data to principal component analysis (Fig. 5B). The tumor xenograft and 3D coculture models were separated by PC1, as expected. Given that PC2 reflected the direction of phenotypic change from solid to acinar in both models (Fig. 5B), we focused on genes contributing to PC2, which may be specifically associated with SAT induced by CAFs. With this approach, we identified 25 genes, which included those related to the cytoskeleton or to TGFβ or NF-κB signaling (Fig. 5C). RT and real-time PCR analysis confirmed that the expression of CCN2 and PMEPA1, both of which are related to the TGFβ signaling pathway, was significantly higher in acinar-type than in solid-type tumor cells in both the in vivo and in vitro models (Fig. 5D and E). In addition, the expression of TGFBI, which is induced by activation of TGFβ signaling (27, 28), was increased in the acinar-type tumor cells (Fig. 5F). Immunofluorescence (Fig. 5G) and immunohistochemical (Fig. 5H) analysis revealed that the amount of phosphorylated Smad3 (p-Smad3) was increased in CAFs and in cancer cells with neighboring fibroblasts of acinar-type tumors compared with cancer cells of solid-type tumors. These results thus indicated that the TGFβ pathway was activated in association with SAT.
Figure 5.
TGFβ signaling is activated in acinar-type tumor cells. A, Experimental scheme for RNA-seq analysis of solid- or acinar-type tumor cells derived from pleural tumors formed by A549 cells in immunodeficient mice or from tumor colonies formed by A549 cells cultured with (acinar) or without (solid) CAFs in vitro. B, PCA plots for the transcriptomes of solid- or acinar-type tumor cells determined as in A. Blue dots, cells isolated by laser microdissection (LMD) from the mouse tumor model; yellow dots, cells sorted from the 3D coculture model. C, Genes contributing to PC2 that are positively related to the change from solid-to-acinar type in the PCA analysis. Those related to the cytoskeleton (blue), to TGFβ signaling (red), or to NF-κB signaling (green) are highlighted. D and E, RT and real-time PCR analysis of genes related to TGFβ signaling (CCN2, PMEPA1, and CDH2) or to NF-κB signaling (RELB, TNFAIP3, and TNFRSF12A) in cells isolated as in A from the mouse model (D) or from the 3D coculture model (E). F, RT and real-time PCR analysis of TGFBI expression in cells as in D and E. Data in D through F were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for solid-type tumor cells or single-cultured tumor cells, and are means + SD of triplicates. Each sample is pooled from three mice examined in one experiment. **, P < 0.01; ****, P < 0.0001 (unpaired two-tailed Student t test). G, Immunofluorescence staining for GFP (green) and p-Smad3 (red) in solid or acinar tumors formed after subrenal capsule injection of A549-GFP cells in immunodeficient mice. Nuclei were stained with Hoechst 33342 (blue). Scale bars, 50 μm. The images are representative of two independent experiments. H, Immunohistochemical staining for Smad2, p-Smad2, Smad3, and p-Smad3 in solid- or acinar-type tumors as in G. Scale bars, 50 μm. The images are representative of two independent experiments.
TGFβ1 released by CAFs contributes to SAT induction in lung cancer cells
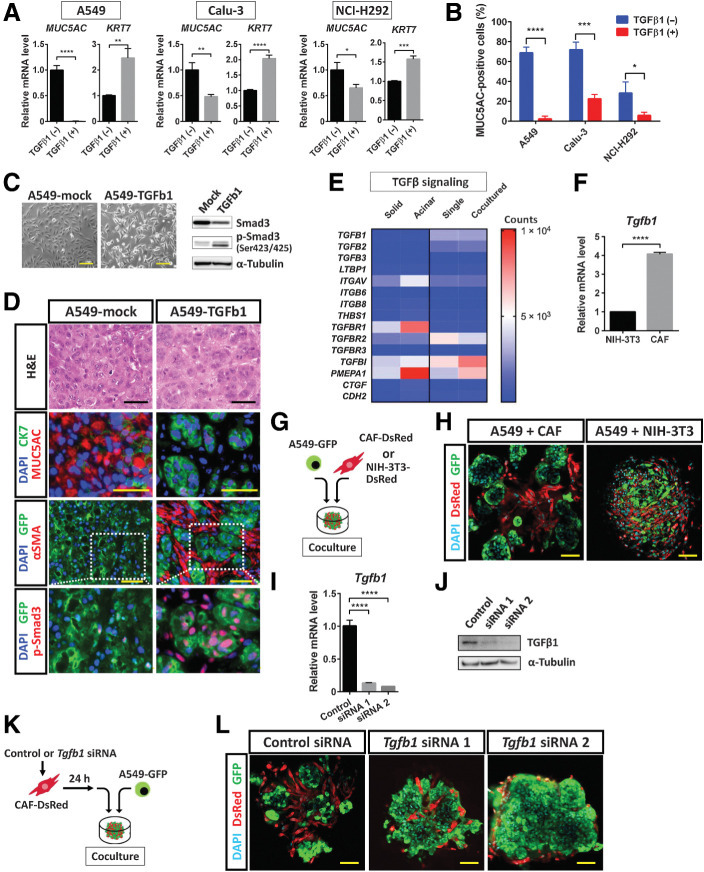
Exogenous TGFβ1 increased the expression of KRT7 and reduced that of MUC5AC in A549, Calu-3, and NCI-H292 cells (Fig. 6A). In addition, mucinous differentiation was suppressed by exogenous TGFβ1 in these three cell lines (Fig. 6B, Supplementary Fig. S5A). We next established A549-TGFb1 cells, which were engineered to express an active form of human TGFβ1 as well as GFP. Immunoblot analysis revealed a marked increase in Smad3 phosphorylation in these cells compared with the corresponding A549-mock control cells, confirming that the TGFβ signaling pathway was activated (Fig. 6C). Subcutaneous injection of A549-mock cells into immunodeficient mice resulted in the formation of solid-type tumors, whereas that of A549-TGFb1 cells gave rise to tumors with acinar-like morphology, a MUC5AClow/CK7high protein expression pattern, and the presence of αSMA+ CAFs (Fig. 6D). Comparison of gene expression levels, including those for genes related to TGFβ signaling, showed upregulation of TGFβ–responsive genes (TGFBR1, TGFBI, and PMEPA1) in the acinar-type tumor cells or 3D coculture cells (Fig. 6E). Overexpression of wild-type TGFβ1 in A549 cells did not result in the formation of acinar-type tumors after subcutaneous injection of the cells in immunodeficient mice (Supplementary Fig. S5B), suggesting that activation of TGFβ signaling is required for acinar tissue formation. Our results were thus consistent with the notion that paracrine TGFβ signaling mediated by CAFs rather than autocrine TGFβ signaling in tumor cells contributes to SAT induction. Indeed, in the absence of CAFs, autocrine activation of TGFβ signaling in A549 cells was insufficient to induce morphological changes in 3D culture (Supplementary Fig. S5C). Attenuation of MUC5AC expression by exogenous TGFβ1 in A549 cells in 3D culture without CAFs was also insufficient for the induction of morphological changes associated with SAT (Supplementary Fig. S5D), indicating that direct interaction between lung cancer cells and CAFs is important for SAT induction. Of note, washout of exogenous TGFβ1 in such 3D cultures attenuated CK7 expression and restored MUC5AC expression at both mRNA and protein levels, suggesting that SAT is a reversible phenomenon, at least with regard to gene and protein expression (Supplementary Fig. S5E and S5F).
Figure 6.
TGFβ1 signaling mediated by CAFs contributes to SAT induction in lung cancer cells. A, RT and real-time PCR analysis of MUC5AC and KRT7 expression in A549, Calu-3, and NCI-H292 lung cancer cells cultured in the absence or presence of TGFβ1 (2 ng/mL) for 24 hours. Data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for cells cultured in the absence of TGFβ1, and are means + SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (unpaired two-tailed Student t test). B, Flow cytometric analysis of MUC5AC expression in A549, Calu-3, and NCI-H292 cells cultured under mucinous differentiation conditions in the absence or presence of TGFβ1 (2 ng/mL) for 48 hours. Data are means + SD for the percentage of MUC5AC-positive cells from three independent experiments (a representative experiment is shown in Supplementary Fig. S5A). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (unpaired two-tailed Student t test). C, Phase-contrast microscopy of (left; scale bars, 50 μm) as well as immunoblot analysis (right) of Smad3 and p-Smad3 in A549-TGFb1 cells (expressing GFP and an active form of TGFβ1) and control (A549-mock) cells. α-Tubulin was examined as a loading control for immunoblot analysis. Data are representative of two independent experiments. D, Hematoxylin and eosin (H&E) staining as well as immunofluorescence staining for MUC5AC (red) and CK7 (green), GFP (green) and αSMA (red), or GFP (green) and p-Smad3 (red), as indicated, in subcutaneous tumors formed by A549-mock or A549-TGFb1 cells in immunodeficient mice. Nuclei were stained with DAPI (blue) in the immunofluorescence images. Scale bars, 50 μm. Data are representative of three independent experiments. E, Heat map of expression levels (counts) of TGFβ signaling–related genes derived from RNA-seq data of solid- and acinar-type tumor cells from both xenograft tumor and 3D coculture models (see Fig. 5A). F, RT and real-time PCR analysis of Tgfb1 expression in NIH3T3 fibroblasts and immortalized mouse CAFs. Data were normalized by the amount of B2 m mRNA, are expressed relative to the value for NIH3T3 cells, and are means + SD of triplicates from one of two similar experiments. ****, P < 0.0001 (unpaired two-tailed Student t test). G, Experimental scheme for 3D coculture of A549-GFP cells with CAF-DsRed or NIH3T3–DsRed cells for 4 days. H, Confocal microscopy of GFP (green) and DsRed (red) immunofluorescence in colonies formed as in G. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The images are representative of two independent experiments. I and J, RT and real-time PCR analysis of Tgfb1 mRNA (I) and immunoblot analysis of TGFβ1 (J) in CAF-DsRed cells transfected with a control siRNA or Tgfb1 siRNAs 1 or 2 for 48 hours. The RT-PCR data were normalized by the amount of B2 m mRNA, are expressed relative to the value for cells transfected with the control siRNA, and are means + SD from three independent experiments. ****, P < 0.0001 (unpaired two-tailed Student t test). The immunoblot data are representative of two independent experiments. K, Experimental scheme for 3D coculture of A549-GFP cells with CAF-DsRed cells transfected with siRNAs as in I and J. L, Confocal microscopy of GFP (green) and DsRed (red) immunofluorescence in colonies formed after 4 days in culture as in K. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The images are representative of two independent experiments.
We found that the relative abundance of Tgfb1 mRNA in immortalized CAFs derived from A549 acinar-type tumor tissue was about four times that in NIH3T3 mouse embryonic fibroblasts (Fig. 6F). NIH3T3 fibroblasts also did not induce the adoption of acinar-type morphology by A549-GFP cells during coculture under 3D conditions (Fig. 6G and H). We then introduced Tgfb1 siRNAs into CAFs to determine the relevance of paracrine TGFβ signaling mediated by these cells (Fig. 6I and J). CAFs transfected with control or Tgfb1 siRNAs were thus cocultured with A549-GFP cells under 3D conditions (Fig. 6K). Acinar formation by A549-GFP cells was induced by CAFs transfected with the control siRNA but not by those transfected with Tgfb1 siRNAs (Fig. 6L). In addition, treatment with TGFBR inhibitors (SB431542 or LY2157299) suppressed acinar formation by A549-GFP cells cultured with CAFs under 3D conditions (Supplementary Fig. S6A). Together, these results indicated that TGFβ signaling mediated by CAFs plays a key role in SAT induction in lung cancer cells.
Immunohistochemical analysis showed that both A549 cells and CAFs in acinar tumor tissue expressed TGFβ1 (Supplementary Fig. S6B). RT and real-time PCR analysis also revealed that the expression of TGFβ1, TGFβ2, and TGFβ3 genes was upregulated in CAFs that had been cocultured with A549 cells compared with those that had been cultured alone (Supplementary Fig. S6C). TGFβ is secreted as a biologically inactive (latent) complex containing a latency-associated peptide and latent TGFβ–binding proteins (LTBP; ref. 29), and this complex is converted to active TGFβ by integrin-, thrombospondin 1 (THBS1)-, or protease-mediated mechanisms (30). To examine the activation mechanism for TGFβ related to cross-talk between cancer cells and CAFs during SAT, we determined gene expression levels for LTBP1, integrins (ITGAV, ITGB6, and ITGB8), THBS1, matrix metalloproteinases (MMP2, MMP3, MMP9, and MMP13), and cathepsin cysteine proteinases (CTSB and CTSD; Supplementary Fig. S6D). The expression of Thbs1, Mmp9, and Mmp13 was markedly upregulated in CAFs under the coculture condition, suggesting that CAFs play a role in the activation of TGFβ.
CXCL8 expression is upregulated by TGFβ-smad3 signaling in acinar-type tumor cells
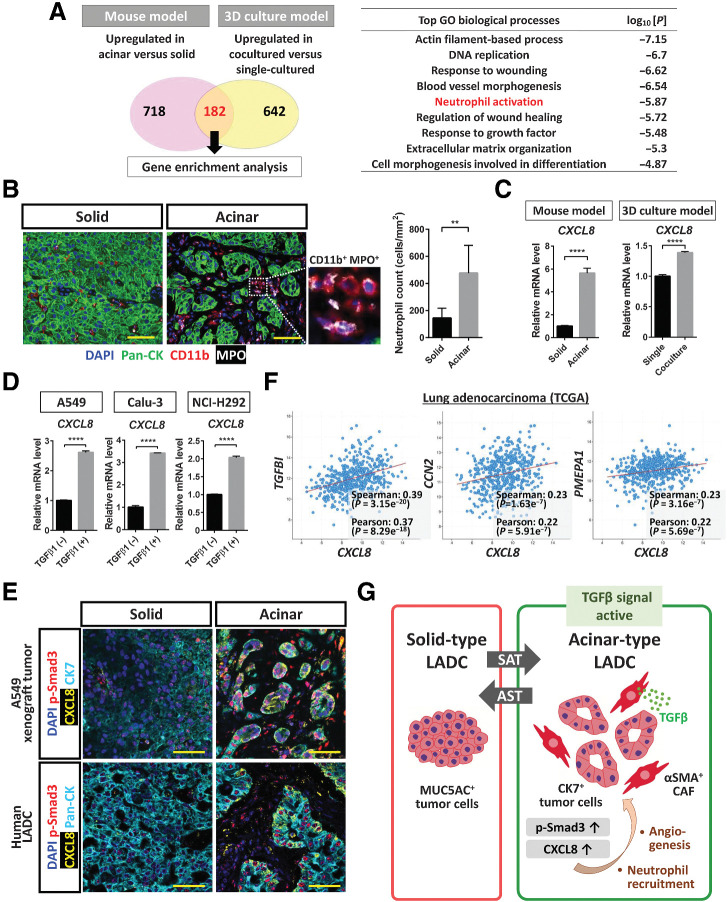
To characterize further the effects of SAT on the biology of tumor tissue, we again examined genes whose expression was upregulated in acinar-type tumor cells compared with solid-type tumor cells of both our in vivo mouse and in vitro 3D culture models. Our RNA-seq analysis identified 182 such genes whose expression was upregulated in association with acinar formation in both models (Fig. 7A). Gene ontology (GO) analysis revealed that neutrophil activation was one of the most significant enriched biological process terms for these 182 genes (Fig. 7A). Indeed, the number of infiltrating neutrophils [CD11b+ myeloperoxidase (MPO)+ immune cells] was significantly increased in acinar-type tumor tissue compared with solid-type tumor tissue (Fig. 7B). CXCL8 was identified both as a key gene contributing to SAT in lung cancer cells (Fig. 5C) as well as one of the 182 genes whose expression was increased in acinar-type tumor cells in both models. CXCL8 encodes C–X–C motif chemokine ligand 8 (CXCL8), also known as IL8, which is a key cytokine in inflammatory responses and serves as a chemotactic factor for recruitment of neutrophils as well as a potent angiogenic factor. Our RNA-seq analysis revealed that GO terms related to angiogenesis were also associated with SAT (Fig. 7A). CXCL8 is released from various cell types, including immune cells, endothelial cells, fibroblasts, and tumor cells, and RT and real-time PCR analysis confirmed that the relative abundance of CXCL8 mRNA was increased in acinar-type tumor cells (Fig. 7C). We also found that TGFβ1 treatment upregulated CXCL8 expression in A549, Calu-3, and NCI-H292 lung cancer cells (Fig. 7D). Furthermore, we performed fluorescent multiplex IHC of A549 xenograft tumors and human LADC tissue, and we found that the expression of CXCL8 was increased in CK7-positive acinar-type tumor tissue compared with solid-type tumor tissue (Fig. 7E). In addition to the clinical sample examined, we identified the SAT phenotype in a mutant KRAS (KRASmut)-driven mouse lung cancer model that we recently established (Supplementary Fig. S7A; ref. 31). Of note, CXCL8 was expressed in p-Smad3+ acinar-type tumor cells of mouse xenograft and human LADC tissue (Fig. 7E) as well as in those of KRASmut-driven mouse lung cancer tissue (Supplementary Fig. S7B). Collectively, these findings suggested that CAF-mediated activation of TGFβ-Smad3 signaling induces CXCL8 secretion in lung cancer cells during SAT.
Figure 7.
CXCL8 expression is upregulated by TGFβ-Smad3 signaling in acinar-type tumor cells. A, Enrichment analysis for genes whose expression was upregulated in acinar-type cells compared with solid-type cells in both the mouse xenograft and 3D culture models as determined from RNA-seq data (see Fig. 5A). The most significantly enriched GO terms for biological processes are listed together with the corresponding P values. B, Immunofluorescence staining for pan-CK (green), CD11b (red), and MPO (gray) in solid and acinar components of a pleural tumor formed after injection of A549 cells in immunodeficient mice. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The number of neutrophils (CD11b+MPO+ cells) in solid or acinar components of the tumor tissue was also quantified, with the presented data being means + SD from three independent experiments. **, P < 0.01 (unpaired two-tailed Student t test). C, RT and real-time PCR analysis of CXCL8 expression in solid- or acinar-type tumor cells isolated from A549 pleural tumor tissue or from A549 colonies cultured in the absence or presence of CAFs. Data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for solid-type tumor cells or single-cultured tumor cells, and are means + SD of triplicates. Each sample was pooled from three mice in one experiment. ****, P < 0.0001 (unpaired two-tailed Student t test). D, RT and real-time PCR analysis of CXCL8 expression in A549, Calu-3, and NCI-H292 lung cancer cells cultured in the absence or presence of TGFβ1 (2 ng/mL) for 48 hours. Data were normalized by the amount of HPRT1 mRNA, are expressed relative to the corresponding value for cells cultured in the absence of TGFβ1, and are means + SD from three independent experiments. ****, P < 0.0001 (unpaired two-tailed Student t test). E, Immunofluorescence staining for CK7 or pan-CK (cyan), for p-Smad3 (red), and for CXCL8 (yellow) in solid and acinar components either of an A549 pleural tumor (top) or of a human LADC tumor (bottom). Nuclei were stained with DAPI (blue). Scale bars, 50 μm. The image of the A549 xenograft tumor is representative of three independent experiments, and one human LADC tumor was examined. F, Relationship between CXCL8 and TGFBI, CCN2, or PMEPA1 expression levels determined from RNA-seq data for 566 LADC tumors in TCGA. Spearman and Pearson correlation coefficients are indicated. G, LADC cells form tumors with distinct histological features, including MUC5AC-expressing solid-type and CK7-expressing acinar-type tumors. In a manner dependent on the tumor microenvironment, these two types of tumor can undergo interconversion. αSMA-positive CAFs surrounding LADC cells induce the formation of acinar tumor tissue by eliciting TGFβ–Smad3 signaling and consequent SAT in the tumor cells. Activation of TGFβ signaling also contributes to upregulation of CXCL8 expression in LADC cells, which has the potential to lead to changes in the tumor microenvironment, including the induction of angiogenesis and the recruitment of neutrophils into the tumor tissue.
We performed an apoptosis assay in A549 cells treated with the anticancer agent cisplatin to investigate the effect of SAT on drug sensitivity. The 3D culture of A549 cells with CAFs or treatment with TGFβ1 showed that the acinar-type A549 cells are more sensitive to cisplatin than the solid-type A549 cells (Supplementary Fig. S8A–S8D). However, therapeutic resistance and patient outcomes in clinical settings are influenced not only by cellular sensitivity to anticancer drugs but also by multiple environmental factors such as immune response and drug delivery. For instance, CXCL8 may contribute to the SAT-associated microenvironmental changes because it plays a role in inflammatory responses such as angiogenesis and neutrophil recruitment, and intratumor neutrophils contribute to the immune suppression that can underlie resistance to immune checkpoint therapy in patients with cancer (32, 33). Further experiments using experimental models that recapitulates in in vivo microenvironments, including neutrophils, other immune cells, and endothelial cells, are necessary to determine the differences in the biological properties of solid tumors and acinar-type tumors.
The NF-κB and JNK signaling pathways have previously been implicated as key regulators of CXCL8 expression (34). Given that genes related to NF-κB signaling were also activated in acinar-type tumor cells (Fig. 5C), both TGFβ and NF-κB pathways may contribute to the expression of CXCL8 in these cells. Indeed, we found that the relative abundance of CXCL8 mRNA was positively correlated with the expression of TGFβ target genes (TGFBI, CCN2, and PMEPA1) as well as with that of NF-κB target genes (JUNB, RELB, TNFAIP3, TNFAIP6, and TNFRSF12A) in LADC tissue of patients in The Cancer Genome Atlas (TCGA) database (Fig. 7F, Supplementary Fig. S8E).
Discussion
We have here described a potential mechanism underlying the generation of LADC tumor heterogeneity. We found that human A549 cells form tumors with distinct histological features in immunodeficient mice. In particular, the cells form MUC5AC-expressing solid-type or CK7-expressing acinar-type tumors depending on the tumor microenvironment. We found that αSMA-positive CAFs surrounding cancer cells induce acinar tissue formation via activation of TGFβ-Smad3 signaling and consequent induction of SAT in the cancer cells (Fig. 7G). SAT was found to be characterized by changes in morphology (solid to acinar), gene expression (in particular, expression of MUC5AC and KRT7), and TGFβ signaling. Our data thus show that TGFβ signaling mediated by CAFs induces SAT in LADC cells, and that SAT may contribute to tumor histological heterogeneity in LADC.
More than 80% to 90% of human LADC tumors comprise several histological subtypes, with such intratumoral histological heterogeneity being thought to contribute to tumor malignancy and therapeutic resistance. However, the mechanisms underlying the histological heterogeneity of LADC have remained unclear. No definitive studies have shown a direct relation between the histological subtypes of LADC and specific genetic alterations. Multiregion analysis of genetic mutations in individual patients with LADC has revealed the same EGFR mutations in spatially distinct locations within a single tumor (35, 36). Furthermore, components of LADC tumors corresponding to distinct histological subtypes within individual patients have been found to harbor the same driver gene mutation (37), suggesting that extrinsic factors contribute to the determination of tumor histology. In the present study, we examined three human lung cancer cell lines and found that they all manifested histological plasticity, including the ability to undergo SAT. A549 cells are wild-type for EGFR but have a mutant KRAS (G12S) allele; Calu-3 cells manifest amplification of ERBB2, are wild type for EGFR, and possess a mutant KRAS (G13D) allele; and NCI-H292 cells are wild-type for both EGFR and KRAS. Our data therefore also suggest that histological phenotype is regulated by the tumor environment rather than being determined by driver gene mutations.
We have shown that cross-talk between lung cancer cells and CAFs contributes to changes in tumor histology—in particular, to SAT. We established a line of αSMA-expressing CAFs and found that these cells interact directly with LADC cells and that TGFβ signaling was activated in both interacting cell types. CAFs that exist in close proximity to tumor cells have previously been shown to express αSMA at a high level as a result of TGFβ signaling in pancreatic cancer (38). Our data further suggest that TGFβ signaling contributes to the induction of CXCL8 expression in LADC cells. Tumor-associated neutrophils have recently been implicated in a shift in the histological features of lung tumors from adenocarcinoma to squamous cell carcinoma (39). Given that neutrophils have recently been shown to produce MMP9 and to activate latent TGFβ in colon cancer (40), neutrophils in LADC might also play a role in the activation of TGFβ and thereby complete a positive feedback loop involving TGFβ signaling, CXCL8 expression, neutrophil recruitment, and TGFβ activation. Further studies are thus warranted to clarify the role of the interaction between CAFs and such neutrophils in determination of the histological subtype of LADC.
Our RNA-seq analysis suggested that NF-κB signaling might also contribute to SAT, with orchestration between TGFβ and NF-κB signaling pathways, therefore, possibly playing an important role in determination of the histological features of lung tumor tissue. In vitro experiments have shown that transcription of CXCL8 is regulated by multiple stimulus-responsive transcription factors—in particular, by NF-κB, activator protein–1(AP-1), and C/EBPβ (34). We found that the activation of TGFβ signaling increased the expression of CXCL8 in lung cancer cells, and analysis of RNA-seq data for patients with LADC in TCGA also revealed a correlation between CXCL8 expression and the expression of genes related to TGFβ signaling or to NF-κB signaling. Regulation of CXCL8 expression by TGFβ and NF-κB signaling pathways may therefore be a key determinant of SAT.
CAFs and CAF-derived TGFβ contribute to tumor development by modulating aspects of the tumor microenvironment, including the production of ECM and growth factors, as well as by promoting tumor cell proliferation. In addition, evidence suggests that CAFs may promote tumor growth by targeting the immune system (41). Differences in the tumor immune microenvironment have been found to be dependent on tumor histological subtype (42, 43), with solid-type LADC tumors having been shown to express programmed death–ligand 1 (PD-L1) at a higher level compared with other subtypes (44). Moreover, the serum level of CXCL8 allowed prediction of responsiveness to immune checkpoint inhibitors in patients with LADC (45, 46). Determination of the relevance of regulation of CXCL8 expression by TGFβ and NF-κB signaling during SAT to therapeutic resistance, including immunosuppression may provide a basis for the development of new treatment strategies for and consequent improvement in the prognosis of patients with lung cancer.
Our data thus show that interference with cross-talk between LADC cells and CAFs via suppression of TGFβ signaling may influence not only tumor heterogeneity but also the tumor immune microenvironment. Given that neutrophils have been shown to suppress tumor-infiltrating T cells via MMP-mediated activation of TGFβ in the tumor microenvironment (40), targeting of TGFβ signaling might increase the efficacy of T-cell–based immunotherapies. Furthermore, p-Smad3 rather than p-Smad2 can be a potential biomarker for the TGFβ–targeting therapy. In our model, we found that the A549 acinar-type tumor cells highly expressed CK7, CK8, and CK18 genes. Cytokeratins (CK) have become an important diagnostic marker for a disease state of cancer, and CK7, CK8, CK18, and CK19 are known to be preferentially expressed in non–small cell lung cancer (47). Therefore, change of the CKs expression is a potential diagnostic marker for SAT.
Authors' Disclosures
R. Sato reports grants from Japan Society for the Promotion of Science during the conduct of the study. Y. Tomita reports grants from JSPS KAKENHI grant number JP18K15928 and grants from Takeda Science Foundation outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Fig
Supplementary Info
Acknowledgments
This work was supported by Japan Society for the Promotion of Science KAKENHI grants 17H06971 and 19K17677 (to R. Sato) by Translational Research Network Program, Research on Applying Health Technology, and Research on Rare and Intractable Diseases grants from the Japan Agency for Medical Research and Development (AMED) grant 22130007 (to H. Saya) and by grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (KAKENHI 22130007 to H. Saya; KAKENHI 20K08968 to Y. Arima). The authors thank T. Kitamura, H. Seimiya, and E. Batlle for retroviral plasmids; E. Sugihara, K. Suina, A. Kasuga, R. Harigai, I. Ishimatsu, S. Hayashi, M. Sato, H. Kohrogi, H. Ichiyasu, M. Takahashi, K. Emoto, and other laboratory members for technical assistance and discussion; and Collaborative Research Resources, Keio University School of Medicine, for technical support.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
R. Sato: Conceptualization, resources, data curation, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. K. Imamura: Conceptualization, data curation, investigation, visualization, methodology, writing–review and editing. T. Semba: Conceptualization, resources, data curation, investigation, methodology, writing–review and editing. Y. Tomita: Conceptualization, data curation, validation, investigation, visualization, methodology, writing–review and editing. S. Saeki: Project administration, writing–review and editing. K. Ikeda: Project administration, writing–review and editing. Y. Komohara: Data curation, investigation, project administration, writing–review and editing. M. Suzuki: Resources, project administration, writing–review and editing. T. Sakagami: Resources, project administration, writing–review and editing. H. Saya: Conceptualization, resources, data curation, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. Y. Arima: Conceptualization, resources, data curation, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu Y-L, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299–311. [DOI] [PubMed] [Google Scholar]
- 2. Warth A, Muley T, Meister M, Stenzinger A, Thomas M, Schirmacher P, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438–46. [DOI] [PubMed] [Google Scholar]
- 3. Cadioli A, Rossi G, Costantini M, Cavazza A, Migaldi M, Colby TV. Lung cancer histologic and immunohistochemical heterogeneity in the era of molecular therapies. Am J Surg Pathol 2014;38:502–9. [DOI] [PubMed] [Google Scholar]
- 4. Solis LM, Behrens C, Raso MG, Lin HY, Kadara H, Yuan P, et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer 2012;118:2889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TB, Veeriah S, et al. Tracking the evolution of non–small cell lung cancer. N Engl J Med 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 6. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato R, Semba T, Saya H, Arima Y. Stem cells and epithelial–mesenchymal transition in cancer: biological implications and therapeutic targets. Stem Cells 2016;34:1997–2007. [DOI] [PubMed] [Google Scholar]
- 8. Sharma A, Merritt E, Hu X, Cruz A, Jiang C, Sarkodie H, et al. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep 2019;29:2164–74.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer 2020;20:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli K, Andersen S, Sirera R, et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non–small cell lung cancer. J Thorac Oncol 2011;6:209–17. [DOI] [PubMed] [Google Scholar]
- 11. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer 2019;19:9–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014;211:1503–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shangguan C, Gan G, Zhang J, Wu J, Miao Y, Zhang M, et al. Cancer-associated fibroblasts enhance tumor 18F-FDG uptake and contribute to the intratumor heterogeneity of PET-CT. Theranostics 2018;8:1376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Pilato MD, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell 2019;178:160–75.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyazono K, Katsuno Y, Koinuma D, Ehata S, Morikawa M. Intracellular and extracellular TGFβ signaling in cancer: some recent topics. Front Med 2018;12:387–411. [DOI] [PubMed] [Google Scholar]
- 17. Otsuki Y, Saya H, Arima Y. Prospects for new lung cancer treatments that target EMT signaling. Dev Dyn 2018;247:462–72. [DOI] [PubMed] [Google Scholar]
- 18. Colak S, ten Dijke P. Targeting TGFβ signaling in cancer. Trends Cancer 2017;3:56–71. [DOI] [PubMed] [Google Scholar]
- 19. Syed V. TGFβ signaling in cancer. J Cell Biochem 2016;117:1279–87. [DOI] [PubMed] [Google Scholar]
- 20. Real FX, Egea G, Francí C, Schüssler MH, Xu M, Welt S. Mucin production by colon cancer cells cultured in serum-free medium. Int J Cancer 1991;49:787–95. [DOI] [PubMed] [Google Scholar]
- 21. Iwashita J, Hongo K, Ito Y, Abe T, Murata J. Regulation of MUC5AC mucin production by the cell attachment dependent pathway involving integrin β1 in NCI-H292 human lung epithelial cells. Adv Biol Chem 2013;3:1–10. [Google Scholar]
- 22. Lakshmanan I, Ponnusamy MP, Macha MA, Haridas D, Majhi P, Kaur S, et al. Mucins in lung cancer: diagnostic, prognostic, and therapeutic implications. J Thorac Oncol 2015;10:19–27. [DOI] [PubMed] [Google Scholar]
- 23. Warth A, Muley T, Herpel E, Meister M, Herth FJ, Schirmacher P, et al. Large-scale comparative analyses of immunomarkers for diagnostic subtyping of non–small cell lung cancer biopsies. Histopathology 2012;61:1017–25. [DOI] [PubMed] [Google Scholar]
- 24. Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, et al. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest 2009;119:2914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, et al. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells 2009;27:623–33. [DOI] [PubMed] [Google Scholar]
- 26. Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int J Cancer 2020;146:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol 1992;11:511–22. [DOI] [PubMed] [Google Scholar]
- 28. Yokobori T, Nishiyama M. TGFβ signaling in gastrointestinal cancers: progress in basic and clinical research. J Clin Med 2017;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robertson IB, Rifkin DB. Regulation of the bioavailability of TGFβ and TGFβ–related proteins. Cold Spring Harb Perspect Biol 2016;8:a021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayashi H, Sakai T. Biological significance of local TGFβ activation in liver diseases. Front Physiol 2012;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Semba T, Sato R, Kasuga A, Suina K, Shibata T, Kohno T, et al. Lung adenocarcinoma mouse models based on orthotopic transplantation of syngeneic tumor-initiating cells expressing EpCAM, SCA-1, and Ly6d. Cancers 2020;12:3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schalper KA, Carleton M, Zhou M, Chen T, Feng Y, Huang SP, et al. Elevated serum interleukin-8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune-checkpoint inhibitors. Nat Med 2020;26:688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. David JM, Dominguez C, Hamilton DH, Palena C. The IL-8/IL-8R axis: a double agent in tumor immune resistance. Vaccines 2016;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol 2002;72:847–55. [PubMed] [Google Scholar]
- 35. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol 2011;29:2972–7. [DOI] [PubMed] [Google Scholar]
- 36. Suda K, Sakai K, Obata K, Ohara S, Fujino T, Koga T, et al. Inter- and intratumor heterogeneity of EGFR compound mutations in non–small cell lung cancers: analysis of five cases. Clin Lung Cancer 2020;S1525–7304:30273–4. [DOI] [PubMed] [Google Scholar]
- 37. Schneider F, Derrick V, Davison JM, Strollo D, Incharoen P, Dacic S. Morphological and molecular approach to synchronous non–small cell lung carcinomas: impact on staging. Mod Pathol 2016;29:735–42. [DOI] [PubMed] [Google Scholar]
- 38. Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mollaoglu G, Jones A, Wait SJ, Mukhopadhyay A, Jeong S, Arya R, et al. The lineage-defining transcription factors SOX2 and NKX2–1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity 2018;49:764–79.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Germann M, Zangger N, Sauvain M, Sempoux C, Bowler AD, Wirapati P, et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFβ. EMBO Mol Med 2020;12:e10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jaeghere EA, Denys HG, Wever O. Fibroblasts fuel immune escape in the tumor microenvironment. Trends Cancer 2019;5:704–23. [DOI] [PubMed] [Google Scholar]
- 42. Saruwatari K, Ikemura S, Sekihara K, Kuwata T, Fujii S, Umemura S, et al. Aggressive tumor microenvironment of solid predominant lung adenocarcinoma subtype harboring with epidermal growth factor receptor mutations. Lung Cancer 2016;91:7–14. [DOI] [PubMed] [Google Scholar]
- 43. Nagaraj AS, Lahtela J, Hemmes A, Pellinen T, Blom S, Devlin JR, et al. Cell of origin links histotype spectrum to immune microenvironment diversity in non–small cell lung cancer driven by mutant Kras and loss of Lkb1. Cell Rep 2017;18:673–84. [DOI] [PubMed] [Google Scholar]
- 44. Dong Z-Y, Zhang C, Li Y-F, Su J, Xie Z, Liu S-Y, et al. Genetic and immune profiles of solid predominant lung adenocarcinoma reveal potential immunotherapeutic strategies. J Thorac Oncol 2018;13:85–96. [DOI] [PubMed] [Google Scholar]
- 45. Sanmamed MF, Perez-Gracia JL, Schalper KA, Fusco JP, Gonzalez A, Rodriguez-Ruiz ME, et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non–small cell lung cancer patients. Ann Oncol 2017;28:1988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oyanagi J, Koh Y, Sato K, Mori K, Teraoka S, Akamatsu H, et al. Predictive value of serum protein levels in patients with advanced non–small cell lung cancer treated with nivolumab. Lung Cancer 2019;132:107–13. [DOI] [PubMed] [Google Scholar]
- 47. Kanaji N, Bandoh J, Fujita J, Ishii T, Ishida T, Kubo A. Compensation of type I and type II cytokeratin pools in lung cancer. Lung Cancer 2007;55:295–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig
Supplementary Info