Abstract
Testing peripheral blood for circulating tumor DNA (ctDNA) offers a minimally invasive opportunity to diagnose, characterize, and monitor the disease in individual cancer patients. ctDNA can reflect the actual tumor burden and specific genomic state of disease and thus might serve as a prognostic and predictive biomarker for immune checkpoint inhibitor (ICI) therapy. Recent studies in various cancer entities (e.g., melanoma, non–small cell lung cancer, colon cancer, and urothelial cancer) have shown that sequential ctDNA analyses allow for the identification of responders to ICI therapy, with a significant lead time to imaging. ctDNA assessment may also help distinguish pseudoprogression under ICI therapy from real progression. Developing dynamic changes in ctDNA concentrations as a potential surrogate endpoint of clinical efficacy in patients undergoing adjuvant immunotherapy is ongoing. Besides overall ctDNA burden, further ctDNA characterization can help uncover tumor-specific determinants (e.g., tumor mutational burden and microsatellite instability) of responses or resistance to immunotherapy. In future studies, standardized ctDNA assessments need to be included in interventional clinical trials across cancer entities to demonstrate the clinical utility of ctDNA as a biomarker for personalized cancer immunotherapy.
Introduction
Over the past decade, the identification of the molecular mechanisms by which tumor cells hamper immunity marked the coming of a new era in the management of cancer patients. Since first immune checkpoint inhibitor (ICI) approval in unresectable malignant melanoma (1), up to 15 different clinical entities, comprising both solid and hematologic malignancies, currently benefit from an FDA-approved indication for ICI-based treatment (2) and the field of applications is rapidly evolving. Notably, the repertoire of immune-oncology (IO) therapeutic options is constantly expanding by targeting additional immune checkpoints or costimulatory molecules, combining ICI with other therapeutic strategies (3, 4) and introducing innovative approaches based on T-cell bioengineering (5).
Early identification of relapse and early therapeutic intervention are essential determinants for improved overall survival. However, an objective biomarker associated with the efficacy of IO drugs is an urgent but still unmet clinical need.
The past decade has also seen the advent of liquid biopsy (6, 7). Contrary to tumor tissue biopsy, liquid biopsy gives access to tumor material in a minimally invasive way, therefore offering the patient a more acceptable, safer, and easily repeatable option to monitor tumor response. Liquid biopsy applies to detecting tumor cells or tumor-derived products like tumor DNA (referred to as circulating tumor DNA, ctDNA) mainly shed in peripheral blood and other body fluids. The field of ctDNA clinical applications is mainly based on mutation detection and has greatly benefited from significant improvements of detection methods in terms of sensitivity and multiplexing. The utility of monitoring tumor genomics through plasma ctDNA analysis has been widely investigated over the past years in diverse clinical settings (8, 9).
This review will present the different clinical applications of ctDNA analysis in the specific context of IO. We will discuss the capability of ctDNA, quantified either before or during therapy, to identify patients who will benefit from the treatment. We will finally describe ctDNA as a privileged substrate to study and monitor the genetic determinants of immunotherapy response, such as tumor mutation burden or microsatellite instability and underline the value of ctDNA-based decision-making in cancer treatments.
Pretreatment Levels of ctDNA as a Prognostic Biomarker in IO
Clinical value of pretreatment ctDNA levels in metastatic patients
Supplementary Table S1 recapitulates the studies investigating the correlation of ctDNA measured before the treatment with the primary clinical endpoints. Most of the studies were conducted on melanoma and NSCLC populations who received ICI either as a first or later-line therapy, according to the timing of drug approval. Recent pan-cancer studies and hematologic malignancies, implementing new IO strategies, highlight the advantage of ctDNA to be implemented agnostic to cancer types and independent from a specific ICI treatment, as long as one mutation can be detected. There is a high level of heterogeneity between the studies about the number of included subjects, the types of clinical cohorts, and the methodology adopted to measure ctDNA, including the detection of a single mutation—usually the driver—versus multiple with gene panels, different sensitivity thresholds, and quantification strategies. Notably, most of the studies so far have only demonstrated the clinical validity of ctDNA as a biomarker (10). The use of pretreatment ctDNA value as a biomarker in the clinic will therefore necessitate establishing precise pretreatment ctDNA cutoff points for each particular assay and for each particular tumor type. Moreover, interventional studies are needed to demonstrate the clinical utility of ctDNA measurements. Nevertheless, several investigations have identified a congruent association between undetectable ctDNA or low ctDNA levels [inferior to the cohort's median variant allele frequency (VAF)] and a longer progression-free survival (PFS) and overall survival (OS) in univariate analyses (11–18). Owing to the close relationship between ctDNA and tumor burden, well established in NSCLC (19–21) and melanoma (11, 12, 15, 22–26), the underlying influence of anatomic tumor disease burden in the duration of response to ICI therapy might partially explain the pretreatment ctDNA association to PFS or OS (27, 28). However, in the up-to-now most extensive study encompassing 16 different tumor types in 790 patients, ctDNA association with OS after adjustment for Eastern Cooperative Oncology Group (ECOG) performance status, baseline liver metastases, baseline lymph node metastases, smoking status, tumor burden, and tumor PD-L1 score suggests that ctDNA is not simply a surrogate marker for baseline tumor burden (29). In this line, the mutation selected to quantify ctDNA could also play a role in associating ctDNA levels to the clinical outcome since specific mutations identified in tumor tissues could have different prognostic values (refs. 30, 31; cf. chapter 4.1). As an additional confounding factor, the capability to equally detect all different mutations in ctDNA remains unclear, as reported in melanoma (17).
Contrary to PFS and OS, the association to objective response rate (ORR) is poorly reported. When mentioned, ORR and pretreatment ctDNA levels association was not significant (Supplementary Table S1). This observation rather confers to ctDNA pretreatment levels a prognostic value than a direct link to clinical efficacy. Of note, the pretreatment ctDNA levels discrimination of patients with durable or nondurable clinical benefit reported by Nabet and colleagues can be explained by a different evaluation model of clinical response from immune Response Evaluation Criteria In Solid Tumors (RECIST) criteria (32).
Pretreatment ctDNA levels associated with PFS and OS should also be differently examined between the first- or the second-line treatment setting. For instance in melanoma, pretreatment ctDNA levels are only associated with clinical outcome in patients receiving ICI therapy as a first line (13, 17). Brain metastasis development in patients who relapsed after first-line therapy might be one potential explanation for the limited discriminative capacity in the second-line setting due to an insufficient ctDNA detection. This observation merits further clinical investigations, notably in other tumor types where ICI therapy can be proposed in the second line.
Clinical value of pretreatment ctDNA levels in adjuvant immunotherapy
Adjuvant immunotherapy, by definition, is being applied to tumor-resected patients. Several reports in melanoma (33–36) or lung (20) and colorectal cancer (37) have shown that the prevalence of ctDNA-positive patients after resection is low despite the use of highly sensitive digital PCR techniques. To increase the sensitivity of ctDNA testing, one could particularly recommend the interrogation of multiple mutations with personalized gene panels based on the primary tumor sequencing, the analysis of higher volumes of plasma, and repeated sampling to increase the sensitivity of mutation detection (38). However, in the adjuvant setting, the low quantities of ctDNA and sequencing artifacts currently limit the usage of large sequencing panel assays. Error suppression strategies to reduce background error rate will be necessary to improve the analytical specificity of ctDNA assays (39). In this line, ctDNA detection via personalized profiling by cancer personalized profiling by deep sequencing (CAPP-seq) was associated with a better outcome in a cohort of 28 locally advanced NSCLC patients receiving ICI as consolidation therapy after adjuvant chemoradiotherapy (40). In a clinical trial comparing adjuvant administration of the anti-PDL1 antibody atezolizumab versus observation in operable urothelial cancer patients, ctDNA positivity (detected by patient-specific mutation) at the beginning of the treatment identified a high-risk population who will benefit from adjuvant ICI therapy (41). This study design paves the way for additional high level of evidence studies in other clinical entities aimed to achieve clinical utility of ctDNA testing in the adjuvant setting.
On-treatment ctDNA Measurement to Predict Clinical Outcome
ctDNA measurements can easily be repeated throughout therapy. On-treatment levels of ctDNA either were used to calculate ctDNA changes by comparison with ctDNA levels at baseline or were directly associated with clinical outcome. Supplementary Table S1 also details the corresponding studies.
Early ctDNA dynamics after the onset of systemic therapy
The terms ctDNA “dynamics,” “kinetics,” and “variations” denote changes in VAF or concentration measured between before the first and before subsequent treatment infusions. It is worth mentioning here that, besides tumor driver mutations, those encoding for neoantigens (42) or even chromosomal number aberrations (CNA; refs. 43, 44) were used to quantify ctDNA changes.
ctDNA decrease is associated with a higher ORR, PFS, and OS. However, studies significantly differ by the ctDNA change threshold (20%, 50%, nonspecified increase or decrease, complete clearance) and time point (after one infusion or more, between 4 and 8 weeks) to assess molecular response. In the future, it will be critical to harmonize the strategy to adopt by a precise definition of a cutoff and of the time point to compare with baseline. In addition, a better knowledge of ctDNA intraday variation (45, 46) and the reproducibility of the methods is necessary to identify actual biological ctDNA variations correctly. Again, the reported studies have provided evidence for the clinical validity of ctDNA monitoring while demonstration of its clinical utility is still pending (10).
In the metastatic setting, the superior association of early on-treatment ctDNA changes to clinical efficacy over baseline ctDNA values is noteworthy (32, 47). Indeed, as a direct reflection of tumor burden (11, 12, 22–26, 48–54), ctDNA changes would encompass all variables that contribute to overall tumor response.
CtDNA variations evaluated in the early course of therapy correlated to radiographic best response evaluated 5 to 12 weeks later, suggesting an exciting capacity to anticipate tumor response in NSCLC or metastatic melanoma (55–58). However, this conclusion can be inherently biased by the study's design (i.e., most studies report on radiologic evaluations performed in daily clinical routine later than ctDNA sampling). Anticipating tumor response presents several advantages for the clinician, notably in case of treatment interruption due to severe side effects or in patients presenting with stable disease at their first assessment, to identify those who will finally go in clinical response (29, 32, 55). Nevertheless, the agreement between the first radiologic evaluation of tumor response and ctDNA evolution profile is not total (59, 60). For instance, 23% of the patients present discordant ctDNA kinetics from the first RECIST evaluation (59). Pseudoprogression, defined as a radiologic finding of disease progression before response caused by various immune cells infiltrating the tumor mass, thus contributing to increased tumor volume, can be one source of discordance (61). Although not frequent (incidence range, 0% to 9.7%), pseudoprogression is a specific challenge associated with ICI treatment. One study in 29 metastatic melanoma patients treated with PD-1 antibodies has demonstrated that decreasing ctDNA profiles can accurately differentiate pseudoprogression from a proper disease progression (62). First RECIST evaluation cannot be considered as an accurate predictor of clinical outcome for ICI (32, 55), mandating additional response assessment at later time points during tumor evolution. Therefore, it will be necessary to explore early ctDNA variations with clinical benefit determined several months later to better understand its potential to guide clinician decisions. In this line, Nabet and colleagues acknowledge that ctDNA early kinetics misclassified 25% of NSCLC patients for durable clinical benefit (32), highlighting the need for continuous monitoring of ctDNA throughout the therapy. The limited value of ctDNA as a biomarker of intracranial response suggests that ctDNA measurements and clinical imaging are not redundant but rather complementary. In metastatic melanoma, intracranial disease control did not associate with on-treatment ctDNA favorable profiles or undetectability (11, 12). Properly designed studies with simultaneous assessment of tumor response by both methods would provide interesting hints to understand this complementarity better and build more accurate models to predict clinical outcome (32, 55, 59). Exploring the cerebrospinal fluid as another compartment for liquid biopsy would also be a good alternative for clinicians (63).
Measuring ctDNA variations could also be applied to predict other immunotherapy regimes' efficacy. CtDNA clearance after the first cycles of treatment identified responders to adjuvant therapies in urothelial carcinoma or NSCLC (40, 41).
Recent advancements in immunotherapy have allowed treatment of relapsed or refractory diffuse large B-cell lymphoma thanks to CD19-targeted chimeric antigen receptor T cells (CAR T cells; ref. 64). In a pilot study of six patients, investigating specific clonotypic V(D)J rearrangements in ctDNA through the treatment could predict patient response to CAR-T cell therapy (65). In a subcohort of patients with metastatic cervical cancer treated with tumor-infiltrating lymphocyte therapy (TIL), a solid but transient HPV peak detected in cfDNA, immediately after the TIL therapy start was preferentially observed in patients with a complete and long-term response to TIL therapy (66). A similar post-TIL ctDNA “flair” was also observed in melanoma patients (67).
Association between on-treatment ctDNA concentrations and clinical outcome
Among the aforementioned studies, some have also directly correlated the ctDNA levels after the first cycles of ICI therapy to a clinical endpoint, with notable superiority to predict clinical efficacy over pretreatment levels of ctDNA (cf. Supplementary Table S1; refs. 11, 29, 47, 59, 68). Whether on-treatment levels or ctDNA variations is the most accurate way to predict the clinical outcome is still an open question. The disadvantage of ctDNA dynamics could be that, depending on its calculation mode, it can equalize patients presenting low pretreatment ctDNA and significant decrease with patients presenting high pretreatment ctDNA and smaller decrease. As such, an integrated metric defined as the ratio of on-treatment VAF to pretreatment VAF had a superior association with immunotherapy outcomes than on-treatment levels (29). On the other hand, ctDNA clearance was associated with the most favorable outcome profile (14, 29, 55, 59, 69), and conversely, detection of high levels of ctDNA is associated with future progression (14). Therefore, stratifying patients by both pretreatment and on-treatment levels and distinguishing ctDNA clearance should result in the most accurate evaluation of patient outcome, as initially suggested by Lee and colleagues in melanoma patients (12) or more recently by Zhang and colleagues (29). More studies comparing on-treatment ctDNA with on-treatment RECIST tumor evaluation would also be necessary to understand the complementarity between the two approaches better (70).
Even with favorable ctDNA kinetics, some of these patients will ultimately progress, and early ctDNA variations might not be able to discriminate long-term responders. Some studies have then evaluated ctDNA level at later time points of therapy. In NSCLC, 31 blood samples from patients achieving long-term benefit were collected at a median of 26.7 months after initiation of therapy (71). At this surveillance timepoint, 25/27 patients with undetectable ctDNA remained progression-free while all four patients with detectable ctDNA eventually progressed. A similar observation was reported in 38 melanoma patients evaluated after cessation of ICI therapy (72). Of the 28 patients with no progression, ctDNA was undetectable in 27 patients, and among the ten patients who progressed, four had detectable ctDNA at the time of treatment cessation. These independent observations corroborate the hypothesis raised by Bratman and colleagues in which ctDNA clearance at any time point during the therapy is associated with long-term survival (59). Concerning the lack of knowledge in the optimal treatment duration (and its consequences in terms of potentially severe side-effects exposure and financial costs), both studies pave the way for additional ctDNA evaluation later in therapy to better discriminate patient personal benefit. In this setting also, the usage of highly sensitive methods to detect ctDNA will be necessary to reduce the probability of false-negative results.
Genetic Determinants of Response to IO Therapies Assessed on ctDNA
In addition to a quantitative assessment, other genetic determinants of ICI therapy response can also be measured on ctDNA, such as the association of specific mutations to ICI therapy outcome, the assessment of tumor mutational burden and microinstability phenotype.
Status of specific cancer mutations relevant to therapy
As a surrogate of tumor tissue, plasma genotyping could also be used to directly evaluate the association of tumor-specific molecular alterations with response to ICI therapy or with the onset of immune-related adverse events. After excluding patients with no detectable ctDNA, Guibert and colleagues confirmed a better prognosis in patients harboring TP53 or KRAS mutations and the detrimental effect of STK11 mutations and loss of PTEN compared with wild-type patients (30). Similarly, in 38 metastatic gastric cancer patients, the mutation status of TGFBR2, RHOA, and PREX2 in ctDNA at baseline negatively influenced the PFS (31). In the same metastatic gastric cohort, patients with alterations in CEBPA, FGFR4, MET, or KMT2B detected in plasma at baseline had a greater likelihood of experiencing irAEs (31). In classic Hodgkin lymphoma (cHL), CHD8 mutation in ctDNA was only detected in patients with the longest PFS (73).
Repetitive sampling throughout therapy is the main advantage offered by ctDNA analysis. Large gene panels or whole-exome sequencing (WES) analysis on ctDNA depicting tumor clonal evolution can lead to identifying specific mutations implicated in resistance to immunotherapy. Mutation in FOXL2 and RHOA genes and copy-number variation of FGFR2 gene were identified as candidate resistant mechanisms after plasma analysis of 13 metastatic gastric cancer patients who had initially benefited from the treatment (31). Serial sequencing of ctDNA with a 329 pan-cancer‐-related gene panel and WES identified mutations in PTCH1 and B2M genes in two out of four NSCLC patients with progressive disease (74). WES on ctDNA performed on eight different NSCLC patients reported alterations of Wnt-signaling pathway-related genes, an increase of copy-number aberrations in cancer-related genes, and loss of PTEN or B2M as molecular mechanisms associated with late progression (i.e., progression observed after six months of treatment) to ICI therapy (75). Considering the broader usage of comprehensive genome sequencing in the near future, one could strongly emphasize the need for additional studies across different clinical entities with regular plasma sampling to decipher the tumor molecular landscape at the onset of resistance to immunotherapy.
Tumor mutational burden
Following the hypothesis that the more nonsynonymous mutations are present in the tumor DNA, the more neoantigens will be presented at the surface of the tumor, tumor mutational burden (TMB; i.e., the number of somatic mutations per megabase of interrogated genomic sequence) has been extensively explored as an additional predictor of clinical benefit in ICI therapies. However, the correlation between a high TMB and better response to ICI therapy is still not completely established, varying between cancer entities (76–78). If WES would be the most accurate way to assess TMB in tumor tissue (named tTMB hereafter), panel sequencing-based estimates of TMB were mainly used in the clinic so far. Nevertheless, a lack of standardization in TMB score determination due to technical features (i.e., location and size of the sequenced regions, types of mutation detected, differences in the germline mutations filtering methods, and mode of calculation of TMB score) prevents TMB score comparison across platforms and tumor types (76, 77, 79–81) and has led to the recent initiative of establishing harmonization guidelines (82). Moreover, tTMB determination on a single biopsy can also be affected by intratumor heterogeneity and might evolve with treatment.
As an alternative to tissue determination, blood-based determination of TMB (bTMB) could overcome the double problem associated with repeated access to tumor material and tumor heterogeneity (83–86). However, bTMB assays face specific challenges, such as tumor-derived molecules' input varying upon cancer type and clonal hematopoiesis (87, 88). Importantly, standardization in bTMB assays is also currently lacking but should rapidly benefit from the harmonization efforts currently ongoing for tTMB determination.
Nevertheless, bTMB via ctDNA analysis with multiple gene panels was first evaluated as a surrogate for tTMB. Despite the use of different gene panels and independent cohorts of patients, a similar level of correlation (R around 0.6) between tissue and plasma was reported (89–91) The absence of a higher correlation between tTMB and bTMB could originate from the intratumor heterogeneity. However, a low VAF and an extended time interval between blood and tissue collection in some cases could also explain the reported level of correlation (90).
bTMB was then evaluated as a predictor of ICI therapy outcome. Like for tTMB, there is an association between a high bTMB score and a better ORR and improved PFS and OS in NSCLC patients (90, 92, 93). However, no association with OS was reported by several studies (94, 95), leading Wang and colleagues to question ctDNA-based TMB determination rationale. Patients with the highest amount of ctDNA have the highest number of mutations and the highest tumor burden, and both situations result in a contradictory effect on OS. Upon adjustment by VAF, bTMB-high eventually associated with improved ORR, PFS, and OS in uni- but also multivariate analysis (96). Still, prospective studies are needed to validate the predictive efficacy of low allele frequency bTMB. Interestingly, Nabet and colleagues recently addressed this issue by defining normalized bTMB as the ratio of bTMB and ctDNA level. Normalized bTMB was superior to both individual metrics (bTMB and ctDNA levels) for predicting durable clinical benefit (32).
Microsatellite instability
In colorectal cancer, microsatellite instability (MSI) was associated with a high Th1/CTL infiltration and upregulation of immune-checkpoint proteins, suggesting a link between MSI and response to ICI (97). Like for TMB, minimally invasive determination of MSI is highly desirable in a context of a constantly expanding usage of ICI therapy.
Next-generation sequencing (NGS)-based approaches can nowadays determine MSI by measuring the length of altered microsatellites sequences (98–101). Several NGS-based assays were recently developed on cfDNA to determine tumor MSI status by overcoming the technical challenges associated with detecting low-level allele length polymorphisms in coexisting excessive amounts of wild-type DNA and PCR originating errors on long mononucleotides repeats (98, 102–104). Despite a lack of consensus on the selected loci number and nature, the different NGS assays had a sensitivity around 0.1%–1% tumor fraction and presented a high concordance with tissue MSI status (102–104). Landscape studies performed in large plasma samples sets from cancer patients reported an MSI-high prevalence among tumor types similar to the one observed with tissue-based analyses (102, 104). This approach paves the way for a pragmatic strategy to identify better the subset of patients who might benefit from ICI therapies, especially in tumor types where the benefit of the IO treatment is not yet fully established. In small cohorts of gastrointestinal cancers treated by ICI therapy, patients detected with an MSI phenotype had significantly prolonged PFS (98, 102, 104), demonstrating clinical validity of the developed assays.
NGS-based methodologies present the advantage to enable simultaneous determination of the MSI status of the tumor together with detection of other genomic determinants of response to ICI therapy like TMB. The European Society for Medical Oncology (ESMO) recommendations on MSI tissue testing for immunotherapy in cancer stated that the relationships between MSI and TMB are complex and differ according to tumor types (105). Studies exploring the complementarity between these two biomarkers are needed to predict the outcome of ICI more finely. In this line, Willis and colleagues observed a significantly superior number of SNV in MSI-high than in microsatellite stable (MSS) patients (102). Wang and colleagues, in a pan-cancer plasma analysis, questioned this putative complementarity by dichotomizing the bMSS patient's cohort into bTMB-high and bTMB-low subsets. bMSS-TMB-high and the bMSI-high groups collectively predicted significantly improved outcome, indicating that bMSI combined with bTMB may maximize the scope of ICB therapy (104).
General Conclusions
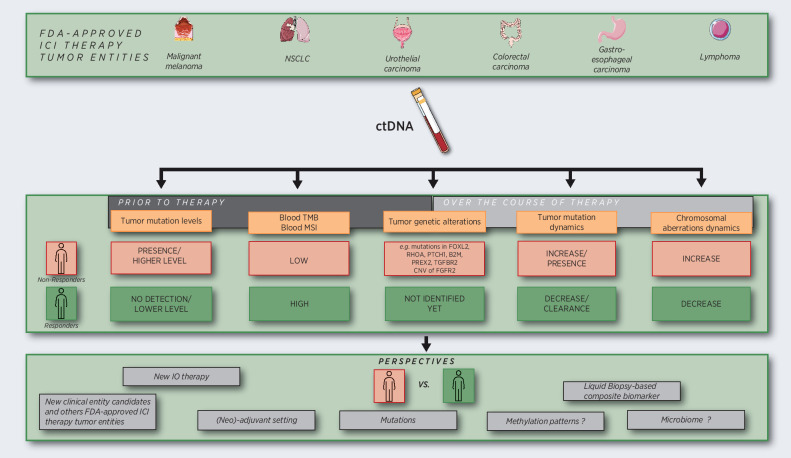
The last years witnessed a growing body of evidence supporting the use of ctDNA's multiple features (e.g., ctDNA levels, mutations, bTMB, bMSI) for discrimination of patient response to ICI therapy (Table 1 and Fig. 1).
Table 1.
Clinical applications of ctDNA in IO.
| Biomarker | Clinical entities | Biomarker study type | Clinical trial number and ICI therapy evaluated | Refs. |
|---|---|---|---|---|
| Quantitative analysis of ctDNAa | Melanoma—metastatic | Evaluated in IO clinical trialb | NCT02374242 + NCT02089685 (nivolumab, pembrolizumab) | (12) |
| Standard-of-care cohorts | Pembrolizumab/ipilimumab/nivolumab | (11, 13–18, 22, 58, 121, 122) | ||
| NSCLC—metastatic | Evaluated in IO clinical trial | NCT01693562 + NCT02087423 (durvalumab) | (123) | |
| NCT01903993 + NCT02008227 (atezolizumab) | (32) | |||
| NCT02475382 (nivolumab) | (124) | |||
| Standard-of-care cohorts | Nivolumab, pembrolizumab | (30, 42, 47, 55, 57, 125, 126) | ||
| NSCLC—localized | Evaluated in IO clinical trial | Durvalumab + NCT02525757 (atezolizumab) | (40) | |
| Gastric cancer—metastatic | Standard-of-care cohorts | Nivolumab, pembrolizumab, toripalimab, sintilimab | (31) | |
| Evaluated in IO clinical trial | NCT02589496 (pembrolizumab) | (89) | ||
| Biliary tract—metastatic | Evaluated in IO clinical trial | SHR1210-GEMOX-BTC-IIT03 | (95) | |
| Classic Hodgkin lymphoma | Evaluated in IO clinical trial | NCT03114683 (sintilimumab) | (73) | |
| Urothelial cancer—metastatic | Evaluated in IO clinical trial | NCT01693562 + NCT02087423 (durvalumab) | (123) | |
| Urothelial cancer – localized | PRCT designed to address clinical utility | NCT02450331 (atezolizumab) | (41) | |
| Multiple clinical entities—metastatic | Evaluated in IO clinical trial | NCT02644369 (pembrolizumab) | (59) | |
| NCT01693562 (durvalumab), NCT02087423 (durvalumab), NCT02261220 (durvalumab + tremelimumab) | (29) | |||
| Tumor mutational burden estimation | NSCLC—metastatic | Evaluated in IO clinical trials | NCT01903993 + NCT02008227 (atezolizumab) | (90, 91, 96) |
| NCT02453282 | (93) | |||
| NCT02478931 | (92) | |||
| NCT02848651 | (89) | |||
| Biliary tract—metastatic | Evaluated in IO clinical trial | SHR1210-GEMOX-BTC-IIT03 | (95) | |
| Microsatellite instability estimation | Gastric cancer—metastatic | Evaluated in IO clinical trials | NCT02589496 (pembrolizumab) | (102) |
| Gastrointestinal cancer—metastatic | Standard-of-care cohorts | (104) | ||
| Prostate cancer—metastatic | Standard-of-care cohorts | (103) | ||
| Multiple clinical entities—metastatic | Standard-of-care cohorts | NCT01876511 (pembrolizumab) | (98) |
Note: This table summarizes the main clinical applications of ctDNA sorted by disease severity (localized or metastatic) and tumor type. For further details on the cited studies, please refer to Supplementary Table S1.
Abbreviation: PRCT, prospective randomized controlled trial.
aPlease note that ctDNA quantification refers to both quantification before the treatment and early on treatment.
bThe clinical trial was initially designed to measure drug efficiency/safety and ctDNA was measured as an observational parameter.
Figure 1.
Current strategies and perspectives of clinical applications of ctDNA analysis in IO. Besides the detection and quantification of tumor mutations, other features like chromosomal aberrations can be quantified as a score, with the advantage of not requiring any prior knowledge of tumor mutation in an individual patient. Performing low-coverage genome-wide sequencing of cfDNA may also present interests in comparison with high-depth sequencing of targeted panels, in terms of cost and time. As a future perspective, the same kind of ctDNA analysis could be applied to new clinical entities where ICI therapy will be approved, or in new clinical therapeutic settings (neo/adjuvant). Other fascinating perspectives offered by studying ctDNA will be to identify other determinants of response to ICI therapy such as tumor-derived methylation patterns and the signatures of the host microbiome.
Most data have been obtained in metastatic patients with different types of solid tumors in the context of IO clinical trials, establishing the clinical validity of ctDNA quantification (before and early on-treatment) as prognosticator for response to therapy. The few reports on patients with localized disease in NSCLC and urothelial cancers suggest the capacity of ctDNA measurement to discriminate response from failure to therapy also in the adjuvant setting. The standardization of tests including the harmonization of cutoff points to discriminate ctDNA responders from nonresponders is now the priority task of international consortia like the European Liquid Biopsy Consortium (www.elbs.eu) or the International Alliance of Liquid Biopsy Standardization (ILSA; ref. 106). Indeed, most of the work so far reported was performed on patients included in standard-of-care cohorts or in the frame of a clinical trial initially designed to measure drug efficiency/safety. To introduce ctDNA measurements into clinical practice, interventional ctDNA-based clinical decision trials need to be designed to demonstrate the clinical utility of this biomarker. In this context, it is worth to highlight the pioneering clinical trial in localized urothelial cancers where ctDNA detection was used to personalize treatment selection for patients. In the same line, several clinical trials in early-stage NSCLC or triple-negative breast cancer are currently ongoing, in which adjuvant or neoadjuvant treatment choice is based on ctDNA positivity status after surgery (NCT04966663, NCT04849364, and NCT04585490). To better predict clinical benefit, ctDNA monitoring of tumor response could also open new avenues in the management of side effects and treatment costs. In the metastatic setting, such monitoring could also help to determine the best time point for switching from first- to second-line treatment. The CAcTUS trial in metastatic melanoma (NCT03808441) is a good example of this strategy; based on the determination of BRAF-mutant ctDNA levels patients receiving targeted therapy as first-line therapy are switched to immunotherapy as second-line therapy. In future studies, one could also imagine trials where increasing ctDNA kinetics will guide a switch from PD-1 monotherapy to a more aggressive PD-1 and CTLA-4 combination therapy while decreasing ctDNA will guide a deescalation from combination to the less aggressive monotherapy. Finally, medico-economic comparison with conventional radioimaging technologies is now also needed.
Despite the current technical challenges discussed above, ctDNA can also be used to estimate bTMB and bMSI, two genetic determinants of ICI therapy response. However, the overall response to immunotherapy is not solely dependent on tumor genomics. Tumor escape mechanisms driven at the transcriptional level and host immune system features have been highlighted as additional parameters involved in treatment efficacy (107–109). Therefore, it is very likely that multicomposite biomarkers capable of integrating several metrics will present the highest accuracy to predict tumor response to ICI. Thus, peripheral blood, including circulating tumor cells, circulating cytokines, peripheral T cells population profiles, and extracellular vesicles could be an ideal source to encompass simultaneously all parameters involved in tumor immune response, and that have already been separately demonstrated as a candidate biomarker of clinical efficacy (110–114). Likewise, Nabet and colleagues have recently developed the DIREct-On score (Durable Immunotherapy Response Estimation by immune profiling and ctDNA) to predict the response of NSCLC patients receiving ICI-based therapies that incorporates three pretreatment biomarkers (ctDNA-normalized TMB, PDL1 tissue expression, circulating CD8 T-cell fraction) but also ctDNA levels after a single cycle of ICI therapy. This score outperformed each metric on the clinical classification accuracy and prognostic value and was the only feature independently associated with PFS in the multivariate Cox proportional model comprising age, ECOG, and line of therapy (32).
Besides mutations, other valuable information like methylation of specific loci or methylation patterns could be extracted from ctDNA analysis (115). Recently, the EPIMUNNE signature based on methylome analysis of the tumor tissue was successfully correlated to the clinical outcome of NSCLC patients treated by immunotherapy (116). Moreover, with thousand copies per cell, mitochondrial DNA in plasma represents an abundant source to exploit, potentially providing valuable information on both tumor and microenvironment (117, 118). Other exciting perspectives of exploiting plasma information could come from the emergent possibility to dissect the microbiome in peripheral blood that would make sense in this context owing to the putative role of intratumor bacteria in response to ICI therapy (119, 120). Thus, liquid biopsy analysis expands the offer to interrogate several features originating from both the host and tumor in a minimally invasive way, leading to the development of a personalized biomarker of response to ICI therapy.
Disclaimer
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, and approval of the manuscript.
Authors' Disclosures
J.C. Stadler reports personal fees from Mildred Scheel Cancer Career Center HaTriCS4, University Medical Center Hamburg- Eppendorf, Hamburg, Germany, during the conduct of the study. B. Deitert reports other support from IPRime Scholarship outside the submitted work. C. Gebhardt reports personal fees from Amgen, personal fees and other support from BioNTech, Bristol-Myers Squibb, Immunocore, MSD Sharp & Dohme, other support from Dermagnostix, grants, personal fees, and other support from Novartis, personal fees and other support from Pierre-Fabre, Roche, Sanofi Genzyme, grants from Sciomics, and grants and personal fees from Sysmex outside the submitted work. No disclosures were reported by the other authors.
Acknowledgments
J.C. Stadler was financially supported by Mildred Scheel clinician scientist program. B. Deitert was financially supported by IPRime Scholarship, UKE, Hamburg. L. Keller and K. Pantel were financially supported by KMU-innovativ-23 n 031B0843D. Y. Belloum and K. Pantel were financially supported by the European Union's Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement No 765492. K. Pantel received funding from the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Program (grant agreement No 834974).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. [DOI] [PubMed] [Google Scholar]
- 2. Vaddepally RK, Kharel P, Pandey R, Garje R, Chandra AB. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers 2020;12:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanna NH, Schneider BJ, Temin S, Baker S Jr, Brahmer J, Ellis PM, et al. Therapy for stage IV non-small-cell lung cancer without driver alterations: ASCO and OH (CCO) joint guideline update. J Clin Oncol 2020;38:1608–32. [DOI] [PubMed] [Google Scholar]
- 4. Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 5. Ellis GI, Sheppard NC, Riley JL. Genetic engineering of T cells for immunotherapy. Nat Rev Genet 2021;22:427–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med 2010;16:398–406. [DOI] [PubMed] [Google Scholar]
- 7. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297–312. [DOI] [PubMed] [Google Scholar]
- 8. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. [DOI] [PubMed] [Google Scholar]
- 9. De Mattos-Arruda L, Siravegna G. How to use liquid biopsies to treat patients with cancer. ESMO Open 2021;6:100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayes DF. Biomarker validation and testing. Mol Oncol 2015;9:960–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee JH, Menzies AM, Carlino MS, McEvoy AC, Sandhu S, Weppler AM, et al. Longitudinal monitoring of ctDNA in patients with melanoma and brain metastases treated with immune checkpoint inhibitors. Clin Cancer Res 2020;26:4064–71. [DOI] [PubMed] [Google Scholar]
- 12. Lee JH, Long GV, Boyd S, Lo S, Menzies AM, Tembe V, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol 2017;28:1130–36. [DOI] [PubMed] [Google Scholar]
- 13. Marsavela G, Lee J, Calapre L, Wong SQ, Pereira MR, McEvoy AC, et al. Circulating tumor DNA predicts outcome from first-, but not second-line treatment and identifies melanoma patients who may benefit from combination immunotherapy. Clin Cancer Res 2020;26:5926–33. [DOI] [PubMed] [Google Scholar]
- 14. Seremet T, Jansen Y, Planken S, Njimi H, Delaunoy M, El Housni H, et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J Transl Med 2019;17:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McEvoy AC, Warburton L, Al-Ogaili Z, Celliers L, Calapre L, Pereira MR, et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. BMC Cancer 2018;18:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015;6:42008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herbreteau G, Vallée A, Knol AC, Théoleyre S, Quéreux G, Frénard C, et al. Circulating tumour DNA is an independent prognostic biomarker for survival in metastatic BRAF or NRAS-mutated melanoma patients. Cancers 2020;12:1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forschner A, Battke F, Hadaschik D, Schulze M, Weißgraeber S, Han CT, et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma: results of a prospective biomarker study. J Immunother Cancer 2019;7:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv 2020;6:eabc4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braune J, Keller L, Schiller F, Graf E, Rafei-Shamsabadi D, Wehrle J, et al. Circulating tumor DNA allows early treatment monitoring in BRAF- and NRAS-mutant malignant melanoma. JCO Precis Oncol 2020;4:20–31. [DOI] [PubMed] [Google Scholar]
- 23. Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer 2018;88:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wong SQ, Raleigh JM, Callahan J, Vergara IA, Ftouni S, Hatzimihalis A, et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Prec Oncol 2017;1:1–14. [DOI] [PubMed] [Google Scholar]
- 25. Chang GA, Tadepalli JS, Shao Y, Zhang Y, Weiss S, Robinson E, et al. Sensitivity of plasma BRAFmutant and NRASmutant cell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol Oncol 2016;10:157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito K, Schöder H, Teng R, Humm JL, Ni A, Wolchok JD, et al. Prognostic value of baseline metabolic tumor volume measured on (18)F-fluorodeoxyglucose positron emission tomography/computed tomography in melanoma patients treated with ipilimumab therapy. Eur J Nucl Med Mol Imaging 2019;46:930–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joseph RW, Elassaiss-Schaap J, Kefford R, Hwu WJ, Wolchok JD, Joshua AM, et al. Baseline tumor size is an independent prognostic factor for overall survival in patients with melanoma treated with pembrolizumab. Clin Cancer Res 2018;24:4960–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020;10:1842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guibert N, Jones G, Beeler JF, Plagnol V, Morris C, Mourlanette J, et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 2019;137:1–6. [DOI] [PubMed] [Google Scholar]
- 31. Jin Y, Chen DL, Wang F, Yang CP, Chen XX, You JQ, et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer 2020;19:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nabet BY, Esfahani MS, Moding EJ, Hamilton EG, Chabon JJ, Rizvi H, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 2020;183:363–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee JH, Saw RP, Thompson JF, Lo S, Spillane AJ, Shannon KF, et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann Oncol 2019;30:815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan L, Sandhu S, Lee RJ, Li J, Callahan J, Ftouni S, et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann Oncol 2019;30:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McEvoy AC, Pereira MR, Reid A, Pearce R, Cowell L, Al-Ogaili Z, et al. Monitoring melanoma recurrence with circulating tumor DNA: a proof of concept from three case studies. Oncotarget 2019;10:113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee RJ, Gremel G, Marshall A, Myers KA, Fisher N, Dunn JA, et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann Oncol 2018;29:490–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moding EJ, Liu Y, Nabet BY, Chabon JJ, Chaudhuri AA, Hui AB, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat Cancer 2020;1:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021;595:432–37. [DOI] [PubMed] [Google Scholar]
- 42. Jia Q, Chiu L, Wu S, Bai J, Peng L, Zheng L, et al. Tracking neoantigens by personalized circulating tumor DNA sequencing during checkpoint blockade immunotherapy in non-small cell lung cancer. Adva Sci 2020;7:1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss GJ, Beck J, Braun DP, Bornemann-Kolatzki K, Barilla H, Cubello R, et al. Tumor cell-free DNA copy number instability predicts therapeutic response to immunotherapy. Clin Cancer Res 2017;23:5074–81. [DOI] [PubMed] [Google Scholar]
- 44. Jensen TJ, Goodman AM, Kato S, Ellison CK, Daniels GA, Kim L, et al. Genome-wide sequencing of cell-free DNA identifies copy-number alterations that can be used for monitoring response to immunotherapy in cancer patients. Mol Cancer Ther 2019;18:448–58. [DOI] [PubMed] [Google Scholar]
- 45. Hojbjerg JA, Madsen AT, Schmidt HH, Sorensen SF, Stougaard M, Meldgaard P, et al. Intra-individual variation of circulating tumour DNA in lung cancer patients. Mol Oncol 2019;13:2098–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Bai H, Hong C, Wang J, Mei TH. Analyzing epidermal growth factor receptor mutation status changes in advanced non-small-cell lung cancer at different sampling time-points of blood within one day. Thoracic Cancer 2017;8:312–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leprieur GE, Herbretau G, Dumenil C, Julie C, Giraud V, Labrune S, et al. Circulating tumor DNA evaluated by next-generation sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. Oncoimmunology 2018;7:e1424675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang EW, Dagogo-Jack I, Kuo A, Rooney MM, Shaw AT, Digumarthy SR. Association between circulating tumor DNA burden and disease burden in patients with ALK-positive lung cancer. Cancer 2020;126:4473–84. [DOI] [PubMed] [Google Scholar]
- 49. Fernandez-Cuesta L, Perdomo S, Avogbe PH, Leblay N, Delhomme TM, Gaborieau V, et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 2016;10:117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Du M, Thompson J, Fisher H, Zhang P, Huang CC, Wang L. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer 2018;120:113–21. [DOI] [PubMed] [Google Scholar]
- 51. Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 2018;9:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maia MC, Bergerot PG, Dizman N, Hsu J, Jones J, Lanman RB, et al. Association of circulating tumor DNA (ctDNA) detection in metastatic renal cell carcinoma (mRCC) with tumor burden. Kidney Cancer 2017;1:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 2014;16:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaira K, Higuchi T, Naruse I, Arisaka Y, Tokue A, Altan B, et al. Metabolic activity by (18)F-FDG-PET/CT is predictive of early response after nivolumab in previously treated NSCLC. Eur J Nucl Med Mol Imaging 2018;45:56–66. [DOI] [PubMed] [Google Scholar]
- 55. Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res 2019;79:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herbreteau G, Langlais A, Greillier L, Audigier-Valette C, Uwer L, Hureaux J, et al. Circulating tumor DNA as a prognostic determinant in small cell lung cancer patients receiving atezolizumab. J Clin Med 2020;9:3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 2018;24:1872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Váraljai R, Wistuba-Hamprecht K, Seremet T, Diaz JMS, Nsengimana J, Sucker A, et al. Application of circulating cell-free tumor DNA profiles for therapeutic monitoring and outcome prediction in genetically heterogeneous metastatic melanoma. JCO Precis Oncol 2020;3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 2020;1:873–81. [DOI] [PubMed] [Google Scholar]
- 60. Ricciuti B, Jones G, Severgnini M, Alessi JV, Recondo G, Lawrence M, et al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J Immunother Cancer 2021;9:e001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frelaut M, du Rusquec P, de Moura A, Le Tourneau C, Borcoman E. Pseudoprogression and hyperprogression as new forms of response to immunotherapy. BioDrugs 2020;34:463–76. [DOI] [PubMed] [Google Scholar]
- 62. Lee JH, Long GV, Menzies AM, Lo S, Guminski A, Whitbourne K, et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol 2018;4:717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mouliere F, Smith CG, Heider K, Su J, van der Pol Y, Thompson M, et al. Fragmentation patterns and personalized sequencing of cell-free DNA in urine and plasma of glioma patients. EMBO Mol Med 2021;13:e12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med 2017;377:2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hossain NM, Dahiya S, Le R, Abramian AM, Kong KA, Muffly LS, et al. Circulating tumor DNA assessment in patients with diffuse large B-cell lymphoma following CAR T-cell therapy. Leuk Lymphoma 2019;60:503–06. [DOI] [PubMed] [Google Scholar]
- 66. Kang Z, Stevanović S, Hinrichs CS, Cao L. Circulating Cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin Cancer Res 2017;23:6856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xi L, Pham TH, Payabyab EC, Sherry RM, Rosenberg SA, Raffeld M. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res 2016;22:5480–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern MH, et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol 2018;15:639–50. [DOI] [PubMed] [Google Scholar]
- 69. Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017;28:1996–2001. [DOI] [PubMed] [Google Scholar]
- 70. Marsavela G, McEvoy AC, Pereira MR, Reid AL, Al-Ogaili Z, Warburton L, et al. Detection of clinical progression through plasma ctDNA in metastatic melanoma patients: a comparison to radiological progression. Br J Cancer 2021. doi: 10.1038/s41416-021-01507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hellmann MD, Nabet BY, Rizvi H, Chaudhuri AA, Wells DK, Dunphy MPS, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L)1 blockade in NSCLC. Clin Cancer Res 2020;26:2849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Warburton L, Calapre L, Pereira MR, Reid A, Robinson C, Amanuel B, et al. Circulating tumour DNA in advanced melanoma patients ceasing PD1 inhibition in the absence of disease progression. Cancers 2020;12:3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shi Y, Su H, Song Y, Jiang W, Sun X, Qian W, et al. Circulating tumor DNA predicts response in Chinese patients with relapsed or refractory classical Hodgkin lymphoma treated with sintilimab. EBioMedicine 2020;54:102731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li L, Wang Y, Shi W, Zhu M, Liu Z, Luo N, et al. Serial ultra-deep sequencing of circulating tumor DNA reveals the clonal evolution in non-small cell lung cancer patients treated with anti-PD1 immunotherapy. Cancer Med 2019;8:7669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giroux Leprieur E, Hélias-Rodzewicz Z, Kamga TP, Costantini A, Julie C, Corjon A, et al. Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J Immunother Cancer 2020;8:e000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov 2020;10:1808–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Strickler JH, Hanks BA, Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res 2021;27:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yakirevich E, Patel NR. Tumor mutational burden and immune signatures interplay in renal cell carcinoma. Ann Transl Med 2020;8:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Buchhalter I, Rempel E, Endris V, Allgäuer M, Neumann O, Volckmar AL, et al. Size matters: dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer 2019;144:848–58. [DOI] [PubMed] [Google Scholar]
- 80. Valero C, Lee M, Hoen D, Wang J, Nadeem Z, Patel N, et al. The association between tumor mutational burden and prognosis is dependent on treatment context. Nat Genet 2021;53:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Merino DM, McShane LM, Fabrizio D, Funari V, Chen SJ, White JR, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the friends of cancer research TMB harmonization project. J Immunother Cancer 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marusyk A, Janiszewska M, Polyak K. Heterogeneity: The rosetta stone of therapy resistance. Cancer Cell 2020;37:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cresswell GD, Nichol D, Spiteri I, Tari H, Zapata L, Heide T, et al. Mapping the breast cancer metastatic cascade onto ctDNA using genetic and epigenetic clonal tracking. Nat Commun 2020;11:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 2019;25:1415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 2018;24:4437–43. [DOI] [PubMed] [Google Scholar]
- 88. Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 2017;21:374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kim ST, Cristescu R, Bass AJ, Kim K-M, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. [DOI] [PubMed] [Google Scholar]
- 90. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441–48. [DOI] [PubMed] [Google Scholar]
- 91. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small Cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019;5:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Khagi Y, Goodman AM, Daniels GA, Patel SP, Sacco AG, Randall JM, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res 2017, 23:5729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rizvi NA, Cho BC, Reinmuth N, Lee KH, Luft A, Ahn MJ, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol 2020;6:661–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chae YK, Davis AA, Agte S, Pan A, Simon NI, Iams WT, et al. Clinical implications of circulating tumor DNA tumor mutational burden (ctDNA TMB) in non-small cell lung cancer. Oncologist 2019;24:820–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen X, Wu X, Wu H, Gu Y, Shao Y, Shao Q, et al. Camrelizumab plus gemcitabine and oxaliplatin (GEMOX) in patients with advanced biliary tract cancer: a single-arm, open-label, phase II trial. J Immunother Cancer 2020;8:e001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang Z, Duan J, Wang G, Zhao J, Xu J, Han J, et al. Allele frequency-adjusted blood-based tumor mutational burden as a predictor of overall survival for patients with NSCLC treated with PD-(L)1 inhibitors. J Thorac Oncol 2020;15:556–67. [DOI] [PubMed] [Google Scholar]
- 97. Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Georgiadis A, Durham JN, Keefer LA, Bartlett BR, Zielonka M, Murphy D, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin Cancer Res 2019;25:7024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res 2016;22:813–20. [DOI] [PubMed] [Google Scholar]
- 100. Ladas I, Yu F, Leong KW, Fitarelli-Kiehl M, Song C, Ashtaputre R, et al. Enhanced detection of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic Acids Res 2018;46:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192–9. [DOI] [PubMed] [Google Scholar]
- 102. Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 2019;25:7035–45. [DOI] [PubMed] [Google Scholar]
- 103. Barata P, Agarwal N, Nussenzveig R, Gerendash B, Jaeger E, Hatton W, et al. Clinical activity of pembrolizumab in metastatic prostate cancer with microsatellite instability high (MSI-H) detected by circulating tumor DNA. J Immunother Cancer 2020;8:e001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang Z, Zhao X, Gao C, Gong J, Wang X, Gao J, et al. Plasma-based microsatellite instability detection strategy to guide immune checkpoint blockade treatment. J Immunother Cancer 2020;8:e001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol 2019;30:1232–43. [DOI] [PubMed] [Google Scholar]
- 106. Connors D, Allen J, Alvarez JD, Boyle J, Cristofanilli M, Hiller C, et al. International liquid biopsy standardization alliance white paper. Crit Rev Oncol Hematol 2020;156:103112. [DOI] [PubMed] [Google Scholar]
- 107. Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med 2018;24:1545–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cordonnier M, Nardin C, Chanteloup G, Derangere V, Algros MP, Arnould L, et al. Tracking the evolution of circulating exosomal-PD-L1 to monitor melanoma patients. J Extracell Vesicles 2020;9:1710899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yamauchi T, Hoki T, Oba T, Jain V, Chen H, Attwood K, et al. T-cell CX3CR1 expression as a dynamic blood-based biomarker of response to immune checkpoint inhibitors. Nat Commun 2021;12:1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Janning M, Kobus F, Babayan A, Wikman H, Velthaus JL, Bergmann S, et al. Determination of PD-L1 expression in circulating tumor cells of NSCLC patients and correlation with response to PD-1/PD-L1 inhibitors. Cancers 2019;11:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Guibert N, Delaunay M, Lusque A, Boubekeur N, Rouquette I, Clermont E, et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer 2018;120:108–12. [DOI] [PubMed] [Google Scholar]
- 115. Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 2021;124:345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Duruisseaux M, Martínez-Cardús A, Calleja-Cervantes ME, Moran S, Castro de Moura M, Davalos V, et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis. Lancet Respir Med 2018;6:771–81. [DOI] [PubMed] [Google Scholar]
- 117. Brinker AE, Vivian CJ, Beadnell TC, Koestler DC, Teoh ST, Lunt SY, et al. Mitochondrial haplotype of the host stromal microenvironment alters metastasis in a non-cell autonomous manner. Cancer Res 2020;80:1118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mair R, Mouliere F, Smith CG, Chandrananda D, Gale D, Marass F, et al. Measurement of plasma cell-free mitochondrial tumor dna improves detection of glioblastoma in patient-derived orthotopic xenograft models. Cancer Res 2019;79:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021;592:138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Herbreteau G, Vallee A, Knol AC, Theoleyre S, Quereux G, Varey E, et al. Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget 2018;9:25265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Keller L, Guibert N, Casanova A, Brayer S, Farella M, Delaunay M, et al. Early circulating tumour DNA variations predict tumour response in melanoma patients treated with immunotherapy. Acta Derm Venereol 2019;99:206–10. [DOI] [PubMed] [Google Scholar]
- 123. Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res 2018;24:6212–22. [DOI] [PubMed] [Google Scholar]
- 124. Alama A, Coco S, Genova C, Rossi G, Fontana V, Tagliamento M, et al. Prognostic relevance of circulating tumor cells and circulating cell-free DNA association in metastatic non-small cell lung cancer treated with nivolumab. J Clin Med 2019;8:1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Iijima Y, Hirotsu Y, Amemiya K, Ooka Y, Mochizuki H, Oyama T, et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. Eur J Cancer 2017;86:349–57. [DOI] [PubMed] [Google Scholar]
- 126. Passiglia F, Galvano A, Castiglia M, Incorvaia L, Calo V, Listi A, et al. Monitoring blood biomarkers to predict nivolumab effectiveness in NSCLC patients. Ther Adv Med Oncol 2019;11:1758835919839928. [DOI] [PMC free article] [PubMed] [Google Scholar]