Abstract
A recombinant Newcastle Disease Virus (NDV), encoding either a human (NDVhuGM-CSF, MEDI5395) or murine (NDVmuGM-CSF) GM-CSF transgene, combined broad oncolytic activity with the ability to significantly modulate genes related to immune functionality in human tumor cells. Replication in murine tumor lines was significantly diminished relative to human tumor cells. Nonetheless, intratumoral injection of NDVmuGM-CSF conferred antitumor effects in three syngeneic models in vivo; with efficacy further augmented by concomitant treatment with anti–PD-1/PD-L1 or T-cell agonists. Ex vivo immune profiling, including T-cell receptor sequencing, revealed profound immune-contexture changes consistent with priming and potentiation of adaptive immunity and tumor microenvironment (TME) reprogramming toward an immune-permissive state. CRISPR modifications rendered CT26 tumors significantly more permissive to NDV replication, and in this setting, NDVmuGM-CSF confers immune-mediated effects in the noninjected tumor in vivo. Taken together, the data support the thesis that MEDI5395 primes and augments cell-mediated antitumor immunity and has significant utility as a combination partner with other immunomodulatory cancer treatments.
Introduction
Treatment of cancer has been revolutionized by the development of immune checkpoint blockade (ICB) therapies; which confer durable clinical benefit in a subset of patients (1, 2). Defining predictive biomarkers of response to ICB therapies has become a priority, with significant progress made with respect to genetic signatures (e.g., microsatellite instability) and inhibitory receptor ligand expression patterns (e.g., PD-L1) linked to treatment outcomes (3, 4). Clinical response to ICB is more frequently observed in “inflamed” tumors, that is, those often associated with high mutational burden and/or high PD-L1 expression and higher densities of cytotoxic CD8+ T lymphocytes (CTLs) and NK cells. Conversely, “cold” tumors have a paucity of these effector populations and tend to be unresponsive to ICB (5). In some cancers, T cells may be present but either “excluded” from the tumor nests or actively suppressed by the local tumor microenvironment (TME; ref. 6). Identifying treatments that transform TMEs from an ICB resistant to sensitive phenotype is a priority to enable rational immunotherapy combinations with enhanced clinical efficacy.
Oncolytic viruses (OVs) could potentiate immunologic responses in patients for whom ICB therapy is currently not effective (7). OVs selectively infect and replicate in tumor cells, leading to oncolysis, an inherently immunogenic process, with release of tumor-associated antigens, damage-associated molecular patterns (DAMPs), and proinflammatory cytokines culminating in the activation of antigen-presenting cells, recruitment of immune effector cells (e.g., T and NK cells) and de novo priming of T-cell–mediated antitumor immunity (8). The intrinsic oncolytic activity and immunomodulatory properties of some viruses can be further augmented by introduction of immunomodulatory transgenes; with expression limited to the TME, due to tumor-restricted viral replication (8, 9).
Newcastle Disease Virus (NDV) has a number of distinguishing characteristics that make it an attractive OV platform: (i) low seroprevalence, (ii) precedence of safe use in humans (10), (iii) ability to infect a wide variety of tumor types through binding to sialic acids on the tumor cell surface, (iv) potential for intravenous administration, and (v) option to include therapeutic transgenes with tumor-restricted expression linked to viral replication (11).
MEDI5395, is a genetically modified NDV that retains oncolytic activity while also engineered to express human GM-CSF from infected tumor cells (12). Infection of human peripheral blood mononuclear cells (PBMCs) with MEDI5395 leads to polarization of monocyte populations and an enhancement in cell surface molecules and cytokine profiles associated with an antitumor phenotype. NDV-mediated expression of GM-CSF results in enhancement of monocyte activation compared with NDV lacking the transgene (13).
In this study, we demonstrate that MEDI5395 has broad oncolytic and proinflammatory activity across a range of preclinical human tumor models. We extend these observations to immune competent murine syngeneic models using a surrogate NDV (NDVmuGM-CSF) where we show that treatment leads to repolarization of the TME and to robust antitumor activity. Efficacy was further potentiated when NDVmuGM-CSF was combined with ICB antibodies or T-cell agonists. NDVmuGM-CSF treatment induced immune-mediated anenestic (abscopal) effects in CT26 tumors modified to facilitate enhanced viral replication through CRISPR-mediated disruption of Rig-I and Ifnar1; consistent with induction of a systemic adaptive immune response. These results support the development of MEDI5395 as a broad immuno-oncology–based therapeutic and combination partner with T-cell–targeted immunotherapies. The data also provide important insight into potential patient stratification strategies, and early biomarkers of response that could be used in the clinic.
Materials and Methods
In vitro tumor cell infectivity assay
A total of 10,000 cells per well were plated in a 96-well plate in cell-specific full-growth medium and allowed to adhere for 4 hours. Cells were subsequently treated with a dose range of virus [multiplicity of infection (MOI) 10–0.00001]. Staurosporine (5 μmol/L; Sigma) was used as a positive death-inducing control. At 72 hours after infection, the supernatant from cells infected at MOI 0.1 was used to assess GM-CSF expression levels by electrochemiluminescent immunoassay (Meso Scale Diagnostics), according to the manufacturer's instructions, and virus titer via plaque assay (12). Cell viability at each MOI was measured with CellTiter Glo (Promega). All experiments were performed in triplicate and were repeated at least four times. Details of all cell lines used can be found in Supplementary Table S1.
Transcriptomic analysis and cytokine measurement of virus-infected cells
NDV-GFP was used to determine the MOI that resulted in 100% infectivity of 6 × 105 cells in 6-well plates at 24 hours after infection for each cell line. For human cell lines, the following MOIs were used: HT1080, MOI 0.1; DU145, MOI 0.5; NCIH358, MOI 0.5; OVCAR4, MOI 0.5; CAL27, MOI 2.0; CT26, MOI 10; 4T1, MOI 5.0; and B16F10, MOI 0.5. Cells were incubated with virus for 24 hours, after which supernatant was removed and chemokine and cytokine levels were analyzed with Luminex-based assays (eBioscience).
Cells were either harvested for FACS analysis or used to prepare RNA according to the manufacturer's instructions (Qiagen). For microarrays, 100 ng of RNA was used and biological replicates were run for each condition. Whole-transcriptome amplification, reverse transcription and DNA labeling, fragmentation, and array hybridization were performed according to the manufacturer's instructions for Affymetrix HTA 2.0 arrays. Arrays were hybridized, washed, and scanned in randomized batches of eight to minimize batch effects. Files were processed with Affymetrix Expression Console, and differential expression was determined with Affymetrix Transcriptome Analysis Console.
Animal husbandry and in vivo experiments
In vivo experiments used 8- to 10-week-old female C57BL/6J or BALB/c mice (Charles River Laboratories) for syngeneic models. All experiments in the United Kingdom were conducted under a U.K. Home Office Project Licence in accordance with the U.K. Animals (Scientific Procedures) Act 1986 and in accordance with EU Directive EU 2010/63/EU. All experiments in the United States were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)–accredited and United States Department of Agriculture (USDA)–licensed facility. All mice were housed in sterile and standardized environmental conditions. Mice received autoclaved food and bedding and drinking water ad libitum. Mice were shaved on the right flank and anesthetized with isoflurane, and all tumor cells were implanted subcutaneously into the right flank. For the rechallenge or dual-flank model, the left flank was also used as an implantation site.
Group sizes for all studies were powered on the basis of existing tumor growth data to test for more than 50% tumor growth inhibition. Power calculation was conducted with a one-sided, two-sample t test at 5% significance. Animals were randomized to treatment groups on the basis of tumor volume before the first treatment. Tumors were routinely measured with calipers, and data were automatically captured with the dasHost system (Microsoft). A partial response to therapy was deemed to occur when median survival of that animal was greater than two SDs from the median survival of the control group. A complete response to therapy was deemed to occur when tumors had five sequential measurements below 200 mm3 following cessation of treatment.
NDVmuGM-CSF was administered intratumorally (i.t.) at 20 μL. Antibodies to PD-1 (clone RMP4–14; Bio X Cell), PD-L1 (mouse IgG1; AstraZeneca), GITRL-FP (mouse IgG2a; AstraZeneca), and OX40L-FP (mouse IgG1; AstraZeneca) were all administered intraperitoneally. For live imaging with NDV-luciferase, animals were injected with 100 μL of 33-mg/mL d-Luciferin (Perkin Elmer). Under recoverable isoflurane anesthesia, animals were imaged with the IVIS Bioluminescence System (Perkin Elmer) 10 minutes after intraperitoneal (i.p.) injection.
ELISpot detection of antigen-reactive T cells
Spleens were harvested from control or previously “cured” NDVmuGM-CSF–treated animals following CT26 tumor rechallenge. Spleens were dissociated to a single-cell suspension through a 70-μm nylon mesh and depleted of red blood cells with lysing buffer (Sigma) before being resuspended in media. As per manufacturer's instructions (MAB-TECH), 3 × 105 cells were plated into 96-well ELISpot PVDF plates (Millipore), which had been coated with anti-IFNγ capture antibody. Cells were pulsed with 10 μmol/L control (β-galactosidase peptide) or AH-1 peptide (both from MBL International). The assays were incubated at 37°C for 36 hours, and spots were detected as per the manufacturer's directions. Spots were enumerated with an ImmunoSpot Plate Reader (CTL Europe).
T-cell receptor sequencing from CT26 tumors
CT26 tumors were excised and DNA was prepared according to the manufacturer's instructions (Qiagen) and shipped to Adaptive Biotechnologies for T-cell receptor (TCR)-β sequencing and analysis with the ImmunoSeq platform.
Clustergram and abacus plot
Genes that were differentially expressed after NDV-GFP infection were identified by volcano plot analysis (twofold change, P < 0.05) and used for pathway analysis. Data for each cell line were submitted separately to the Reactome pathway engine, and all pathway results were merged in a single output file that was subsequently ranked by the FDR for each pathway. Abacus plots represent the median expression levels of the entire pathway members, which can be either up- or downregulated, and the clustergram highlights genes that were upregulated after NDV infection. All expression data can be found in Supplementary Materials.
CRISPR-Cas9 engineering and screening of mutated murine cancer cell lines
Rig-I (Ddx58) and Ifnar1 genes were disrupted with double-guided wild-type (WT) Cas9. Guide RNAs were designed against critical exons within each gene with proprietary software (AstraZeneca). Both guides were synthesized and cloned into DNA 2.0 vectors, one containing WT Cas9–2A-GFP (Rig-I) and the other WT Cas9–2A-RFP (Ifnar1). The two plasmids were transfected as single complexes into CT26 cells (ATCC) with Lipofectamine LTX (Life Technologies), and after 3 days, single cells were sorted on a FACSAria Sorter (Becton Dickinson) for GFP–red fluorescent protein (RFP) double-positive cells into 96-well plates. Colonies were imaged by whole-well imaging on an IncuCyte Zoom (Essen BioScience), and colonies with average growth rates were selected for target disruption screening. Gene disruption was assessed by either Western blotting or FACS expression, and respective target loss of function was confirmed by DNA sequencing.
Flow cytometry on in vitro and ex vivo samples
Animals were euthanized and spleens were removed and dissociated to a single-cell suspension through a 70-μm nylon mesh. Tumors were dissociated with a commercial kit (Miltenyi Biotec). Cells were stained with a fixable live/dead dye (Life Technologies), followed by blocking with anti-CD16/32 antibody (eBiosciences) and staining with cocktails of fluorescence-conjugated antibodies before fixation in 1% formaldehyde–PBS. Intracellular nuclear staining was carried out on fixed and permeabilized cells (FoxP3/Transcription Factor Staining Buffer Set; eBioscience). Stained cells were analyzed by flow cytometry (LSR Fortessa; BD Biosciences) and data analysis was performed in FlowJo software (Tree Star). Positive-staining gates were identified by comparison with cells stained with the full antibody panel minus the antibody of interest (FL-1 control). In vitro cells were washed once in PBS and then incubated in nonenzymatic cell dissociation solution for 10 minutes before being neutralized in relevant media, washed, and resuspended in PBS before staining was performed as described above. A representative gating strategy is shown in Supplementary Fig. S6.
Live imaging of NDV-GFP–infected cells
After infection with NDV-GFP at MOIs described above, cells were imaged with one field per well every 3 hours on an IncuCyte Zoom (Essen BioScience), and viral GFP expression was quantified in each cell line in real time and at various times over a period of 96 hours.
Statistical analysis
All in vivo data were collated in Microsoft Excel and transferred to Prism (GraphPad Software) for graphical representation. Tumor volumes were log transformed before statistical analysis. For comparison of tumor growth inhibition between groups, significant P values, if any, were obtained from two-way ANOVA corrected with the Tukey test for multiple comparisons. Results from survival studies were analyzed with a log-rank (Mantel–Cox) test, comparing only two survival curves at a time. P values were not adjusted for multiple testing. Where means of two groups were compared, then statistical analysis was performed using a two-tailed, unpaired t test with Welch correction.
Results
NDV drives a potent tumor-intrinsic inflammatory phenotype
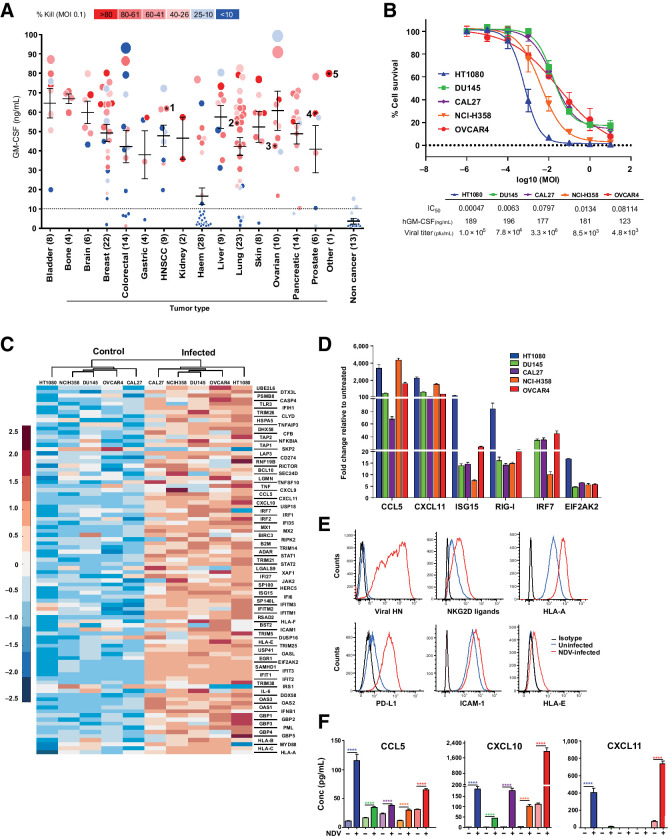
To test the oncolytic potency of MEDI5395, virus replication and oncolysis was measured across 176 human tumor cell lines, of which 82 (46%) showed more than 40% cytotoxicity/cell death 72 hours after infection (Fig. 1A). Using GM-CSF levels as a surrogate of virus replication (>10 ng/mL), we found that MEDI5395 infected 146 of 176 (83%) cell lines (Fig. 1A). To investigate the contribution of the GM-CSF transgene to oncolysis, we compared the oncolytic activity of MEDI5395 with a murine surrogate (NDVmuGM-CSF) because human GM-CSF is not bioactive in mice (14). NDVmuGM-CSF exhibited equivalent oncolytic potency across all cell lines tested, indicating that GM-CSF did not contribute to oncolytic activity (Supplementary Fig. S2C and S2D). To further demonstrate the oncolytic potency of MEDI5395, repeat local i.t. and systemic administration regimens of MEDI5395 were tested in vivo in a patient-derived triple-negative breast cancer (TNBC) xenograft model (Supplementary Fig. S1). Regardless of the route of administration, both MEDI5395 and NDVmuGM-CSF demonstrated potent oncolytic activity that depended on the virus's ability to replicate.
Figure 1.
MEDI5395 combines broad oncolytic activity with induction of immunologic changes within tumor cells in vitro. A, Output from >100 human cell lines infected with MEDI5395 at MOI 0.1. Potency of virus classified as percent kill (symbol colour) at 72 hours after infection and GM-CSF levels in supernatant (symbol size), which was used as a surrogate of susceptibility to virus. 1 = CAL27, 2 = NCI-H358, 3 = OVCAR4, 4 = DU145, and 5 = HT0180 cell lines. Each data point is an average of three individual replicates. B, Fifty percent inhibitory concentration (IC50) curves from the five sensitive cell lines identified in A at 72 hours after infection with MEDI5395. Data represent the mean of four biological replicates for each cell line. C, Clustergram analysis of RNA expression of 96 genes involved in immune-cell interactions at 24 hours after infection at MOIs that resulted in 100% infection. D, RT-PCR analysis of gene expression expressed as fold change relative to untreated levels at 24 hours after NDV-GFP infection at MOIs that resulted in 100% infection; n = 6 per group. Data are mean ± SEM. E, Representative FACS analysis histogram plots of cell surface marker expression at 24 hours after NDV-GFP infection from one of the five sensitive cell lines (CAL27). F, Levels of CCL5, CXCL10, and CXCL11 protein in supernatants from the same NDV-GFP–infected cells shown in D at 24 hours after infection. Statistical analysis was performed with an unpaired t test with Welch correction; data are mean ± SEM. *, P < 0.05; ****, P < 0.0001.
Five of the most sensitive cell lines (HT1080, DU145, CAL27, NCI-H358, and OVCAR4) were selected to further understand pathways that are altered after NDV infection (Fig. 1B).
These cells were infected with an NDV that can express GFP (NDV-GFP) at MOIs resulting in 100% infection, then assessed 24 hours postinfection for alterations in gene expression by transcriptomic microarray analysis (Affymetrix). More than 50% (11/20) of the most significantly altered pathways were involved in immune regulation. Hierarchical clustering of the 90 genes involved in these immune regulatory pathways segregated the cell lines into uninfected and infected groups (Fig. 1C). Gene expression changes in infected cells were confirmed by RT-PCR. As expected, genes involved in RNA virus–sensing, (RIG-I and IRF7) were upregulated alongside genes known to be downstream of a type I IFN (IFN-I) response ISG15, CCL5, and CXCL11, which have previously been demonstrated to drive a proinflammatory response in vivo (Fig. 1D; refs. 15, 16). Further analysis of cell surface and secreted proteins by flow cytometry and protein arrays, respectively, demonstrated that alterations in gene expression were matched by changes in protein levels. NDV-GFP–infected cells upregulated HLA-A, HLA-E, PD-L1, ICAM, and NKG2D ligands, all of which have been implicated in modulating the immune system (Fig. 1E; refs. 17, 18). Upregulation of the chemokines CCL5 (P < 0.0001 in all five cell lines), CXCL10 (P < 0.0001 in all five cell lines), and CXCL11 (P < 0.0001 in HT1080 and OVCAR4 cell lines) was also confirmed following analysis of cell supernatants collected from NDV-GFP–infected tumor cells (Fig. 1F). These data suggest that, in addition to a direct cytolytic effect, infection of tumor cells by MEDI5395 may lead to immune modulation through multiple pathways.
To further investigate the hypothesis that MEDI5395 exerts a proinflammatory effect in tumors, sensitivity to MEDI5395 infection was profiled in 11 syngeneic mouse cell lines. Sensitivity to MEDI5395 was greatly reduced in murine tumor cell lines compared with human cell lines; in line with data reported for other OVs including Maraba and Vesicular Stomatitis Viruses (19). Nonetheless, transgene expression could be detected in most murine tumor cell lines 72 hours after infection at MOI 0.1 (Supplementary Fig. S2A). Infection of these syngeneic mouse cell lines with MEDI5395 at MOI of 10 conferred broadly equivalent levels of oncolytic activity to that detected following infection of the human HT1080 cell line (Supplementary Fig. S2B). No difference in sensitivity to MEDI5395 or NDVmuGM-CSF, was observed in murine tumor cell lines (Supplementary Fig. S2E).
Despite reduced susceptibility, 100% infectivity of syngeneic murine cells could be achieved with NDV-GFP 24 hours postinfection by using higher MOIs than in the human tumor cell studies. Gene expression analysis of infected and uninfected tumor cells revealed that of the 15 differentially regulated pathways identified in syngeneic cell lines, 12 matched those identified in human tumor cell lines, and all were immune modulatory pathways (Supplementary Fig. S4). Expression levels of these immune pathway genes were again segregated into uninfected and infected cells after cluster analysis (Supplementary Fig. S3A). Immune cell surface markers PD-L1 and MHC-I were also upregulated after infection of these murine cell lines (Supplementary Fig. S2B). Although the pattern of altered gene expression associated with the antiviral response was similar between human and murine tumor cell lines, the absolute levels of changes in gene and protein expression between uninfected and infected murine tumor cells were several orders of magnitude larger than those observed in human cell lines. Potentially, this may be one mechanism by which murine tumor cell lines suppress replication and virus spread to neighboring cells (Supplementary Fig. S3D). Therefore, although sensitivity to MEDI5395 differed between mouse and human tumor cell lines, activation of similar pathways and cellular responses resulted at matched effective MOIs.
NDVmuGM-CSF modulates the TME resulting in antitumor activity in vivo
Three murine syngeneic tumor models, that differed in both their in vitro response to MEDI5395 and their in vivo immune TME profiles, were chosen to evaluate the antitumor efficacy of NDV. (i) CT26 model, characterized as immune responsive (inflamed TME, but low sensitivity to NDV); (ii) the B16F10 model (cold TME, most sensitive mouse cell line to NDV); and (iii) the 4T1 model (“immunosuppressed” TME, insensitive to NDV; ref. 20; Supplementary Fig. S2A). To reproduce the high MOIs required to observe any lytic activity in the mouse tumor cell lines in vitro, direct delivery of NDVmuGM-CSF into subcutaneously implanted tumors via i.t. route was utilized.
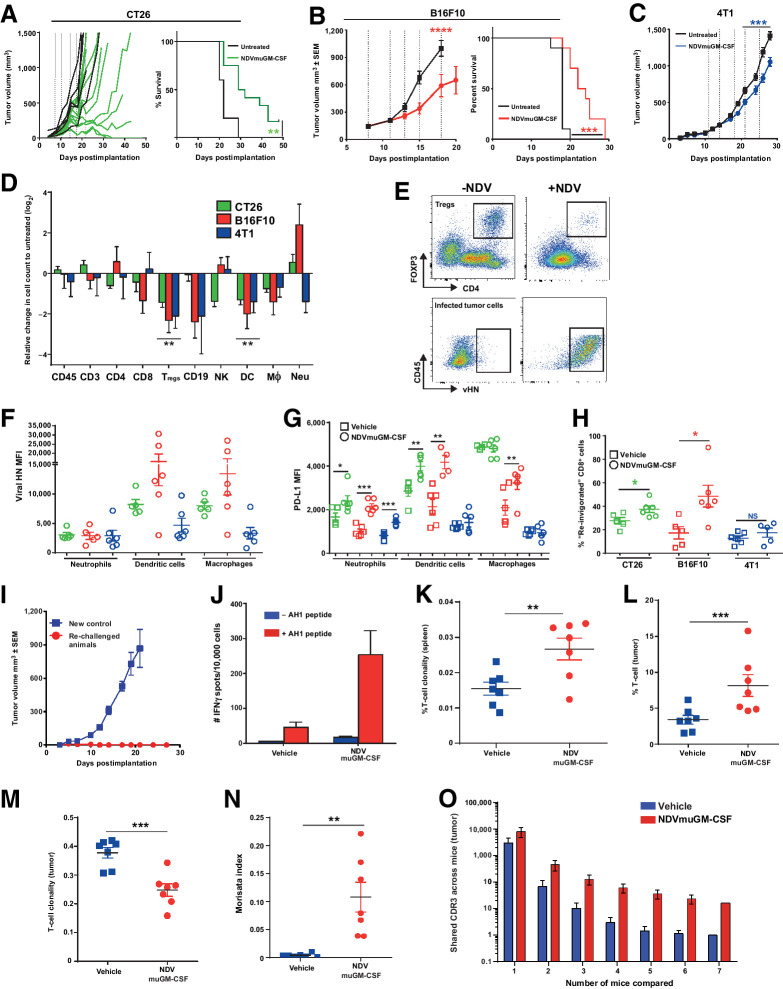
In the CT26 model, despite limited replication, NDVmuGM-CSF treatment resulted in significant antitumor activity and an increase in median survival times (P = 0.006), from 22 days in untreated controls to 30.5 days in NDVmuGM-CSF–treated animals; two of 12 animals had complete tumor regression, compared with no animals in the untreated control group (Fig. 2A). In the B16F10 model, tumor growth inhibition was observed in all treated animals during treatment, but the benefit was rapidly lost after treatment cessation. Antitumor activity resulted in a significant increase in median survival times (P = 0.0002), from 18 days in untreated controls to 23 days in NDVmuGM-CSF–treated animals, but no complete responses were observed (Fig. 2B). Finally, in the 4T1 model, the least sensitive cell line, a modest but reproducible antitumor effect (25% reduction, P = 0.0071) was achieved (Fig. 2C). Similar antitumor activity in these models was observed following treatment with MEDI5395 (Supplementary Fig. S3D and S3E).
Figure 2.
NDV infection remodeling of TME leading to immune-mediated antitumor activity. A, Tumor volumes after i.t. dosing with NDVmuGM-CSF in CT26 (n = 12), B, B16F10 (n = 10), and C, 4T1 (n = 10) syngeneic tumor models. Dashed vertical lines represent days of NDVmuGM-CSF injections (5 × 108 pfu). CT26 tumor growth data (mean ± SEM) shown as spider plots demonstrating heterogeneous antitumor response. D, Representative immunophenotyping of tumor immune infiltrate from three syngeneic models at 24 hours after third i.t. dose of 5 × 108 pfu at 8, 11, and 14 days postimplantation; n = 6 vehicle and 6 treated animals for each model. Data are mean ± SEM log2 values relative to untreated cell counts. NK = natural killer cells; CD19 = B cells; Mθ = macrophage. E, Representative pseudocolour dot plots showing Treg proportions in treated tumors and presence of NDV vHN protein on infected tumor cell surfaces. F, vHN protein expression (MFI) on cell surface of myeloid cell subsets in TME. Data are mean ± SEM. G, PD-L1 expression (MFI) on the surface of myeloid cell subsets in TME. Data are mean ± SEM. H, Detection of “reinvigorated” (PD-1+, Eomes+, Ki67+, Gzmb+) CD8+ T cells in TME. Data are mean ± SEM; *, P < 0.05. I, Tumor growth plots from control CT26 tumor-bearing animals or rechallenged CT26 “cured” animals after NDVmuGM-CSF treatment with CT26 cells on opposite flank. Data represent two separate experiments (controls, n = 3; rechallenged, n = 6). J, Detection of AH1 reactive T cells from splenocytes isolated from control or previously “cured” NDVmuGM-CSF treated animals following CT26 tumor rechallenge as judged by increased number of IFNγ-positive “spots” following stimulation with (red) or without (blue) AH1 peptide ex vivo. Data are mean ± SEM (n = 2 per group). K and L, Clonality of TCR sequencing isolated from spleen (K) and tumor (L). Values approaching 1 indicate a nearly monoclonal population of TCRs (controls, n = 8; virus treated, n = 7). M, Comparison of T-cell clonality from vehicle- and NDVmuGM-CSF–treated tumors. N, Morisita index quantitates the similarity between TCR repertoires in spleen and tumors of vehicle- and NDVmuGM-CSF–treated mice. O, Comparison of shared TCR sequences between individual animals in untreated and treated groups. For all comparisons between two data sets, statistical analysis was performed with unpaired t test with Welch correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Flow cytometric immune-phenotyping of resected tumors, 24 hours after the third i.t. administration of NDVmuGM-CSF revealed that several immune-cell populations present in the TME were differentially modulated; in a model-dependent manner (i.e., significantly altered in only one or two of the three models) following NDVmuGM-CSF treatment. Absolute numbers of regulatory T cells (Tregs; CD4+, Foxp3+) and dendritic cells (DC; CD11b+, CD11c+, MHC-II+) within the TME were significantly reduced by 1.8- to threefold and 2.2- to 2.5-fold, respectively, compared with untreated controls (Fig. 2D and E) across all three models after NDVmuGM-CSF treatment.
NDV also modulated the tumor resident/infiltrating T-cell compartment. NDVmuGM-CSF treatment in CT26 and B16F10 tumor-bearing mice conferred a significant increase in the proportion of “reinvigorated” T cells (CD8+, PD-1+, Eomes+, Ki67+, and Granzyme B+; ref. 21; 1.4- and 2.8-fold, respectively) observed in the TME compared with untreated controls (P = 0.0359 and 0.0192, respectively; Fig. 2H). Notably, rechallenge of NDVmuGM-CSF–treated CT26 tumor-bearing mice with a second inoculation of CT26 cells on the opposite flank, failed to induce tumor development (Fig. 2I); reflective of protective immunologic memory. In addition, animals who were rechallenged with CT26 had higher number of T cells that reacted to the dominant CT26 peptide AH1 (22) as judged by increased number of IFNγ-positive “spots” (Fig. 2J), indicating that a proportion of T cells now recognize tumor antigens.
Virally infected cells were identified by FACS using an anti-NDV hemagglutinin-neuraminidase (vHN) antibody. FACS analysis identified not only vHN-positive tumor cells but also vHN-positive myeloid populations (macrophages, DCs, and neutrophils) in all three models (Fig. 2E and F). Virus infection was associated with a significant upregulation of PD-L1 on the surface of vHN+ myeloid populations within the TME compared with untreated animals (Fig. 2G).
To better understand the adaptive immune response observed in CT26 tumors, we explored the TCR repertoire following viral infection in tumor-bearing mice. Mice received three i.t. administrations of 5 × 108 plaque-forming units (pfu) of NDVmuGM-CSF, and the TCR repertoire from tumors and spleens were analyzed 7 days after the final injection. Enhanced TCR clonality was only observed in spleens of CT26 tumor–bearing mice treated with NDVmuGM-CSF; indicating a systemic effect to local administration of NDVmuGM-CSF (P = 0.011; Fig. 2K). Using the number of TCR copies as a surrogate for number of resident/infiltrating T cells revealed a significant increase in T-cell number within NDVmuGM-CSF–infected tumors compared with controls (P = 0.02; Fig. 2L), This correlates with a significantly reduced clonal TCR repertoire within NDVmuGM-CSF–treated tumors (P = 0.007; Fig. 2M), suggesting that the T-cell response may be focused against specific antigens (virus or tumor derived) present within the TME. There was a significant overlap in TCR sequences isolated from tumor and spleens indicating that T-cell clones from the periphery are infiltrating infected tumors (P = 0.0081; Fig. 2N). Comparison of shared sequences between NDVmuGM-CSF–treated individuals was indicative of an adaptive immune response that was conserved between individual animals (Fig. 2O).
NDV sensitizes tumors to immune checkpoint blockade and T-cell agonist agents
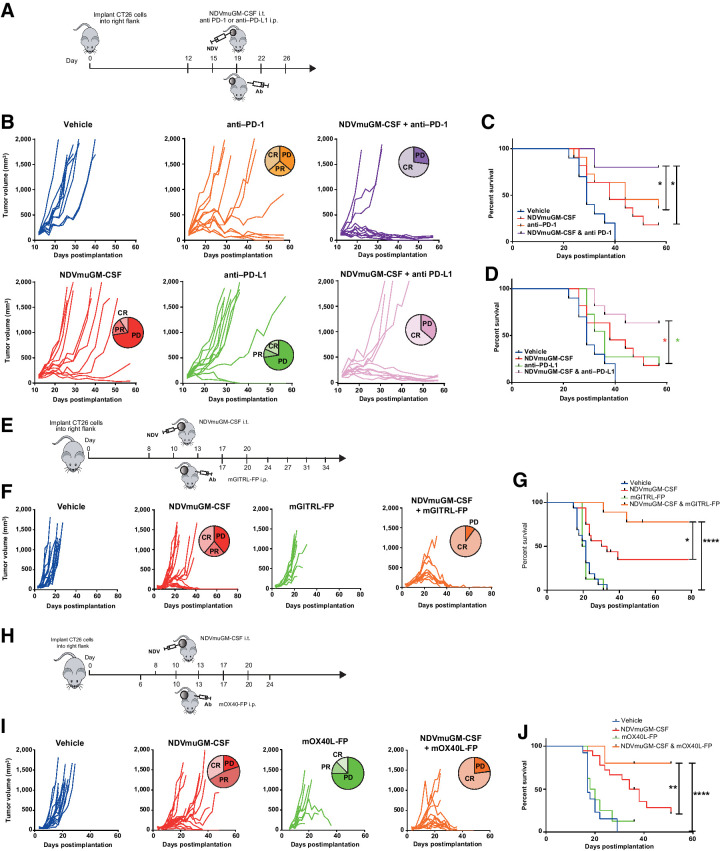
Given the immune-modulatory effects observed with NDVmuGM-CSF, we next explored whether the antitumor response could be augmented by combination with either anti-PDX (PD-L1 or PD-1) antibodies or T-cell agonists. To test this hypothesis, NDVmuGM-CSF was combined with either an anti–PD-1 or an anti–PD-L1 antibody (Fig. 3A). NDVmuGM-CSF monotherapy significantly increased median survival time to 38 days compared with 29 days in vehicle control group (P = 0.277) while anti–PD-1 and PD-L1 treatment alone increased median survival time to 44 days and 36 days, respectively (P = 0.005 and P = 0.077). The combination of NDVmuGM-CSF with anti–PD-1 resulted in a significant increase in median survival time compared with either PD-1 (P = 0.0499) or NDVmuGM-CSF (P = 0.007) monotherapy. The combination of NDVmuGM-CSF with anti–PD-L1 resulted in a significant increase in median survival time compared with either PD-L1 (P = 0.025) or NDVmuGM-CSF (P = 0.041) monotherapy. No significant difference in efficacy was observed between the NDVmuGM-CSF and PD-L1 or NDVmuGM-CSF and PD-1 combination groups (Fig. 3B–D).
Figure 3.
Combination of NDV with immune checkpoint blockade or T-cell agonists in preclinical models. A, Representation of study design combining local NDVmuGM-CSF administration with antibodies to PD-1 or PD-L1 in the CT26 model. B, Spider plots showing tumor growth kinetics in individual animals in each group (n = 10 per group), data are representative of three separate experiments. C and D, Kaplan–Meier survival analyses of NDVmuGM-CSF plus C, anti–PD-1 or D, PD-L1 antibody combination therapy compared with monotherapies. E, Schematic representation of study design combining NDVmuGM-CSF administration with mGITRL-FP agonist in the CT26 model. F, Spider plots showing tumor growth kinetics of the individual animals in each group (n = 10 per group except NDVmuGM-CSF, n = 18). G, Kaplan–Meier survival analysis of NDVmuGM-CSF plus mGITRL-FP agonist combination therapy compared with vehicle control and NDVmuGM-CSF and mGITRL-FP monotherapies. H, Schematic representation of study design combining local administration of NDVmuGM-CSF with mOX40L-FP agonist in CT26 model. I, Spider plots showing tumor growth kinetics of the individual animals in each group (n = 10 per group except NDVmuGM-CSF and NDVmuGM-CSF + OX40L-FP, n = 18). J, Kaplan–Meier survival analysis of NDVmuGM-CSF plus mOX40L-FP agonist combination therapy compared with monotherapy. CR, complete response; PD, progressive disease; PR, partial response. Significance in survival was assessed by log–rank (Mantel–Cox) test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To examine the combination potential of NDV with T-cell agonists; NDVmuGM-CSF was combined with a murine GITR ligand fusion protein (mGITRL-FP) or a murine OX40 ligand fusion protein (mOX40L-FP; refs. 23, 24). Combining NDVmuGM-CSF with mGITRL-FP, which had no antitumor activity as a monotherapy (Fig. 3E and F) resulted in a significant increase in median survival times compared with vehicle (22 days, P < 0.001) and either mGITRL-FP (21 days, P = 0.0018) or NDVmuGM-CSF (32.5 days, P = 0.0300) monotherapy. Notably, treatment with the combination of NDVmuGM-CSF and mGITRL-FP resulted in complete regression of tumors as large as approximately 700 mm3 (Fig. 3G). NDVmuGM-CSF combined with mOX40L-FP resulted in a significant increase in median survival times compared with vehicle (17 days, P < 0.0001) or either mOX40L-FP (19 days, P < 0.0001) or NDVmuGM-CSF (36 days, P = 0.0075) monotherapy (Fig. 3H–J). These data indicate that NDVmuGM-CSF can enhance the activity of multiple immunotherapeutic agents and support the clinical evaluation of several IO combination strategies with MEDI5395.
Enhanced viral replication delivers enhanced antitumor activity in vivo
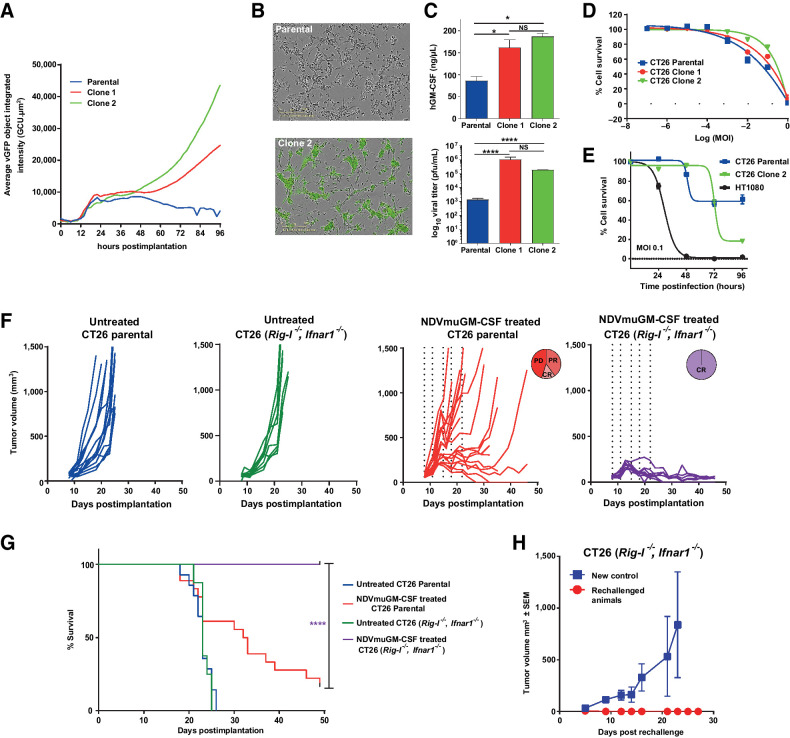
Because of the intrinsic differences between murine and human tumor cells to support NDV replication and oncolysis, currently there is no suitable preclinical model that can be used to explore oncolytic activity or sustained expression of the therapeutic GM-CSF transgene (which is driven by virus replication) in the presence of a fully intact immune system. In an effort to enhance virus susceptibility in murine cell lines, the retinoic acid–inducible gene I (Rig-I) and IFN-α/β receptor subunit 1 (Ifnar1) genes were deleted in a CT26 syngeneic tumor line using CRISPR gene editing. Successful targeting of both loci and complete loss of protein expression were determined by Western blot and flow cytometry, respectively (Supplementary Fig. S5A and S5B). CT26 Rig-I−/−, Ifnar1−/− clones were functionally screened with an NDV-GFP virus for evidence of enhanced sensitivity to NDV infection at MOI 0.1. CT26 Rig−/−, Ifnar1−/− cells were able to support greater levels of virus replication compared with the parental cell line for at least 96 hours, as measured by GFP intensity on a per-cell basis (Fig. 4A). Furthermore, virus spread appeared to be enhanced in CT26 Rig-I−/−, Ifnar1−/− cells at 72 hours (Fig. 4B). At 72 hours after infection with MEDI5395, there were significantly higher levels of infectious virus particles (>2 log) and GM-CSF (GM-CSF) transgene protein (>twofold value) in CT26 Rig-I−/−, Ifnar1−/− supernatants than in the parental cell line (Fig. 4C). This enhanced virus replication did not result in significantly increased oncolytic activity at 72 hours (Fig. 4D). However, a decrease in cell survival was observed at 96 hours after infection, demonstrating that the engineered cell lines remained less sensitive than the control human HT1080 cell line to NDV oncolysis (Fig. 4E). In vivo validation of CT26 Rig-I−/−, Ifnar1−/− cells with an NDV-luciferase virus revealed that enhanced susceptibility to NDV infection was retained in vivo after a single i.t. administration (Supplementary Fig. S5F and S5G). An enhancement in levels of viral replication combined with a limited impact on sensitivity to oncolysis was observed when these two deletions were introduced into the B16F10 syngeneic cell lines (Supplementary Fig. S5C–S5E).
Figure 4.
Genetic modifications of syngeneic tumor cells modulates susceptibility to NDV infection in vitro and in vivo. A, Quantitative measurement of GFP intensity per individual cell over time after infection of CT26 Rig-I−/−, Ifnar1−/− cells with NDV-GFP. B, Representative Incucyte images of CT26 parental and Rig-I−/−, Ifnar1−/− (clone 2) cells at 72 hours after infection with NDV-GFP at MOI 0.1. C, hGM-CSF transgene expression levels (top) and virus titers (bottom) at 72 hours after infection at MOI 0.1 from parental CT26 cell line and a CT26 Rig-I−/−, Ifnar1−/− clones. Data represent four separate experiments. *, P = 0.05; ****, P < 0.0001; NS, not significant. D, Oncolytic potency of MEDI5395 (percent cell survival) in CT26 parental and CT26 Rig-I−/−, Ifnar1−/− clones. Data represent four separate experiments. E, Comparison of percent cell death observed over 96 hours after MEDI5395 infection at MOI 0.1 in CT26 and CT26 Rig-I−/−, Ifnar1−/− clone 2 versus HT1080 human xenograft cell line. Data represent two separate experiments. F, Representative spider plots showing tumor growth kinetics of individual animals implanted with CT26 parental cells (blue lines) or CT26 Rig-I−/−, Ifnar1−/− cells (green lines), alongside growth kinetics from CT26 parental tumors (red lines) and CT26 Rig-I−/−, Ifnar1−/− tumors (purple lines) treated with i.t. NDVmuGM-CSF at 5 × 108 pfu. Vertical dashed lines represent days of dosing. CR = complete response; PD = progressive disease; PR = partial response. G, Kaplan–Meier survival analysis of untreated and NDVmuGM-CSF–treated animals implanted with the indicated tumor. H, Tumor growth kinetics after rechallenge of “cured” CT26 Rig-I−/−, Ifnar1−/− tumor-bearing animals with new CT26 Rig-I−/−, Ifnar1−/− cells on the opposite flank (red lines; n = 6), compared with tumors grown in naïve animals (blue lines; (n = 3). Significance in survival was assessed by log–rank (Mantel–Cox) test.
In vivo, the CT26 Rig-I−/−, Ifnar1−/− modified model had similar tumor growth kinetics and survival times to those of the CT26 parental tumors (Fig. 4F and G). NDVmuGM-CSF i.t. treatment resulted in a significant increase in median survival time in CT26 Rig-I−/−, Ifnar1−/− tumors compared with CT26 parental tumors (P < 0.0001). Complete responses were observed in 100% (8/8) of CT26 Rig-I−/−, Ifnar1−/− tumor-bearing animals compared with 22% (4/18) in CT26 parental tumors (Fig. 4F and G). Enhanced viral replication within these cells did not negatively affect the host's ability to mount an effective antitumor immune response against rechallenge (Fig. 4H). These data indicate that increased virus replication is associated with significant improvement in antitumor activity in the immunocompetent setting.
Potentiation of systemic antitumor effects by increased susceptibility to NDVmuGM-CSF
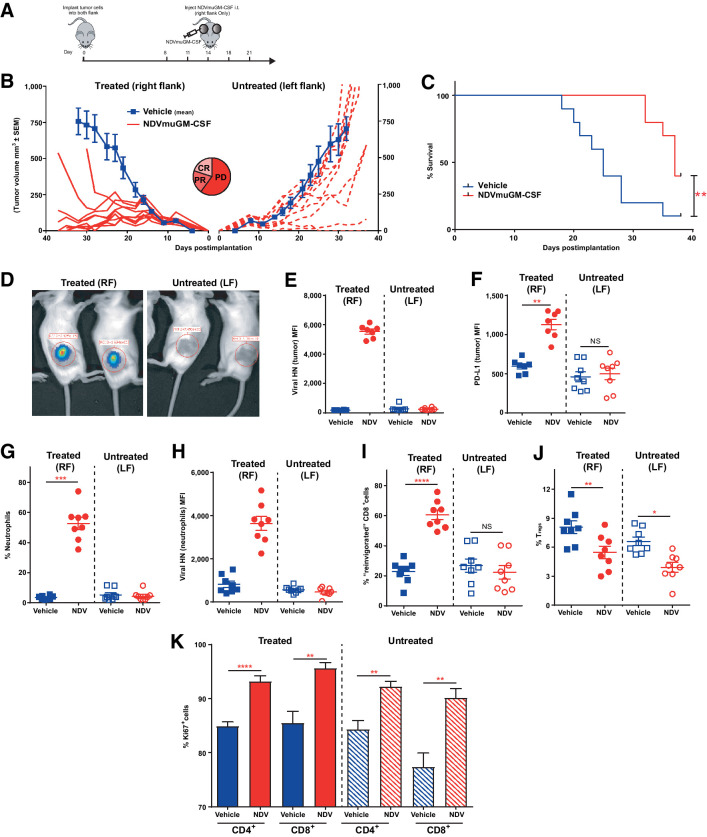
To assess the ability of local NDVmuGM-CSF treatment to generate an immune response capable of leading to systemic therapeutic responses including in noninjected “anenestic” tumors and understand whether this was affected by the level of virus replication, CT26 parental or CT26 Rig-I−/−, Ifnar1−/− cells were implanted and tumors established on both flanks of BALB/c mice. NDVmuGM-CSF was then administered i.t. only in the right-flank tumor (Fig. 5A). In the parental CT26 model, a small proportion of treated right-flank tumors responded to treatment, but untreated left-flank tumors grew unimpeded and no survival benefit was observed (Supplementary Fig. S5I). All CT26 Rig-I−/−, Ifnar1−/− treated tumors responded to local administration of NDVmuGM-CSF, and complete regression of untreated left-flank tumors in 20% (2/10) of animals was also observed (Fig. 5B). Median survival in the NDVmuGM-CSF–treated tumors were also significantly increased, compared with the vehicle control group (P = 0.0031; Fig. 5C). Similar response rates and activity was observed with MEDI5395 (Supplementary Fig. S5K).
Figure 5.
Increased susceptibility to NDV was required to generate potent anenestic antitumor responses, which were entirely dependent on modulation of the host immune system. A, Schematic representation of study design with NDVmuGM-CSF administered in a dual-flank CT26 Rig-I−/−, Ifnar1−/− tumor model. B, Mean tumor growth in untreated animals (blue lines) and spider plots from individual NDVmuGM-CSF–treated animals (red lines) in both treated tumors (solid lines) and untreated tumors (dashed lines; n = 10 animals per group). CR = complete response; PD = progressive disease; PR = partial response. C, Kaplan–Meier plot of survival proportions in dual flank model. D, Representative image of bioluminescent signal detected in NDV-luciferase–treated animals at 48 hours after a single i.t. administration of virus at 5 × 108 pfu (n = 10). Immunophenotyping of treated and untreated tumors with three i.t. doses of NDVmuGM-CSF was performed on days 8, 11, and 14 after implantation and then analyzed 24 hours later on day 15. E–J, Detection of E, vHN protein and F, PD-L1 from NDVmuGM-CSF on surface of CD45− tumor cells. F, Detection of PD-L1 on surface of tumor cells. G, Detection of neutrophils (CD11b+, Ly6Ghi) within tumors and H, analysis of vHN levels on the surface of tumor-infiltrating neutrophils. I, Detection of “reinvigorated” T cells and J, Tregs within tumors. Blue solid squares, vehicle-treated tumors; blue open squares, untreated tumors; red solid circles, NDVmuGM-CSF–treated tumors; red open circles, untreated tumors. MFI = mean fluorescence intensity. K, Quantification of Ki67+, CD4+, and CD8+ T-cell populations in vehicle-treated (solid blue bars) and untreated (shaded blue bars) tumors and in NDVmuGM-CSF–treated (solid red bars) and untreated (shaded red bars) tumors (n = 8 animals per group). Statistical analyses were performed with an unpaired t test with Welch correction; data are mean ± SEM. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significance in survival benefit was assessed by log–rank (Mantel–Cox) test. RF, right flank; LF, left flank.
In the dual flank CT26 Rig-I−/−, Ifnar1−/− model no evidence of virus replication was found at any time point in the left flank (untreated, anenestic) tumor by bioluminescence imaging following i.t. injection of the contralateral tumor with NDV-luciferase (Fig. 5D). The absence of viral infection in untreated/uninjected tumors was further confirmed with flow cytometry 24 hours after the third dose. NDV vHN protein was detected only on the surface of treated (right-flank enestic) tumors and was associated with a significant upregulation of PD-L1 on the surface of infected cells (P = 0.001), whereas no alterations in PD-L1 levels was observed in vHN-negative, untreated left-flank tumors (Fig. 5E and F). Increased numbers of infiltrating neutrophils were detected in the treated right-flank tumors (P = 0.0005), and these polymorphonuclear cells tended to be positive for vHN protein on their cell surface (Fig. 5G and H). CD8+ T cells with a “reinvigorated” phenotype were also significantly upregulated, but only in the treated right-flank tumors (P < 0.0001; Fig. 5I). However, Treg proportions were significantly reduced in both treated right-flank (P = 0.001) and untreated left-flank tumors (P = 0.013; Fig. 5J). This reduction in immune-suppressive potential within the TMEs of both tumors may be responsible for the significant increase in the proliferative potential of both CD4+ and CD8+ T cells observed both in treated (P < 0.0001 and 0.0017, respectively) and untreated (P = 0.0015 and 0.0016 respectively) tumors (Fig. 5K). The data suggest that the anenestic effect is not due to the ability of virus to replicate at distal uninjected sites after local administration but rather the ability of NDV to prime and activate a systemic antitumor immune response at the site of the injected tumor.
Discussion
The paradigm underlying the mechanism of OVs in the clinical setting has undergone a radical transformation over the last decade. Historically, interactions between OV vectors and host immunity were thought to be antithetical to the antitumor activity of OVs. This opinion has altered, with the advent of ICB therapy demonstrating that modulation of the host immune system results in durable antitumor responses in some patients. It is now understood that the ability of OVs to combine potent immune activation and modulation with direct tumoricidal activity is one of the key properties of this class of immunotherapy agents. Therefore, achieving the optimal balance between these two distinct mechanisms of actions is the greatest challenge for successful clinical development (9).
We show that MEDI5395 has potent and broad oncolytic activity across a range of solid human tumor types in vitro. Cell lines from hematologic malignancies appear comparably insensitive to the oncolytic action of MEDI5395, recapitulating findings made with other OVs including pexastimogene-devacirpvec (25). Further stratification of the hematologic cell lines used in the screen revealed that sensitivity to MEDI5395 appears restricted to myeloid lineage malignancies, a phenomenon previously reported for other viruses (26).
Despite the limitations of in vitro screening for estimating efficacy with an OV, this can be used to identify potential mechanisms of insensitivity/sensitivity to OV replication and oncolysis. Interestingly, we found that several colorectal cancer cell lines were resistant to MEDI5395-induced cell lysis but were still able to support high levels of viral replication. Extending the assay beyond 72 hours led to increased cell death (Supplementary Fig. S2F), suggesting that these cells may have delayed cell death kinetics rather than intrinsic resistance to MEDI5395-mediated oncolysis, perhaps as a consequence of defects in apoptosis/necroptosis pathways that are required for efficient NDV-induced death (27).
Furthermore, infected tumor cells are likely to be immunologically “visible” and susceptible to immune-mediated clearance. Therefore, a simplified cell screen, that measures only tumor cell death and only over a short time period in vitro, may substantially underestimate actual antitumor activity in the clinical setting.
RNA viruses, such as NDV, are potent inducers of IFN-I (11). Accordingly, we show that MEDI5395 infection led to increased expression of gene signatures associated with IFN-I pathway activation and release of additional proinflammatory cytokines and chemokines. Transcriptomic data indicated that the inflammatory phenotype induced in tumor cells after MEDI5395 infection is similar to that engendered by viral pathway mimetics or agonists of RIG-I and STING (28, 29). These mimetic strategies can produce potent antitumor responses in preclinical models through DC activation and enhanced tumor-specific CD8+ T-cell responses (30). Consistent with this, an attenuated NDV has previously demonstrated strong antitumor efficacy in preclinical models, driven by its ability to modulate the TME and synergise with ICB to drive potent anenestic responses (31, 32). MEDI5395 also induced significant antitumor activity in vivo; even in murine tumor models, where virus replication is relatively restricted (Fig. 2A–C). Importantly, we clearly demonstrated that MEDI5395 generated potent adaptive immune responses, as indicated by the following: (i) protective effect against rechallenge; (ii) increased TCR diversity in the tumor; (iii) reinvigoration of tumor infiltrating CD8+ T cells; (iv) systemic anenestic effect in the dual-flank model.
We also demonstrated that NDVmuGM-CSF infection modulated the immune contexture of the TME in vivo, as evidenced by the pronounced effect on Treg and DC infiltrate densities relative to tumors derived from vehicle-treated animals (Fig. 2D). This does not appear to be a direct effect of Treg infection and lysis by NDV (13) but possibly by an altered cytokine milieu within the TME (33).
ICB therapy with patient-derived xenografts (PDXs) has transformed the standard of care across a range of cancers. However, although effective for some patients, many patients either fail to respond to PDX therapy or suffer from disease relapse. In preclinical models, we show that NDVmuGM-CSF infection leads to systemic T-cell priming, as measured by an increased TCR clonality in the spleen, expansion of tumor-infiltrating T cells commensurate with increased IFN-I and IFN-II production, and increased PD-L1 expression on tumor and myeloid cells within the TME. Moreover, our data demonstrate that the capacity of NDVmuGM-CSF infection to alter the TME through conversion of an immunologically cold to an inflamed tumor may be used to increase both the frequency of patients responding to PDX blockade and depth of response.
Alongside ICB agents, T-cell agonists also continue to be investigated in the clinic, although initial clinical response has been disappointing (34). Combining T-cell agonists with OVs could simultaneously reduce immune-suppressive mechanisms within the TME and prime T cells to new tumor-associated antigens, which may result in improved clinical benefit. Not all OVs appear to synergise with T-cell agonists; VSV-IFNβ has been shown to have no activity when combined with an OX40 L agonist in preclinical models, because OX40 L primes an antiviral rather than an antitumor response (35). This is in contrast to our findings with MEDI5395 where combinations of NDVmuGM-CSF and GITRL or OX40 L agonists were significantly more effective than monotherapies in syngeneic tumor models.
Modeling the therapeutic activities of these OVs in preclinical tumor models remains a key challenge in the field, owing to the naturally variable tropism across virus species and the lack of relevant preclinical pharmacology animal models for OV investigations. Enhanced susceptibility to MEDI5395 (driven by loss of Rig-I and Ifnar1 genes involved in antiviral response) through CRISPR engineering resulted in CT26 Rig-I−/−, Ifnar1−/− cells that were able to support significantly greater levels of virus replication than the parental cell line in vitro and in vivo resulted in significantly improved antitumor efficacy. This enhanced activity of NDVmuGM-CSF in engineered cell lines was not entirely dependent on oncolysis.
In the dual-flank tumor setting, antitumor activity in untreated (anenestic) lesions was observed only when tumor cells had a higher capacity to support virus replication. This activity was accompanied by an apparent enhanced antitumor immune response that resulted in changes within the immune contexture of the untreated lesion in the absence of any evidence of virus replication.
In summary, MEDI5395 demonstrated potent oncolytic properties across a broad spectrum of human and murine cancer cell lines and PDX models. The ability of MEDI5395 to lyse cancer cells and simultaneously modulate the immune system results in significant antitumor activity in vivo, which can be further augmented when the virus is administered in combination with immune-modulating antibodies. These preclinical data support clinical development of MEDI5395; both as a monotherapy and in combination with other immuno-oncology agents.
Authors' Disclosures
J. Harper reports other support from AstraZeneca outside the submitted work. S. Burke reports other support from AstraZeneca during the conduct of the study. N. Rath reports other support from AstraZeneca during the conduct of the study. A. Leinster reports other support from AstraZeneca during the conduct of the study. S.J. Dovedi reports personal fees from AstraZeneca during the conduct of the study and personal fees from AstraZeneca outside the submitted work. N.M. Durham reports other support from AstraZeneca outside the submitted work. N.M. Durham is an employee of AstraZeneca and receives salary and stock from AstraZeneca. H. Jin reports other support from AstraZenca employee outside the submitted work; in addition, H. Jin has a patent for WO 2015/032755 AI issued. J. Eyles reports other support from AstraZeneca Stock outside the submitted work. R.W. Wilkinson reports other support from AstraZeneca outside the submitted work. D. Carroll reports other support from AstraZeneca during the conduct of the study; in addition, D. Carroll has a patent for US 2020/0181581A1 issued. No disclosures were reported by the other authors.
Acknowledgments
We thank Margaret Blundy, Alison Keen, and staff in the Core Tissue Culture laboratory for their assistance in providing cell lines and the Animal Science Team for their assistance in providing in vivo support.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
J. Harper: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. S. Burke: Resources, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. J. Travers: Resources, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. N. Rath: Formal analysis, validation, investigation, methodology, writing–original draft. A. Leinster: Investigation, methodology, writing–original draft. C. Navarro: Investigation, methodology. R. Franks: Investigation. R. Leyland: Investigation. K. Mulgrew: Investigation. K. McGlinchey: Investigation. L. Brown: Investigation, visualization. S.J. Dovedi: Visualization, writing–review and editing. J.O. Koopmann: Investigation, methodology, writing–review and editing. N.M. Durham: Investigation, methodology, writing–review and editing. X. Cheng: Resources. H. Jin: Resources. J. Eyles: Writing–review and editing. R.W. Wilkinson: Funding acquisition, writing–review and editing. D. Carroll: Conceptualization, formal analysis, supervision, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borghaei H, Brahmer J. Nivolumab in nonsquamous non-small-cell lung cancer. N Engl J Med 2016;374:493–4. [DOI] [PubMed] [Google Scholar]
- 3. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang A, Wang HY, Liu Y, Zhao MC, Zhang HJ, Lu ZY, et al. The prognostic value of PD-L1 expression for non-small cell lung cancer patients: a meta-analysis. Eur J Surg Oncol 2015;41:450–6. [DOI] [PubMed] [Google Scholar]
- 5. Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight 2019;4:e126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumors with combination immunotherapies. Nat Rev Drug Discov 2019;18:197–218. [DOI] [PubMed] [Google Scholar]
- 7. Breitbach CJ, Lichty BD, Bell JC. Oncolytic viruses: therapeutics with an identity crisis. EBioMedicine 2016;9:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov 2015;14:642–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrington K, Freeman DJ, Kelly B, Harper J, Soria JC. Optimizing oncolytic virotherapy in cancer treatment. Nat Rev Drug Discov 2019;18:689–706. [DOI] [PubMed] [Google Scholar]
- 10. Lorence R, Scot Roberts M, O'Neil J, Groene W, Miller J, Mueller S, et al. Phase 1 clinical experience using intravenous administration of PV701, an oncolytic Newcastle disease virus. Curr Cancer Drug Targets 2007;7:157–67. [DOI] [PubMed] [Google Scholar]
- 11. Zamarin D, Palese P. Oncolytic Newcastle disease virus for cancer therapy: old challenges and new directions. Future Microbiol 2012;7:347–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng X, Wang W, Xu Q, Harper J, Carroll D, Galinski MS, et al. Genetic modification of oncolytic newcastle disease virus for cancer therapy. J Virol 2016;90:5343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burke S, Shergold A, Elder MJ, Whitworth J, Cheng X, Jin H, et al. Oncolytic Newcastle disease virus activation of the innate immune response and priming of antitumor adaptive responses in vitro. Cancer Immunol Immunother 2020;69:1015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaushansky K, Shoemaker SG, Alfaro S, Brown C. Hematopoietic activity of granulocyte/macrophage colony-stimulating factor is dependent upon two distinct regions of the molecule: functional analysis based upon the activities of interspecies hybrid growth factors. Proc Natl Acad Sci U S A 1989;86:1213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 2012;86:2900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res 2014;2:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 2006;6:836–48. [DOI] [PubMed] [Google Scholar]
- 18. Aquino-Lopez A, Senyukov VV, Vlasic Z, Kleinerman ES, Lee DA. Interferon gamma induces changes in natural killer (NK) cell ligand expression and alters NK cell-mediated lysis of pediatric cancer cell lines. Front Immunol 2017;8:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bourgeois-Daigneault M-C, Roy DG, Aitken AS, El Sayes N, Martin NT, Varette O, et al. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple-negative breast cancer to immune checkpoint therapy. Sci Transl Med 2018;10:eaao1641. [DOI] [PubMed] [Google Scholar]
- 20. Mosely SIS, Prime JE, Sainson RCA, Koopmann J-O, Wang DYQ, Greenawalt DM, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res 2017;5:29–41. [DOI] [PubMed] [Google Scholar]
- 21. Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2016;520:373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci USA 1996;93:9730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leyland R, Watkins A, Mulgrew KA, Holoweckyj N, Bamber L, Tigue NJ, et al. A novel murine GITR ligand fusion protein induces antitumor sctivity as a monotherapy that is further enhanced in combination with an OX40 agonist. Clin Cancer Res 2017;23:3416–27. [DOI] [PubMed] [Google Scholar]
- 24. Oberst MD, Augé C, Morris C, Kentner S, Mulgrew K, McGlinchey K, et al. Potent immune modulation by MEDI6383, an engineered human OX40 ligand IgG4P Fc fusion protein. Mol Cancer Ther 2018;17:1024–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee NH, Kim M, Oh SY, Kim S-G, Kwon H-C, Hwang T-H. Gene expression profiling of hematologic malignant cell lines resistant to oncolytic virus treatment. Oncotarget 2017;8:1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bais S, Bartee E, Rahman MM, McFadden G, Cogle CR. Oncolytic virotherapy for hematological malignancies. Adv Virol 2012;2012:186512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Yuan J, Zhang HY, Simayi D, Li PD, Wang YH, et al. Natural resistance to apoptosis correlates with resistance to chemotherapy in colorectal cancer cells. Clin Exp Med 2012;12:97–103. [DOI] [PubMed] [Google Scholar]
- 28. van den Boorn JG, Hartmann G. Turning tumors into vaccines: co-opting the innate immune system. Immunity 2013;39:27–37. [DOI] [PubMed] [Google Scholar]
- 29. Woo S-R, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014;41:830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med 2015;7:283ra252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med 2014;6:226ra232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zamarin D, Ricca JM, Sadekova S, Oseledchyk A, Yu Y, Blumenschein WM, et al. PD-L1 in tumor microenvironment mediates resistance to oncolytic immunotherapy. J Clin Invest 2018;128:5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fournier P, Arnold A, Wilden H, Schirrmacher V. Newcastle disease virus induces pro-inflammatory conditions and type I interferon for counter-acting Treg activity. Int J Oncol 2012;40:840–50. [DOI] [PubMed] [Google Scholar]
- 34. Mayes PA, Hance KW, Hoos A. The promise and challenges of immune agonist antibody development in cancer. Nat Rev Drug Discov 2018;17:509–27. [DOI] [PubMed] [Google Scholar]
- 35. Durham NM, Mulgrew K, McGlinchey K, Monks NR, Ji H, Herbst R, et al. Oncolytic VSV primes differential responses to immuno-oncology therapy. Mol Ther 2017;25:1917–32. [DOI] [PMC free article] [PubMed] [Google Scholar]