Abstract
Porphyromonas gingivalis, an important periodontal disease pathogen, forms black-pigmented colonies on blood agar. Pigmentation is believed to result from accumulation of iron protoporphyrin IX (FePPIX) derived from erythrocytic hemoglobin. The Lys-X (Lys-gingipain) and Arg-X (Arg-gingipain) cysteine proteases of P. gingivalis bind and degrade erythrocytes. We have observed that mutations abolishing activity of the Lys-X-specific cysteine protease, Kgp, resulted in loss of black pigmentation of P. gingivalis W83. Because the hemagglutinating and hemolytic potentials of mutant strains were reduced but not eliminated, we hypothesized that this protease played a role in acquisition of FePPIX from hemoglobin. In contrast to Arg-gingipain, Lys-gingipain was not inhibited by hemin, suggesting that this protease played a role near the cell surface where high concentrations of hemin confer the black pigmentation. Human hemoglobin contains 11 Lys residues in the α chain and 10 Lys residues in the β chain. In contrast, there are only three Arg residues in each of the α and β chains. These observations are consistent with human hemoglobin being a preferred substrate for Lys-gingipain but not Arg-gingipain. The ability of the Lys-gingipain to cleave human hemoglobin at Lys residues was confirmed by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry of hemoglobin fragments resulting from digestion with the purified protease. We were able to detect several of the predicted hemoglobin fragments rendered by digestion with purified Lys-gingipain. Thus, we postulate that the Lys-gingipain of P. gingivalis is a hemoglobinase which plays a role in heme and iron uptake by effecting the accumulation of FePPIX on the bacterial cell surface.
Iron is an indispensable nutrient for growth of most living organisms. In humans, most of the iron is present intracellularly as hemoglobin (76%) and ferritin (23%). Extracellular iron is rapidly bound by transferrin in serum and lactoferrin at mucosal surfaces (38). Because the in vivo concentration of free iron is too low (10−18 M) to support the growth of microorganisms (20), specialized iron acquisition mechanisms are needed for bacterial colonization of the human host. Many pathogenic bacteria produce siderophores capable of chelating iron from transferrin and lactoferrin (8). Also, in Neisseria gonorrhoeae siderophore-independent acquisition of iron from transferrin and lactoferrin through direct binding to a specific receptor has been observed (33).
Hemoglobin, the largest reservoir of iron in the human adult (38), is a tetramer composed of two α and two β polypeptide chains. Each polypeptide chain has a noncovalently bound heme group consisting of a porphyrin ring and a ferrous atom. The heme group is another nutrient used by bacteria as a cofactor for cytochromes and catalase (52, 56). However, heme and heme-associated iron are not normally available to pathogenic organisms. Hemoglobin released by lysis of erythrocytes is rapidly bound by haptoglobin and transported to the liver. Also, heme released from hemoglobin is bound by hemopexin. Although several gram-negative bacteria such as Neisseria species, Haemophilus influenzae, Vibrio species, Bacteroides fragilis, and Escherichia coli are known to use heme and heme iron from hemoglobin (38, 39), the mechanisms of heme extraction from hemoglobin remain poorly understood.
Porphyromonas gingivalis, an obligately anaerobic bacterium, is recognized as an etiologic agent of adult periodontitis (29, 46, 47). It forms black-pigmented colonies resulting from accumulation of hemin (oxidized form of heme) on the cell surface and within the bacterial cell when grown on blood agar (44, 50). Hemin has been shown to be an important supplement for P. gingivalis growth (19, 44). Since this organism is unable to synthesize protoporphyrin IX (42) and siderophores have not been reported in this organism (2, 16), hemin may serve a dual nutritional function as an iron and protoporphyrin IX source. At the site of infection, hemoglobin derived from lysed erythrocytes abundant in periodontal pockets is the most probable source of this compound. Although P. gingivalis is capable of utilizing various hemin-containing compounds (2, 16), Shizukuishi et al. have shown that this organism utilizes hemoglobin more efficiently than other iron-containing compounds (45).
The mechanism of hemin acquisition from erythrocytes involves hemagglutination, hemolysis, binding, and degradation of the hemoglobin molecule. P. gingivalis produces large quantities of cysteine proteases with either arginine (Arg-gingipain; rgpA and rgpB genes) or lysine (Lys-gingipain; kgp gene) specificity. The hemagglutinin/adhesin domain of these enzymes, which is similar to the P. gingivalis HagA hemagglutinin (21), has been shown to possess hemagglutinin activities (9, 40). P. gingivalis is also able to lyse erythrocytes (5). Although Arg-gingipain has been observed to possess hemolytic activity (43), the role of Lys-gingipain in hemolysis remains unknown. Recently, a hemoglobin binding protein called HbR, intragenically encoded by the rgpA, kgp, and hagA genes, has been identified (35). In addition, the Lys-gingipain has shown hemoglobin binding activity (28). Last, although the hemoglobinolytic potential of P. gingivalis has been demonstrated (14, 45), the molecular mechanisms of acquiring hemin and iron from hemoglobin have not been elucidated.
We have constructed mutants in Lys-gingipain gene, kgp, of P. gingivalis W83 (30). Using the mouse abscess model, we demonstrated that Kgp played a role in virulence of P. gingivalis (30). We have since observed that when grown on blood agar plates these mutants appear white, suggesting a major role of this protease in acquisition of hemin from erythrocytes. Because neither the hemagglutinating nor the hemolytic potentials of the mutant strains were totally eliminated, we reasoned that degradation of hemoglobin resulting in release of hemin might be involved. To test this hypothesis, we have compared the hemoglobinolytic activities of the mutant and parent strains. These results have been confirmed in studies using a P. gingivalis strain with restored Kgp activity and purified Kgp protein. Our results indicate that the Lys-gingipain is a hemoglobinase. This observation suggests that this enzyme plays a role in heme and iron acquisition by efficient extraction of iron protoporphyrin from hemoglobin that in turn is accumulated on the bacterial cell surface.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. gingivalis strains used in this study are described in Table 1. W83 was obtained from H. A. Schenkein, VCU Clinical Research Center for Periodontal Diseases. P. gingivalis V2543 was a naturally occurring variant of P. gingivalis W83 carrying an insertion sequence (IS)-like element, IS195 inserted into prtP (30). However, we now have abandoned this gene designation in favor of kgp, indicating the conserved lysine-specific cysteine protease (Lys-gingipain) typically found in P. gingivalis (10). We use that genotypic designation hereafter in this report, with Kgp used to designate the Lys-gingipain protease. P. gingivalis V2577 was an allelic exchange mutant of W83 carrying the ermF-ermAM cassette inserted into the prepropeptide domain of the kgp gene (30). P. gingivalis V2602 was constructed in this study by restoration of the V2543 mutation via recombination (Fig. 1). V2383 was a double mutant (RgpA and Kgp deficient), V2448 was a HagA mutant, and V2546 was an RgpA mutant.
TABLE 1.
Strains of P. gingivalis used in this study
| Strain | Characteristics | Comment |
|---|---|---|
| W83 | RgpA+ Kgp+ | Wild type |
| V2383 | RgpA− Kgp− | Allelic exchange mutanta in rgpA, IS195 insertional mutant in kgp (30) (unpublished data) |
| V2448 | RgpA+ Kgp+ HagA− | Allelic exchange mutanta in hagA (unpublished data) |
| V2543 | RgpA+ Kgp− | IS195 insertional mutant in kgp (30) |
| V2546 | RgpA− Kgp+ | Allelic exchange mutanta in rgpA (unpublished data) |
| V2577 | RgpA+ Kgp− | Allelic exchange mutant in kgp (30) |
| V2602 | RgpA+ Kgp+ | V2543 with restored kgp gene constructed in this study |
Constructed by using the ermF-ermAM cassette described and used as previously published (13).
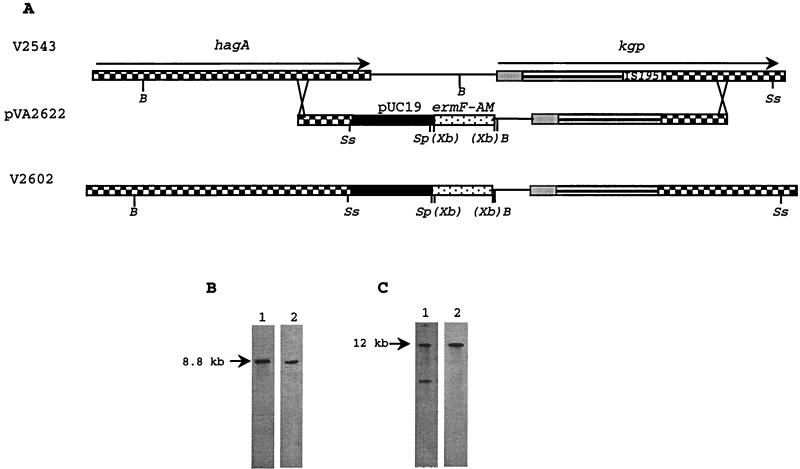
FIG. 1.
Restoration of the kgp gene in P. gingivalis V2543. (A) Maps of hagA and kgp on the chromosome of V2543. The hagA and kgp open reading frames are indicated by rightward arrows. Different regions of the kgp gene encoding different domains are indicated by various patterns: prepropeptide (grey), protease domain (parallel lines), and hemagglutinin domain (checkerboard). Shadings and patterns indicate similar sequences. Relevant restriction sites: B, BamHI; Xb, inactive XbaI; Sp, SphI; Ss, SstI. (B) Southern blot analysis of SstI/SphI-digested chromosomal DNA of V2602, probed with the kgp fragment encoding the prepropeptide domain (lane 1) and ermF-ermAM cassette (lane 2). (C) Southern blot analysis of BamHI-digested chromosomal DNA of V2602 probed with a 1.2-kb fragment encoding hemagglutinin (lane 1) and ermF-ermAM cassette (lane 2).
E. coli TB1 and pUC18 were used in all cloning work. pVA2622 was a suicide vector constructed by cloning of the ermF-ermAM cassette into XbaI site of a recombinant plasmid consisting of pUC18 and 6.8-kb fragment carrying the entire kgp gene from P. gingivalis W12 (obtained from A. Progulske-Fox, Department of Oral Biology, University of Florida, Gainesville) (Fig. 1). The kgp gene from P. gingivalis W12 was 99.9% identical to the kgp homolog from P. gingivalis W83 (28).
Growth media and conditions.
P. gingivalis was grown in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) supplemented with hemin (5 μg/ml), vitamin K3 (0.5 μg/ml), and cysteine (1%). Agar (2% [wt/vol]) was added when solid medium was required. Cultures were incubated at 37°C in an anaerobic chamber (Coy Manufacturing, Ann Arbor, Mich.) in 80% N2–10% CO2–10% H2. To evaluate black pigment formation, P. gingivalis strains were inoculated onto blood agar plates and incubated for 4 days anaerobically at 37°C and then for 10 days anaerobically at room temperature. E. coli strains were cultivated aerobically at 37°C in Luria-Bertani (LB) broth (Gibco, BRL Inc., Gaithersburg, Md.). Solid LB medium was obtained by addition of 2% (wt/vol) agar. Antibiotic concentrations used for selection were 0.5 μg/ml for clindamycin (P. gingivalis) and 300 μg/ml for erythromycin (E. coli).
Restoration of Kgp activity in Kgp mutant.
The genetic strategy for restoration of the kgp defective gene is depicted in Fig. 1. Briefly, recombinant pUC18 carrying the ermF-ermAM cassette and the entire kgp gene was electroporated into P. gingivalis V2543 (carrying the inactivated kgp gene due to insertion of IS195) (30) according to the protocol of Fletcher et al. (13). Clindamycin-resistant colonies testing positive for Lys-gingipain activity were analyzed by Southern blotting to verify the presence and correct chromosomal location of the intact kgp gene.
Protein preparation.
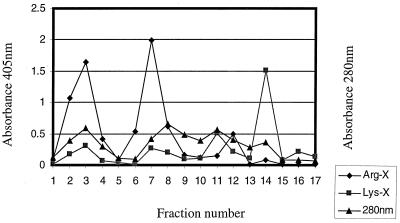
Extracellular membrane vesicles were prepared as described previously (30). Partially purified Kgp protease from V2546 was prepared by a combination of ammonium sulfate saturation followed by a hemoglobin affinity column with the buffer system of Fujimura et al. (15). Bacteria (2 liters) were grown for 72 h and harvested by centrifugation (10,000 × g, 30 min, 4°C). Extracellular vesicles and proteins present in the culture supernatant were precipitated by addition of ammonium sulfate (40 to 60% saturation), harvested by centrifugation (20,000 × g, 40 min, 4°C), suspended in 120 ml of Tris-HCl buffer (50 mM, pH 9.5) and dialyzed overnight against the same buffer. Vesicular proteins were harvested by centrifugation (15,000 × g, 40 min, 4°C) and stored in 20 ml of sodium acetate buffer (50 mM, pH 5.5). Vesicular proteins were solubilized by addition of octylthioglucoside (OTG) (54) to a final concentration of 0.6% followed by overnight gentle mixing at 4°C. The mixture was centrifuged at 150,000 × g for 1 h to remove insoluble material, and the supernatant was applied to hemoglobin-conjugated agarose (Sigma Chemical Co., St. Louis, Mo.) column (1 by 5 cm) equilibrated with 50 mM sodium acetate buffer (pH 5.5) containing 0.6% OTG. The column was washed with 3 volumes of the same buffer followed by a wash with 50 mM sodium phosphate buffer (pH 7.2) containing 300 mM sodium chloride and 0.6% OTG. Hemoglobin-bound proteins were eluted with 50 mM Tris-HCl buffer (pH 9.0) containing 0.6% OTG. Affinity column fractions (5 ml each) were analyzed for protein content by monitoring absorbance at 280 nm (A280). Protease activity was determined with the chromogenic substrates 1 mM N-α-benzoyl-dl-arginine-p-nitroanilide (BApNA) (Sigma) and 1 mM N-α-benzyloxycarbonyl-l-lysine-p-nitroanilide (Z-Lys-pNA) (Novabiochem, La Jolla, Calif.) in 50 mM Tris-HCl buffer–10 mM l-cysteine–1 mM CaCl2 (for Arg-X activity) and pH 7.5 (for Arg-gingipain activity) or 8.0 (for Lys-gingipain activity); 20 μl of eluted fractions and 10 μl of starting material were used in enzyme activity assays (Fig. 5).
FIG. 5.
Hemoglobin-agarose chromatography. The 40 to 60% ammonium sulfate fraction from P. gingivalis culture supernatant solubilized with 0.6% (wt/wt) OTG was subjected to hemoglobin-agarose chromatography. Fractions (5 ml) were collected and assayed for protein content (A280) and protease activity (A405) as described in Materials and Methods.
Protease activity.
Extracellular vesicular protease activity was assayed as described previously (30). Reaction mixtures of 500 μl containing 36 μg of extracellular vesicular proteins were incubated at 37°C in either 50 mM Tris-HCl (pH 7.2)–1 mM BApNA in the presence of 2 mM dithiothreitol (DTT) or 50 mM Tris-HCl (pH 8.0)–1 mM Z- Lys-pNA in the presence of 2 mM DTT. Hydrolysis was measured by monitoring A405. The effect of hemin and imidazole on protease activity was determined by using 10 μg of extracellular vesicular proteins in reaction mixture as described above supplemented with either 15 μM hemin or 20 mM imidazole.
Lysis of erythrocytes.
Sheep defibrinated blood obtained from BBL (Becton Dickinson Microbiology Systems, Cockeysville, Md.) was washed in phosphate-buffered saline (PBS) until the supernatant was free of red pigment. Bacterial cells were also washed in PBS and adjusted to an optical density at 660 nm (OD660) of 1. Vesicles were prepared as described above. Reaction mixtures were prepared by mixing 1 ml of 2.5% (vol/vol in PBS) sheep erythrocytes containing 2 mM DTT with 50 μl of bacterial cells (OD = 1) or 36 μg of vesicular proteins. After 8 and 15 h of incubation with gentle mixing at 37°C, intact erythrocytes were removed by centrifugation 10,000 × g for 5 min. The hemolytic activity was determined by monitoring absorbance for red pigment released from lysed cells at 520 nm.
Hemagglutination assays.
Hemagglutination activity was determined by using bacterial cells (50 μl, OD = 1) and extracellular vesicles (18 μg) as described previously (4).
Hemoglobinase activity.
For assay of hemoglobinase activity of vesicular proteins, 500-μl reaction mixtures containing 72 μg of extracellular proteins and 4 μg of bovine hemoglobin (Sigma) per μl (2 mg) in 50 mM Tris-HCl (pH 8.0) were incubated 15 h at 37°C in the presence of 2 mM DTT; 20 μl of this mixture was added to an equal volume of reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (Laemmli sample buffer; Bio-Rad, Hercules, Calif.), boiled for 10 min, and electrophoresed on a 15% polyacrylamide gel. Hemoglobin fragments were visualized by staining with Coomassie blue (0.2% Coomassie blue R-250 in destaining solution) and destaining in destaining solution (45% ethanol, 10% actetic acid). For assay of hemoglobinase activity of purified Kgp protease, a 200-μl reaction mixture containing 150 μg of pure Kgp protease and 600 μg of human hemoglobin (Sigma) in 50 mM Tris-HCl (pH 8.0) was incubated for 15 h at 37°C in the presence of 2 mM DTT; 15 μl of this mixture was analyzed on a 15% polyacrylamide gel. Proteolysis of hemoglobin was visualized as described above.
Microdialysis.
The development of a microdialysis approach to prepare oligonucleotides, peptides, and proteins has been reported elsewhere in detail (23) and is described here briefly. The protein digests contained >10 mM Tris-HCl as well as additional nonvolatile, monatomic ionic species (e.g., Na+). To ensure that the sample was compatible with the electrospray ionization (ESI) process, a rapid (<3 min), single-step microdialysis approach was used. The microdialysis system employed a 13,000-molecular-weight-cutoff (MWCO) conical cellulose acetate dialysis fiber with a countercurrent buffer of 10 mM ammonium acetate with a gravity-induced flow rate of 1.2 ml/min (23). Typically, 5 μl of sample was injected into the microdialysis system at a flow rate of 2 μl/min; desalting efficiencies greater than 99.9% were routinely achieved with the dialysate collected in a microcentrifuge tube (23).
Mass spectrometry.
Mass spectrometric data for hemoglobin fragments rendered by digestion with purified Kgp were obtained using an ESI (12)-Fourier transform ion cyclotron resonance (FTICR) mass spectrometer (7) modified to accept micro- and nanoelectrospray sources. Electrospray emitters were 50-μm fused-silica capillaries pulled to ca. 10 to 20 μm as previously reported (22). Electrosprayed ions entered into the dielectric capillary of the electrospray ionization source (Analytica of Branford, Inc., Branford, Conn.) coupled to an ESI-FTICR mass spectrometer (Ionspec, Irvine, Calif.). The Ionspec Omega 586 data station was used for processing and signal generation, and the broad-band pulse sequence was employed. The instrument utilized a 4.7-T horizontal bore superconducting magnet with a 128-mm bore (Cryomagnetics, Inc., Oak Ridge, Tenn.). Mass calibration of the FTICR was accomplished by using the isotopically resolved multiply charged ions of bovine ubiquitin as the external standard. The calibration parameters were subsequently verified by analysis of the 5+ charge state of bovine insulin, which resulted in a mass error of less than 10 ppm (0.001%). The dialyzed samples were microsprayed from a solvent of 49/49/2 (vol %) methanol–10 mM ammonium acetate–acetic acid with a flow rate of 0.5 μl/min. Ions were injected for 2 s, and all spectra obtained were single acquisitions.
RESULTS
Restoration of the defective kgp gene.
To determine whether inabilities to both accumulate black pigment and degrade hemoglobin resulted from inactivation of the kgp gene, we restored the Kgp activity in Kgp-deficient mutant. Two types of Kgp mutants (V2543 and V2577) used in this study were described and characterized previously (28). While V2543 was a naturally occurring mutant resulting from insertion of the IS-like element IS195 into the kgp gene, V2577 was an allelic exchange mutant of the kgp gene of P. gingivalis W83 constructed by insertion of the ermF-ermAM gene cassette into the prepropeptide region of kgp. The genetic structure of V2543 mutant is shown in Fig. 1A. We chose the Kgp mutant V2543 as a starting point for the restoration of kgp gene because it contained neither suicide vector nor resistance cassette integrated into the chromosome.
Following electroporation of P. gingivalis V2543 with a suicide vector containing an intact copy of the kgp gene and ermF-ermAM gene cassette, pVA2622, six clindamycin-resistant colonies were detected after a 10-day incubation period on brain heart infusion plates containing clindamycin. Only one of the six colonies exhibited Kgp activity. This colony was characterized by Southern blot analysis, and the hybridization pattern of SphI-SstI-digested genomic DNA probed with a fragment of kgp encoding for the propeptide sequence revealed the presence of a complete copy of the kgp gene (Fig. 1A and B, lane 1). These results were confirmed by probing the same blot with the ermF-ermAM cassette (Fig. 1A and B, lane 2) (13). Further analyses using V2602 BamHI-digested genomic DNA probed with a 1.2-kb fragment internal to hagA and the ermF-ermAM cassette indicated that the insertion of the suicide vector pVA2622 occurred by a double crossover between the hemagglutinin-encoding portion of the kgp gene present in the suicide vector and the homologous sequences present in kgp and hagA genes resident on the chromosome of V2543 (Fig. 1A and C). The recombination that regenerated the kgp gene in V2602 was evidently accompanied by a 3-kb deletion upstream of the kgp sequence (Fig. 1A). This deletion was replaced by ermF-ermAM gene and pUC18 sequence present in the suicide vector. This event may explain the decreased Lys-X proteolytic activity of the strain with restored kgp gene compared to the wild-type strain.
Growth of P. gingivalis strains on blood agar plates.
P. gingivalis grown on blood agar plates forms black colonies after several days of incubation. This black colony pigmentation has been shown to be a result of accumulation of hemin derived from erythrocytes on a surface and within bacterial cells (44, 50). All strains tested were able to grow on blood agar plates; however, mutants containing a disrupted kgp gene lost the ability to accumulate black pigment (Fig. 2). This ability was restored in V2602 strain containing the restored kgp gene (Fig. 2).
FIG. 2.
Accumulation of black pigment by P. gingivalis strains grown on blood agar. P. gingivalis strains were inoculated onto blood agar medium and incubated for 4 days anaerobically at 37°C followed by 10 days anaerobically at room temperature. Examples of the two colonial phenotypes are shown. To obtain the photograph, P. gingivalis cultures of W83 and V2577 were mixed, diluted, and spread on blood agar plates. Well-isolated colonies thus allowed the simultaneous viewing of both pigment phenotypes. Black-pigmented colonies were formed by the wild-type strain (W83). The mutation abolishing the Lys-gingipain activity (V2577) resulted in loss of pigmentation (nonpigmented colony in the photograph). Strains examined in this study are described at the right.
Protease activity.
Both cells and extracellular vesicles were assayed for protease activity. Examination of extracellular vesicles of P. gingivalis strains for protease activity revealed that Lys-gingipain activity was sixfold lower in both V2543 and V2577 than in the wild-type strain (Table 2). This activity was largely restored (70% of wild-type activity) in the strain V2602 containing the restored kgp gene. The Lys-gingipain activity of cells of the Kgp-deficient strains (V2543 and V2577) was fivefold lower than that of the parental W83 and V2602 strains (Table 2). While Arg-gingipain activities were similar for all strains tested, the activity of extracellular vesicles was reduced in the insertion mutant (V2543) and in the strain with restored Lys-gingipain activity (V2602) (results not shown).
TABLE 2.
Lys-gingipain proteolytic activities of extracellular vesicles and cells of P. gingivalis strains
| Strain |
A405a
|
|
|---|---|---|
| Vesicles | Cells | |
| W83 | 2.8, 3.0 | 1.0, 1.0 |
| V2543 | 0.5, 0.5 | 0.2, 0.2 |
| V2577 | 0.5, 0.5 | 0.2, 0.2 |
| V2602 | 2.1, 2.2 | 0.9, 0.9 |
Proteolytic activity was measured spectrophotometrically by monitoring the release of p-nitroanilide resulting from hydrolysis of Z-Lys-pNA detectable at A405. Results for two obtained in two independently performed assays are presented.
Effect of hemin and imidazole on proteolytic activity.
Production of white colonies by Kgp-deficient strains indicated that there was a link between Kgp and accumulation of hemin from erythrocytes present on blood agar plates. This prompted our interest in the effect of hemin on Kgp activity. We believed that there may be some hemin present in vesicular preparations and so used imidazole, which interacts with the iron atom present in the hemin molecule, thus distorting the hemin structure (1, 57) and resulting in loss of its ability to bind protein. Arg-gingipain activity was lower in the presence of hemin and higher in the presence of imidazole compared to the control reaction (Table 3). These results indicated the inhibitory effect of hemin on Arg-gingipain and confirmed the results of U et al. (55). Lys-X activity was not affected by the presence of either hemin or imidazole (Table 3).
TABLE 3.
Effects of hemin and imidazole on protease activity of extracellular vesicles of P. gingivalis W83
| Addition to reaction |
A405a
|
|||||
|---|---|---|---|---|---|---|
| Arg-gingipain activity
|
Lys-gingipain activity
|
|||||
| 15 min | 30 min | 60 min | 15 min | 30 min | 60 min | |
| None | 0.4 | 0.6 | 0.9 | 0.5 | 0.8 | 1.2 |
| Hemin | 0.1 | 0.1 | 0.1 | 0.5 | 0.7 | 1.2 |
| Imidazole | 1.7 | 2.2 | 2.6 | 0.5 | 0.8 | 1.2 |
Protease activity was monitored by change of A405 resulting from release of p-nitroanilide from chromogenic substrates.
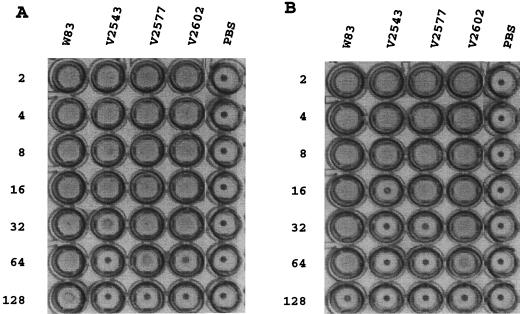
Hemagglutination activity.
Since spontaneous mutants deficient in the ability to attach to erythrocytes also have shown loss of pigmentation (4), we wondered whether similar results would be obtained for our Kgp-deficient strains. P. gingivalis W83, V2543, V2577, and V2602 were assessed for hemagglutination ability. The hemagglutinating activity of cells of the strain containing the restored kgp gene, V2602, was comparable to that of the wild type. However, the Kgp-deficient strains showed significantly reduced hemagglutination activity (eightfold for V2543 and fourfold for V2577 [Fig. 3A]). The activity of extracellular vesicles of the wild-type strain exceeded that of the insertion mutant strain V2543 (fourfold) and the complemented strain V2602 (fourfold) but was similar to that of the allelic exchange mutant V2577 (Fig. 3B).
FIG. 3.
Hemagglutinating activities of P. gingivalis strains. Hemagglutinating activities of 18 μg of extracellular vesicles (A) and 50 μl of cells (B) were analyzed as described in Materials and Methods. The reciprocals of serial twofold dilutions are shown at the left; the strains tested are given at the top. PBS was used as a control.
Hemolytic activity.
Hemolytic activities of extracellular vesicles and cells of the Kgp-deficient mutants V2543 and V2577 was lower than hemolytic activities of vesicles and cells of the wild-type strain (Table 4). Hemolytic activity was partially restored in cells of the P. gingivalis strain with a restored kgp gene (V2602) (Table 4).
TABLE 4.
Hemolytic activities of extracellular vesicles and cells of P. gingivalis strains
| Strain |
A520a
|
|||
|---|---|---|---|---|
| Vesicles
|
Cells
|
|||
| 8 h | 15 h | 8 h | 15 h | |
| W83 | 0.70 | 0.97 | 0.40 | 0.62 |
| V2543 | 0.27 | 0.58 | 0.24 | 0.33 |
| V2577 | 0.24 | 0.67 | 0.22 | 0.32 |
| V2602 | 0.30 | 0.51 | 0.28 | 0.54 |
Hemolysis of erythrocytes was monitored by the change of absorbance for red pigment resulting from release of hemoglobin from the lysed cells at 520 nm in assays using 36 μg of extracellular vesicles or 50 μl of cells (OD = 1).
Hemoglobinase activity.
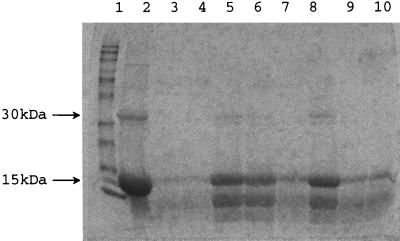
To assess the hemoglobinase activity of Kgp, we examined bovine hemoglobin degradation by extracellular vesicular proteins from P. gingivalis W83, V2543, V2577, V2602, V2383, V2448, and V2546. Hydrolytic activities of Kgp-deficient strains V2543 and V2577 were significantly lower than that of wild-type strain W83 (Fig. 4, lanes 3 to 6). While in the case of the wild-type strain almost all hemoglobin used for the reaction was degraded into fragments that were not detectable on an SDS–15% polyacrylamide gel (Fig. 4, lanes 3 and 4), Kgp mutants degraded only part of the hemoglobin used for the reaction and created at least one detectable fragment (approximately 6 to 8 kDa) (Fig. 4, lanes 5, 6, and 8). The ability to degrade hemoglobin into fragments no longer detectable on SDS-polyacrylamide gels was restored in the strain containing the restored kgp gene, V2602 (Fig. 4, lane 7). The hemoglobin degradation pattern by RgpA− (V2546 [Fig. 4, lane 10]) and HagA− (V2448 [Fig. 4, lane 9]) strains was similar to that of the wild-type strain. The double mutant V2383 (RgpA− Kgp−) displayed a digestion pattern similar to that of the Kgp-deficient strains (Fig. 4, lanes 5, 6, and 8).
FIG. 4.
Hemoglobinase activities of extracellular proteins of P. gingivalis strains. Samples of 500 μl containing 72 μg of extracellular vesicular protein and 4 μg of bovine hemoglobin (Sigma) per μl in 50 mM Tris-HCl (pH 8.0) were incubated 15 h at 37°C in the presence of 2 mM DTT; 20 μl of this mixture was added to equal volume of reducing SDS-PAGE sample buffer (Laemmli sample buffer; Bio-Rad), boiled for 10 min, and electrophoresed on a 15% polyacrylamide gel. Proteolysis of hemoglobin was visualized as described in Materials and Methods. Lanes: 1, BenchMark prestained protein ladder (Gibco, BRL); 2, hemoglobin only; 3, hemoglobin, W83, and DTT; 4, hemoglobin and W83; 5, hemoglobin, V2577, and DTT; 6, hemoglobin, V2543, and DTT; 7, hemoglobin, V2602, and DTT; 8, hemoglobin, V2383 (RgpA− Kgp−), and DTT; 9, hemoglobin, V2448 (HagA−), and DTT; 10, hemoglobin, V2546 (RgpA−), and DTT.
Purification of Kgp from P. gingivalis.
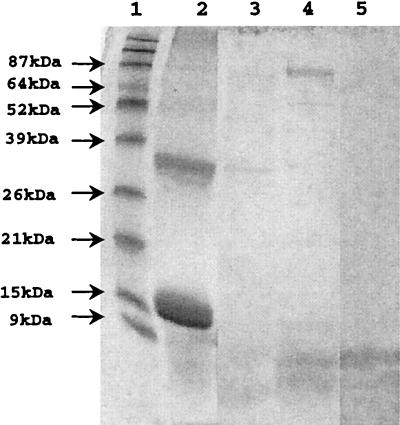
Hemoglobinolytic activities for vesicles indicated that although both Arg-gingipain and Lys-gingipain were capable of degrading hemoglobin, the latter protease was far more efficient because it degraded hemoglobin into many fragments undetectable by SDS-PAGE. For further analysis of hemoglobin fragments rendered by Lys-gingipain, we separated the Arg-gingipain and Lys-gingipain activities. Since Lys-gingipain, but not Arg-gingipain, has been shown to bind hemoglobin (28), we chose hemoglobin affinity chromatography to isolate the Lys-gingipain. We obtained one fraction exhibiting Lys-gingipain activity but not Arg-gingipain activity (Fig. 5, fraction 14) and considered it a partially purified Kgp. The hemoglobinase activity of partially purified Kgp was comparable to that obtained with extracellular vesicles from wild-type strain W83 (Fig. 6).
FIG. 6.
SDS-PAGE analysis of hemoglobinase activity of purified Kgp. A 200-μl reaction mixture containing 600 μg of human hemoglobin and 2 mM DTT in 50 mM Tris-HCl (pH 8.0) was incubated 15 h at 37°C in the presence of either 150 μg of pure Kgp protease or 30 μg of vesicular proteins from W83; 15 μl of this mixture was analyzed on a 15% polyacrylamide gel. Protein fragments were visualized by staining with Coomassie blue R-250. Lanes: 1, BenchMark prestained protein ladder; 2, hemoglobin; 3, hemoglobin plus Kgp; 4, Kgp; 5, hemoglobin plus W83 vesicles.
Hemoglobinase activity of Kgp.
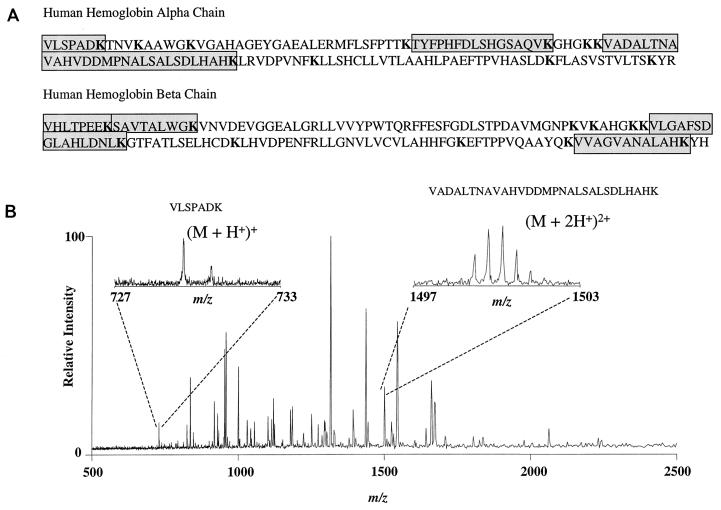
We used mass spectrometry to demonstrate fragments of the hemoglobin α and β chains predicted to occur following Kgp digestion. Figure 7B shows a typical positive-ion ESI-FTICR mass spectrum of the hemoglobin fragments rendered by Kgp. The fragments in the protease digest were observed as protonated species, (M + nH)n+, where n is the number of protons. Direct charge state determination was facilitated by the isotopic resolution obtained over the wide m/z range where the fragments reside which is unique to FTICR; the charge (z) was determined as the reciprocal of the carbon-13 isotopic spacing (24). The two insets in Fig. 7B illustrate the isotopic resolution that was achieved for two different fragments varying in intensity. Mass accuracy for the observed digestion fragments were determined from the monoisotopic peak, with errors typically being less than 60 ppm (0.006%) compared to the theoretical masses. Figure 7A illustrates the sequences of the Kgp peptides demonstrated by ESI-FTICR.
FIG. 7.
Mass spectrometric analysis of Kgp-digested hemoglobin fragments. (A) Amino acid sequence of human α and β hemoglobin chains. Lysine residues are indicated in boldface; amino acid sequences of peptides demonstrated by ESI-FTICR spectroscopy are shaded. (B) ESI-FTICR mass spectrum of a human hemoglobin sample digested by the lysine-specific cysteine protease Kgp. The hemoglobinolytic potential of Kgp is verified by the observance of expected hemoglobin digestion fragments. The resolved carbon-13 isotopic distribution is evident in the two insets for two of the hemoglobin fragments selected as examples. Additional peaks in the spectrum not related to hemoglobin are suggestive of autodigestion of the protease.
Table 5 summarizes the observed Kgp digestion fragments observed in the ESI-FTICR mass spectrum (Fig. 7) in addition to the theoretical monoisotopic masses. Digest fragments of less than 700 Da, which account for 9 of the 23 possible digestion fragments, were not observed or expected due to the essential usage of the microdialysis purification step. Microdialysis of protein standards of various molecular masses (1.2 to 29 kDa) using the same 13,000-MWCO dialysis fiber has been shown to filter out low-molecular-weight proteins in addition to the buffer and salt (23). The observance of proteins less than 13,000 Da is not uncommon, and it is also speculated to be related to the initial concentration of the peptide or protein (23). The observed hemoglobin fragments verify Kgp digestion at 5 of the 10 expected cleavage sites for the α chain and 6 of the 9 for the β chain. The demonstration of several of the predicted Kgp-generated peptides argues strongly against the possibility of hemoglobin degradation by other proteases or peptidases that might be present in our partially purified enzyme preparation. Additional peaks in the spectrum not related to hemoglobin suggests autodigestion of Kgp.
TABLE 5.
Comparison of theoretical and experimental monoisotopic masses of human hemoglobin fragments
| Fragment | Monoisotopic mass (Da)
|
|
|---|---|---|
| Theoretical | Measured | |
| α chain | ||
| VLSPADK | 728.4066 | 728.4255 |
| TYFPHFDLSHGSAQVK | 1,832.8844 | 1,833.0992 |
| VADALTNAVAHVDDMP-NALSALSDLHAHK | 2,995.4820 | 2,995.6590 |
| β chain | ||
| SAVTALWGK | 931.5100 | 931.5587 |
| VHLTPEEK | 951.5600 | 951.5587 |
| VVAGVANALAHK | 1,148.6665 | 1,148.7779a |
| VLGAFSDGLAHLDNLK | 1,668.8830 | 1,668.9468 |
Charge-state determination is tentative due to observation of overlapping isotopic distributions.
DISCUSSION
The existence of nonpigmented variants of P. gingivalis has been reported, but the mechanism of pigment accumulation from erythrocytes has not been previously explained (18, 25, 37). Although other work (35) has suggested that pigmentation may be related to Kgp activity, we have demonstrated here that the black pigmentation of P. gingivalis colonies observed on blood agar plates is dependent on Kgp activity. We have shown that kgp mutants obtained naturally or by allelic exchange were nonpigmented. We also have restored by recombination the kgp gene of a strain that contained an insertionally inactivated allele of this gene. This strain expressed Kgp activity and formed pigmented colonies when grown on blood agar plates. There also were striking differences between the interactions of our P. gingivalis strains and sheep erythrocytes. P. gingivalis W83 was able to mediate hemagglutination and to effect hemolysis (Table 4; Fig. 3). Both of these properties were greatly reduced in kgp mutants of this strain (Table 4; Fig. 3). Finally, our biochemical and biophysical data clearly supported the notion that hemoglobin was a preferred substrate for Kgp (Table 5; Fig. 4, 6, and 7). These observations lead us to conclude that Kgp production is necessary for the provision of large amounts of hemin from hemoglobin. This hemin accumulates on the cell surface, ultimately leading to pigment formation (44). However, other factors, including surface binding components, may be involved in the pigmentation process. For example, hemin binding proteins as well as the hemin binding capability of lipopolysaccharide may be important (3, 11, 27).
Kgp is a multidomain polypeptide composed of a signal peptide, prepropeptide, protease domain, and hemagglutinin/adhesin domain. The hemagglutinin/adhesin domain, also associated with the Arg-gingipain protease RgpA and the hemagglutinin HagA (9, 21, 41, 53), has been shown to contain determinants which bind to erythrocytes (9). To determine the involvement of Kgp in hemin acquisition, we analyzed our mutants for the ability to hemagglutinate and lyse erythrocytes, and for the ability to degrade hemoglobin. Our results with cell fractions indicated that Kgp contributes to hemagglutination of P. gingivalis (Fig. 3). However, as previously reported (30), its role was less significant in the vesicle fraction. The reason for this is unknown. Our general lack of knowledge about the formation of vesicles precludes a ready answer to this question, but several possibilities, including the uneven partitioning of membrane associated proteins during vesicle formation, exist.
Although, the ability of Rgp to lyse erythrocytes has been observed (43), our study provides the first evidence that Kgp also contributes to this process (Table 4). Kgp-deficient strains were still capable of degrading erythrocytes but not to the same extent as the wild type.
Our analysis of hemoglobinase activities for wild-type, mutant, and kgp-restored variants of W83 was instructive. We found that the Kgp-deficient strain had reduced ability to degrade both α and β chains of hemoglobin. Because hemoglobin contains numerous Lys residues (i.e., human hemoglobin contains 11 Lys residues in the α chain and 10 Lys residues in the β chain [Fig. 7]), it was reasonable to postulate that Kgp specificity qualified it as a hemoglobinase, degrading hemoglobin into small peptides and effecting the efficient release of hemin. Although the hemoglobinolytic activity of Kgp explains the generation of high amounts of hemin from hemoglobin, the mechanism of storage of hemin on a surface of P. gingivalis remains to be elucidated. However, the reports of hemin binding surface components (3, 11, 27) are in keeping with our recent preliminary observation that the Lys-gingipain protease was able to bind hemin (data not shown).
Although Fujimura et al. have reported that Arg-gingipain is also capable of degrading hemoglobin (14), its role is likely to be minor on both theoretical and practical grounds (Fig. 4). First, there are only three Arg residues in either α or β chain of human hemoglobin molecule (Fig. 7). Therefore, this protease presumably degrades hemoglobin into three (α chain) or four (β chain) fragments. Second, the role of Arg-gingipain may be insignificant since its activity is inhibited by hemin (55), and so the degradation of hemoglobin and subsequent release of hemin by the action of the enzyme would be self-limiting. On the other hand, our results have shown that Lys-gingipain activity is not affected by the presence of hemin (Table 3). Because high concentrations of hemin favor binding of both hemin and hemoglobin by P. gingivalis (17, 49) Lys-gingipain activity in high concentrations of hemin is an important strategy in initiating the process of iron acquisition. The phenotypic manifestation of this activity is the accumulation of black pigment when cells are grown on blood agar. Our observed Kgp-related hemoglobinase activity is in contrast to the report of Fujimura et al. (14), who failed to see substantial degradation of hemoglobin by this activity. The reason for this inconsistency is not known but may be attributable to differences in strain origin, enzyme purification methods, or other technical factors.
In light of the results obtained by other laboratories and results presented here, a plausible explanation for the contribution of Kgp to provision of hemin from erythrocytes abundant in the periodontal pocket (34) can be proposed. First, Kgp degradation of fibrinogen deregulates the clotting cascade, maximizing the availability of free erythrocytes (6, 26, 37, 40, 53). Second, Kgp binds erythrocytes and degrades them, releasing hemoglobin. Third, the 51-kDa protein encompassing the catalytic domain of Kgp binds hemoglobin (28). Taken together, our results suggest that Kgp is a principal protein involved in acquisition of hemin from hemoglobin.
We recently showed that Kgp-deficient strains had reduced virulence in a mouse abscess model (30). Multiple possibilities may explain this. Kgp has been shown to degrade fibrinogen (26, 37, 40, 53). We have observed that this activity also correlates with the number of Lys residues present in this protein. The activity was shown to be very high for the γ chain, which contains 32 Lys residues and 10 Arg residues. The α and β chains, which have been shown to be equally degraded by Arg-gingipains and Lys-gingipains (35), contain a similar number of Arg and Lys residues. The fibrinogenolytic activity may serve to diminish the clotting process followed by prolonged bleeding providing the bacteria with erythrocytes. The erythrocytes, in turn, may be degraded by Lys-gingipain, resulting in release of hemoglobin. Hemoglobin is then bound and degraded by Lys-gingipain, resulting in the provision of the essential nutrient, heme.
Heme has a profound effect on virulence of P. gingivalis (31, 32). Bacteria grown under hemin limitation conditions were less virulent than their counterparts grown in hemin excess. Heme can contribute to virulence in several ways. First, heme can serve as an iron source. Second, the oxidation-reduction potential of heme, required as a prosthetic group of cytochrome b, allows it to mediate electron transfer with generation of cellular energy that is required for growth and propagation of P. gingivalis. Third, hemin has a regulatory effect on expression of protease activity in P. gingivalis (31, 48). In addition, heme could be detrimental to mammalian cells because of the combined lipophilic and oxidative nature of the molecule (51), resulting in lipid peroxidative catalysis of cellular membranes and constituents. The black pigmentation of P. gingivalis cells may be a form of sequestering and detoxifying of hemin that in high concentrations (10 to 20 μg/ml) has been shown to have antibacterial effects on P. gingivalis as well as other gram-positive cocci and gram-negative rods. Hemin has been proposed to inactivate bacterial cells through an oxidation-reduction process related to peroxide (36). The deteriorating effect of hemin on bacteria can be abrogated in the presence of thiol reagents. An alternative way is formation of black pigmentation composed of μ-oxo dimer, [Fe(III)PPIX]2O (50), serving not only as a scavenger of hemin but also binding free oxygen and thereby reducing the hemin-mediated oxygen radical cell damage as well as protecting from reactive oxidants generated by neutrophils (50). Therefore, interference with mechanisms involved in accumulation of black pigmentation may be significant in controlling the pathogenic potential of P. gingivalis.
ACKNOWLEDGMENTS
This work was supported by USPHS grants DE04224 (to F.L.M.) and DE07606 (to H. A. Schenkein).
We thank K. R. Jones for expert assistance with enzyme purification and with hemolysis assays.
REFERENCES
- 1.Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93→Gly. Biochemistry. 1994;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 2.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramanti T E, Holt S C. Heminoptake in Porphyromonas gingiralis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandad F, Mayrand D, Grenier D, Hinode D, Mouton C. Selection and phenotypic characterization of nonhemagglutinating mutants of Porphyromonas gingivalis. Infect Immun. 1996;64:952–958. doi: 10.1128/iai.64.3.952-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu L, Bramanti T E, Ebersole J L, Holt S C. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect Immun. 1991;59:1932–1940. doi: 10.1128/iai.59.6.1932-1940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciborowski P, Nishikata M, Allen R D, Lantz M S. Purification and characterization of two forms of a high-molecular-weight cysteine proteinase (porphypain) from Porphyromonas gingivalis. J Bacteriol. 1994;176:4549–4557. doi: 10.1128/jb.176.15.4549-4557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comisarow M B, Marshall A G. Fourier transform ion cyclotron resonance mass spectrometry. Chem Phys Lett. 1974;25:282–283. [Google Scholar]
- 8.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis M A, Aduse-Opoku J, Slaney J M, Rangarajan M, Booth V, Cridland J, Shepherd P. Characterization of an adherence and antigenic determinant of the ArgI protease of Porphyromonas gingivalis which is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. Submitted for publication. [DOI] [PubMed]
- 11.Cutler C W, Eke P I, Genco C A, Van Dyke T E, Arnold R R. Hemin-induced modifications of the antigenicity and hemin-binding capacity of Porphyromonas gingivalis lipopolysaccharide. Infect Immun. 1996;64:2282–2287. doi: 10.1128/iai.64.6.2282-2287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimura S, Hirai K, Shibata Y, Nakayama K, Nakamura T. Comparative properties of envelope-associated arginine-gingipains and lysine-gingipain of Porphyromonas gingivalis. FEMS Microbiol Lett. 1998;163:173–179. doi: 10.1111/j.1574-6968.1998.tb13042.x. [DOI] [PubMed] [Google Scholar]
- 15.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalis and isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genco C A. Regulation of hemin and iron transport in Porphyromonas gingivalis. Adv Dent Res. 1995;9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 17.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genco C A, Simpson W, Forng R Y, Egal M, Odusanya B M. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for the coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons R J, Macdonald J B. Hemin and vitamin K compounds as required factors for the cultivation of certain strains of Bacteroides melaningogenicus. J Bacteriol. 1960;80:164–170. doi: 10.1128/jb.80.2.164-170.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths E. The iron-uptake of pathogenic bacteria. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological and clinical aspects. Chichester, United Kingdom: John Wiley & Sons; 1987. pp. 69–138. [Google Scholar]
- 21.Han N, Whitlock J, Progulske-Fox A. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect Immun. 1996;64:4000–4007. doi: 10.1128/iai.64.10.4000-4007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannis J C, Muddiman D C. Nanoelectrospray mass spectrometry using non-metalized, tapered (50-10 μm) fused-silica capillaries. Rapid Commun Mass Spectrom. 1998;12:443–448. [Google Scholar]
- 23.Hannis, J. C., and D. C. Muddiman. Characterization of a microdialysis approach for the preparation of PCR products for ESI-MS using ICP-AES and on-line UV-vis detection. Rapid Commun. Mass Spectrom., in press.
- 24.Henry K D, McLafferty F W. Electrospray ionization with Fourier-transform mass spectrometry. Charge state assignment from resolved isotopic peaks. Org Mass Spectrom. 1990;25:490–492. [Google Scholar]
- 25.Hoover C I, Yoshimura F. Transposon-induced pigment-deficient mutants of Porphyromonas gingivalis. FEMS Microbiol Lett. 1994;124:43–48. doi: 10.1111/j.1574-6968.1994.tb07259.x. [DOI] [PubMed] [Google Scholar]
- 26.Imamura T, Potempa J, Pike R N, Moore J N, Barton M H, Travis J. Effect of free and vesicle-bound cysteine proteinases of Porphyromonas gingivalis on plasma clot formation: implications for bleeding tendency at periodontitis sites. Infect Immun. 1995;63:4877–4882. doi: 10.1128/iai.63.12.4877-4882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S J, Chu L, Holt S C. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 28.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 29.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis J P, Macrina F L. IS195, an insertion sequence-like element associated with protease genes in Porphyromonas gingivalis. Infect Immun. 1998;66:3035–3042. doi: 10.1128/iai.66.7.3035-3042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh P D, McDermid A S, McKee A S, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology. 1994;140:861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 32.McKee A S, McDermid A S, Baskerville A, Dowsett A B, Ellwood D C, Marsh P D. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect Immun. 1986;52:349–355. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna W R, Mickelsen P A, Sparling P F, Dyer D W. Iron uptake from lactoferrin and transferrin by Neisseria gonorrhoeae. Infect Immun. 1988;56:785–791. doi: 10.1128/iai.56.4.785-791.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S. The role of crevicular fluid iron in periodontal disease. J Periodontol. 1998;56:22–27. doi: 10.1902/jop.1985.56.11s.22. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama K, Ratnayake D, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 36.Nitzan Y, Wexler H M, Finegold S M. Inactivation of anaerobic bacteria by various photosensitized porphyrins or by hemin. Curr Microbiol. 1994;29:125–131. doi: 10.1007/BF01570752. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 38.Otto B R, Verweij-van Vught A M, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 39.Payne S M. Iron and virulence in the family enterobacteriaceae. Crit Rev Microbiol. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 40.Pike R N, Potempa J, McGraw W, Coetzer T H, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rangarajan M, Aduse-Opoku J, Slaney J M, Young K A, Curtis M A. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 42.Schifferle R E, Shostad S A, Bayers-Thering M T, Dyer D W, Neiders M E. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J Endod. 1996;22:352–355. doi: 10.1016/S0099-2399(96)80216-0. [DOI] [PubMed] [Google Scholar]
- 43.Shah H N, Gharbia S E. Lysis of erythrocytes by the secreted cysteine protease of Porphyromonas gingivalis W83. FEMS Microbiol Lett. 1989;61:213–218. doi: 10.1016/0378-1097(89)90199-7. [DOI] [PubMed] [Google Scholar]
- 44.Shah H N, Bonnett R, Mateen B, Williams R A. The porphyrin pigmentation of subspecies of Bacteroides melaninogenicus. Biochem J. 1979;180:45–50. doi: 10.1042/bj1800045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;131:313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 46.Slots J, Bragd L, Wikstrom M, Dahlen G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 47.Slots J, Genco R J. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984;63:412–421. doi: 10.1177/00220345840630031101. [DOI] [PubMed] [Google Scholar]
- 48.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-restriction influences haemin-binding, haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalis W50. FEMS Microbiol Lett. 1991;69:63–67. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 49.Smalley J W, Birss A J, McKee A S, Marsh P D. Hemin regulation of hemoglobin binding by Porphyromonas gingivalis. Curr Microbiol. 1998;36:102–106. doi: 10.1007/s002849900287. [DOI] [PubMed] [Google Scholar]
- 50.Smalley J W, Silver J, Marsh P J, Birss A J. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith A. Transport of tetrapyrroles: mechanisms and biological and regulatory consequences. In: Dailey H A, editor. Biosynthesis of heme and chlorophylls. New York, N.Y: McGraw-Hill Publishing Company; 1990. pp. 435–490. [Google Scholar]
- 52.Thony-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodont Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 54.Tsuchiya T, Saito S. Use of n-octyl-beta-d-thioglucoside, a new nonionic detergent, for solubilization and reconstitution of membrane proteins. J Biochem (Tokyo) 1984;96:1593–1597. doi: 10.1093/oxfordjournals.jbchem.a134989. [DOI] [PubMed] [Google Scholar]
- 55.U S, Harper F, Curtis M A. Haemin inhibits the trypsin-like enzyme activity of Porphyromonas gingivalis W83. FEMS Microbiol Lett. 1990;60:169–172. doi: 10.1111/j.1574-6968.1990.tb03883.x. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg E D. Iron withholding: a defense against infection and neoplasia. Physiol Rev. 1984;64:65–102. doi: 10.1152/physrev.1984.64.1.65. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]