Abstract
Arginyltransferase (ATE1) plays critical roles in many biological functions including cardiovascular development, angiogenesis, adipogenesis, muscle contraction, and metastasis of cancer. However, the role of ATE1 in hepatocellular carcinoma (HCC) remains unknown. In this study, we find that ATE1 plays an essential role in growth and malignancy of liver cancer. ATE1 expression is significantly reduced in human HCC samples compared with normal liver tissue. In addition, low ATE1 expression is correlated with aggressive clinicopathologic features and is an independent poor prognostic factor for overall survival and disease-free survival of patients with HCC. Lentivirus-mediated ATE1 knockdown significantly promoted liver cancer growth, migration, and disease progression in vitro and in vivo. Opposing results were observed when ATE1 was upregulated. Mechanistically, ATE1 accelerated the degradation of β-catenin and inhibited Wnt signaling by regulating turnover of Regulator of G Protein Signaling 5 (RGS5). Loss- and gain-of-function assays confirmed that RGS5 was a key effector of ATE1-mediated regulation of Wnt signaling. Further studies indicated that RGS5 might be involved in regulating the activity of GSK3-β, a crucial component of the cytoplasmic destruction complex. Treatment with a GSK inhibitor (CHIR99021) cooperated with ablation of ATE1 or RGS5 overexpression to promote Wnt/β-catenin signaling, but overexpression of ATE1 or RGS5 knockdown did not reverse the effect of GSK inhibitor.
Implications:
ATE1 inhibits liver cancer progression by suppressing Wnt/β-catenin signaling and can serve as a potentially valuable prognostic biomarker for HCC.
Introduction
Hepatocellular carcinoma (HCC) is the sixth-most common cancer type and the fourth leading cause of cancer-related death worldwide in 2018, with about 841,000 new cases and 782,000 deaths annually (1). As the treatments for HCC have been improved notably in recent years, the prognosis of HCC is gaining unsatisfactory rate of improvement (2–4). Therefore, a thorough understanding of the underlying mechanisms regarding tumor growth and metastasis is critical for developing effective therapeutic targets for patients with HCC.
Protein arginylation is a posttranslational modification mediated by ATE1 that transfers Arg from tRNA directly to protein targets. It has been previously believed that ATE1 regulated the degradation of RGS4, RGS5, and RGS16, which contained the Met-Cys- N-terminal motif through N-end rule pathway (5–7). Recent studies showed that the acidic side chains of Asp and Glu can also serve as targets for arginylation in intact proteins (8, 9). ATE1 was proved to play an important role in many biological functions including cardiovascular development (10), angiogenesis (11), adipogenesis (12), cell migration (13), muscle contraction (14), neurodegeneration (15), and neurite outgrowth during brain development (16), but the understanding about the precise role of ATE1 in cancer remains insufficient. Previous studies have proved that ATE1 was a suppressor and a potential prognostic indicator which inversely correlated with metastatic progression and survival in human cancer (17, 18). Despite these intriguing observations, no direct functional and pathway mechanism studies have ever been reported in liver cancer.
We studied and indicated the suppressor role of ATE1 in liver cancer. Specifically, we first expounded the functional and molecular mechanism of ATE1′s regulatory function on the proliferation, migration, and invasion of HCC cells to inhibit the progression for a patient with HCC.
Materials and Methods
HCC samples and patients
This study was approved by the Ethics Committee of Xiangya Hospital at Central South University (Hunan, China). All patients and their families provided written informed consent and agreed to the use of their tissue samples in the study in accordance with the Declaration of Helsinki. A total of 120 HCC specimens collected from January 2010 to December 2013 were randomly selected from the Department of Surgery, Xiangya Hospital of Central South University (Hunan, China). Another 80 HCC specimens collected from January 2010 to December 2013 were randomly selected from the Department of Abdominal Surgical Oncology, Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University (Hunan, China). The patient demographics and clinicopathologic variables of the two cohorts are described in Supplementary Tables S1 and S2. The diagnosis of HCC in all patients was confirmed by two independent histopathologists. Follow-up procedures were conducted as described in our previous study (19). The complete clinicopathologic features of these patients were collected and stored in our database. Research protocols followed the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) recommendations for reporting prognostic biomarkers in cancer (20).
Cell lines and cell culture
Human liver cancer cell lines, HepG2 (RRID: CVCL_0027) and Hep3B (RRID: CVCL_0326) cell lines were purchased from the ATCC. MHCC97-L (RRID: CVCL_4973), MHCC97-H (RRID: CVCL_4972), and HCCLM3 (RRID: CVCL_6832) were kindly provided by the Liver Cancer Institute of Fudan University, Shanghai, China, where these cell lines were established. Huh7 (RRID: CVCL_0336) and LO2 (RRID: CVCL_6926) were purchased from the Cell Bank of Typical Culture Preservation Committee of Chinese Academy of Science, Shanghai, China. The cell lines have been authenticated using short tandem repeat profiling within the last three years. The cell lines also have been regularly tested for Mycoplasma contamination using the Universal Mycoplasma Detection Kit obtained from ATCC. Cells were cultured in equal mix of DMEM (high glucose with Glutamax, Biological Industries), supplemented with antibiotics and 10% FBS (Biological Industries, catalog no. 04–001–1A). Cells were maintained in 5% CO2 at 37°C, according to the manufacturer's protocol.
Quantitative real-time PCR
Quantitative real-time PCR assay were performed and analyzed for this study as described in our previous work (19). GAPDH was used as endogenous control. The experiments were performed in triplicate. The primers were all synthesized by and purchased from Sangon Biotech (China). Specific primers for each gene were as follows: ATE1 (forward primer: CAGTTCCTAAGCCAGGCAAA, reverse primer: AGCCTGGAAACCCTCAAGTT), RGS5 (forward primer: AAGCCACCTGCCAAAATGTGC, reverse primer: AGCGAGGTTTTCTGGGTCTTG), RGS4 (forward primer: TACAGGACGCAGGCATGTGA, reverse primer: GTCTCCACGCAGTGATTTGC), RGS16 (forward primer: GAATCCTCACGACCCTGCCT, reverse primer: GGTGGCGCTGTGTTCTTTAG), β-catenin (forward primer: TGGATTGATTCGAAATCTTGCC, reverse primer: GAACAAGCAACTGAACTAGTCG), GAPDH (forward primer: CAACGTGTCAGTGGTGGACCTG, reverse primer: GTGTCGCTGTTGAAGTCAGAGGAG)
Protein extraction and Western blot analysis
Tissues or cells were lysed with RIPA buffer (Pierce) supplemented with protease inhibitors. Protein concentration was measured using a BCA Protein Assay (Thermo Scientific). Protein lysates, suspended in loading buffer, were separated on 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore). Then these membranes were blocked with 5% skim milk at room temperature for 1 hour, and incubated with primary antibodies at 4°C overnight. After washed, they were incubated with suitable HRP-conjugated secondary antibody at room temperature for 30 minutes and detected using an enhanced chemiluminescence (ECL) kit (Thermo Scientific). The antibodies used are listed as followed: anti-Ubiquitin, Abcam (ab7780); APC, Cell Signaling Technology (2504); ATE1, Abcam (ab199423); Axin1, Cell Signaling Technology (2087); CCND2, Cell Signaling Technology (3741); CK1, Cell Signaling Technology (2655); c-MYC, Abcam (ab32072); GSK3-β, Cell Signaling Technology (9315); MMP16, Abcam (ab73877); MMP7, Cell Signaling Technology (71031); p-GSK3-β (Tyr216), Abcam (ab75745); p-β-catenin (Ser33/37/Thr41), Cell Signaling Technology (9561); RGS4, Abcam (ab97307); RGS5, LifeSpan BioSciences (LS-C162581); RGS16, Abcam (ab119424); β-Actin, Affinity (T0022); β-catenin, Cell Signaling Technology (9562); β-TrCP, Cell Signaling Technology (4394); goat anti-mouse IgG (H+L) secondary antibody, Multi Sciences (GAM0072); goat anti-rabbit IgG(H+L) secondary antibody, Multi Sciences(GAR0072); goat anti-rabbit IgG(H+L) secondary antibody, Thermo Scientific (35560).
IHC
IHC staining on formalin-fixed, paraffin-embedded tissue sections 4-μm in thickness was performed using the polymer HRP detection system (Zhongshan Goldenbridge Biotechnology). IHC experiments were conducted as described previously (19). The antibodies used are listed in Supplementary Table S4. The IHC staining intensity was scored as negative (−,0), weak (+,1), moderate (++,2), or strong (+++,3), and the percentage of positive cells was scored as <5% (0), 6%–25% (1), 26%–75% (2), or >75% (3). The final scores were calculated by multiplying the intensity and percentage values (range: 0–9). The HCC specimens were divided on the basis of protein expression into a low expression group (<4) and a high expression group (≥4) for further analysis (21).
Immunofluorescence
Immunofluorescence assay were performed and analyzed for the present study as described in our previous work (19).
Establishment of lentiviral-transfected cells
The ectopic expression and knockdown lentivirus as well as control lentivirus for ATE1 (carrying purinomycin resistance) and RGS5 (carrying hygromycin resistance) were all purchased from Vigene Bioscience. Transfection was performed according to the manufacturer's instructions. Puromycin (2 μg/mL) and hygromycin (200 μg/mL) were used to select stable clones. The sequences of the shRNA and cDNA clones were as follows: ATE1-shRNA-1, CGGGTGACTTTGCATTGATAA; ATE1-shRNA-2, TGTACTACGATCCTGATTATT; ATE1-shRNA-3, GATGACATCAAAGAGAGTTTA; RGS5-shRNA-1, GCTCCTAAAGAGGTGAATATT; RGS5-shRNA-2, GACCTTGTCATTCCGTACAAT; RGS5-shRNA-3, CAAGGAGATTAAGATCAAGTT; The accession of ATE1 Nucleotide Sequence, NM_001288736.1; The accession of RGS5 Nucleotide Sequence, AB008109.1
MTT and colony formation assays
MTT assay was performed using CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega, catalog no. G3582). MTT assay and colony formation assay were performed and analyzed for the present study as described in our previous work (19).
Wound healing and transwell invasion assays
Wound healing and transwell invasion assays were performed and analyzed for this study as described in our previous work (19).
Cell-cycle analysis
Cell-cycle analysis were performed using a Cell Cycle Staining Kit (Multi Sciences, catalog no. CCS012) following the manufacturer's protocol. A total of 5 × 105 cells were seeded in 6-well plates and incubated for 24 hours. Then cells were harvested and fixed with cold 70% ethanol at − 20°C overnight. After washing, cells were stained in a solution containing PI (0.5 mg/mL) and RNase A (10 mg/mL). Then cells were filtered through a 70-μm cell strainer immediately prior to flow cytometry, which was carried out on a FACS caliber flow cytometer (BD Biosciences).
Cignal Finder Cancer 10-Pathway reporter arrays
A Cignal Finder 10-Pathway Reporter Array (Qiagen, catalog no. CCA-101L) was performed to explore the signaling pathways that were regulated by ATE1 in liver cancer cells. The assay was conducted according to the manufacturer's protocol. Relative firefly luciferase activity was calculated and normalized to the constitutively expressed Renilla luciferase. Experiments were done in triplicates. The data were presented as the mean ± SD.
Coimmunoprecipitation assay and cycloheximide chase assay
Coimmunoprecipitation (co-IP) was performed using a Thermo Scientific Pierce Co-IP kit (Thermo Scientific, catalog no. 88804) following the manufacturer's protocol. The cells were treated with 5 μmol/L Mg132 (Abcam, catalog no. ab141003) for 6 hours, followed collected and lysed. Primary antibody was first immobilized for 2 hours using AminoLink Plus coupling resin, which was washed and incubated with whole-cell lysates overnight. After incubation, the resin was washed, and the protein was eluted using elution buffer. A negative control that was provided in the IP kit to assess nonspecific binding received the same treatment as the co-IP samples, including the primary antibody. In this control, the coupling resin was not amine-reactive, thus preventing covalent immobilization of the primary antibody on to the resin. Then, samples were analyzed by Western blot. A cycloheximide (Abcam, catalog no. ab120093) chase assay was used to determine the half-lives of β-catenin. Liver cancer cells with aberrant ATE1 and/or RGS5 expression were treated with cycloheximide (10 μg/mL) for the indicated times, then the cells were collected, lysed, and followed by Western blot assay with the indicated antibodies.
Mouse models
Xenograft experiments were conducted with 6-week-old male BALB/c nude mice. A total of 5 × 106 of the indicated liver cancer cells were injected subcutaneously into the right upper flank regions of BALB/c nude mice (6 mice per group). After 4 weeks, the mice were killed. The tumors were excised, cut into pieces approximately 1 mm3 in size, and implanted into the livers of BALB/c nude mice in each group (3 mice per group; ref. 19). At 6 weeks after implantation, the mice were killed, and the livers were harvested, imaged, and processed for histopathologic examination. All animal studies were conducted at the Animal Institute of CSU according to the protocols approved by the Medical Experimental Animal Care Commission of CSU.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software (SPSS Inc.) and GraphPad Prism 7. Data are presented as the mean ± SD from three repeated experiments. Quantitative data between groups were compared using Student t test. The χ2 test was applied to examine the associations between ATE1 expression and clinicopathologic parameters. Spearman rank analysis was performed to determine the correlations between the different protein levels. Overall survival (OS) and disease-free survival (DFS) curves were obtained by the Kaplan–Meier method, and differences were compared by the log-rank test. Univariate and multivariate analyses were performed with the Cox proportional hazard regression model to verify the independent risk factors. All differences were deemed statistically significant at P < 0.05.
Results
ATE1 is frequently low expression and predicts poor prognosis in HCC
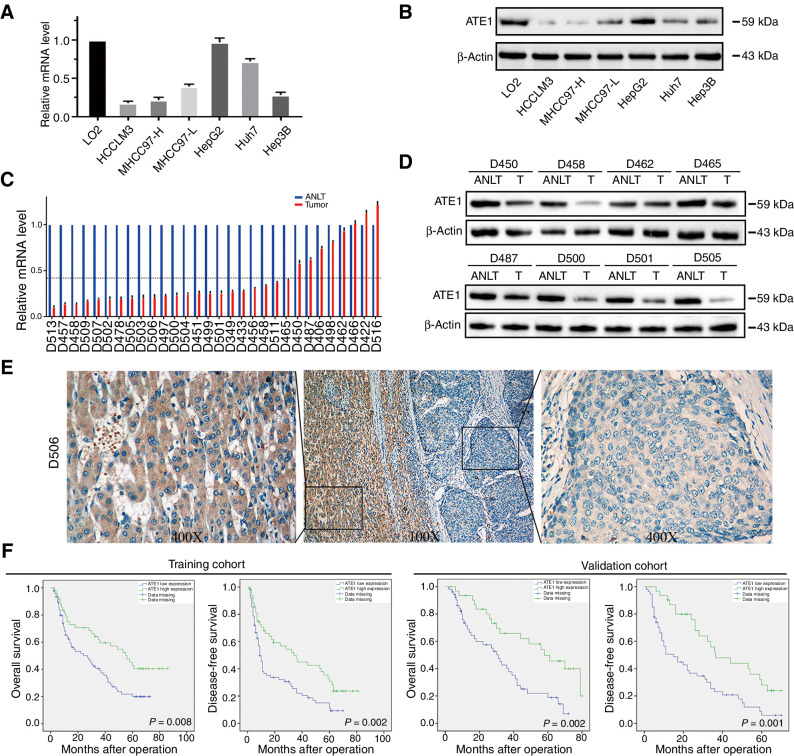
We firstly evaluated the expression of ATE1 in human liver cancer cell lines, which indicated that ATE1 expression in HCCLM3, MHCC97-H, MHCC97-L, Huh7, and Hep3B cell lines were lower than L02, an immortalized human liver cell line (Fig. 1A and B). Moreover, to verify ATE1 expression in human HCC samples, we randomly selected 30 paired fresh HCC samples for qRT-PCR analysis and 8 paired fresh samples for Western blot analysis, the results showed that the expression level of ATE1 in HCC tissues was lower than in their matched adjacent nontumor liver tissues (Fig. 1C and D). Accordingly, IHC analysis in HCC samples also showed that ATE1 was highly expressed in adjacent-tumor liver cells (magnified image shown on the left) and lowly expressed in tumor cells (magnified image shown on the right; Fig. 1E).
Figure 1.
ATE1 is frequently downregulated and predicts poor prognosis in HCC. ATE1 expression was downregulated in HCC cell lines analyzed by real-time PCR (A) and Western blot (B). C, ATE1 mRNA level was detected in 30 pairs of HCC tissues and their adjacent nontumor liver tissues (ANLT) by real-time PCR. Imaginary line indicates the mean fold change. D, 8 of the 30 pairs of HCC tissues and their ANLTs were selected randomly and ATE1 expression was detected by Western blot analysis. E, Representative IHC images of ATE1 expression also showed that ATE1 expression was lower in tumor tissues than in ANLTs. Magnification, 100×, inset magnification, 400×. F, Kaplan–Meier curves for OS and DFS according to ATE1 expression in the training cohort (n = 120) and validation cohort (n = 80).
To explore the association between ATE1 expression and clinicopathologic features of patients with HCC, we dichotomized patients with HCC into two subgroups according to the ATE1 expression in tumor tissues, namely high ATE1 expression and low ATE1 expression. In training cohort, it showed that low ATE1 expression was significantly associated with tumor size (>5 cm, P < 0.001), multiple tumor nodules (≧2, P = 0.027), microvascular invasion (P = 0.008), advanced TNM stage (P = 0.026), advanced BCLC stage (P = 0.003), and advanced Edmonson–Steiner grade (P < 0.001, Supplementary Table S1), which was further confirmed in the validation cohort (Supplementary Table S4). Patients with HCC in low ATE1 expression group exhibited shorter OS and worse DFS than those in high ATE1 expression group (Fig. 1F). Multivariate analysis further revealed that low ATE1 expression was an independent risk factor for both OS and DFS of patients with HCC after liver resection, and the HR for OS and DFS in the ATE1-low group were 1.973 [95% confidence interval (CI), 1.134–3.430; P = 0.009] and 1.744 (95% CI, 1.105–2.752; P = 0.017) respectively (Supplementary Tables S2 and S3). The prognostic value of ATE1 expression in HCC was further verified in the validation cohort (Supplementary Tables S5 and S6). Thus, these results suggested that low ATE1 expression in patients with HCC predicts poor prognosis and may contribute to HCC progression. We then deduced that ATE1 may play as a tumor suppressor in HCC.
ATE1 inhibits the proliferation, migration, and invasion of liver cancer in vitro
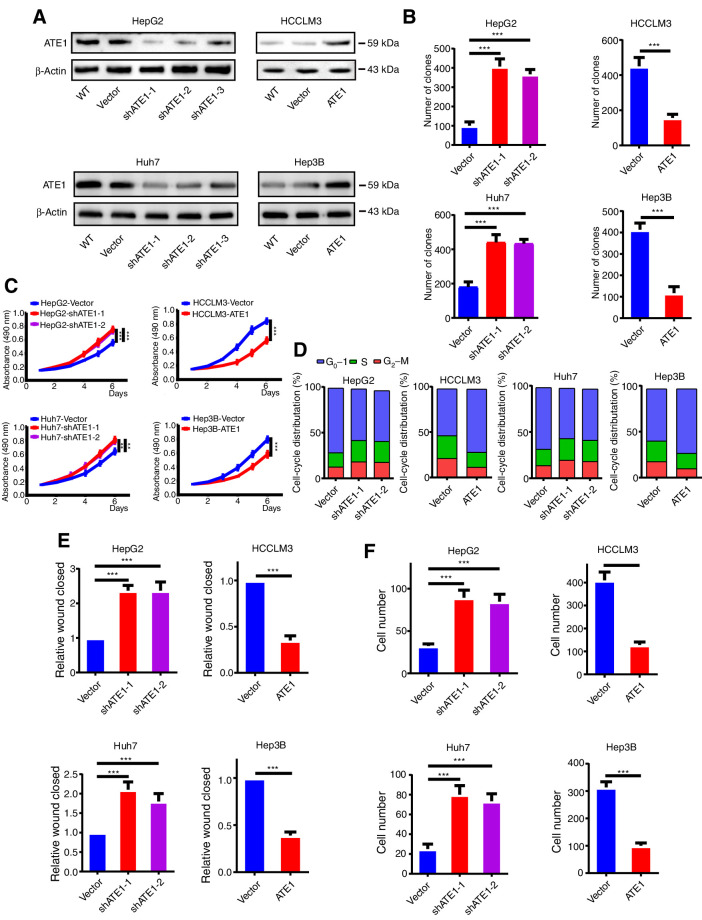
To further study the role of ATE1 in development of liver cancer, we chose high ATE1 expression liver cancer cell line HepG2 and Huh7 for stably downregulated ATE1; and chose low ATE1 expression HCCLM3 and Hep3B cell lines for stably overexpression through lentivirus transfection (Fig. 2A; Supplementary Fig. S1A). We performed clone formation assay to investigate the effect of ATE1 on cell proliferation. The clones of HepG2-Vector, Huh7-Vector, HCCLM3-ATE1, and Hep3B-ATE1 groups were significantly less than that of HepG2-shATE1, Huh7-Vector, HCCLM3-Vector, and Hep3B-Vector groups (Fig. 2B; Supplementary Fig. S1B). The similar results were shown by MTT assays (Fig. 2C). Furthermore, cell-cycle assay showed that much more cells were blocked in the G0–G1 phase in HepG2-Vector and Huh7-Vector cell lines than that in HepG2-shATE1 and Huh7-shATE1 cells, whereas more cells entered into S and G2 phase in HCCLM3-Vector and Hep3B-Vector cells than that in HCCLM3-ATE1 and Hep3B-ATE1 cells (Fig. 2D). The wound-healing and transwell assays showed that HepG2-shATE1 and Huh7-shATE1 cells had a faster wound closure rate and more invasion cells than HepG2-Vector and Huh7-Vector cells, whereas HCCLM3-ATE1 and Hep3B-ATE1 cells had markedly reduced migratory and invasive capacity (Fig. 2E and F; Supplementary Fig. S1C and S1D). It verifies that ATE1 inhibits liver cancer cell lines proliferation, migration, and invasion capacity in vitro.
Figure 2.
ATE1 inhibits the proliferation, migration, and invasion of liver cancer in vitro. A, HepG2 and Huh7 cell lines were infected by shATE1 lentivirus. HCCLM3 and Hep3B cell lines were infected by ATE1-overexpressing lentivirus, and the expression of ATE1 was determined by Western blot analysis, respectively. B, Quantifications of cell colonies in indicated liver cancer cell lines, as determined by colony formation assay. C, MTT analysis of the proliferation ability of HepG2, Huh7, HCCLM3, and Hep3B transfectants. D, DNA-content staining–based cell-cycle analysis in HepG2, Huh7, HCCLM3, Hep3B, and their ATE1-intervened cell lines. DNA was stained by propidium iodide. Wound-healing assays (E) and transwell invasion assays (F) were subjected to detect the migration and invasion capacity of ATE1-interfered cell lines. The histograms showed the statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001 based on the Student t test. Error bars, SD.
ATE1 inhibits the growth and correlates with low potential malignancy of liver cancer xenografts in vivo
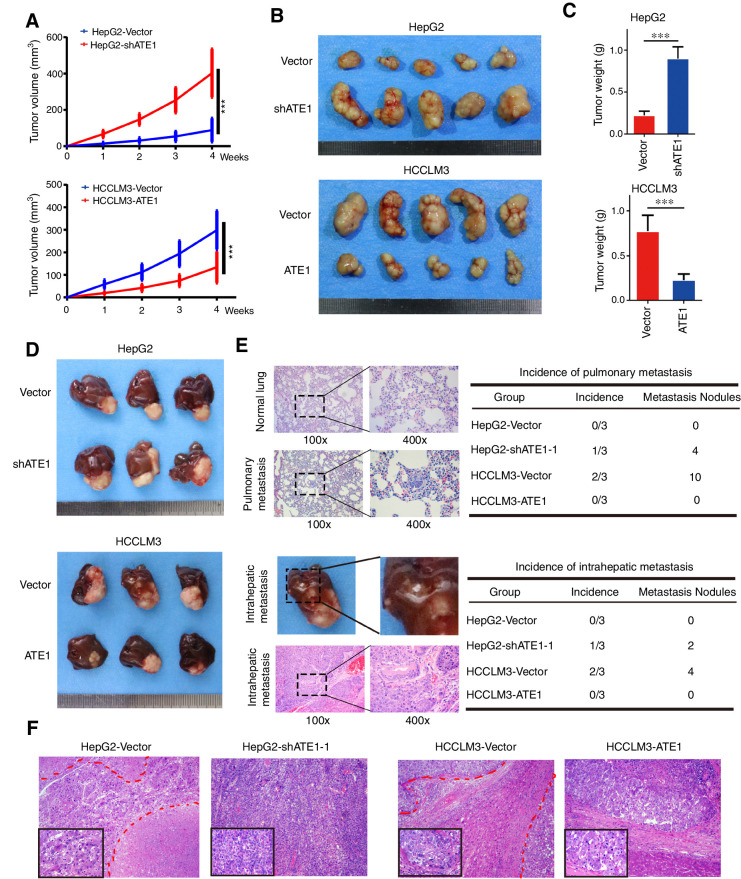
To study the role of ATE1 in vivo, we established subcutaneous xenograft tumor models in nude mice as previously described (22). As ATE1 expression of HepG2-shATE1–1 cell line was reduced more significantly, we selected HepG2-shATE1–1 cell line for further study and shortly named as HepG2-shATE1. The size of subcutaneous tumors were measured and calculated every week. The growth curve of subcutaneous tumors were shown (Fig. 3A). After 4 weeks, the subcutaneous tumors were removed and weighed (Fig. 3B). HepG2-Vector cell-derived tumors at the subcutaneous implantation sites were smaller and grew more slowly than HepG2-shATE1 cell–derived tumors, whereas HCCLM3-Vector cell–derived tumors were larger and grew more rapidly than HCCLM3-ATE1 cell–derived tumors implantation. The weights of each group tumors also showed the significant differences (Fig. 3C). To further understand the influence of ATE1 for liver cancer cell lines in the liver environment, we established orthotopic xenograft tumor models. After 8 weeks of implantation, the mice were sacrificed and the orthotopic xenograft tumors were removed and shown (Fig. 3D; Supplementary Fig. S1E). Consistently, the liver orthotopic xenograft tumor also showed that ATE1 inhibits tumor growth, whereas ATE1 knockdown promoted tumor growth in vivo. We also collected the lung and the liver orthotopic xenograft tumors, which were sectioned and subjected to histopathologic analysis subsequently by hematoxylin and eosin staining. We analyzed the incidence of pulmonary metastasis and intrahepatic metastasis. Furthermore, we also counted the metastasis nodules of and pulmonary metastasis and intrahepatic metastasis (Fig. 3E). The results suggested that ATE1 knockdown increased the incidence of pulmonary/intraheptatic metastasis and the number of pulmonary/intraheptatic metastasis nodule, whereas ATE1 overexpression reduced the incidence of pulmonary/intraheptatic metastasis and the number of pulmonary/intraheptatic metastasis nodule. As the representative images shown in Fig. 3F, all the orthotopic tumors presented significant atypia. HepG2-Vector cell–derived orthotopic tumor showed lower tumor cell density and more focal necroses (marked with red lines) than HepG2-shATE1 cell–derived orthotopic tumor whose nucleus were bigger, more pleomorphism, and hyperchromatic. Meanwhile, HCCLM3-Vector cell–derived orthotopic tumor presented intrahepatic metastasis (marked with red lines) and the nucleus of tumor cells were more pleomorphism and hyperchromatic than HCCLM3-ATE1 cell–derived orthotopic tumor, which also showed more lymphocytes infiltration. More pleomorphism, hyperchromatic nucleus, higher tumor cell density, and more intrahepatic metastasis predict higher malignancy potential and poorer prognosis. In consequence, we suggested that ATE1 inhibited the growth and correlated with low potential malignancy of liver cancer in vivo.
Figure 3.
ATE1 inhibits the growth and correlates with low-grade malignancy of liver cancer in vivo. A, Subcutaneous xenograft tumors from HepG2-shATE1 and HCCLM3-ATE1 cells and their control cells were established in nude mice. The volumes of subcutaneous tumors were measured per week and calculated according to the following equation: V (mm3) = width2 (mm2) × length (mm)/2. The growth curves are shown. B, Subcutaneous tumors from HepG2-shATE1 and HCCLM3-ATE1 cells and their control cells were removed and shown. C, The histograms showed the weight and statistical analysis of subcutaneous tumors. D, Orthotopic tumors were established using subcutaneous tumors and removed after 6 weeks. Each indicated groups are shown. Tumor volumes of tumors are shown in the right panels. *, P < 0.05; **, P < 0.01; ***, P < 0.001 based on the Student t test. Error bars, SD. E, Representative images of H&E staining in normal lung tissue and pulmonary metastasis sections from orthotopic tumor models were shown (top left). The incidence and the total metastasis nodules of pulmonary metastasis were shown (top, right). Representative images of intrahepatic metastasis and H&E staining in intrahepatic metastasis tissues (bottom left). The incidence and the total metastasis nodules of intrahepatic metastasis were shown (bottom, right). F, Representative images of H&E staining in tissue sections from orthotopic tumors were shown. Magnification, 100×; inset magnification, 400×.
ATE1 inhibits Wnt signaling pathway by accelerating the degradation of β-catenin
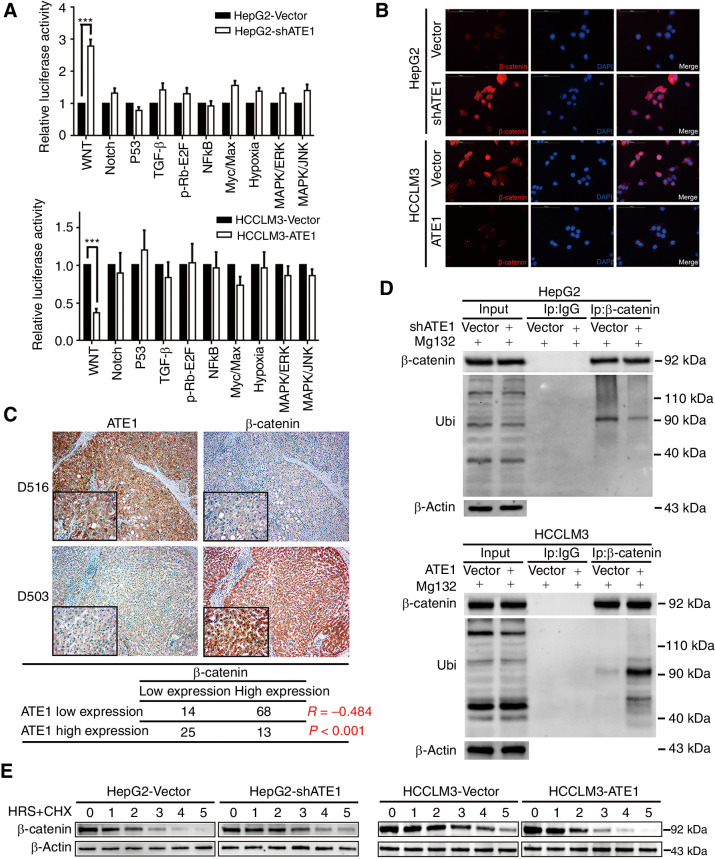
Here, we have confirmed the effect of ATE1 on the proliferation, migration, invasion and malignant degree of liver cancer in vitro and in vivo. But the question is how does ATE1 play the role in liver cancer cell lines. Cignal Finder Cancer 10-Pathway Reporter Array was used to scan and assess the activity of the key transcription factors of 10 signaling pathway by dual-luciferase reporter systerm. ATE1 knockdown significantly enhanced the activity of Wnt signaling pathway, which was presented by β-catenin in HepG2 cell line and ATE1 overexpression attenuated Wnt signaling activity in HCCLM3 cell line (Fig. 4A). To further study the mechanism, we examine the manipulation of β-catenin by immunofluorescence in HepG2-Vector/HepG2-shATE1 and HCCLM3-Vector/HCCLM3-ATE1 cell lines. Results showed that knockdown ATE1 increased β-catenin expression in HepG2 cell line, whereas ectopic expression ATE1 reduced β-catenin expression in HCCLM3 cell line (Fig. 4B). Western blot assay also verified the expression alteration of β-catenin and its downstream proteins (Supplementary Fig. S2A). Consistent with this, IHC for HCC consecutive sections revealed that high ATE1 expression was associated with low β-catenin expression level (Fig. 4C), which suggested a negative correlation between the two variables. Interestingly, intervening ATE1 expression in HepG2 and HCCLM3 cell lines did not affect the β-catenin mRNA level (Supplementary Fig. S2B). Here, we inferred that ATE1 regulates β-catenin steady-state level through degradation pathway which depends on ubiquitination and proteasome, not transcription. To confirm it, we immunoprecipitated β-catenin from lysates of HepG2-Vector/HepG2-shATE1 and HCCLM3-Vector/HCCLM3-ATE1 cell lines, which were treated with MG132. Ubiquitination was confirmed using an anti-ubiquitin antibody. There was much weaker presence of ubiquitinated β-catenin in HepG2-shATE1 cell line, meanwhile there were no macroscopic difference in input lane. The opposite results were observed in HCCLM3 cell lines (Fig. 4D). Furthermore, we performed a time course experiment for cycloheximide treatment. HepG2, HCCLM3 cell lines and their ATE1-intervened cells were treated with cycloheximide at the interval of 1 hour. We found that the half-life period of β-catenin in HepG2-Vector cells was about 1.5–2 hours, which was extended to almost 3 hours in HepG2-shATE1 cells. Meanwhile, the half-life period of β-catenin in HCCLM3-Vector and HCCLM3-ATE1 cells was shorten from 3–4 hours to 2 hours (Fig. 4E). The observation suggested that ATE1 accelerated the degradation of β-catenin through ubiquitination pathway.
Figure 4.
ATE1 inhibits Wnt signaling pathway by accelerating the deregulation of β-catenin. A, The 10-Pathway Reporter Array showed the signaling change in ATE1-interfered cells. B, IF assays to show the expression and distribution of β-catenin in liver cancer cell lines with ATE1 knockdown or ectopic expression. C, Representative IHC images of ATE1 and β-catenin expression in HCC serial slices. Magnification, 100×; inset magnification, 400×. The correlation analysis between ATE1 and β-catenin was shown in the table below. D, Liver cancer cells were treated with MG132. Samples were immunoprecipitated with β-catenin antibody and IgG as control followed by Western blot analysis using the indicated antibodies. E, Liver cancer cells were treated with cycloheximide at different time points as indicated followed by Western blot analysis using the indicated antibodies.
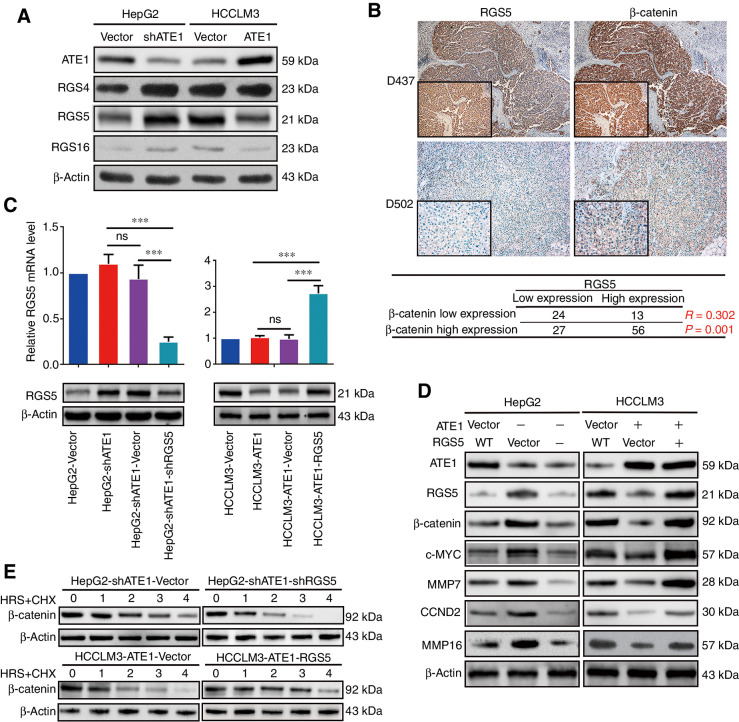
RGS5 is the key downstream target of ATE1 to regulate the degradation of β-catenin
We then intend to understand the pathway that ATE1 regulated the degradation of β-catenin. We first focused on the signaling protein which most possibly and directly took part in the degradation of β-catenin. According to the mechanism of ATE1-dependent N-terminal pathway rule and ubiquitination-dependent degradation, RGS4, RGS5, and RGS16 are the three downstream signaling proteins of ATE1 that have been discerned (5–7). To verify that ATE1 negatively regulated the steady-state level of RGS4, RGS5 and RGS16, we collected the expression level of RGS4, RGS5, and RGS16 in ATE1-intervened cell lines respectively by Western blot analysis (Fig. 5A). Here, we hope that at least one of RGS4, RGS5, and RGS16 had a proven correlation with β-catenin degradation. To determine it, we monitored the expression of RGS4, RGS5, RGS16, and β-catenin, respectively, in serial slices from 120 HCC samples by IHC. As representative images and correlation analysis were shown in Fig. 5B, RGS5 expression, instead of RGS4 or RGS16, had a strongly positive correlation with β-catenin expression (P = 0.001, R = 0.302). Consequently, we supposed that RGS5 was the key downstream of ATE1 to regulate the β-catenin degradation. To further confirm it, we downregulated RGS5 expression in HepG2-shATE1(named as HepG2-shATE1-shRGS5–1, HepG2-shATE1-shRGS5–2 and HepG2-shATE1-shRGS5–3) cell line while upregulated it in HCCLM3-ATE1 (named as HCCLM3-ATE1-RGS5) cell line through lentivirus transfection and examined the infection efficiency, respectively, by Western blot analysis and qRT-PCR, HepG2-shATE1-shRGS5–2 was the most effective one and shortly named as HepG2-shATE1-shRGS5 for further study (Fig. 5C, Supporting Supplementary Fig. S2C). Interestingly, β-catenin protein level was reduced again after downregulating RGS5 in HepG2-shATE1 cell line, whereas β-catenin expression reverted after overexpression RGS5 in HCCLM3-ATE1 cell line. The similar observations of the Wnt signaling pathway downstream proteins were also monitored by Western blot analysis (Fig. 5D). Furthermore, we tested the effect of RGS5 on the β-catenin degradation efficiency. Similarly, the β-catenin degradation efficiency increased in knockdown RGS5 HepG2-shATE1 cell line, whereas the β-catenin degradation efficiency decreased in ectopic RGS5 expression HCCLM3-ATE1 cell line (Fig. 5E). Together with the observations in Fig. 4D, we confirmed that ectopic overexpression of RGS5 reversed the effect of ATE1 on accelerating β-catenin degradation. Here, we have shown that RGS5 is the key downstream target of ATE1 to regulate the degradation of β-catenin.
Figure 5.
RGS5 is the key downstream of ATE1 to regulate the deregulation of β-catenin. A, The expressions of ATE1 and its downstream proteins RGS4, RGS5, RGS16 were detected by indicated antibodies. B, Representative IHC images of RGS5 and β-catenin expression in HCC serial slices. Magnification, 100×; inset magnification, 400×. The correlation analysis between RGS5 and β-catenin was shown in the table below. C, The expression of ATE1 was determined by real-time PCR and Western blot analysis, respectively in RGS5-intervened HepG2-shATE1 and HCCLM3-ATE1 cell lines. D, The expression of β-catenin and its representative downstream proteins were monitored in with or without RGS5 intervening cell lines by Western blot analysis using the indicated antibodies. E, HCC cells were treated with cycloheximide at different time points as indicated followed by Western blot analysis using the indicated antibodies.
The effect of ATE1 on proliferation, migration, and invasion of liver cancer was reversed by RGS5 in vitro.
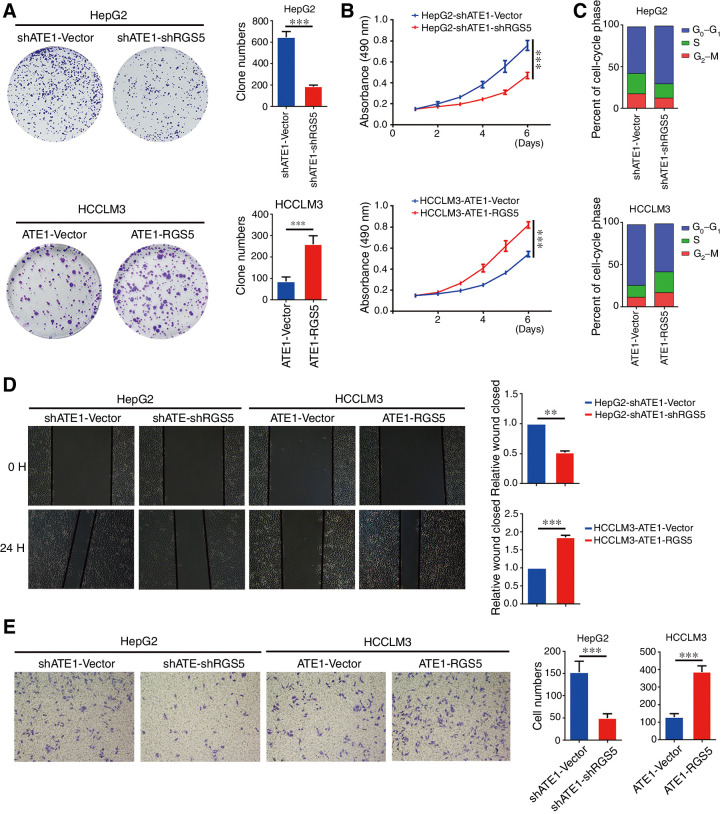
To further understand the effect of RGS5 in liver cancer cell lines, we tested the proliferation, migration, and invasion capacity in RGS5-intervened HepG2-shATE1 and HCCLM3-ATE1 cell lines. Clone formation and MTT assay as shown in Fig. 6A and B, RGS5 increased the proliferation capacity in liver cancer cell lines while it had been reduced by ATE1 (Fig. 2A and B). Cell-cycle assay showed that knockdown RGS5 in HepG2-shATE1 cells blocked the cells in the G0–G1 phase, whereas ectopic RGS5 in HCCLM3-ATE1 cells promoted the cells to enter into S and G1–G2 phase (Fig. 6C). We investigated the migration and invasion capacity of RGS5-intervened liver cancer cells through wound-healing and transwell assays. Results showed that knockdown RGS5 in HepG2-shATE1 cells decreased the migration and invasion capacity. In contrast, ectopic expression of RGS5 in HCCLM3-ATE1 cells had the inverse effect (Fig. 6D and E). Together, with the previously mentioned observations (Fig 2E and F), we suggested that RGS5 reversed the effect of ATE1 on the proliferation, migration, and invasion capacity in liver cancer cell lines. Therefore, we confirmed again that RGS5 was the key downstream target of ATE1 to regulate the degradation of β-catenin.
Figure 6.
The effect of ATE1 on proliferation, migration, and invasion of liver cancer was reversed by RGS5 in vitro. A, Representative micrographs (left) and quantifications (right) of cell colonies in indicated liver cancer cell lines, as determined by colony formation assay. B, The proliferation ability of RGS5-intervened HepG2-shATE1 and HCCLM3-ATE1 cell lines was monitored by MTT assay. C, DNA-content staining based cell-cycle analysis in RGS5-intervened HepG2-shATE1 and HCCLM3-ATE1 cell lines. DNA was stained by Propidium iodide. Wound-healing assays (D) and transwell invasion assays (E) were subjected to detect the migration and invasion capacity of RGS5-intervened HepG2-shATE1 and HCCLM3-ATE1 cell lines (left). The histograms (right) of all images showed the statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001 based on the Student t test. Error bars, SD.
RGS5 reduces the phosphorylation of β-catenin through GS3K-β
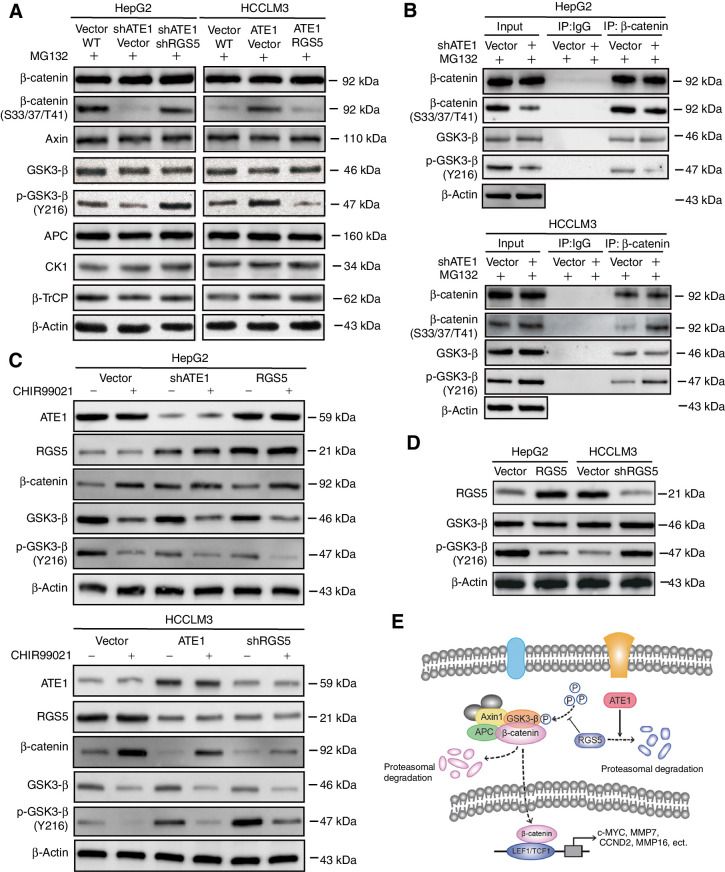
We have shown that ATE1 accelerated the degradation of β-catenin in liver cancer cell lines through regulating RGS5 degradation. However, how RGS5 regulates the degradation of β-catenin remains unknown. Previous studies have shown that β-catenin degradation mainly depends on the cytoplasmic destruction complex, which consists of Axin, APC, GSK3-β, and CKI (23, 24). The destruction complex phosphorylates β-catenin at N-terminal Thr41, Ser37, and Ser33 residues. Phosphorylated β-catenin is subsequently ubiquitinated by the F-box-containing protein β-TrCP ubiquitin E3 ligase (25–27). In consequence, it is necessary to determine whether potential change has happened to the destruction complex and β-TrCP in ATE1- and RGS5-intervened HCC cell lines. We treated HCCLM3, HepG2, and their lentivirus-infected cells with MG132. Then, the expression of the destruction complex members and β-TrCP were monitored. As predicted, the phosphorylation of β-catenin (S33/37/T41) was reduced after ATE1 knockdown in HepG2 cell line, while it was reverted as RGS5 knockdown in HepG2-shATE1 cell line. The opposite results were shown in HCCLM3 and its ATE1- and RGS5-intervened cell lines (Fig. 7A). It was verified again that ATE1 accelerated the degradation of β-catenin depending on phosphorylation of β-catenin through regulating RGS5 degradation, and RGS5 decreased the phosphorylation of β-catenin (S33/37/T41). Interestingly, we also found that the phosphorylation of GSK3-β (Y216) had a negative correction with RGS5 expression after ATE1 and/or RGS5 expression intervened in HepG2 and HCCLM3 cell lines, but the total GSK3-β expression and the other members of the destruction complex kept stable. To confirm that p-GSK3-β (Y216) was the key regulator, which directly participates in the phosphorylation of β-catenin (S33/37/T41), and not the side effect, HCCLM3, HepG2, and their lentivirus-infected cells were treated with MG132 and subjected to β-catenin immunoprecipitation. The total β-catenin, p-β-catenin (S33/37/T41), total GSK3-β, and p-GSK3-β (Y216) expression levels were monitored by Western blot analysis. Similarly, p-GSK3-β (Y216) expression decreased in HepG2-shATE1 cells and it increased in HCCLM3-ATE1 cells, whereas the total β-catenin and GSK3-β expression kept stable (Fig. 7B). The p-β-catenin (S33/37/T41) expression had a positive correlation with p-GSK3-β (Y216) expression in live cancer cell lines. For further affirming that the activity of GSK3-β was regulated by RGS5 rather than the other arginylation substrates of ATE1, we treated each cell lines with or without CHIR99021 (a GSK3 inhibitor, 5 μmol/L). Then we monitored the expression of β-catenin, GSK3-β, and p-GSK3-β (Y216), respectively. The results showed that CHIR99021 did not affect the expression of ATE1 or RGS5. Moreover, CHIR99021 can cooperate with shATE1 or RGS5 to promote Wnt/β-catenin signaling pathway, but overexpression of ATE1 or shRGS5 did not entirely reverse the effect of GSK3 inhibitor. These results suggested that ATE1-RGS5 may serve as the upstream regulator of GSK3 (Fig. 7C). Furthermore, we upregulated RGS5 solely in HepG2 cell line and downregulated it solely in HCCLM3 cell line. Expression level of RGS5 was identified by qRT-PCR and Western blotting (Fig. 7D; Supplementary Fig S2D). Then we determined GSK3-β and p-GSK3-β (Y216) expression. As predicted, p-GSK3-β (Y216) expression was reduced in HepG2-RGS5 cell line and enhanced in HCCLM3-shRGS5 cell line. The total GSK3-β expression kept stable as usual.
Figure 7.
RGS5 reduces the phosphorylation of β-catenin through GS3K-β. A, HepG2, HCCLM3, and their lentivirus-infected cells were treated with proteasome inhibitor MG132 for 6 hours. Treated cells were collected, lysed, the expression of β-catenin destruction complex members was monitored by Western blot analysis using the indicated antibodies. B, HepG2, HCCLM3 and their lentivirus-infected cells were treated with proteasome inhibitor MG132 for 6 hours. Treated cells were collected, lysed, and used for β-catenin immunoprecipitation, The expression of β-catenin, phosphorylation β-catenin, GSK3-β and phosphorylation GSK3-β were monitored by Western blot analysis using the indicated antibodies. C, HepG2, HCCLM3, and their lentivirus-infected cells were treated with or without CHIR99021 (5 μmol/L), the expression of β-catenin, GSK3-β, and phosphorylation GSK3-β (Y216) were monitored by Western blot analysis using the indicated antibodies. D, Phosphorylation level of GSK3-β was detected by Western blot analysis using the indicated antibodies in RGS5-intervened or RGS5-controled HepG2 and HCCLM3 cells. E, The schematic diagram of ATE1 suppresses WNT signaling pathway through degradation of β-catenin induced by RGS5.
Taken together, as shown in Fig. 7E, we indicated that ATE1 influenced the steady-state level of β-catenin through regulating RGS5 degradation, which was likely to regulate the phosphorylation of GSK3-β.
Discussion
The high recurrence and metastasis rates after liver resection are the main cause of death in patients with HCC (4). In this study, we found that a downregulation of ATE1 was commonly detected in HCC, and was correlated with poor prognosis of patients with HCC. We further showed that downregulation of ATE1 promoted the proliferation, invasion, and migration capacity in liver cancer by inhibiting the degradation of β-catenin to activate Wnt signaling pathway. Moreover, we demonstrated the relationship between RGS5 as a downstream key of ATE1 and the activity of cytoplasmic destruction complex, which dominated the degradation of β-catenin. Thus, ATE1 is a novel suppressor in liver cancer and could serve as a potentially valuable prognostic biomarker.
ATE1 is evolutionarily conserved in eukaryotes and in large subset bacteria (28). Hundreds of arginylation substrates have been identified and these substrates include cytoskeletal, chromosomal, and signaling proteins, transcription and translation factors, metabolic enzymes, and molecular chaperones and so on (5–9). However, the understanding of the molecular mechanism underlying the role of ATE1 in physiology and pathology is still limited, especially in cancer development. Previous reports already found that the target proteins of ATE1 involved in mitosis, chromatin structure, apoptosis (29–31), and ATE1 was essential for stress-induced cell-cycle checkpoints and/or apoptosis in mouse fibroblasts (17). It was also shown that silencing or inhibiting ATE1 disrupted E-cadherin–mediated cell–cell contacts in MCF-7 human breast carcinoma which probably promoted cell migration (32). Consistently, in our study, we observed that ATE1 inhibited the proliferation, migration and invasion of HCC in vitro and in vivo. A recent study whose observation had a slight difference from ours reported that no effects of ATE1 on the growth of prostate cancer cells were observed in nonstressed, nonconfluent conditions, but reduction of ATE1 retains cellular viabilities under chemical stressors or radiation (18). We attribute this to the tumor specificity that HCC is a kind of continuous enduring hypoxia condition tumor but not a hormone-derived tumor type. We first observed that ATE1 had low expression in HCC patients' samples and was correlated with aggressive clinicopathologic features. Low ATE1 expression predicted shorter OS and worse DFS than high ATE1 expression in patients with HCC. We also found that low ATE1 expression in liver cancer cell–derived orthotopic tumor showed more malignant pathologic feature than high ATE1 expression. These observations confirmed the suppressor and potential prognostic indicator role of ATE1 in liver cancer.
Wnt signaling pathway is critical for embryo development, angiogenesis, stem cell differentiation, and cancer development (33, 34). We first found the regulatory relationship between ATE1 and Wnt signaling pathway by using Cignal Finder Cancer 10-Pathway Reporter Array, which detected the transcriptional activity of β-catenin. Moreover, we observed that ATE1 inhibited the transcriptional activity of β-catenin by accelerating its degradation. Here, previous studies did not provide any evidence or possibility that β-catenin was an arginylation substrate of ATE1. It means an important key intermediate must exist between ATE1 and β-catenin. So we focused on the signaling protein of ATE1 arginylation substrates. The regulator of G-protein signaling (RGS) proteins negatively regulate G-protein–mediated receptor signaling pathways by acting as GTPase-activating proteins (GAP) and stimulate the hydrolysis of the Gα-bound GTP back to GDP (35). Researchers have demonstrated that RGSs played a critical role in the formation of blood vessels, in blood pressure regulation, and in tumors and inflammatory diseases (36–38). Interestingly, RGS4, RGS5, and RGS16 were the signaling proteins of ATE1 arginylation substrates and were degraded through ATE1-dependent N-terminal pathway rule and ubiquitination. We first affirmed that RGS5 expression, but not RGS4 or RGS16, got a strongly positive correlation with β-catenin expression. Then, we observed that the phosphorylation of GSK3-β reduced in ATE1 knockdown liver cancer cells. This observation could be reversed by RGS5 knockdown.
These results indicated an important signaling pathway mechanism of ATE1 regulating the proliferation, migration, and malignancy degree in liver cancer. However, the exact mechanism how RGS5 regulates the activity of GSK3-β remains unknown, which requires further research.
Authors’ Disclosures
C. Xu reports grants from National Natural Science Foundation of China during the conduct of the study. Y. Li reports grants from National Natural Science Foundation of China during the conduct of the study. B. Sun reports grants from National Natural Science Foundation of China during the conduct of the study. F. Zhong reports grants from National Natural Science Foundation of China during the conduct of the study. L. Yang reports grants from National Natural Science Foundation of China during the conduct of the study. No other disclosures were reported.
Acknowledgments
This work was supported by the Key Project of the National Natural Science Foundation of China (grant number 81330057); the National Natural Science Foundation of China (grant number 81773139); the National Science and Technology Major Project (grant number 2017ZX10203207–002–003). The authors thank Prof. Qiong-Qiong He and Prof. Geng-Qiu Luo (Department of Pathology, Xiangya Hospital of Central South University, Hunan, China) for the help of pathologic diagnoses and guidance.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Contributions
C. Xu: Conceptualization, formal analysis, methodology, writing–original draft, writing–review and editing. Y.-M. Li: Software, methodology. B. Sun: Investigation, methodology. F.-J. Zhong: Formal analysis, investigation. L.-Y. Yang: Resources, data curation, supervision, project administration.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301–14. [DOI] [PubMed] [Google Scholar]
- 3. Vogel A, Saborowski A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat Rev 2019;82:101946. [DOI] [PubMed] [Google Scholar]
- 4. Finn RS, Zhu AX, Farah W, Almasri J, Zaiem F, Prokop LJ. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: a systematic review and meta-analysis. Hepatology 2018;67:422–35. [DOI] [PubMed] [Google Scholar]
- 5. Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102:15030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu RG, Sheng J, Qi X, Xu Z, Takahashi TT, Varshavsky A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 2005;437:981–6. [DOI] [PubMed] [Google Scholar]
- 7. Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem 2000;275:22931–41. [DOI] [PubMed] [Google Scholar]
- 8. Saha S, Kashina A. Posttranslational arginylation as a global biological regulator. Dev Biol 2011;;358:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J, Han X, Wong CC, Cheng H, Aslanian A, Xu T, et al. Arginyltransferase ATE1 catalyzes midchain arginylation of proteins at side chain carboxylates in vivo. Chem Biol 2014;;21:331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rai R, Wong CC, Xu T, Leu NA, Dong DW, Guo C, et al. Arginyltransferase regulates alpha cardiac actin function, myofibril formation and contractility during heart development. Development 2008;135:3881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, et al. An essential role of N-terminal arginylation in cardiovascular development. Science 2002;297:96–9. [DOI] [PubMed] [Google Scholar]
- 12. Verma K P Barah, Saha S. Arginylation regulates adipogenesis by regulating expression of PPARγ at transcript and protein level. Biochim Biophys Acta Mol Cell Biol Lipids 2019;;1864:596–607. [DOI] [PubMed] [Google Scholar]
- 13. Batsios P, Ishikawa-Ankerhold HC, Roth H, Schleicher M, Wong CCL, Müller-Taubenbergera A. Ate1-mediated posttranslational arginylation affects substrate adhesion and cell migration in Dictyostelium discoideum. Mol Biol Cell 2019;30:453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cornachione AS, Leite FS, Wang J, Leu NA, Kalganov A, Volgin D, et al. Arginylation of myosin heavy chain regulates skeletal muscle strength. Cell Rep 2014;8:470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, Han X, Leu NA, Sterling S, Kurosaka S, Fina M, et al. Protein arginylation targets alpha synuclein, facilitates normal brain health, and prevents neurodegeneration. Sci Rep 2017;7:11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Pavlyk I, Vedula P, Sterling S, Leu NA, Dong DW, et al. Arginyltransferase ATE1 is targeted to the neuronal growth cones and regulates neurite outgrowth during brain development. Dev Biol 2017;430:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rai R, Zhang F, Colavita K, Leu NA, Kurosaka S, Kumar A, et al. Arginyltransferase suppresses cell tumorigenic potential and inversely correlates with metastases in human cancers. Oncogene 2016;35:4058–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Birnbaum MD, Zhao N, Moorthy BT, Patel DM, Kryvenko ON, Heidman L, et al. Reduced Arginyltransferase 1 is a driver and a potential prognostic indicator of prostate cancer metastasis. Oncogene 2019;38:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao S, Chang RM, Yang MY, Lei X, Liu X, Gao WB, et al. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology 2016;63:1256–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst 2018;110:803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu L, Dai Y, Chen J, Zeng T, Li Y, Chen L, et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3β/Snail signaling. Hepatology 2014;59:531–43. [DOI] [PubMed] [Google Scholar]
- 22. Kapanadze T, Gamrekelashvili J, Ma C, Chan C, Zhao F, Hewitt S, et al. Regulation of accumulation and function of myeloid derived suppressor cells in different murine models of hepatocellular carcinoma. J Hepatol 2013;59:1007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012;149:1245–56. [DOI] [PubMed] [Google Scholar]
- 24. Roberts DM, Pronobis MI, Poulton JS, Waldmann JD, Stephenson EM, Hanna S, et al. Deconstructing the ßcatenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Mol Biol Cell 2011;22:1845–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002;108:837–47. [DOI] [PubMed] [Google Scholar]
- 26. Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 1997;16:3797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, et al. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J 1999;18:2401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balzi E, Choder M, Chen WN, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem 1990;265:7464–71. [PubMed] [Google Scholar]
- 29. Wong CC, Xu T, Rai R, Bailey AO, Yates JR, Wolf YI, et al. Global analysis of posttranslational protein arginylation. PLoS Biol 2007;5:e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. PNAS 2012;109:E1839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saha S, Wong CC, Xu T, Namgoong S, Zebroski H, Yates JR, et al. Arginylation and methylation double up to regulate nuclear proteins and nuclear architecture in vivo. Chem Biol 2011;18:1369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eisenach PA, Schikora F, Posern G. Inhibition of arginyltransferase 1 induces transcriptional activity of myocardin-related transcription factor A (MRTF-A) and promotes directional migration. J Biol Chem 2014;289:35376–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006;127:469–80. [DOI] [PubMed] [Google Scholar]
- 34. MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 2002;54:527–59. [DOI] [PubMed] [Google Scholar]
- 36. Squires KE, Montañez-Miranda C, Pandya RR1, Torres MP, Hepler JR. Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease. Pharmacol Rev 2018;70:446–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie Z, Chan EC, Druey KM. R4 Regulator of G Protein Signaling (RGS) Proteins in Inflammation and Immunity. AAPS J 2016;18:294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther 2007;116:473–95. [DOI] [PMC free article] [PubMed] [Google Scholar]