This study characterizes a previously unexplored role of ncNATs in regulation of isoform expression of associated sense genes, highlighting a mechanism of alternative promoter usage in cancer.
Abstract
Multiple noncoding natural antisense transcripts (ncNAT) are known to modulate key biological events such as cell growth or differentiation. However, the actual impact of ncNATs on cancer progression remains largely unknown. In this study, we identified a complete list of differentially expressed ncNATs in hepatocellular carcinoma. Among them, a previously undescribed ncNAT HNF4A-AS1L suppressed cancer cell growth by regulating its sense gene HNF4A, a well-known cancer driver, through a promoter-specific mechanism. HNF4A-AS1L selectively activated the HNF4A P1 promoter via HNF1A, which upregulated expression of tumor suppressor P1-driven isoforms, while having no effect on the oncogenic P2 promoter. RNA-seq data from 23 tissue and cancer types identified approximately 100 ncNATs whose expression correlated specifically with the activity of one promoter of their associated sense gene. Silencing of two of these ncNATs ENSG00000259357 and ENSG00000255031 (antisense to CERS2 and CHKA, respectively) altered the promoter usage of CERS2 and CHKA. Altogether, these results demonstrate that promoter-specific regulation is a mechanism used by ncNATs for context-specific control of alternative isoform expression of their counterpart sense genes.
Significance:
This study characterizes a previously unexplored role of ncNATs in regulation of isoform expression of associated sense genes, highlighting a mechanism of alternative promoter usage in cancer.
Introduction
Although treatment options are steadily improving, hepatocellular carcinoma (HCC) remains the fourth most common cause of cancer-related deaths worldwide (1, 2). To better understand the molecular nature of HCC, international studies have profiled genetic, epigenetic, and transcriptional changes in hundreds of patient samples (3, 4). Although genetic driver mutations are the underlying cause of cancer development, they are often rare, affecting on average 4.6 genes per tumor, with different patients harboring a distinct mutational profile (5). In contrast, transcriptional aberrations are widespread and recently considered as nongenetic drivers for cancer (6). Transcriptional controls in cancer cells can be disrupted via multiple mechanisms, such as alterations in promoter sequence (7), DNA methylation (8), transcription factor binding (9) as well as dysregulation of important, yet poorly studied noncoding natural antisense transcripts (ncNAT; ref. 10).
ncNATs are a specific class of noncoding RNA sequences that are transcribed on the opposite strand relative to a protein-coding or non-coding transcript (11). It has been reported that ncNATs can regulate the expression of their counterpart sense genes through diverse mechanisms, including competition for transcription factor binding to the shared promoter(s), collision with RNA polymerase, induction of DNA and histone modifications, modulation of mRNA stability, degradation, and translation (2), among others. ncNATs have been found to regulate key biological events such as cell differentiation and carcinogenesis (12–15).
Most human protein-coding genes have multiple promoters that direct transcription of promoter-specific gene isoforms. The choice of promoter has been shown to be a major influence on the cancer transcriptome (16, 17), as these promoters are deregulated across tissues and cancer types, affecting cancer-related genes. When these promoters are differentially activated within a particular gene and derive isoform-specific protein variants with distinct functions, it is much more informative and accurate to use promoter activity than gene expression for survival analysis (17). Given the proximity of ncNATs to sense transcripts, ncNATs are potential modulators of promoter selection and activity of their counterpart sense genes. However, as antisense RNAs are often expressed at very low abundance, they tend to be poorly annotated and scarcely studied. Therefore, their real impact on promoter regulation in cancer remains largely unexplored.
Here, we analyzed publicly available RNA-seq data from The Cancer Genome Atlas (TCGA), the Pan-Cancer-Analysis of Whole Genomes (PCAWG), and the Genotype-Tissue Expression (GTEx) project to study deregulated ncNATs in HCC. We identified a previously undescribed isoform of HNF4A antisense RNA that acts as a promoter-specific regulator of its counterpart sense gene HNF4A, a well-characterized cancer driver in HCC (18). Our results suggest that ncNATs participate in promoter selection, modulating the expression of different promoter-specific gene isoforms that, as observed for HNF4A, may exert distinct, or even opposed, cancer-related functions. More importantly, our further pan-cancer analysis revealed approximately a hundred ncNATs whose expression correlates specifically with the activity of only one promoter of its counterpart sense gene. This association was observed in multiple types of cancers, suggesting a broaden role for ncNATs in regulation of isoform expression across tissues and cancers.
Materials and Methods
Analysis of RNA-seq data downloaded from TCGA and GTEx
Tumor and nontumor (NT) normal RNA-seq samples were obtained from GTEx (19) and TCGA (20). Transcript expression and promoter quantification were performed using Kallisto (21) and proActiv (17). For the analysis of differential gene expression, TCGA and GTEx samples were matched on the basis of their derived tissues and divided between tumor and NT samples (tumor state). TCGA datasets that were derived from the same tissue, but different tumor types were considered separately from one another. We conducted a batch effect correction for the samples based on their derived project using the removeBatchEffect function in R from the limma package (22), and confirmed clustering of samples based on tissue type and tumor state via principal component analysis. Differential analysis was performed using the DESeq2 package in R for all annotated ncNATs (23). For the identification of differentially expressed ncNATs in HCC, our criteria for candidates require an absolute fold change greater than 2 between the tumorous/NT conditions and a Benjamini-Hochberg-adjusted P value of <0.01. To determine the most likely associated gene for each ncNAT, we looked for the following factors in genes encoded on the opposite strand in order of priority; overlapping gene regions with similar gene name, overlapping gene region size, and chromosome distance up to a maximum of 10,000 bp. Approximately 1.88% were assigned on the basis of name, 83.14% due to overlapping, 10.18% by general proximity and 4.80% could not be assigned an associated gene with this method. Cancer driver annotations were retrieved from the PCAWG study (5) hosted by the ICGC Data Portal.
To identify ncNAT potentially implicated in the promoter selection of their sense genes (ncNATs-sense gene pairs), we limited our search to ncNATs with an associated sense gene controlled by two active promoters. The list of considered promoters and their genomic coordinates are specified in Supplementary Table S1. Both mean ncNAT expression and promoter activity of the sense genes promoters were required to be >1. NT or tumor samples with the 10% lowest and highest ncNAT expression were categorized in "low-ncNAT" and "high-ncNAT" groups for each ncNAT, respectively. This 10% threshold ensures a dramatic difference in ncNAT expression between the low/high ncNAT groups while keeping the statistical power of the analysis. To classify ncNAT-sense gene pairs as significant positive or negative correlation between ncNAT expression and promoter activity of its associated sense gene, we applied two criteria, including (i) the promoter (promoter a or b) activity of sense gene must be significantly higher or lower in the “high-ncNAT” than that in “low-ncNAT group” [fold change (high/low) >1.5 or <0.67; P < 0.001, a single-sided Wilcoxon test], and (ii) promoter activity of sense gene is correlated with the expression level of its associated ncNAT among all NT or tumor samples (Pearson correlation score is >0.5 or <−0.5), respectively.
To identify potential promoter switch ncNATs, we applied two additional criteria: (i) It must demonstrate a significant positive or negative correlation with either promoter a or b of its sense gene in NT or tumor samples (defined as "pos and null" and "neg and null" in the column Q of Supplementary Table S2 and S3); and (ii) if both promoters (promoter a and b) pass the Wilcoxon test (P < 0.001), this ncNAT will be excluded.
Cell culture
SNU-398 (CRL-2233, RRID: CVCL_0077, Homo sapiens, male, Asian, 42-years-old, anaplastic HCC), PLC/PRF/5 (CRL-8024, RRID: CVCL_0485, Homo sapiens, hepatoma) and HEK 293T (CRL-11268, RRID: CVCL_0063, Homo sapiens, fetus, kidney) cell lines were purchased from the ATCC. Huh7 (JCRB0403, RRID: CVCL_0336, Homo sapiens, male, Asian, 57-years-old, differentiated hepatoma) was obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB). Cells were tested for Mycoplasma and characterized using STR profiling by the ATCC and JCRB, respectively. Cells were used within six months of resuscitation. Unless otherwise stated, HEK 293T, Huh7, and PLC/PRF/5 were grown in DMEM High Glucose (biowest) supplemented with 10% FBS (biowest) and SNU398 cells were cultured in RPMI-1640 medium (biowest) also supplemented with 10% FBS. Transfections were carried out in Opti-MEM (Gibco, Thermo Fisher Scientific) in combination with lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). All the cell lines were cultured in a humidified incubator at 37°C, with 5% CO2.
Cell fractionation
Cell fractionation was carried out following the protocol of the PARIS Kit (Life Technologies), to isolate pure nuclear and cytosolic fractions of Huh7 cells. Each fraction was subjected to RNA extraction and equivalent volumes of nuclear and cytosolic RNA were converted into cDNA. The purity of these fractions was assessed through the quantification of a well-known nuclear RNA MALAT1 and a cytoplasmatic RNA GAPDH. Sequences of RT-qPCR primers are listed in Supplementary Table S4.
Western blot
Protein lysates were quantified using Bradford assay (Bio-Rad), resolved on a 10% SDS-PAGE gel and subsequently transferred onto a polyvinylidene difluoride membrane. The following antibodies were used: mouse-anti HNF4A-P1 isoforms (1:1,500, 3 hours, room temperature; R&D Systems PP-K9218–00, RRID: AB_1964277), mouse anti-GAPDH (1:5,000, 3 hours, room temperature; Santa Cruz Biotechnology sc-59540, RRID: AB_631587), anti-mouse IgG, horseradish peroxidase–linked Antibody 7076P2 (1:10,000, 1 hour; room temperature; Cell Signaling Technology 7076p2, RRID: AB_330924). Amersham ECL Western Blotting Detection Reagents (GE Healthcare Life Sciences), an enhanced luminol-based detection system, was used for luminescent signal generation.
Luciferase assay
Promoter regions of HNF4A P1 and HNF4A P2 were amplified by PCR using the high-fidelity PrimeSTAR Max DNA Polymerase (Takara) and cloned into the pGL3-basic luciferase reporter vector. Different promoter length constructs were generated by deletion following a directed-mutagenesis strategy. Sequences of used primers are listed in Supplementary Table S4. Absence of mutations was carefully confirmed by Sanger Sequencing in all the derivative constructs. For luciferase assay, Huh7, PLC/PRF/5 or SNU398 cells were plated in 96-well plates and incubated for 48–72 hours with the corresponding reporter constructs and the appropriate treatment, using lipofectamine 2000 as transfection reagent. The pRL vector encoding the Renilla protein (Rluc) was also cotransfected in every well as an internal control to compensate the variability in both transfection and harvest efficiencies. Luciferase and renilla protein production was revealed using the Dual-Luciferase Reporter Assay System (Promega). Signal luminosity was measured in the EnVision 2105 Multimode Plate Reader (PerkinElmer).
shHNF4A-AS1L stable cell lines generation and in vitro tumorigenicity assays
For virus packaging, HEK293T cells were cotransfected with pLenti6-V5 packaging plasmids and the shHNF4A-AS1L/Scramble pLKO.1-puro constructs using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) in Opti-MEM I Reduced Serum Media for 24 hours. Transfection media were replaced with DMEM supplemented with 10% FBS and left incubating for additional 24–48 hours. Virus-containing media were collected and kept at −80 °C for subsequent use. Huh7 cells in 10 cm plates were transduced in DMEM supplemented with 10% FBS by adding 1 mL of lentivirus of interest in the presence of polybrene for 6 hours, when media are replaced. 24 hours after transduction, selection was carried out by adding puromycin to the media at a final concentration of 0.65 μg/mL. A nontransduced huh7 cell plate is used as a reference to monitor the selection process. For the foci formation assay, 5 × 103 cells were seeded in a 6-well plate. After culturing for 14 days, surviving colonies (>50 cells per colony) were counted and stained using crystal violet solution (0.1% crystal violet, 25% methanol in water). Three independent experiments were performed. For colony formation in soft agar, 5 × 103 cells in 0.4% bacto agar were seeded on top of a solidified layer of 0.6% bacto agar in 6-well plates. Colonies consisting of more than 50 cells were counted after 3 weeks. Three independent experiments were conducted.
In vivo tumorigenicity assay
We subcutaneously injected approximately 1 × 106 cells into the right flank of 4- to 5-week-old male SCID mice. We monitored tumor formation in the SCID mice approximately a 4-week period and calculated the tumor volume weekly by the formula V (volume) = 0.5 × L (length) × W (width) × W (24). All animal experiments were approved by and performed in accordance with the Institutional Animal Care and Use Committees of National University of Singapore (NUS, Singapore).
Data and materials availability
Strand-specific RNA-seq data from six pairs of HCC and normal adjacent liver tissues have been deposited in Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE174338. Codes for TCGA and GTEx data analysis are available at https://github.com/GoekeLab/ncNAT_promoter_switch_code. All other data files supporting the findings of this study are available from the corresponding author upon reasonable request.
Results
Identification of differentially expressed ncNATs in HCC
First, to identify ncNATs contributing to transcriptional regulation in HCC, we retrieved publicly available The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) RNA-seq data (https://www.cancer.gov/tcga) of 374 tumors and 50 NT liver samples as well as RNA-seq data of 136 NT liver samples from the GTEx database (https://gtexportal.org/; ref. 19). We scrutinized the expression of 6,748 currently annotated ncNAT transcripts to identify those differentially expressed in HCC. Our analysis revealed 85 ncNATs fulfilling our filter criteria: Fold change of ncNAT expression (tumor/NT) >2 or <0.5 and Padj < 0.01 (Fig. 1A; Supplementary Table S5) and the differential expression of six randomly selected ncNATs could be experimentally validated in six matched pairs of HCC and their adjacent NT liver samples (Fig. 1B). In-house strand-specific RNA-seq data from these six pairs of HCC and NT liver tissues supported the existence of these ncNATs (Fig. 1C). Among our list of 85 ncNATs deregulated in HCC, a high proportion (49/85, 57.6%) had not been linked to cancer, whereas some (21/85, 24.71%) had been previously related to HCC (Supplementary Table S5), supporting the robustness of our analysis. From the list of 85 ncNATs, 15 (18%) have an associated sense gene abundantly expressed in the liver, and 7 out of these 15 ncNAT-sense gene pairs demonstrated a strong correlation (r > 0.40) between expression of ncNAT and its corresponding sense gene in both normal and tumor samples (Fig. 1A; Supplementary Table S6), indicating a potential regulatory role of these ncNATs in modulating the expression of their associated sense genes. Next, we assessed whether any of these 85 ncNATs had a counterpart sense gene previously reported as cancer driver (4), and identified two such ncNAT-sense gene pairs: HNF4A-AS1-HNF4A and ENSG00000255224-RUNX1 (Fig. 1A), among which, HNF4A was specifically associated with HCC (18). HNF4A encodes a transcription factor that participates in a wide array of hepatic functions and is considered a master regulator of liver differentiation (25–27). Considering the relevance of HNF4A in liver development and HCC, we selected HNF4A-AS1 for further study.
Figure 1.
Identification of differentially expressed ncNATs in HCC. A, Heatmap displaying the expression (Z-score) of 85 differentially expressed ncNATs [fold change (tumor/nontumor) >2 or <0.5, and Padj < 0.01] sorted by fold change of gene expression. ncNATs names are shown on the right side following the Ensembl nomenclature, whereas the top bar indicates the nature of the samples (tumoral or NT liver tissues). The three bars (1–3) on the left side relate to the ncNAT-associated sense gene, as determined by the opposite strand proximity and encoding capability. The bar 1 reflects the expression level (Z-score) of each ncNAT counterpart sense gene; the bar 2 indicates the expression correlation between each ncNAT and its sense gene; and the bar 3 specifies whether the associated sense gene has been identified as a cancer-driver gene in the PCAWG study (Pan-cancer Analysis of Whole Genomes, 2020). Seven ncNAT-sense gene pairs demonstrating a strong correlation (r > 0.40) between expression of ncNAT and its corresponding sense gene in both NT and tumor samples are indicated by an asterisk (*). B, qPCR analysis of six randomly selected ncNATs (highlighted in red in A) was conducted in six matched pairs of HCC tumors and their NT liver samples. The expression of ncNATs transcripts in each pair of samples was calculated by the formula 2−ΔCt [ΔCt = Ct(ncNAT) – Ct(β-actin)]. Bar indicates the fold change of ncNAT expression in tumors, relative to their corresponding NT samples (defined as 1.0). Data are presented as mean ± SEM of technical duplicates from a representative experiment. C, An Integrative Genomic Viewer coverage plot of our in-house strand-specific RNA-seq data from one representative HCC or NT liver sample, showing reads from both forward (top) and reverse strand (bottom) for each ncNAT (ENSG00000233554 and DLG5-AS1 from tumor sample of case NT10; ENSG00000263400 and ENSG00000234678 from NT liver sample of case NT10).
HNF4A-AS1L, a previously unannotated isoform of HNF4A-AS1, functions as a tumor-suppressive ncNAT in HCC
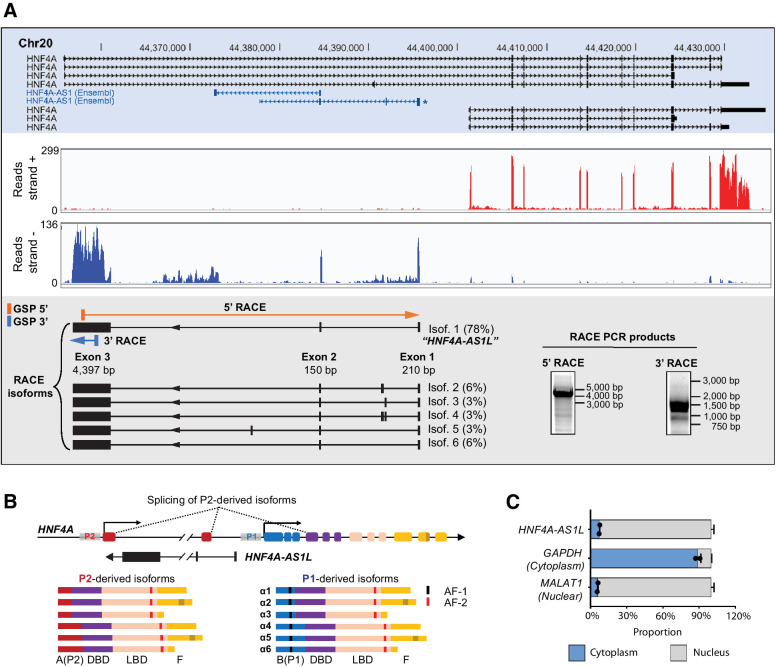
Unlike protein-coding genes, noncoding RNAs are often incompletely annotated in public databases, largely due to their low expression levels. To accurately determine the full-length sequence of HNF4A-AS1 transcript(s), we performed rapid amplification of cDNA ends (RACE) in NT liver samples. From our in-house strand-specific RNA-seq data abovementioned, the HNF4A-AS1 region with the highest read density was selected as “seed sequence” where the RACE gene-specific primers were derived from (Fig. 2A). We identified a previously undescribed 4.8 kb isoform of HNF4A-AS1 (78% of transcripts, namely HNF4A-AS1L; Fig. 2A). HNF4A-AS1L encompasses a large exon 3 (4.4 kb) that is not present in two previously annotated HNF4A-AS1 isoforms in Ensembl (highlighted in blue, Fig. 2A). Moreover, the wide extension of HNF4A-AS1L, residing between two contrasting tumor-suppressor and oncogenic alternative promoters (P1 and P2) of the HNF4A gene (Fig. 2B; refs. 28–33), led us to hypothesize its role in specifically regulating the activities of these promoters. Analysis of nuclear and cytoplasmic RNA fractions revealed that HNF4A-AS1L appeared to predominantly accumulate in the cell nuclei (Fig. 2C), suggesting its potential role in transcriptional regulation (34).
Figure 2.
HNF4A-AS1L, a nuclear ncNAT located between HNF4A P1 and P2 promoters. A, Top, genomic view of HNF4A gene as annotated in the UCSC genome browser. Two HNF4A-AS1 isoforms annotated in Ensembl are highlighted in blue. Middle, an Integrative Genomic Viewer coverage plot of our in-house strand-specific RNA-seq data from the NT liver of case NT10 showing both forward-strand and reverse-strand reads within HNF4A gene locus. Bottom, left, schematic diagram illustrating multiple isoforms of HNF4A-AS1 and their expression abundance, as detected by 5′ and 3′ RACE. Blue and orange rectangles indicate the location of gene-specific primers and arrows denote the amplification direction. The most abundant isoform detected by RACE is named by HNF4A-AS1L. Note that exons 1 and 2 of HNF4A-AS1L overlap the exons 1 and 3 of one of previously annotated HNF4A-AS1 isoforms (*). Bottom right, PCR amplicons obtained with the indicated gene-specific primer were resolved by agarose electrophoresis. B, Top, schematic diagram showing that HNF4A-AS1L resides between HNF4A P1 and P2 promoters. Bottom, structural domains of 12 protein variants encoded from HNF4A P1 or P2-derived isoforms. AF-1, activating function-1; AF-2, activating function-2; DBD, DNA binding domain; LBD, ligand binding domain C, Proportion of GAPDH, MALAT1, or HNF4A-AS1L transcripts in nuclear and cytoplasmic fractions of Huh7 cells, as detected by qPCR. GAPDH and MALAT1 were included as a positive control for the cytoplasmic and nuclear fractions, respectively. Data are presented as mean ± SEM of technical duplicates from a representative experiment.
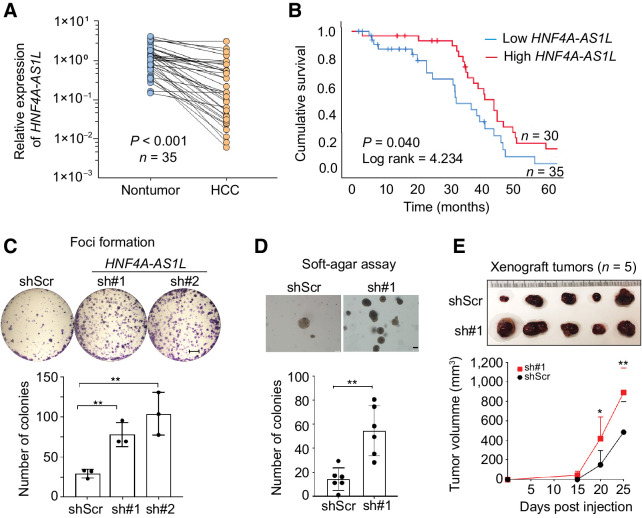
On the basis of the data obtained from our in-house strand-specific RNA-seq and RACE, HNF4A-AS1L is most likely the most abundant HNF4A-AS1 isoform in liver tissues (Fig. 2A). Therefore, we aimed to investigate its functional importance in HCC. We examined the expression of HNF4A-AS1L in a HCC cohort of 89 patients with available follow-up survival data, in which there were 35 matched pairs of HCC tumors and NT liver samples. HNF4A-AS1L was found to be significantly decreased in HCC tumors when compared with their matched NT samples (P < 0.001; Fig. 3A). Patients demonstrating lower tumoral expression of HNF4A-AS1L had worse overall survival than those with high level of HNF4A-AS1L (P = 0.04; Fig. 3B), indicating that this ncNAT may be a favorable HCC prognosis marker. Furthermore, silencing of HNF4A-AS1L by specific shRNAs (sh#1 and/or sh#2) in Huh7 cells significantly promoted tumor aggressiveness, as manifested by in vitro culture-based foci formation and anchorage-independent (soft agar) assays as well as in vivo xenograft assays (Fig. 3C–E; Supplementary Fig. S1). All these data supported that HNF4A-AS1L functions as a tumor-suppressive ncNAT in HCC and that loss of HNF4A-AS1L expression promotes tumorigenesis.
Figure 3.
Silencing of HNF4A-AS1L promotes tumorigenesis in HCC. A, qPCR analysis of HNF4A-AS1L expression was conducted in 35 matched pairs of HCC tumors and their NT liver samples (paired Student t test). The expression of HNF4A-AS1L transcripts in each sample was calculated by the formula described in Fig. 1B and normalized to the average expression in all the NT tissues (defined as 1.0). B, Kaplan–Meier plots for the overall survival rate of patients with HCC regarding their HNF4A-AS1L expression status in tumor. Expression of HNF4A-AS1L was measured in 89 patients by qPCR. The relative expression of HNF4A-AS1L in tumors was normalized to the average relative expression in all tumor samples, which was defined as 1.0. Overall survival rates for the top 40% (high expression, n = 30*) and bottom 40% (low expression, n = 35*) of patients were compared (log rank test). *, only patients with available survival data were included in this analysis. C, Quantification of foci formation induced by the indicated cells. Scale bar, 5 mm. D, Quantification of colonies formed in soft agar induced by the indicated clones. Scale bar, 200 μm. E, Images of xenograft tumors derived from the indicated cells (n = 5 mice per group) at end point (top). Growth curves of tumors derived from the indicated cells (bottom) over a 25-day period. Statistical significance was determined by paired, two-tailed Student t test (*, P < 0.05; **, P < 0.01). C and D, Data are presented as the mean ± SEM of technical triplicates (C) or six replicates (D) from a representative experiment of three independent experiments. Statistical significance was determined by unpaired, two-tailed Student t test (**, P < 0.01).
Silencing HNF4A-AS1L selectively deactivates HNF4A P1-derived isoform expression
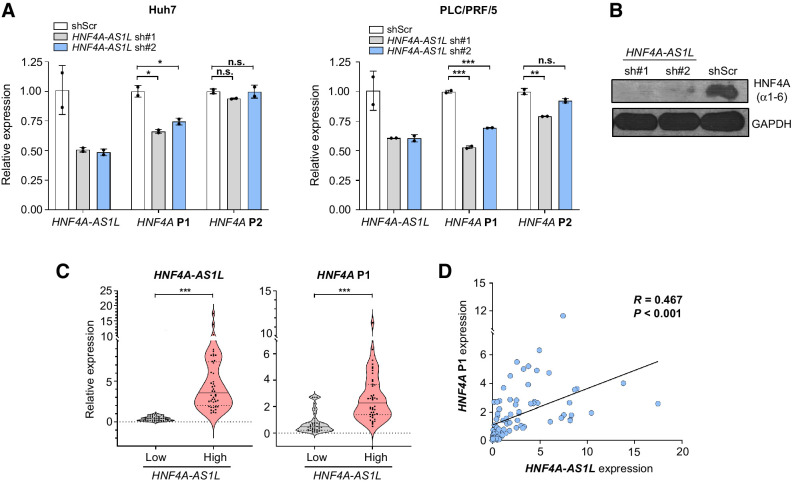
HNF4A encodes up to 12 forms of HNF4A, a key transcription factor for liver differentiation (25–27). HNF4A P1 and P2 promoters, each transcribes six distinct isoforms (HNF4A α1–6 and HNF4A α7–12, respectively; Fig. 2B) that are spatiotemporally expressed in opposite ways: HNF4A-P2 isoform family is prevalent in undifferentiated fetal liver, whereas HNF4A-P1 isoforms are predominantly expressed in adult liver (35). In HCC, HNF4A-P1 isoforms appear downregulated whereas the expression of HNF4A-P2 isoforms is recovered (31, 33). Further studies provided solid evidence supporting the tumor-suppressive and oncogenic roles of P1 and P2-derived isoforms in HCC, respectively (31, 33, 36, 37). Moreover, the choice and activity of HNF4A P1 and P2 promoters appear dysregulated in diverse types of cancer apart from HCC, including renal, colorectal, and gastric (30, 38).
Because HNF4A-AS1L resides between the HNF4A promoters (Fig. 2B) and its transcripts are predominately accumulated in the nucleus, we set out to investigate the regulatory effect of HNF4A-AS1L on HNF4A P1 and/or P2 promoter activity. With this aim, we experimentally evaluated the effect of HNF4A-AS1L knockdown on the expression of HNF4A P1 or P2-derived isoforms in the HCC cell lines Huh7 and PLC/PRF/5. Silencing HNF4A-AS1L by sh#1 and sh#2 led to a pronounced reduction in expression of P1-derived isoforms, whereas no effect or only a subtle decrease was observed for P2-derived isoforms (Fig. 4A). Moreover, a drastic decline in HNF4A protein encoded by P1-derived isoforms (HNF4A α1–6) could also be detected in HNF4A-AS1L–depleted Huh7 cells (Fig. 4B). We could not detect the protein expression of HNF4A P2 isoforms, due to their low basal levels. In our aforementioned HCC cohort (Fig. 3A), significantly reduced expression of HNF4A P1 isoforms was observed in those patients with HCC with relatively low levels of HNF4A-AS1L in tumor (Fig. 4C). This positive relationship between the expression of HNF4A P1 isoforms and HNF4A-AS1L was also confirmed in a case-by-case correlation analysis (R = 0.467 and P < 0.0001; Fig. 4D). All these findings suggested that in normal condition, HNF4A-AS1L selectively activates HNF4A P1 promoter, leading to expression of tumor-suppressive P1-derived isoforms. In HCC, loss of HNF4A-AS1L causes the inactivation of the P1 promoter and the subsequent downregulation of P1 isoforms, eventually driving HCC development.
Figure 4.
Silencing HNF4A-AS1L selectively deactivates expression of HNF4A P1-derived isoforms. A, qPCR analysis of expression levels of HNF4A-AS1L, HNF4A P1, and P2-derived isoforms in Huh7 (left) and PLC/PRF/5 (right) cells treated with shScr, sh#1, or sh#2. Data are presented as the mean ± SEM of duplicates from a representative experiment of three independent experiments (unpaired, two-tailed Student t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant). B, Western blot analysis of protein level of HNF4A P1 isoforms (HNF4A α1–6) in Huh7 cells treated with shScr, sh#1, or sh#2. GAPDH was used as a loading control. C, Violin plots showing the expression of HNF4A-AS1L (left) and HNF4A P1-derived isoforms (right) in HCC tumors divided into two groups (low HNF4A-AS1L, n = 43; high HNF4A-AS1L, n = 44), using the median value as a cutoff (unpaired, two-tailed Student t test; ***, P < 0.001). The solid line indicates the median, and the two lines with shorter dashes on either side indicate the first and third quartiles. D, Scatterplot displaying the correlation between the expression of HNF4A-AS1L and HNF4A P1 isoforms based on the data shown in C. Statistical significance is determined by linear regression.
HNF4A-AS1L specifically activates transcription of HNF4A P1 isoforms through HNF1A
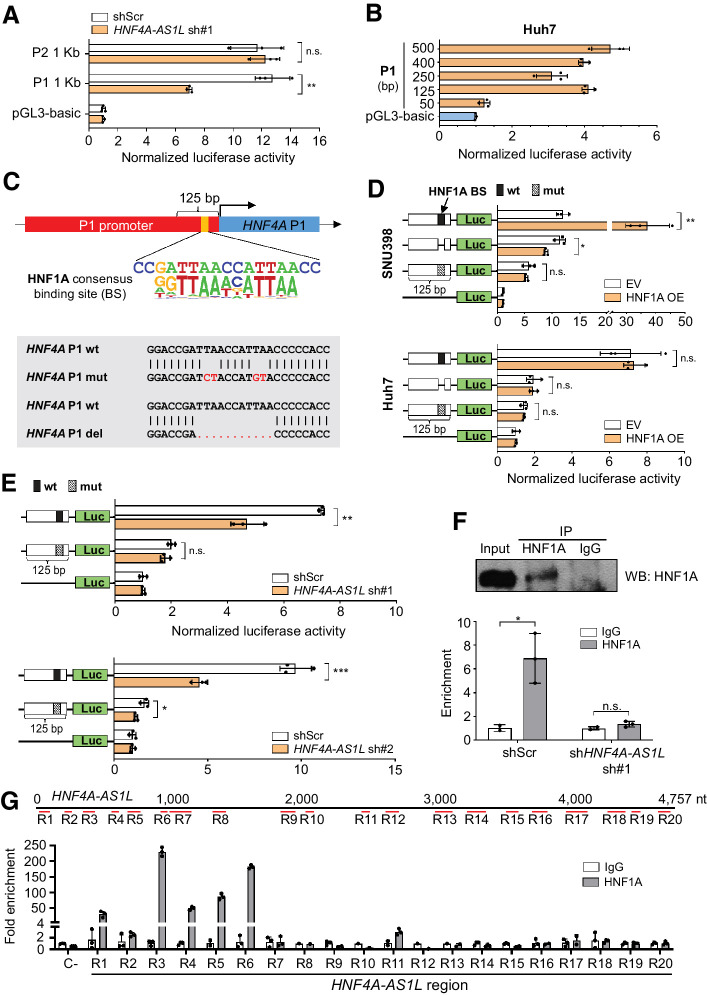
To shed light on the regulatory mechanism of HNF4A-AS1L on HNF4A, we generated reporter constructs by inserting the 1 kb DNA fragment upstream of the transcription starting site (TSS) of HNF4A P1 and P2 isoforms upstream of a luciferase reporter. Upon HNF4A-AS1L knockdown, a significant decrease in luciferase activity of the P1 promoter but not P2, was observed in Huh7 cells (Fig. 5A, Supplementary Fig. S2A).
Figure 5.
HNF4A-AS1L specifically activates transcription of HNF4A P1 isoforms through HNF1A. A, Bar charts demonstrate the luciferase activities associated with a 1-kb promoter sequence upstream of the TSS of HNF4A P1 or P2 isoforms after HNF4A-AS1L knockdown by sh#1 in Huh7 cells. Results for knockdown by sh#2 can be found in Supplementary Fig. S2A. B, Bar charts showing the luciferase activities associated with promoter sequences of the indicated bp upstream of the TSS in Huh7 cells. Results for PLC/PRF/5 cells are described in Supplementary Fig. S2B. C, Top, schematic diagram showing a HNF1A binding site consensus sequence within the proximal promoter sequence 125-bp upstream of the TSS, as predicted by the MatInspector software. Bottom, schematic diagram showing the deletion or point mutations introduced to the predicted HNF1A-binding site within the P1 promoter sequence. wt, wild-type; mut, mutant. D, Bar charts representing the luciferase activities associated with the 125-bp wt or mut HNF4A P1 promoter sequence, upon HNF1A overexpression in SNU398 (top) and Huh7 (bottom) cells. EV, empty vector; OE, overexpression. E, Bar charts representing the luciferase activities associated with the 125-bp wt or mut P1 promoter sequence after stable HNF4A-AS1L knockdown by sh#1 (top) and sh#2 (bottom) in Huh7 cells. F, ChIP-qPCR analysis of the binding of HNF1A protein to the HNF4A P1 promoter in Huh7 cells treated with HNF4A-AS1L sh#1 or shScr (bottom). Top, Western blot analysis of HNF1A protein in ChIPed products. IgG was used as a negative control. G, Top, diagram displaying 20 different regions (R1-R20) that cover the 4.8kb full-length sequence of HNF4A-AS1L and were examined by RNA immunoprecipitation-qPCR assay. Bottom, bar chart representing the fold enrichment of each of the indicated regions of HNF4A-AS1L in the HNF1A pulldown sample compared with IgG. β-Actin transcript was used as a negative control (C−) for HNF1A binding (A and B; D–G). Data are presented as the mean ± SEM of triplicates from a representative experiment of two or three independent experiments (unpaired, two-tailed Student t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant).
After perceiving the capability of HNF4A-AS1L to drive HNF4A P1 activity, we searched for regulatory elements within the P1 promoter. Reporter constructs successively shorter in length were generated for luciferase studies, and when we narrowed down the proximal promoter region from 125 to 50 bp, a dramatic drop in the luciferase signal was observed in both Huh7 and PLC/PRF/5 cells (Fig. 5B, Supplementary Fig. S2B). This indicated that the region 50 to 125 bp upstream of the TSS of P1 isoforms contains the core promoter sequence essential for P1-driven transcription. In silico predictions using the MatInspector software (39) pointed out the existence of a HNF1A consensus–binding site (BS) at 92–99 bp upstream of the HNF4A P1 TSS (top, Fig. 5C; Supplementary Table S7). We disrupted this HNF1A-BS in the P1 reporter constructs by introducing either deletion or point mutations (bottom, Fig. 5C). In the HCC cell line, SNU398 that expresses HNF1A at an approximately 2,500-fold lower level than Huh7 cells (Supplementary Fig. S3), overexpression of HNF1A effectively led to a nearly 3-fold increase in the luciferase activity for the intact P1 reporter construct, whereas no effect was observed for the HNF1A BS-deleted or -mutated construct (top, Fig. 5D). No increase in the luciferase signal upon HNF1A overexpression was observed in Huh7 cells, due to its HNF1A basal saturation (bottom, Fig. 5D). Furthermore, when HNF1A BS was either deleted or mutated, a drastic loss in the luciferase activity occurred in Huh7 cells (bottom, Fig. 5D). All these data demonstrated that HNF1A is indeed a transcription factor strongly involved in the transcriptional activation of HNF4A P1 isoforms. We further investigated whether silencing HNF4A-AS1L could inhibit transcription of HNF4A P1 isoforms via HNF1A. Even though we only included the 125 bp region (−125 to −1) upstream of the HNF4A P1 TSS where the HNF1A BS is located, depletion of HNF4A-AS1L by shRNA-mediated knockdown (sh#1 and sh#2) led to a significant decrease in the luciferase activity for the intact P1 reporter construct (Fig. 5E). On the basis of these observations, we assumed that HNF4A-AS1L might regulate the HNF1A-mediated transcriptional activation of HNF4A P1 isoforms by enhancing the binding of HNF1A to its BS in the P1 promoter. As detected by chromatin immunoprecipitation (ChIP) followed by qPCR (ChIP-qPCR) analysis, silencing HNF4A-AS1L in Huh7 cells led to a reduced occupancy of HNF1A in the HNF4A P1 promoter (Fig. 5F). To further explore how HNF4A-AS1L cooperates with HNF1A to promote HNF4A P1 expression, we performed RNA immunoprecipitation assay to pull down HNF1A-bound RNAs in Huh7 cells, followed by detection of HNF4A-AS1L transcripts using qPCR with primers targeting different regions of HNF4A-AS1L (R1-R20 covering the 4.8 kb full-length sequence of HNF4A-AS1L; Fig. 5G, top). As a result, HNF1A was found to bind to the 5′ end of HNF4A-AS1L, as supported by our observation that four adjacent regions (R3-R6) of HNF4A-AS1L showed significant enrichment in the HNF1A pulldown sample compared with the IgG counterparts (Fig. 5G, bottom). These results indicated the binding of HNF1A to HNF4A-AS1L and suggested that the interaction between HNF4A-AS1L and HNF1A may enhance the binding of HNF1A to the HNF4A P1 promoter, thereby activating the transcription of P1-derived isoforms.
Context-specific regulation of alternative promoters by ncNATs
Alternative promoter switching has been described across all cancer types, yet the precise mechanism of regulation is largely not well understood. After demonstrating HNF4A-AS1L participation in HNF4A P1 selection, we intended to evaluate the global impact of ncNATs on promoter-specific regulation across different tissues and cancer types. With simplification purposes, we focused our analysis on those ncNAT-sense gene pairs where; ncNAT is sufficiently expressed and, its corresponding counterpart sense gene has specifically two active promoters (Fig. 6A). Using RNA-seq datasets of 14,912 cancer and NT normal samples from TCGA and GTEx databases, we identified 521 and 353 unique ncNATs expressed in NT and tumor samples, respectively, following the aforementioned criteria (Supplementary Tables S2, S3, S8, and S9). Strikingly, approximately half of these unique ncNATs (268 out of 521 in NT samples, whereas 156 out of 353 in tumor samples) were specifically identified in only one tissue/cancer type (Supplementary Fig. S4A). There was a lower prevalence of unique ncNATs in tumors compared with NT samples across nearly all tissue/cancer types (Supplementary Fig. S4B), consistent with a previous study where ncNAT expression was found overall downregulated in cancer (40). We further assessed how the expression level of every ncNAT correlates with the promoter (P1 and P2) activity of their corresponding counterpart sense gene. With stringent criteria (Materials and Methods and Fig. 6B), of all the ncNAT-sense gene pairs analyzed, 16.2% and 20.1% of ncNATs exhibited significant correlation with either or both of the two sense gene promoters in NT and tumor samples, respectively (Fig. 6C and D).
Figure 6.
Tissue-wide correlation between ncNAT expression and sense gene promoter activity. A, Schematic representation of ncNAT-sense gene pairs selected for our study. ncNAT-sense gene pairs where expressed ncNATs have a counterpart sense gene with specifically two active promoters were selected for further study. Datasets were independently considered and analyzed for each tissue/tumoral type. B, For those pairs fulfilling the criteria in A, correlation between ncNAT expression and activity of P1 and P2 sense gene promoters was evaluated (see more details in Materials and Methods). Illustration only shows P1 for simplification purposes. FC, fold change. C and D, Doughnut charts depict the percentages of ncNATs showing either no correlation (gray) or a significant correlation (yellow) between its expression and the promoter activity of either one or both promoters of their ncNAT-associated sense gene, as determined in B, in NT (left, C) and tumor (left, D) samples. The 10 × 10 dot plots illustrate the proportions of ncNAT-sense gene pairs, demonstrating either no correlation (null) or any direction of significant correlation [positive (Pos) or negative (Neg)] between ncNAT expression and the promoter activity of each promoter of the counterpart sense gene in NT (right, C) and tumor (right, D) samples. “Pos and Pos” group, activities of both sense gene promoters are significantly positively correlated with expression of the corresponding ncNAT. “Pos and null,” positive correlation is found for one sense gene promoter, but not for the other; “Pos and Neg,” positive correlation is found for one sense gene promoter, but negative correlation for the other; “Neg and null,” negative correlation is found for one sense gene promoter, but not for the other; and “Neg and Neg,” negative correlation is found for both sense gene promoters.
Next, we aimed to identify those ncNATs that, like HNF4A-AS1L, selectively and specifically regulate the promoter activity of only one promoter of their associated sense gene and defined them as “promoter-switch ncNATs.” With this purpose, first, we selected all ncNAT-sense gene pairs where ncNAT expression correlated with only one sense promoter but not with the other (Fig. 6C and D; “positive and null” and “negative and null” groups). And subsequently, we refined this selection to those pairs with the best promoter-specific association (Materials and Methods). As a result, we obtained a list of 90 unique “promoter-switch ncNATs” (Supplementary Table S10). Of these, 65 and 35 ncNATs were positive hits in NT and tumor samples, respectively, with 10 ncNATs fulfilling our selection criteria in both NT and tumor groups (Fig. 7A; Supplementary Figs. S5A–S5C and S6A–S6B). To further prove the true “promoter-switch” capacity of these candidate ncNATs, we designed shRNAs against six of them, including all four ncNATs candidates detected in the NT liver (antisense to, and theoretically regulating SH3GL1, FAM92A1, CHKA, and CLSTN3; Fig. 7A) and two additional ncNATs (antisense to SUN2 and CERS2) that do not fulfill our stringent requirements for promoter-switch ncNATs, but whose expression was notably correlated with the promoter activity of their corresponding sense gene in both NT and liver tumor samples (Supplementary Tables S2 and S3). Because the majority of these six ncNATs was expressed at extremely low level, we only managed to knockdown ENSG00000255031 (antisense to CHKA) and ENSG00000259357 (antisense to CERS2) in Huh7 cells (Fig. 7B). Upon knockdown of ENSG00000255031 or ENSG00000259357, the promoter usage of CHKA and CERS2 was skewed toward the P2 and P1 promoter, respectively (Fig. 7C). This observation was indeed matched with the change in the promoter usage of CHKA and CERS2 between the high-ncNAT and low-ncNAT groups from the analysis of the TCGA LIHC datasets (Supplementary Tables S2 and S3, Supplementary Fig. S7A and S7B). Altogether, these findings suggest a context-dependent regulation of sense gene expression by their associated ncNATs through modulating the selection and activity of the sense gene promoters.
Figure 7.
Context-specific regulation of alternative promoters by ncNATs. A, Bubble plots illustrating all promoter-switch ncNATs identified for each indicated NT tissue (right) or cancer types (left). Qualifying requirements to be classified as a promoter-switch ncNAT include: (i) ncNAT must demonstrate a significant positive or negative correlation with either promoter a or b of its sense gene in NT or tumor samples (defined as "pos and null" and "neg and null," see Fig. 6); and (ii) if both promoters (promoter a and b) pass the Wilcoxon test (P < 0.001), the ncNAT is excluded. Magnitude of difference in promoter activity of the indicated sense gene between high-ncNAT and low groups [log2(fold change)] is proportional to the dot size; pink, positive correlation; blue, negative correlation. #, promoter-switch ncNATs identified in both NT tissue and cancer. *2 ncNATs for CHKA (NT): ENSG00000255031 and ENSG00000255236; (ii) ncNATs for SLC25A25 (tumor): ENSG00000234771 and ENSG00000269988. Left axis lists all the included cancer types with following TCGA abbreviations: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma. B, Bar charts representing the expression levels of ENSG00000255031 (antisense to CHKA) and ENSG00000259357 (antisense to CERS2) in Huh7 cells after treatment with shScr control or shRNAs against the indicated ncNATs. C, Bar charts representing the relative P1 or P2 promoter activity of CHKA or CERS2 in the shRNA-treated cells described in B. qPCR specifically amplifying P1 and P2-derived isoforms were performed for promoter usage measurement. CHKA P1, prmtr.17341; P2, prmtr.17345. CERS2 P1, prmtr.35422; P2, prmtr.35424. Exact genomic coordinates for each promoter are specified in Supplementary Table S1. B and C, Data are presented as the mean ± SEM of triplicates from a representative experiment of two independent experiments (unpaired, two-tailed Student t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Discussion
Most genes have at least two distinct promoters that control the production of discrepant RNA and protein isoforms (41). Promoter regulation can become particularly relevant when distinct promoters within the same gene originate functionally diverse or even opposed products. Consequently, it is not surprising that survival of patients with cancer is more accurately predicted by promoter activity than by gene expression (17). However, how alternative promoters are regulated is often unknown. Up to date, diverse studies evidence that ncNATs can modulate the overall expression of their overlapping sense genes (14, 42, 43). Our analysis identified a novel ncNAT HNF4A-AS1L that, in liver, regulates the expression of HNF4A isoforms through a highly promoter-specific mechanism. HNF4A-AS1L, through its binding to HNF1A, facilitates an independent control of the tumor-suppressive HNF4A P1-derived isoforms, demonstrating that this mechanism can functionally impact hepatocellular tumorigenesis. Although ncNATs have been reported to modulate sense gene expression, this is the first time where an ncNAT was found to participate in the promoter selection of its associated sense gene.
Encouraged by this finding, we examined how expression of multiple ncNATs correlates with the promoter activity of their associated sense genes in numerous tissues and cancer types. Our analysis of the TCGA and GTEx data suggests that this highly promoter-specific regulation might be a mechanism occurring across many tissue and cancer types, thereby enabling context-specific regulation of alternative isoform expression. We centered our efforts in identifying those ncNATs that can regulate the activity of specifically one promoter of their associated sense gene, and called them “promoter-switch” ncNATs. Notably, the majority of these “promoter-switch” ncNATs were detected in a tissue or tumor type–specific manner, suggesting a context-dependent regulation of sense gene expression by associated ncNATs through modulating the selection and activity of sense gene promoters. Of our list of 90 “promoter-switch” ncNATs candidates, some are antisense to known cancer drivers, such as VEGFA and FGFR1. Focusing our efforts on the liver, we managed to validate two additional ncNATs apart from HNF4A-AS1L, as promoter activity regulators of their counterpart sense genes CHKA and CERS2. Interestingly, CHKA and CERS2 have been reported to modulate carcinogenesis in diverse types of cancer (44–47), including HCC (48, 49). Isoform-specific functional characterization for these two genes will help to understand the impact of their associated ncNATs in cancer progression. Overall, these experimental results confirm the validity of our analysis and pave the way for further explorations of this promising field.
Most of the annotated antisense RNAs are believed to be non-coding; however, they might still contain open reading frames or encode for small proteins. Although in silico analysis of protein-coding capability of HNF4A-AS1L, ENSG00000255031 and ENSG00000259357 indicated a very low coding potential, there are several open reading frames potentially encoding small (<110 aa) proteins. Further studies will be needed to fully understand the regulatory mechanism of ncNATs-mediated regulation of sense gene promoters.
It is also feasible that functionally characterized ncNATs can be targeted by antisense oligonucleotides, to disrupt the interaction of ncNAT-sense transcripts via degradation or transcriptional derepression at the chromatin level (50), suggesting the clinical utility of targeting oncogenic ncNATs for cancer treatment.
Altogether, we demonstrate an unexplored role of ncNATs in the regulation of their counterpart sense gene expression through controlling alternative promoter usage. This regulative ability has an impact on hepatocellular tumorigenesis. We proved that HNF4A-AS1L can modulate the expression of HNF4A promoter-specific isoforms that exert distinct cancer-related functions.
Our pan-cancer analysis provided a valuable list of ninety promoter-switch ncNATs potentially exerting a promoter-specific regulation of their associated sense gene in multiple types of tissues and cancers. Further investigation of those inferred promoter-switch ncNATs that are deregulated in tumor will elucidate their functional implications in cancer development as well as their mechanisms of action.
Authors' Disclosures
F. Bellido Molias reports grants from Singapore Ministry of Education and grants from NMRC Clinician Scientist-Individual Research Grant during the conduct of the study. M.W.J. Lim reports grants from Singapore Ministry of Education and NMRC Clinician Scientist-Individual Research Grant during the conduct of the study. J.X.J. Teo reports grants from Singapore Ministry of Education and NMRC Clinician Scientist-Individual Research Grant during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Data - Main File
Exhaustive list of sense-gene promoters considered in our analysis.
List of ncNATs expressed in TCGA tumors, for which their associated sense genes have two active alternative promoters
List of ncNATs expressed in TCGA and GTEx non-tumor samples, for which their associated sense genes have two active alternative promoters
List of 521 unique ncNATs expressed in non-tumor samples of one or multiple cancer/tissue types from TCGA and GTEx.
List of 353 unique ncNATs expressed in tumors of one or multiple tumor types from TCGA.
Acknowledgments
National Research Foundation Singapore; Singapore Ministry of Education under its Research Centers of Excellence initiative; Singapore Ministry of Education's Tier 2 grants (MOE2018-T2–1-005 and MOE2019-T2–2-008); NMRC Clinician Scientist-Individual Research Grant (CS-IRG, project ID: MOH-000214); and Singapore Ministry of Education's Tier 3 grants (MOE2014-T3–1-006).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
F. Bellido Molias: Data curation, validation, investigation, methodology, writing–original draft, writing–review and editing. A. Sim: Formal analysis, investigation, methodology, writing–review and editing. K.W. Leong: Investigation. O. An: Formal analysis. Y. Song: Investigation. V.H.E. Ng: Investigation. M.W.J. Lim: Investigation. C. Ying: Formal analysis. J.X.J. Teo: Investigation. J. Göke: Conceptualization, resources, data curation, supervision, funding acquisition, visualization, methodology, writing–review and editing. L. Chen: Conceptualization, resources, data curation, supervision, funding acquisition, visualization, methodology, project administration, writing–review and editing.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog 2017;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016;48:500–9. [DOI] [PubMed] [Google Scholar]
- 4. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Consortium ITP-CAoWG. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Group PTC, Calabrese C, Davidson NR, Demircioglu D, Fonseca NA, He Y, et al. Genomic basis for RNA alterations in cancer. Nature 2020;578:129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959–61. [DOI] [PubMed] [Google Scholar]
- 8. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- 9. Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 2015;47:818–21. [DOI] [PubMed] [Google Scholar]
- 10. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science 2005;309:1564–6. [DOI] [PubMed] [Google Scholar]
- 12. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishizawa M, Ikeya Y, Okumura T, Kimura T. Post-transcriptional inducible gene regulation by natural antisense RNA. Front Biosci 2015;20:1–36. [DOI] [PubMed] [Google Scholar]
- 14. Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013;14:880–93. [DOI] [PubMed] [Google Scholar]
- 15. Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci 2015;16:3251–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reyes A, Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res 2018;46:582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demircioglu D, Cukuroglu E, Kindermans M, Nandi T, Calabrese C, Fonseca NA, et al. A Pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell 2019;178:1465–77. [DOI] [PubMed] [Google Scholar]
- 18. Taniguchi H, Fujimoto A, Kono H, Furuta M, Fujita M, Nakagawa H. Loss-of-function mutations in Zn-finger DNA-binding domain of HNF4A cause aberrant transcriptional regulation in liver cancer. Oncotarget 2018;9:26144–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 22. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao ZA, Daniel D, Hanahan D. Sub-lethal radiation enhances anti-tumor immunotherapy in a transgenic mouse model of pancreatic cancer. BMC Cancer 2002;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol 2014;20:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 2008;23:2–7. [DOI] [PubMed] [Google Scholar]
- 27. Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4alpha in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol 2013;304:G26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briancon N, et al. Opposing roles of nuclear receptor HNF4alpha isoforms in colitis and colitis-associated colon cancer. Elife 2016;5:e10903.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vuong LM, Chellappa K, Dhahbi JM, Deans JR, Fang B, Bolotin E, et al. Differential effects of hepatocyte nuclear factor 4alpha isoforms on tumor growth and T-cell factor 4/AP-1 Interactions in human colorectal cancer cells. Mol Cell Biol 2015;35:3471–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Babeu JP, Jones C, Geha S, Carrier JC, Boudreau F. P1 promoter-driven HNF4alpha isoforms are specifically repressed by beta-catenin signaling in colorectal cancer cells. J Cell Sci 2018;131;jcs21473. [DOI] [PubMed] [Google Scholar]
- 31. Cai SH, Lu SX, Liu LL, Zhang CZ, Yun JP. Increased expression of hepatocyte nuclear factor 4 alpha transcribed by promoter 2 indicates a poor prognosis in hepatocellular carcinoma. Therap Adv Gastroenterol 2017;10:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oshima T, Kawasaki T, Ohashi R, Hasegawa G, Jiang S, Umezu H, et al. Downregulated P1 promoter-driven hepatocyte nuclear factor-4alpha expression in human colorectal carcinoma is a new prognostic factor against liver metastasis. Pathol Int 2007;57:82–90. [DOI] [PubMed] [Google Scholar]
- 33. Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol 2006;208:662–72. [DOI] [PubMed] [Google Scholar]
- 34. Zhao S, Zhang X, Chen S, Zhang S. Natural antisense transcripts in the biological hallmarks of cancer: powerful regulators hidden in the dark. J Exp Clin Cancer Res 2020;39:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang S, Tanaka T, Iwanari H, Hotta H, Yamashita H, Kumakura J, et al. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4alpha (HNF4alpha) isoforms in human and rats. Nucl Recept 2003;1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fekry B, Ribas-Latre A, Baumgartner C, Mohamed AMT, Kolonin MG, Sladek FM, et al. HNF4alpha-deficient fatty liver provides a permissive environment for sex-independent hepatocellular carcinoma. Cancer Res 2019;79:5860–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walesky C, Apte U. Role of hepatocyte nuclear factor 4alpha (HNF4alpha) in cell proliferation and cancer. Gene Expr 2015;16:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartz B, Algamas-Dimantov A, Hertz R, Nataf J, Kerman A, Peri I, et al. Inhibition of colorectal cancer by targeting hepatocyte nuclear factor-4alpha. Int J Cancer 2009;124:1081–9. [DOI] [PubMed] [Google Scholar]
- 39. Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005;21:2933–42. [DOI] [PubMed] [Google Scholar]
- 40. Maruyama R, Shipitsin M, Choudhury S, Wu Z, Protopopov A, Yao J, et al. Altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci U S A 2012;109:2820–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 2007;8:424–36. [DOI] [PubMed] [Google Scholar]
- 42. Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong N, et al. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget 2015;6:484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dallosso AR, Hancock AL, Malik S, Salpekar A, King-Underwood L, Pritchard-Jones K, et al. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA 2007;13:2287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu L, Wang RY, Cai J, Feng D, Yang GZ, Xu QG, et al. Overexpression of CHKA contributes to tumor progression and metastasis and predicts poor prognosis in colorectal carcinoma. Oncotarget 2016;7:66660–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trousil S, Lee P, Pinato DJ, Ellis JK, Dina R, Aboagye EO, et al. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase alpha overexpression and a hyperactivated deacylation pathway. Cancer Res 2014;74:6867–77. [DOI] [PubMed] [Google Scholar]
- 46. Fan SH, Wang YY, Lu J, Zheng YL, Wu DM, Zhang ZF, et al. CERS2 suppresses tumor cell invasion and is associated with decreased V-ATPase and MMP-2/MMP-9 activities in breast cancer. J Cell Biochem 2015;116:502–13. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Q, Wang JY, Yan W, Wang DD, Yang SJ, Zhou SY, et al. Clinical and pathological significance of Homo sapiens ceramide synthase 2 (CerS-2) in diverse human cancers. Biosci Rep 2019;39:BSR20181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin XM, Hu L, Gu J, Wang RY, Li L, Tang J, et al. Choline kinase alpha mediates interactions between the epidermal growth factor receptor and mechanistic target of rapamycin complex 2 in hepatocellular carcinoma cells to promote drug resistance and xenograft tumor progression. Gastroenterology 2017;152:1187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, et al. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J Biol Chem 2009;284:33549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Latge G, Poulet C, Bours V, Josse C, Jerusalem G. Natural antisense transcripts: molecular mechanisms and implications in breast cancers. Int J Mol Sci 2018;19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data - Main File
Exhaustive list of sense-gene promoters considered in our analysis.
List of ncNATs expressed in TCGA tumors, for which their associated sense genes have two active alternative promoters
List of ncNATs expressed in TCGA and GTEx non-tumor samples, for which their associated sense genes have two active alternative promoters
List of 521 unique ncNATs expressed in non-tumor samples of one or multiple cancer/tissue types from TCGA and GTEx.
List of 353 unique ncNATs expressed in tumors of one or multiple tumor types from TCGA.



![Figure 1. Identification of differentially expressed ncNATs in HCC. A, Heatmap displaying the expression (Z-score) of 85 differentially expressed ncNATs [fold change (tumor/nontumor) >2 or <0.5, and Padj < 0.01] sorted by fold change of gene expression. ncNATs names are shown on the right side following the Ensembl nomenclature, whereas the top bar indicates the nature of the samples (tumoral or NT liver tissues). The three bars (1–3) on the left side relate to the ncNAT-associated sense gene, as determined by the opposite strand proximity and encoding capability. The bar 1 reflects the expression level (Z-score) of each ncNAT counterpart sense gene; the bar 2 indicates the expression correlation between each ncNAT and its sense gene; and the bar 3 specifies whether the associated sense gene has been identified as a cancer-driver gene in the PCAWG study (Pan-cancer Analysis of Whole Genomes, 2020). Seven ncNAT-sense gene pairs demonstrating a strong correlation (r > 0.40) between expression of ncNAT and its corresponding sense gene in both NT and tumor samples are indicated by an asterisk (*). B, qPCR analysis of six randomly selected ncNATs (highlighted in red in A) was conducted in six matched pairs of HCC tumors and their NT liver samples. The expression of ncNATs transcripts in each pair of samples was calculated by the formula 2−ΔCt [ΔCt = Ct(ncNAT) – Ct(β-actin)]. Bar indicates the fold change of ncNAT expression in tumors, relative to their corresponding NT samples (defined as 1.0). Data are presented as mean ± SEM of technical duplicates from a representative experiment. C, An Integrative Genomic Viewer coverage plot of our in-house strand-specific RNA-seq data from one representative HCC or NT liver sample, showing reads from both forward (top) and reverse strand (bottom) for each ncNAT (ENSG00000233554 and DLG5-AS1 from tumor sample of case NT10; ENSG00000263400 and ENSG00000234678 from NT liver sample of case NT10).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d263/9397637/1e3b2b1c9a73/5849fig1.jpg)
![Figure 6. Tissue-wide correlation between ncNAT expression and sense gene promoter activity. A, Schematic representation of ncNAT-sense gene pairs selected for our study. ncNAT-sense gene pairs where expressed ncNATs have a counterpart sense gene with specifically two active promoters were selected for further study. Datasets were independently considered and analyzed for each tissue/tumoral type. B, For those pairs fulfilling the criteria in A, correlation between ncNAT expression and activity of P1 and P2 sense gene promoters was evaluated (see more details in Materials and Methods). Illustration only shows P1 for simplification purposes. FC, fold change. C and D, Doughnut charts depict the percentages of ncNATs showing either no correlation (gray) or a significant correlation (yellow) between its expression and the promoter activity of either one or both promoters of their ncNAT-associated sense gene, as determined in B, in NT (left, C) and tumor (left, D) samples. The 10 × 10 dot plots illustrate the proportions of ncNAT-sense gene pairs, demonstrating either no correlation (null) or any direction of significant correlation [positive (Pos) or negative (Neg)] between ncNAT expression and the promoter activity of each promoter of the counterpart sense gene in NT (right, C) and tumor (right, D) samples. “Pos and Pos” group, activities of both sense gene promoters are significantly positively correlated with expression of the corresponding ncNAT. “Pos and null,” positive correlation is found for one sense gene promoter, but not for the other; “Pos and Neg,” positive correlation is found for one sense gene promoter, but negative correlation for the other; “Neg and null,” negative correlation is found for one sense gene promoter, but not for the other; and “Neg and Neg,” negative correlation is found for both sense gene promoters.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d263/9397637/271861c672a5/5849fig6.jpg)
![Figure 7. Context-specific regulation of alternative promoters by ncNATs. A, Bubble plots illustrating all promoter-switch ncNATs identified for each indicated NT tissue (right) or cancer types (left). Qualifying requirements to be classified as a promoter-switch ncNAT include: (i) ncNAT must demonstrate a significant positive or negative correlation with either promoter a or b of its sense gene in NT or tumor samples (defined as "pos and null" and "neg and null," see Fig. 6); and (ii) if both promoters (promoter a and b) pass the Wilcoxon test (P < 0.001), the ncNAT is excluded. Magnitude of difference in promoter activity of the indicated sense gene between high-ncNAT and low groups [log2(fold change)] is proportional to the dot size; pink, positive correlation; blue, negative correlation. #, promoter-switch ncNATs identified in both NT tissue and cancer. *2 ncNATs for CHKA (NT): ENSG00000255031 and ENSG00000255236; (ii) ncNATs for SLC25A25 (tumor): ENSG00000234771 and ENSG00000269988. Left axis lists all the included cancer types with following TCGA abbreviations: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma. B, Bar charts representing the expression levels of ENSG00000255031 (antisense to CHKA) and ENSG00000259357 (antisense to CERS2) in Huh7 cells after treatment with shScr control or shRNAs against the indicated ncNATs. C, Bar charts representing the relative P1 or P2 promoter activity of CHKA or CERS2 in the shRNA-treated cells described in B. qPCR specifically amplifying P1 and P2-derived isoforms were performed for promoter usage measurement. CHKA P1, prmtr.17341; P2, prmtr.17345. CERS2 P1, prmtr.35422; P2, prmtr.35424. Exact genomic coordinates for each promoter are specified in Supplementary Table S1. B and C, Data are presented as the mean ± SEM of triplicates from a representative experiment of two independent experiments (unpaired, two-tailed Student t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d263/9397637/1d14e92c41a6/5849fig7.jpg)