Abstract
Yersinia pestis, which causes bubonic and pneumonic plague, forms pigmented red colonies on Congo red (CR) dye agar. The hmsHFRS genes required for CR binding (Crb+) are genetically linked to virulence-associated genes encoding a siderophore uptake system. These genes are contained in a 102-kb chromosomal pgm locus that is lost in a high-frequency deletion event, resulting in loss of the Crb+ phenotype. We constructed a recA mutant strain of Y. pestis KIM10+ (YPRA) to test whether the high frequency Crb mutants result from a RecA-mediated deletion of the IS100-flanked pgm locus. Two Pgm-associated phenotypes (Crb+ and pesticin sensitivity [Psts]) were used as markers for the presence of the pgm locus in the RecA+ KIM10+ and RecA− YPRA strains. In KIM10+, both phenotypes were lost at a very high (2 × 10−3) frequency, due to the deletion of the entire pgm locus. In YPRA, the Crb+ phenotype was still lost at a high frequency (4.5 × 10−5), although the loss of the Psts phenotype occurred at spontaneous antibiotic resistance mutation frequencies (2 × 10−7). These RecA-independent Crb− mutants were caused by mutations in both the hmsHFRS locus and in a newly identified gene, hmsT. Nonpigmented Yersinia pseudotuberculosis and Escherichia coli strains transformed with both hmsT and hmsHFRS became Crb+. This study demonstrates that in a laboratory culture, the Crb+ phenotype is unstable, independent of the pgm locus deletion. We propose that a lack of selection for the CR-binding ability of Y. pestis in vitro may contribute to the mutation frequencies observed at the hmsHFRS and hmsT loci.
Yersinia pestis, the bacterium that causes bubonic and pneumonic plague, possesses a variety of plasmid- and chromosome-encoded virulence genes. Wild-type Y. pestis strains bind Congo red (CR) dye in agar media and form pigmented (Crb+) colonies at 26°C (but not at 37°C) (21). This phenotype has been historically used as an indicator for the presence of a group of virulence traits related to iron uptake. This correlation between Crb+ and virulence is due to the genetic linkage between the hmsHFRS locus required for the Crb+ phenotype (29) and virulence-associated genes that encode a siderophore-based iron uptake system (1). The combined presence of these traits is referred to as the pigmentation (Pgm+) phenotype, which is spontaneously lost from Y. pestis at a high frequency (2).
The loss of the Pgm+ phenotype results in the loss of a variety of iron-regulated proteins and has pleiotropic effects. Pgm− bacteria are avirulent in a mouse model unless infection occurs by an intravenous route or in the presence of exogenously supplied iron (5, 38). Pgm− bacteria cannot grow at 37°C under iron-poor conditions (10, 31, 32) and are resistant to the bacteriocin pesticin, because of the loss of the pesticin receptor, which functions as a receptor for both a siderophore and the bacteriocin pesticin (10). Pesticin is produced by Y. pestis and is active against bacteria that possess the pesticin receptor but lack the pesticin production and immunity plasmid pPst (33).
Fetherston et al. showed that the spontaneous loss of the Pgm+ phenotype was due to the deletion of a large (102-kbp) chromosomal pgm locus (12), which had been previously observed (25). The pgm locus contains the hmsHFRS genes that are required for the Crb+ phenotype (29) and transmission of Y. pestis by its flea vector (19), as well as the virulence-associated siderophore genes, which are part of the Y. pestis high-pathogenicity island (YP-HPI) (3, 17).
Single, directly repeated copies of IS100 bound the pgm locus (Fig. 1) (11), but only one copy of IS100 remains when the pgm locus is deleted (12). This led to the hypothesis that the pgm locus deletion is caused by a recombination event between these IS100 copies (11, 20). However, one survey of a collection of 43 Y. pestis strains isolated throughout the world found that 27% of the 26 Crb− strains exhibited only a partial loss of Pgm-associated phenotypes (20). These isolates were Crb− but hybridized to a DNA probe derived from the irp2 gene, which is present at the opposite end of the pgm locus (Fig. 1). However, no irp2− and Crb+ bacteria were identified. These strains’ Crb− phenotypes were traced to deletions encompassing the hmsHFRS genes, as well as mutations in both the hmsHFRS genes and in another, unknown gene(s) (4).
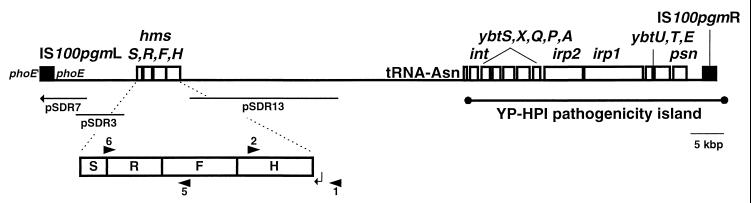
FIG. 1.
The 102-kb pgm locus of Y. pestis KIM10+, showing the hmsHFRS and psn gene locations and the IS100 copies bounding the locus. The inset map of the hmsHFRS genes shows the locations of the PCR primers HMS1, HMS2, HMS5, and HMS6, depicted as arrowheads 1, 2, 5, and 6. The arrow indicates the location and direction of the promoter region of hmsHFRS. The DNA sequences contained in the plasmids pSDR7, pSDR3, and pSDR13 are designated by horizontal lines. The irp2, psn, and other genes at the right end of the pgm locus encode the virulence-associated siderophore production and uptake system (1, 14) encoded in the YP-HPI (3, 17).
These studies indicated that Crb− colonies of Y. pestis bacteria can result from events other than a deletion of the entire pgm locus. The Crb− phenotype could result from deletions at the hmsHFRS end of the pgm locus caused by homologous recombination between appropriately located IS100 copies or other repeated sequences. Alternately, these Crb− mutants could be caused by transposition of IS100 or other insertion sequences into the hmsHFRS locus or may result from point mutations. Our goals in this study were to determine the mechanisms and the relative frequencies of these various deletions and mutations and to identify any other gene(s) required for the Crb+ phenotype.
We investigated the role of RecA-mediated events in causing the high-frequency loss of the Crb+ phenotype in Y. pestis by evaluating the frequency of the loss of two Pgm-associated phenotypes in a recA mutant. We show that at least two separate mechanisms produce the high-frequency Crb− phenotype in Y. pestis and that mutation frequencies at different sites within the pgm locus may vary. RecA-mediated deletion of the entire pgm locus caused the majority of Crb− mutants in RecA+ cells. However, the use of a recA mutant strain, which could not carry out the RecA-mediated pgm locus deletion event, showed that RecA-independent, Crb− mutants occurred at a frequency 100-fold higher than the frequency of spontaneous mutation to drug resistance. These Crb− recA mutants carried mutations in either the previously defined hmsHFRS genes or in a newly identified gene (hmsT), which is located outside the pgm locus. This study defines CR binding as an unstable phenotype of Y. pestis, independent of the high-frequency pgm locus deletion.
MATERIALS AND METHODS
Bacterial strains.
Y. pestis KIM10+ is a pigmented, attenuated strain that was derived from a virulent prototype called KIM (38). KIM10+ is a derivative of KIM6+ (the strain in which the pgm locus and its deletion were initially characterized [12]), which lacks the pesticin production and immunity plasmid (29), making it sensitive to the effects of pesticin. KIM10+ also lacks the 70-kbp calcium dependence plasmid but contains the 100-kbp plasmid and the pgm locus (as denoted by the “+” symbol) (29). Y. pestis and Yersinia pseudotuberculosis strains were grown at 28°C on brain heart infusion (BHI; Difco, Detroit, Mich.) agar or in BHI broth. The Y. pseudotuberculosis strains used in this study were cured of the calcium dependence plasmid to render them avirulent by growth on sodium oxalate plates at 37°C (15). Y. pseudotuberculosis PTB50, PTB51, PTB52, and PTB56 are serotype IB and possess the yersiniabactin siderophore system encoded by the YP-HPI (17). Y. pseudotuberculosis PTB53, PTB54, and PTB55 are serotypes IB, III, and III, respectively, and do not possess the yersiniabactin siderophore system. The KIM10+ recA mutant was named YPRA and is described below. Transforming the cloned, native recA gene back into YPRA generated the RecA+ strain YPRA(pBSrecA).
KIM10+ recA mutant construction.
The Y. pestis recA GenBank entry (accession no. X75336) was used to design PCR primers (forward, 5′-TTAAGTCGACGGTACGTGAAATGGCATTGGG-3′; reverse, 5′-ATTAGGATCCGCAGTTCAGATTCACTTTGG-3′; annealing temperature, 59°C) that would amplify a RecA-expressing cassette. Genomic DNA from Y. pestis 195/P (27) was the DNA template for PCRs (DNA thermal cycler model 480; Perkin-Elmer Cetus, Norwalk, Conn.). The 1.65-kbp Y. pestis recA PCR product was cloned into pBluescript SK(+) (Stratagene, La Jolla, Calif.) and maintained in Escherichia coli JM109 by ampicillin (100 μg/ml).
The cloned recA gene was mutated by cloning a 955-bp kanamycin resistance cassette (aph3′) from Tn903 (8) into unique ClaI and EcoRV sites within the recA gene. This replaced approximately the middle third of the recA coding sequence. This marked recA clone (pBSΔrecA::kan) was maintained in JM109 by kanamycin (50 μg/ml). The deleted 390-bp ClaI-EcoRV recA fragment was cloned into pBluescript SK(+) to form pmidrecA. A 2.2-kbp SacI-SalI fragment of pBSΔrecA::kan containing the marked ΔrecA::kan region was subcloned into the suicide vector pCVD442 to form pCVDΔrecA::kan. pCVD442 has a pir-dependent origin of replication from the plasmid R6K and was maintained in E. coli (λ pir) (9). pCVD442 contains the Bacillus subtilis counterselectable marker sacB, which encodes levan sucrase, an enzyme that is toxic to gram-negative bacteria in the presence of sucrose (13).
pCVDΔrecA::kan was electroporated into Crb+ KIM10+ cells, which were plated on BHI agar containing ampicillin (50 μg/ml) and kanamycin (50 μg/ml) to select for merodiploids possessing the suicide vector construct integrated into native recA. Plasmid absence was verified by performing a plasmid DNA preparation on merodiploid candidates. Direct selection for a second crossover to replace the native recA gene was obtained by plating an overnight culture grown in the absence of drug selection on BHI agar plus 5% sucrose and kanamycin.
Genetic and phenotypic verification of the recA mutant YPRA.
Southern analyses were carried out on the PCR amplification product of the recA genes from the chromosome of the KIM10+ and YPRA strains. Three probes were used: the KIM10+ native recA gene, the kanamycin resistance cassette, and the 390-bp fragment of recA that was replaced in YPRA.
Western blot analyses were carried out by using an anti-E. coli RecA antibody donated by the Charles Radding laboratory. Total cell lysates were obtained by boiling an overnight culture of cells in a standard 1× sodium dodecyl sulfate gel loading buffer (30) for 3 min and were run on a sodium dodecyl sulfate–12.5% polyacrylamide gel. The gel was blotted onto a Zeta-Probe membrane with a TransBlot SD system (both from Bio-Rad Laboratories, Hercules, Calif.). The blot was blocked overnight in Tris-buffered saline (TBS)–0.1% Tween-20–5% nonfat dry milk, washed in TBS–0.1% bovine serum albumin, and incubated with a rabbit anti-E. coli RecA antibody diluted 1:1,000 in TBS-bovine serum albumin-Tween. The blot was then washed and incubated with an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch, West Grove, Pa.). Alkaline phosphatase activity was observed by rinsing the blot in the detection reagents BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium for 4 min, after which the reaction was stopped by rinsing the blot with deionized water.
A UV sensitivity test was carried out by irradiating uncovered BHI plates spread with KIM10+ and YPRA cultures (optical density at 600 nm, 0.2) with 5,000 mJ of UV light per cm2 for 12 s in a UV Stratalinker 1800 (Stratagene) apparatus. These dilutions were also plated on BHI agar plates containing 0.5 μg of mitomycin C (Sigma, St. Louis, Mo.) per ml. Control plates spread with these dilutions were not exposed to UV light or mitomycin C. After 3 days of incubation in the dark at 28°C, viable counts in the presence and absence of UV exposure or mitomycin C were calculated. Colonies were photographed under dark-field conditions on an Olympus SZH10 research stereoscope equipped with an Olympus PM-10ADS automatic photomicrograph system and an Olympus C-335AD-4 camera. Diameters of KIM10+ and YPRA colonies were measured from stereomicrographs at a magnification of ×7.
Phenotype assays.
Pesticin plates were prepared by plating 0.2 ml of an overnight culture of the pesticin-producing Y. pestis strain EV76-x(pKYP1) on BHI agar. After 2 days of incubation at 28°C, 5 ml of molten BHI agar was overlaid on the plates, which were then stored at 4°C for 2 days. CR plates contained heart infusion agar (Difco) plus 0.2% galactose and 0.01% CR dye (Sigma) (36). Surgalla CR plates contained 1% heart infusion broth plus 2% agar, 0.2% galactose, and 0.01% CR dye (36). A single colony of each test strain was inoculated into 3 ml of BHI broth and grown for 24 h at 28°C. The culture was then plated on pesticin and CR plates and incubated at 37°C (pesticin plates) or 28°C (CR plates) for 2 days. The frequency of pesticin resistance was calculated by dividing the CFU per ml for pesticin-resistant colonies by the total CFU per ml for the culture as determined by growth on control BHI agar plates. The Crb− frequency was calculated by dividing the number of Crb− (white) colonies by the total number of colonies observed on the CR plates. Growth at 28°C on BHI plates containing streptomycin (25 μg/ml) or rifampin (20 μg/ml) was used to assay antibiotic resistance mutation frequencies.
Plasmids.
The library and cloning vector pBRΔtet was constructed by removing a 156-bp EcoRV-HindIII fragment of pBR322 to abolish the activity of the tetracycline resistance gene. pHFRS, which expresses the hemin storage proteins HmsHFRS, was constructed by subcloning a 9.65-kbp HindIII-SalI fragment of the plasmid pHMS1 (29) into pBRΔtet. Derivatives of pHFRS were constructed that express a subset of the hemin storage proteins. pFRS is a 9.4-kbp HindIII-SmaI collapse of pHFRS that expresses HmsF, HmsR, and HmsS. pRS is a 7.5-kbp BamHI collapse of pHFRS that expresses HmsR and HmsS. Plasmids are described in Table 1. All plasmids were transformed by electroporation in 0.2-cm gap cuvettes on a Gene Pulser (both from Bio-Rad), set at 2.5 kV, 200 Ω, and 25 μF, and grown for 1 h at 37°C before being plated to the appropriate media.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| pBluescript SK(+) | Apr cloning vector | Stratagene |
| pBSrecA | 1.65-kb Y. pestis recA and promoter in pBluescript SK(+) | This study |
| pBSΔrecA::kan | pBSrecA with 955-bp kanamycin resistance cassette replacing a 390-bp fragment of recA | This study |
| pMidrecA | 390-bp ClaI-EcoRV fragment of the recA coding region in pBluescript SK(+) | This study |
| pCVD442 | Apr suicide vector requiring λ pir for replication | 9 |
| pCVDΔrecA::kan | Kmr Apr; pCVD442 with Kmr-mutated recA insert | This study |
| pHMS1 | KmrhmsHFRS | 29 |
| pHFRS | AprhmsHFRS; 9.65-kb HindIII-SalI insert of pHMS1 into pBRΔtet | This study |
| pFRS | 9.4-kb HindIII-SmaI deletion of pHFRS; hmsFRS | This study |
| pRS | 7.5-kb BamHI deletion of pHFRS; hmsRS | This study |
| pSDR3 | 7.5-kb Sau3A insert from pgm locus; Kmr | 12 |
| pSDR7 | 23-kbp BamHI insert from pgm locus; Apr | 12 |
| pSDR13 | 23.5-kbp Sau3A insert from pgm locus; Apr | 12 |
| pFUS43 | Apr; 18-kb BamHI fusion fragment derived from pgm locus deletion cloned into pHC79 | 12 |
| pBRΔtet | Apr Tets; HindIII-EcoRV deletion of pBR322 | This study |
| pHMSX | Apr; library clone containing ORFA and ORFB | This study |
| pHMSXA | Apr; 6.5-kb EcoRI deletion subclone of pHMSX containing ORFA | This study |
| pHMSXB | Apr; 2.8-kb EcoRI fragment of pHMSX in pBRΔtet, containing ORFB (hmsT) | This study |
KIM10+ library construction and screening.
A genomic library of KIM10+ was constructed by ligating partially Sau3AI-digested KIM10+ genomic DNA into the BamHI site of vector pBRΔtet. We screened the library and identified a complementing clone, pHMSX, which was then digested with EcoRI to yield two DNA fragments suitable for subcloning. A 6.5-kbp EcoRI fragment consisting of both the vector and approximately half of the insert was self-ligated to form pHMSXB, which contained open reading frame B (ORFB). The other, 2.8-kbp EcoRI fragment of pHMSX was ligated into pBRΔtet to form pHMSXA, which contained ORFA.
PCR analyses.
To amplify within hmsH, primers HMS1 (5′-TTCATTGTATCGTAGCC-3′) and HMS2 (5′-CATCAGTCAGTAACTCC-3′) were used (annealing temperature, 44°C). Primers HMS5 (5′-CCCTGAAGTAAGACAGCG-3′) and HMS6 (5′-GATAATGTCAATCCAGCG-3′) were used to amplify part of hmsF, hmsR, and the beginning of hmsS (annealing temperature, 48°C). Primers topgmL (5′-GTCACACCGAATTCAGC-3′) and topgmR (5′-CGCTACCACTGAAATCC-3′) flanking the pgm locus were used to amplify the pgm deletion junction (annealing temperature, 48°C). The pgm deletion junction-containing plasmid pFUS43 (12) was used as a positive control for deletion of the pgm locus.
Southern and colony blot analyses.
Genomic DNA was purified via the Puregene kit (Gentra Systems, Inc., Minneapolis, Minn.). Southern blots were prepared by electrophoresing DNA on a 0.9% agarose gel (Bio-Rad) in 0.5× Tris-borate-EDTA buffer (30) and blotting DNA by capillary action onto a Zeta-Probe nylon membrane (Bio-Rad). The DNA was cross-linked to the membrane by UV light in a UV Stratalinker 1800 (Stratagene). Colony blot preparation, probe preparation, and hybridizations were carried out as previously described (17). All probes were radioactively labeled by random primer labeling (17). The recA probe consisted of the PCR product amplified with the recA forward and reverse primers. A probe for the hmsHFRS locus genes consisted of both a 6-kbp BamHI fragment of pHMS1 containing hmsH and part of hmsF and a PCR product derived from amplification with HMS5 and HMS6 that contained hmsRS. The probe for the ORFA and ORFB region was the purified plasmid pHMSX.
Nucleotide sequence accession number.
The DNA sequence of hmsT is listed in the GenBank database under accession no. AF132982.
RESULTS
Determination of the Crb mutation frequency in Y. pestis KIM10+.
We assayed the RecA+ KIM10+ strain to establish the frequency of the loss of two Pgm-associated phenotypes located at opposite ends of the 102-kbp pgm locus: CR binding and pesticin sensitivity (Psts) (Fig. 1). Loss of the Crb+ phenotype was screened by growing colonies on CR agar; red colonies were Crb+, and white colonies were Crb−. Red colonies with white sectors were occasionally observed but were not used to calculate the Crb mutation frequency, so that only mutations occurring during the initial 24-h growth period were represented in the mutation frequency. The frequency of Crb+ loss was calculated by dividing the number of Crb− colonies by the total number of colonies screened. Loss of Psts was selected by growth on pesticin-containing plates. The frequency of Psts loss was calculated by dividing the CFU per ml of pesticin-resistant (Pstr) colonies by the CFU per ml of cells plated on BHI medium lacking pesticin.
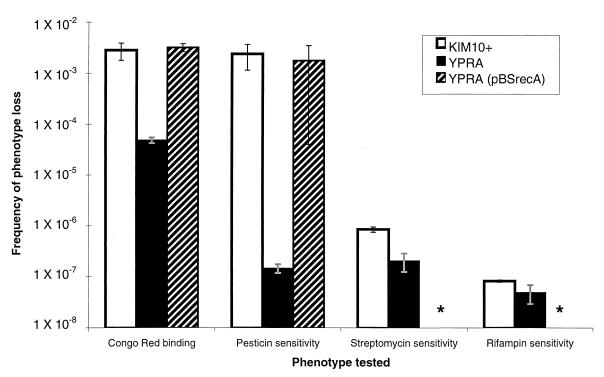
KIM10+ lost both the Psts and Crb+ phenotypes at high frequencies (2.7 × 10−3 and 2.0 × 10−3, respectively) (Fig. 2). We tested some of each type of mutant (Pstr or Crb−) for the other phenotype, because the deletion of the entire pgm locus would result in a simultaneous loss of both markers. All 32 Pstr KIM10+ mutants that we tested were also Crb−, and all of the 20 Crb− KIM10+ colonies that we tested were Pstr, suggesting that these mutants had indeed lost the entire pgm locus.
FIG. 2.
Comparison of the frequencies of loss of various phenotypes in the KIM10+ versus the YPRA strains. The values in the graph are the means ± standard deviations (error bars) for multiple independent experiments. Asterisks indicate that the frequency of resistance to antibiotics was not evaluated in YPRA(pBSrecA).
The deletion of the pgm locus in the Crb− mutants was confirmed by three additional tests. None of the 20 Crb− Pstr mutants hybridized to a hmsHFRS gene probe in colony blot analyses (data not shown). In addition, a 2-kb product was amplified by PCR with primers flanking the pgm locus from all of the five strains that we tested. Finally, DNA sequence analysis of the PCR product from one of these five mutants demonstrated that a single copy of the 2-kb IS100 element remained in place of the pgm locus (data not shown). Transformation of the hmsHFRS-containing plasmid pHMS1 into these Pgm− mutants restored the Crb+ phenotype, as expected and observed previously (30).
The two Pgm-associated phenotypes are lost at different frequencies in YPRA.
We made a Y. pestis KIM10+ recA mutant strain (YPRA) to test the hypothesis that the high-frequency Crb− phenotypes arising in Y. pestis are caused by RecA-mediated deletions, either of the entire IS100-flanked pgm locus or smaller, insertion element-flanked regions. The frequency of spontaneous resistance to two drugs (streptomycin and rifampin) was measured in KIM10+ and YPRA to establish and compare the background mutation frequencies in these strains. Mutation frequencies were similar for each drug in KIM10+ and YPRA (Fig. 2), indicating that YPRA is not a mutator strain (6) that is prone to higher mutation frequencies.
Frequencies of loss of the Crb+ and Psts phenotypes were measured in YPRA to determine whether the high-frequency loss of the Crb+ phenotype occurred by a RecA-dependent process (such as a deletion of the entire 102-kb pgm locus). We reasoned that the high-frequency loss of the Pgm-associated phenotypes would be greatly reduced in YPRA if the pgm locus deletion were RecA mediated. In YPRA, the frequency of Pstr mutants decreased 10,000-fold from RecA+ KIM10+ levels to spontaneous mutation levels (Fig. 2). In contrast, the loss of the Crb+ phenotype was reduced only moderately (40-fold) relative to KIM10+ and remained at a frequency 100- to 1,000-fold higher than the spontaneous mutation frequency measured by resistance to streptomycin and rifampin (Fig. 2). The RecA+ YPRA(pBSrecA) strain lost both the Crb+ and Psts phenotypes at the same high frequency as KIM10+ (Fig. 2), confirming the role of RecA in this process. Twelve Crb− RecA+ YPRA(pBSrecA) mutants were analyzed by PCR and found to have deletions of the entire pgm locus that were indistinguishable from the deletion events observed in the Crb− RecA+ KIM10+ mutants.
In contrast to KIM10+, only 13% of the Pstr YPRA mutants (6 of 46) were also Crb−. Similarly, only 1 of the 18 Crb− YPRA mutants (YPRA11; see below) was also Pstr. These results indicated that in the absence of RecA, the altered phenotypes of most of these YPRA mutants were due to events other than a deletion of the entire pgm locus.
Mutations in the hmsHFRS locus comprise one class of Crb− recA mutants.
In contrast to the loss of Psts, the Crb+ phenotype was lost at a relatively high frequency in YPRA. Eighteen YPRA Crb− mutants were further characterized to learn if the relatively high-frequency loss of this phenotype was due to a common RecA-independent mechanism. The pgm locus was present in nearly all (17 of 18) of these mutants. These mutants remained Psts and contained the hmsHFRS locus as determined by colony blot and PCR analyses of two different regions within the hms locus (data not shown). Southern analysis indicated no obvious insertions, deletions, or rearrangements in the hmsHFRS loci of these strains (Fig. 3, lanes 1 to 9). The remaining mutant (YPRA11) lost the entire pgm locus by a deletion, as indicated by the PCR amplification of a 2-kbp product with primers flanking the pgm locus. This mutant was also Pstr and did not possess DNA from the hmsHFRS locus as measured by the above-mentioned colony, PCR, and Southern analyses (data not shown).
FIG. 3.
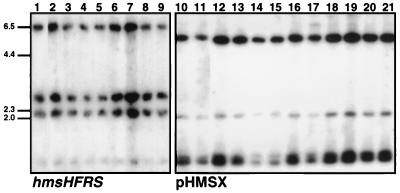
Southern blot analyses of Y. pestis and Y. pseudotuberculosis strains, showing lack of gross chromosomal rearrangements in the hmsHFRS and hmsT regions. Genomic DNA from these strains was digested with EcoRV and probed with the hmsHFRS genes (left panel) and pHMSX (right panel). Lane 1, Y. pestis KIM10+; lane 2, YPRA17; lane 3, YPRA18; lane 4, YPRA19; lane 5, YPRA21; lane 6, YPRA22; lane 7, YPRA23; lane 8, YPRA24; lane 9, YPRA25; lane 10, KIM10+; lane 11, YPRA; lane 12, Y. pseudotuberculosis PTB56; lane 13, PTB52; lane 14, PTB53; lane 15, PTB54; lane 16, Y. pestis YPRA17; lane 17, YPRA18; lane 18, YPRA11; lane 19, YPRA7; lane 20, YPRA22; lane 21, YPRA24. Positions of molecular size markers (in kilobase pairs) for both panels are shown to the left. The sizes of the bands hybridizing to the hmsHFRS genes are as follows: hmsH, 6.5 and 1.2 kb; hmsF, 2.1 kb; hmsRS, 2.7 kb. The 2.0-, 1.3-, and 1.1-kb bands in lanes 10 to 21 hybridize to hmsT DNA sequences.
Plasmid complementation experiments were conducted to identify the mutated gene loci in the YPRA Crb− mutants. Successful complementation was defined as the restoration of red colony growth upon plasmid transformation. Eleven of the 18 mutants were complemented by pHFRS, which encodes the hmsHFRS genes (Table 2). These 11 mutants were further tested with derivatives of pHFRS that expressed only some of the hemin storage proteins to determine which particular hms gene was mutated. Four mutations were localized to hmsH, one mutant was defective in hmsF, five mutants had defects in either hmsR or hmsS, and one mutant (YPRA11) was missing the hmsHFRS genes entirely (Table 2), indicating that these mutants did not carry a common mutation. The control RecA-expressing plasmid pBSrecA did not complement these mutants’ Crb− phenotype.
TABLE 2.
Complementation of Crb− YPRA strains with the hmsHFRS genes
| Y. pestis | Complementation with plasmida
|
Defective inb | |||
|---|---|---|---|---|---|
| pHFRS | pFRS | pRS | pHMSX | ||
| KIM10 | + | ND | ND | ND | HFRS |
| YPRA2 | − | ND | ND | + | T |
| YPRA3 | + | ± | ± | ND | H |
| YPRA4 | + | ± | ± | ND | H |
| YPRA7 | − | ND | ND | + | T |
| YPRA9 | + | + | − | ND | F |
| YPRA11 | + | − | − | ND | HFRS |
| YPRA14 | − | ND | ND | + | T |
| YPRA15 | + | − | − | ND | H |
| YPRA16 | + | + | + | ND | R or S |
| YPRA17 | + | + | + | ND | R or S |
| YPRA18 | + | + | + | ND | R or S |
| YPRA19 | + | + | + | ND | R or S |
| YPRA21 | − | ND | ND | + | T |
| YPRA22 | − | ND | ND | + | T |
| YPRA23 | − | ND | ND | + | T |
| YPRA24 | − | ND | ND | − | ? |
| YPRA25 | + | ± | ± | ND | H |
| YPRA26 | + | + | + | ND | R or S |
Complementation of the Crb− phenotype was either absent (−), fully present (+), or partially present (±). ND, not done; ?, unknown.
Codes indicate presumed location of mutation leading to Crb− phenotype: H, hmsH; F, hmsF; R or S, hmsRS; and T, hmsT (see Results).
A new gene, hmsT, restores the Crb+ phenotype to a second class of Crb− mutants.
The inability of pHFRS to restore the Crb+ phenotype to 7 of the 18 YPRA Crb− mutants is consistent with previous observations suggesting that a second locus was required for the Crb+ phenotype (22, 28). In addition, a recent report suggested that a putative gene needed for the Crb+ phenotype was located in the region of the pgm locus surrounding, but not including, the hmsHFRS genes (4). However, none of the seven YPRA Crb− mutants was complemented with the plasmids pSDR13, pSDR3, and pSDR7, all of which contain DNA from this region of the pgm locus (Fig. 1).
We then screened a Sau3AI library of KIM10+ chromosomal DNA in two of these seven YPRA Crb− mutants (YPRA7 and YPRA22) to identify library clones that would restore the Crb+ phenotype to these mutants. Twelve red colonies derived from four independent library transformation experiments possessed library clones that contained an identical 4.75-kb DNA insert. This library clone (designated pHMSX) complemented the Crb− phenotype in six of the seven YPRA Crb− mutants that were not successfully complemented with pHFRS. The seventh mutant, YPRA24, was not complemented by pHFRS, pHMSX, or both plasmids, and thus the cause of its Crb− phenotype remains unclear. It is possible that this lesion marks another unidentified genetic locus that is involved in the expression of the Crb+ phenotype.
The arrangement of restriction enzyme sites and a partial DNA sequence obtained from pHMSX matched a DNA sequence in the partially completed Y. pestis genome Sanger database (40). The corresponding 4.75-kb DNA sequence in the Y. pestis database contained two ORFs, ORFA (1,440 bp) and ORFB (1,170 bp) (Fig. 4). The predicted protein sequence of ORFA was 67% identical to a hypothetical protein of E. coli (accession no. P39406). The predicted protein sequence of ORFB had a region of 172 amino acids at its C terminus that was 39% identical to a Synechocystis sp. PleD protein of unknown function (accession no. D64005). A PleD homolog in Caulobacter crescentus is a response regulator that is part of a signal transduction pathway involved in motility regulation (7, 18). ORFB’s C-terminal region of homology to PleD contains a 162-amino-acid protein domain of unknown function (DUF9), contained in the Pfam (protein family) database (34). This domain is present in bacterial proteins containing signaling domains (34).
FIG. 4.
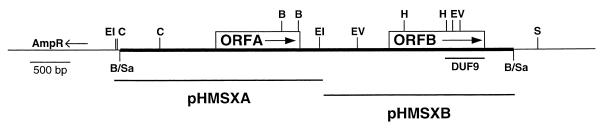
Map of pHMSX. The narrow bar represents library vector pBRΔtet sequences, and the thick bar represents Y. pestis insert DNA containing ORFA and ORFB. The location of a domain of unknown function (DUF9) is shown below ORFB. The Y. pestis insert DNA fragments contained in pHMSXA and pHMSXB are shown as bars below pHMSX. B/Sa indicates the site of Sau3AI-cut Y. pestis DNA ligation into the BamHI site of pBRΔtet. B, BamHI; C, ClaI; EI, EcoRI; EV, EcoRV; H, HindIII; S, SalI.
pHMSX was subcloned to test the abilities of each of the two ORFs to complement the Crb− phenotype of the YPRA Crb− mutants (Fig. 4). pHMSXB (orfB), but not pHMSXA (orfA), restored the Crb+ phenotype to YPRA22 (Fig. 5A). The DNA sequence of ORFB from pHMSX was identical to the ORFB DNA sequence present in the Y. pestis database that was recently described as hmsT (22).
FIG. 5.
CR binding phenotypes of Y. pestis, E. coli, and Y. pseudotuberculosis strains. (A) Y. pestis KIM10+ (Pgm+) and KIM10 (Pgm−), showing Crb+ and Crb− phenotypes, respectively. The YPRA22 strain (Crb−) is shown complemented with the library subclones pHMSXA (Crb−) and pHMSXB (Crb+). (B) E. coli and Y. pseudotuberculosis strains transformed with hmsT and hmsHFRS. Strains were transformed with either pHMS1, pHMSXB, or both plasmids. Only colonies transformed with both plasmids appear red and have a small, wrinkled colony morphology (are Crb+). A partially reddish phenotype is seen in PTB54 and PTB55 on CR agar. (C) An experiment similar to that shown in panel B was done, but cells were plated on slightly different CR agar plates (Surgalla). The PTB55 but not PTB50 Y. pseudotuberculosis strain shows a partially reddish phenotype, to a greater extent than on CR agar. Magnification, ×4. The Crb− colonies appear yellowish in these photos but are whiter when viewed directly on the CR agar plates.
pHMSX was used as a probe in Southern analyses to analyze the YPRA Crb− (hmsT-based) mutants. Wild-type KIM10+, YPRA, and the Crb− (hmsHFRS-based) strains were used as controls. The hmsT-hybridizing DNA bands from both the control and Crb− (hmsT) strains were identical in size (Fig. 3, lanes 10 to 21). This result suggested that no large insertions, deletions, or other rearrangements were present in this region of the chromosome and that point mutations or small lesions might be responsible for the hmsT defects in these strains. The Crb− YPRA11 isolate that lacked the entire pgm locus possessed bands that hybridized to both ORFA and ORFB (hmsT) sequences (Fig. 3, lane 18), indicating that hmsT lies outside the pgm locus.
Y. pseudotuberculosis and E. coli strains transformed with hmsT and hmsHFRS are Crb+.
We transformed Y. pseudotuberculosis PTB55, PTB50, PTB51, and PTB54 and E. coli DH5α with pHMSXB and pHMS1 to determine whether hmsT and/or the hmsHFRS genes could produce a Crb+ phenotype in these normally nonpigmented species (28, 36). Neither pHMSXB nor pHMS1 conferred a Crb+ phenotype on these strains when used alone (Fig. 5B). However, when these strains were transformed with both of these plasmids, they were Crb+ at both 28 and 37°C and acquired a small, wrinkled colony morphology similar to that of Y. pestis (Fig. 5B). This phenotype was apparent when colonies appeared at 20 h of growth.
The presence of both plasmids in these strains also resulted in an unusual, stalactite-like growth in broth. This pattern of broth growth, which is typical of Y. pestis (16), is characterized by reduced turbidity in culture, with cells growing in clumps at the bottom and sides of the tube (Fig. 6). Others have noted a correlation between autoaggregation of Y. pestis cells and pigmentation status consistent with changes in cell surface hydrophobicity upon binding CR or hemin (19, 21). The DH5α, PTB50, and PTB51 strains, all containing both pHMS1 and pHMSXB, repeatedly grew in this stalactite-like fashion after 2 days at 28°C; Y. pseudotuberculosis PTB55 and PTB54 showed fewer of these characteristics (Fig. 6 and data not shown).
FIG. 6.
Broth growth appearance of E. coli and Y. pseudotuberculosis strains transformed with either pHMS1, pHMSXB, or both plasmids. After transformation and growth on CR plates, colonies were cultured in BHI broth for 2 days at 28°C. In the presence of both plasmids, E. coli and Y. pseudotuberculosis PTB51 have a growth pattern similar to that of Y. pestis, with flecks of bacterial growth on the walls and bottom of the tube but little turbidity. PTB55 shows only a few of these growth characteristics. This experiment was repeated three times with similar results.
To determine whether the particular CR agar recipe used affected the requirement for both the hmsT and hmsHFRS genes, we repeated these experiments with the original CR agar recipe developed by Surgalla and Beesley (36). On our CR plates and to a greater extent on Surgalla CR plates, we observed that two of the four Y. pseudotuberculosis strains (PTB55 and PTB54) were pigmented slightly red when transformed with pHMS1 alone (Fig. 5C). However, this partial, reddish phenotype, which developed only after >24 h of growth, was lighter than the dark red Crb+ phenotype that we observed when both pHMS1 and pHMSXB were used. Additionally, neither the wrinkled colony appearance nor the altered broth growth seen in the Crb+ Y. pseudotuberculosis colonies resulted from the transformation of pHMS1 alone. Y. pseudotuberculosis PTB50 and PTB51 did not show this partial phenotype on either type of CR agar.
DISCUSSION
The Pgm+ phenotype is comprised of both the Crb+ phenotype and a group of virulence traits that allow Y. pestis to acquire iron from its mammalian hosts. The original measurement of Pgm+ loss used the Pstr mutation rate as an indicator for the loss of all of the traits that make up the Pgm+ phenotype (2). Another study investigating the effects of a Y. pestis fur mutant found that Crb− colonies were dramatically increased in a fur mutant in the presence of excess iron (35). In this study, we analyzed the variety and frequency of events causing Crb− mutants separately from other Pgm-associated phenotypes. We determined that the instability of the Crb+ phenotype is due to both RecA-mediated deletions and high-frequency RecA-independent mutations.
High-frequency RecA-dependent and -independent mechanisms found for Crb mutants.
This study contains the first description of a recA mutant strain of Y. pestis, which we constructed by replacing the native recA gene of KIM10+ with a mutated, marked recA allele. We used this recA mutant (YPRA) to test whether RecA-mediated homologous recombination caused the deletion of the pgm locus, as others have proposed (12, 20). Three types of events accounted for the high frequency Crb− phenotype we observed in Y. pestis. These included (i) the RecA-dependent deletion of the pgm locus, (ii) mutations in several of the hmsHFRS genes, and (iii) mutations in a previously uncharacterized gene, hmsT.
All Crb− (or Pstr) mutants of the RecA+ KIM10+ strain appeared to result from a pgm locus deletion, similar to previous observations (35). These data confirmed the original observation linking the phenotypes of colony pigmentation and pesticin sensitivity (2) and indicated that a RecA-mediated deletion of the pgm locus is the main source of Crb mutants in RecA+ strains of Y. pestis. Point mutations or small lesions in hmsHFRS and hmsT probably also occur in RecA+ strains, and hmsHFRS mutations have been observed by others (4, 24). However, the RecA-independent mutations we observed occurred 40-fold less frequently than the pgm locus deletions in the RecA+ KIM10+ strain and would be readily masked by the presence of the high-frequency pgm locus deletions.
While a RecA-dependent deletion of the pgm locus was expected to be the predominant mechanism of generating Crb− mutants, we observed that RecA-independent mutations in the hmsHFRS and hmsT loci also occurred at higher frequencies than spontaneous mutation frequencies to antibiotic resistance. Another study also observed losses of the Crb+ phenotype independent of a pgm locus deletion in a collection of Y. pestis strains (20). However, most of these Crb− mutants in the previous study had deletions of the hmsHFRS locus, probably mediated by homologous recombination (4). Furthermore, these mutants were from different isolates in a Y. pestis strain collection and thus represent a range of possible Crb− mutants that can occur and become fixed over time in different Y. pestis populations (4). In contrast, our data were gathered from the events occurring in a particular RecA+ or RecA− Crb+ strain over a 24-h growth period and so directly measure the frequency of Crb− mutants arising in one homogenous population.
The high number of Crb− mutants arising in YPRA was unexpected, given that the levels of spontaneous mutation to Pstr dropped to background antibiotic resistance mutation levels in this strain. The reasons for the higher loss of the Crb− phenotype are not clear. The mutation targets of the hmsHFRS and hmsT loci (8.1 kbp) (reference 24 and this study) are larger than those of the psn gene (2.1 kbp) (10) and could contribute to the unequal mutation frequencies we observed. However, the fourfold difference in the sizes of mutation targets is probably not sufficient to result in a 100-fold increase in the Crb− mutant frequency over background drug resistance levels.
An alternate explanation for this result is the differing amounts of selective pressures that are present when the mutation frequencies of specific genes are measured. The antibiotic resistance assays typically used to calculate spontaneous mutation rates use gene targets that are under selective pressure in the bacterial cell (39). The assays we used, which tested resistance to streptomycin and rifampin, require an altered but functional ribosome structure and RNA polymerase β subunit, respectively (39). Therefore, only a limited number of mutations can be tolerated by these genes and still result in a functional protein. In contrast, genes whose functions are not required for the growth conditions of the assay have fewer restrictions on the types and numbers of mutations they may maintain and may behave as unselected DNA sequences (37).
Mutation frequencies observed for the hmsHFRS and hmsT genes are similar to those reported for E. coli genes in the absence of selective pressure (10−5) (37). The hmsHFRS genes are required for blockage of Y. pestis’ flea vector (19) but not for the pathogenesis of bubonic plague in mammals (23). We observed no difference in the viabilities of Crb+ Y. pestis when grown on CR agar compared to BHI agar (data not shown). Similarly, the apparent size difference in Crb+ versus Crb− colonies was likely due to a difference in colony morphology (Crb+ colonies are wrinkled and domed, while Crb− colonies are flat), rather than the growth inhibition of Crb+ colonies on CR agar. Taken together, these observations suggest that these genes are selected neither for nor against under laboratory growth conditions. Therefore, the seemingly high Crb− mutation frequency in hmsT and hmsHFRS would be predicted by the “selective pressure” argument.
In contrast, the frequency of mutations to Pstr was similar to what we observed with antibiotic resistance markers that are under selective pressure. This result is more difficult to explain in the absence of a known selection for Psn under standard laboratory conditions. Iron acquisition is not likely to be the cause of the selection, as iron is not limiting under laboratory conditions. It is possible that another, unknown function of Psn could limit the accumulation of mutations in psn in the presence of an intact pgm locus.
Role of IS100 in pgm locus deletions.
A deletion of the entire pgm locus likely results from frequent RecA-mediated homologous recombination occurring between the two directly repeated IS100 copies flanking the pgm locus, as predicted from earlier observations (12). The observation that the IS100 sequence formed the exact endpoints of the deletion and the RecA dependence of this deletion process support this conclusion. Previous reports of multiple Y. pestis strains having the same, approximately 102-kbp chromosomal deletion event (12) suggest that the IS100-flanked structure of the pgm locus is highly conserved in Y. pestis strains (4, 12). Other homologous recombination-mediated deletions and inversions may also be occurring elsewhere in the genome at similar frequencies but may not be evident without the loss of an easily observed phenotype such as CR binding.
One YPRA mutant (YPRA11) lacked the entire pgm locus. As RecA is thought to be required for all recombination pathways (26) it is unlikely that this deletion resulted from recombination. An alternate explanation is that this deletion was caused by a transposon-mediated intramolecular deletion event.
A survey of Y. pestis strains collected from clinical cases worldwide found that some Crb− strains had undergone partial deletions of the pgm locus (4, 20), which were either flanked by additional copies of IS100 within the pgm locus or due to undefined causes (4). In contrast, we found no partial deletions of the pgm locus involving the hmsHFRS genes. Presumably this reflects the presence of additional copies of repetitive elements within the pgm locus in these different strains. KIM10+ possesses no additional copies of IS100 within its pgm locus (12).
A second locus is required for the Crb+ phenotype.
By investigating the location of RecA-independent mutations resulting in the Crb− phenotype, we identified a gene (hmsT [22]) that restored the Crb+ phenotype to one class of Y. pestis Crb− mutants. The combined presence of hmsT and the hmsHFRS genes was both necessary and sufficient to confer the Crb+ phenotype upon the normally nonpigmented E. coli and Y. pseudotuberculosis strains (Fig. 6). These results suggest that both of these gene loci are nonfunctional or absent in these species and are consistent with a previous report that E. coli does not become Crb+ when transformed with the hmsHFRS genes alone (28). The changes in colony morphology and growth in liquid culture that occurred in the Crb+ E. coli and Y. pseudotuberculosis strains suggest that the hmsHFRS and hmsT genes contribute to the similar wrinkled colony morphology on plates and stalactite-like growth in liquid culture that is seen in Y. pestis (17, 21). However, unlike in Y. pestis, the Crb+ phenotype in E. coli and Y. pseudotuberculosis was temperature independent. This suggests that another gene, which is present in Y. pestis but absent or nonfunctional in E. coli and Y. pseudotuberculosis, provides the temperature dependence of the Crb+ phenotype. Alternately, it is possible that the temperature-dependent expression of the Crb+ phenotype in Y. pestis could be due to a masking of the heme receptor on the cell surface at 37°C. Both hypotheses are consistent with the observation that the Crb+ phenotype of Pgm− Y. pestis cells remains temperature dependent in the presence of multiple copies of hmsT and hmsHFRS on the plasmids pHMSXB and pHMS1, respectively (data not shown), excluding a simple multicopy effect.
The Crb− Y. pseudotuberculosis strains that we examined contained DNA sequences that hybridized with DNA probes derived from hmsT and hmsHFRS. However, these strains showed no evidence of rearrangements, insertions, or deletions in these loci (Fig. 3 and data not shown), suggesting that the Crb− phenotype of this species may be due to an accumulation of point mutations or small lesions in these genes. As a flea vector does not transmit Y. pseudotuberculosis, these genes may not be selected in this species and may behave as noncoding DNA sequences.
The specific contribution of the hmsT gene product to the Crb+ phenotype and the function of its DUF9 domain, which has been found in bacterial response regulator proteins, are not yet known. The requirement for hmsT in the Crb+ phenotype may indicate a role for it in the flea transmission stage of plague, as is the case for the hmsHFRS genes (19). hmsT is present in the Pgm− strain YPRA11, indicating that it lies outside the pgm locus, and thus is separated from the hmsHFRS genes by at least 15 kbp (11).
In summary, we demonstrated that both RecA-dependent and -independent mechanisms cause the high-frequency loss of the Crb+ phenotype that occurs in Y. pestis. A RecA-mediated deletion of the pgm locus caused the majority of these mutants. However, the RecA-independent mutations in the hmsHFRS and hmsT genes occurred at a frequency 100-fold higher than similar mutations in psn. We propose that this lack of uniformity in mutation frequencies at distinct but genetically linked loci may reflect a smaller amount of selection for the hms genes relative to psn under in vitro conditions. Future studies with additional genes may help clarify the extent to which mutation frequencies can predict gene function under different environmental conditions.
ACKNOWLEDGMENTS
This work was supported by New Investigator Funds to K.A.M. from the Wadsworth Center and National Science Foundation predoctoral fellowship GER9353931 to J.M.H.
We thank the Charles Radding laboratory, for providing us with the anti-E. coli RecA antibody and alkaline phosphatase Western blotting procedure, and the Ontario Ministry of Health Department, for serotyping the Y. pseudotuberculosis isolates. We also thank the Wadsworth Center Molecular Genetics Core Facility, for the synthesis of oligonucleotides and DNA sequencing services, and the photography and illustration units of the Wadsworth Center, for support services. We are also grateful to Keith Derbyshire, Richard Lease, and Dilip Nag for their helpful comments on this manuscript and to Robert Perry for helpful discussions and for sharing unpublished information. Sequence data from the Sanger Y. pestis genome database were produced by the Yersinia pestis Sequencing Group at the Sanger Centre (40).
REFERENCES
- 1.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker R R. Mutation rate to nonpigmentation in Pasteurella pestis. J Bacteriol. 1969;98:1404–1406. doi: 10.1128/jb.98.3.1404-1406.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 4.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrows T W, Jackson S. The virulence enhancing effect of iron on non-pigmented mutants of virulent strains of Pasteurella pestis. Br J Exp Pathol. 1956;37:577–583. [PMC free article] [PubMed] [Google Scholar]
- 6.Cox E C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:133–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- 7.Dennis D T, Meier F A. Plague. In: Horsburgh C R Jr, Nelson A M, editors. Pathology of emerging infections. Washington, D.C: ASM Press; 1997. pp. 21–47. [Google Scholar]
- 8.Derbyshire K M, Hwang L, Grindley N D F. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetherston J D, Lillard J W, Jr, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 12.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 13.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehring A M, DeMoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 15.Gemski P, Lazere J R, Casey T. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect Immun. 1980;27:682–685. doi: 10.1128/iai.27.2.682-685.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray L D. Escherichia, Salmonella, Shigella, and Yersinia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 450–456. [Google Scholar]
- 17.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–304. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 18.Hecht G B, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 20.Iteman I, Guiyoule A, de Almeida A M P, Guilvout I, Baranton G, Carniel E. Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect Immun. 1993;61:2717–2722. doi: 10.1128/iai.61.6.2717-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson S, Burrows T W. The pigmentation of Pasteurella pestis on a defined medium containing hemin. Br J Exp Pathol. 1956;37:570–576. [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, H. A., J. W. Lillard, Jr., and R. D. Perry. HmsT, a protein essential for expression of the hemin storage (Hms+) phenotype of Yersinia pestis. Microbiology, in press. [DOI] [PubMed]
- 23.Lillard J W, Jr, Bearden S W, Fetherston J D, Perry R D. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology. 1999;145:197–209. doi: 10.1099/13500872-145-1-197. [DOI] [PubMed] [Google Scholar]
- 24.Lillard J W, Jr, Fetherston J D, Pedersen L, Pendrak M L, Perry R D. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 25.Lucier T S, Brubaker R R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahajan S K. Pathways of homologous recombination in Escherichia coli. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C: ASM Press; 1988. pp. 87–140. [Google Scholar]
- 27.McDonough K A, Hare J M. Homology with a repeated Yersinia pestis DNA sequence IS100 correlates with pesticin sensitivity in Yersinia pseudotuberculosis. J Bacteriol. 1997;179:2081–2085. doi: 10.1128/jb.179.6.2081-2085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendrak M L, Perry R D. Characterization of a hemin-storage locus of Yersinia pestis. Biol Metals. 1991;4:41–47. doi: 10.1007/BF01135556. [DOI] [PubMed] [Google Scholar]
- 29.Perry R D, Pendrak M L, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sikkema D J, Brubaker R R. Resistance to pesticin, storage of iron, and invasion of HeLa cells by yersiniae. Infect Immun. 1987;55:572–578. doi: 10.1128/iai.55.3.572-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikkema D J, Brubaker R R. Outer membrane peptides of Yersinia pestis mediating siderophore-independent assimilation of iron. Biol Metals. 1989;2:174–184. doi: 10.1007/BF01142557. [DOI] [PubMed] [Google Scholar]
- 33.Sodeinde O A, Goguen J D. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect Immun. 1988;56:2743–2748. doi: 10.1128/iai.56.10.2743-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnhammer E L, Eddy S R, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 35.Staggs T M, Fetherston J D, Perry R D. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol. 1994;176:7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surgalla M J, Beesley E D. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torkelson J, Harris R S, Lombardo M, Nagendran J, Thulin C, Rosenberg S M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Une T, Brubaker R R. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willett H P. Antimicrobial agents. In: Joklik W K, Willett H P, Amos D B, Wilfert C M, editors. Zinsser microbiology. Norwalk, Conn: Appleton & Lange; 1988. pp. 128–160. [Google Scholar]
- 40.Yersinia pestis Sequencing Group. 23 November 1998, posting date. Y. pestis genome database. [Online.] Sanger Centre, Cambridge, England. http://www .sanger.ac.uk/Projects/Y_pestis/. [25 November 1998, last date accessed.]