Abstract
Temozolomide (TMZ) is a DNA-methylating agent used in cancer chemotherapy, notably for glioblastoma multiforme (GBM), where it is applied as a front-line drug. One of the DNA alkylation products of TMZ is the minor lesion O6-methylguanine (O6MeG), which is responsible for nearly all genotoxic, cytotoxic, and cytostatic effects induced in the low-dose range relevant for cancer therapy. Here, we addressed the question of how many O6MeG adducts are required to elicit cytotoxic responses. Adduct quantification revealed that O6MeG increases linearly with dose. The same was observed for DNA double-strand breaks (DSB) and p53ser15. Regarding apoptosis, hockeystick modeling indicated a possible threshold for A172 cells at 2.5 μmol/L TMZ, whereas for LN229 cells no threshold was detected. Cellular senescence, which is the main cellular response, also increased linearly, without a threshold. Using a dose of 20 μmol/L, which is achievable in a therapeutic setting, we determined that 14,000 adducts give rise to 32 DSBs (γH2AX foci) in A172 cells. This leads to 12% cell death and 35% of cells entering senescence. In LN229 cells, 20 μmol/L TMZ induced 20,600 O6MeG adducts, 66 DSBs (γH2AX foci), 24% apoptosis, and 52% senescence. The linear dose response and the genotoxic and cytotoxic effects observed at therapeutically relevant dose levels make it very likely that the TMZ target concentration triggers a significant cytotoxic and cytostatic effect in vivo. Despite a linear increase in the O6MeG adduct level, DSBs, and p53 activation, the low curative effect of TMZ results presumably from the low rate of apoptosis compared to senescence.
Introduction
Temozolomide (TMZ) is a DNA-methylating agent frequently used in cancer therapy (1). It is applied in first-line therapy for high-grade gliomas, including astrocytoma (WHO grade 3) and glioblastoma multiforme (GBM; glioma WHO grade 4; ref. 2) and in some other cancers (3). TMZ is a triazene derivative that does not need metabolic activation. It decomposes spontaneously yielding 3-methyl-(triazen-1-yl)imidazole-4-carboxamide (MTIC) and, in a second step, 5-aminoimidazole-4-carboxamide (AIC) and monomethyl hydrazine, which finally methylates DNA at various sites. As revealed by studies with DNA repair mutants and isogenic cell lines (4, 5), the main genotoxic and cytotoxic target of TMZ is the nuclear DNA. Similar to other SN1 alkylating agents, at least 12 nucleophilic sites are subject of methylation (6). The major methylation products are N-methylpurines such as N7-methylguanine, N3-methylguanine, and N3-methyladenine (comprising about 80% of total alkylation), whereas O-methylpurines are less frequent. Thus, O6-methylguanine (O6MeG) accounts for only 7% of the total DNA methylation (6). Although produced in minor amounts, O6MeG is the main mutagenic, carcinogenic, genotoxic, and cytotoxic lesion (reviewed in ref. 7). It is also responsible for autophagy and cellular senescence, which are endpoints induced by TMZ concomitant to apoptosis (8).
O6MeG is repaired by the suicide enzyme O6-methylguanine-DNA methyltransferase (MGMT) in a damage reversal reaction (9). If cells are repair competent, O6MeG is quickly removed from DNA whereas in repair-incompetent cells, O6MeG persists. Because O6MeG is not eliminated by spontaneous hydrolysis, it can be transmitted even to the next cell generation, exerting long-term effects (10). The MGMT gene is a subject of intensive genetic and epigenetic regulation and therefore cells and tissues vary considerably in their repair capacity (11–13). High expression levels were observed in liver and the lowest in brain (13). Brain tissue of rats is completely devoid of MGMT, leading to persistence of O6MeG, which is the reason for neurotropic carcinogenesis following exposure with SN1 methylating agents such as N-methyl-N-nitrosourea (14). The human brain expresses MGMT, although at low levels compared with other organs (15, 16). About 40% of human brain cancer (glioma grade 3 and 4) show MGMT promoter hypermethylation, which is related to MGMT downregulation, and 17% of GBM are completely devoid of MGMT repair activity as determined in an enzymatic assay (17). Therefore, promoter-methylated GBM are responsive to TMZ therapy (18–20).
In MGMT repair-competent cells, higher doses of TMZ are required to be cytotoxic, which results from non-repaired N-alkylation lesions that interfere with the replication machinery (21). These lesions are repaired by base excision repair (BER) and ALKB homologous proteins (ALKBH; ref. 7) and, therefore, they do not contribute to cytotoxicity if O6MeG is left in the DNA. However, if at high TMZ dose levels the N-alkylation repair system is saturated, these lesions contribute to cell death as well. The doses of TMZ in a therapeutic setting are very likely too low to achieve cell death resulting from N-alkylation products. Therefore, with a serum concentration of ∼50 μmol/L TMZ, the O6MeG adducts are the key players inducing tumor cell death.
The mechanisms of O6MeG triggered cell death responses have been investigated intensively (22). In brief, a widely accepted model claims that O6MeG is a mutagenic mispairing lesion resulting in mismatches with thymine that are subject to mismatch repair (MMR). Reinsertion of thymine during MMR causes a futile MMR cycle, resulting in gaps in the DNA that finally give rise to DNA replication blockage and the formation of replication-mediated DNA double-strand breaks (DSB). This occurs in the posttreatment cell cycle, which is compatible with the time-course of apoptosis and DSB formation (23). Results obtained with synchronized cells confirmed this model (10). It is further known that DSBs induced by the processing of O6MeG/thymine trigger complex DNA damage response pathways, which are activated primarily by ATR, and secondary by ATM, and downstream CHK1 and CHK2, respectively (24) as well as activation of the SIAH1-HIPK2–p53 axis (25). In this scenario, the following factors determine drug resistance: MGMT, MMR, the proliferation level, DNA damage response (DDR) activation, and DSB repair by homologous recombination. O6MeG is not only an apoptosis-inducing damage, it also triggers with high efficiency cellular senescence in glioblastoma cells (8). The pathways involved were described previously (8, 26, 27).
Patients suffering from GBM, which make up ∼70% of high-grade malignant gliomas, have a dismal prognosis (2). Standard therapy includes resection of the tumor followed by radiotherapy and concomitant chemotherapy with TMZ (28). TMZ is also prescribed in the recurrent situation and as maintenance therapy, and is well tolerated even if treatment occurs daily over long periods of time (29). In view of the bad prognosis and weak therapeutic efficiency of TMZ, better knowledge of the action of this anticancer drug is highly desirable. Given the known mechanism of action of TMZ, some questions arise which are a matter of debate (27, 30). First, do GBM cells die (by apoptosis or other forms of cell death) following TMZ treatment or does the drug exert only a cytostatic effect? Second, are the doses used for patient's treatment high enough to kill cancer cells without having toxic side effects on non-cancer cells? This question relates to the paradigm that low doses of a genotoxic anticancer drug are ineffective and that presumably thresholds exist below which cytotoxicity does not occur. Finally, third, how many O6MeG adducts are required for eliciting a specific effect such as apoptosis, necrosis, and senescence and can this level be achieved in vivo?
To address these fundamental questions in the molecular dosimetry of TMZ, we measured the amount of O6MeG induced by TMZ in established GBM cell lines and related this to levels of DSBs, apoptosis, necrosis, overall cell death, and cellular senescence. The data revealed that the amount of O6MeG following TMZ exposure, similar to DSBs, increased linearly with dose. Likewise, cell death, and senescence tracked with the formation of these damage products. Finally, we found that even at low-dose levels genotoxic, cytotoxic, and cytostatic effects were induced by TMZ. The amount of lesions inducing a particular effect was determined.
Materials and Methods
Cell lines and culture conditions
The human glioblastoma cell lines LN229 and A172 were purchased from ATCC, expanded, stored frozen in batches, and kept in culture only for limited time periods (maximally 2 month). LN229-MGMT (clone 12) was generated by stable transfection of LN229 with a human MGMT cDNA expression vector as described previously (8, 31). Cells were cultured in DMEM with Glutamax (Gibco, Life Technologies Corporation) and 10% fetal calf serum at 37°C in a humidified 5% CO2 atmosphere and routinely checked for mycoplasma contamination using a Kit (Venor GeM Classic) from Minerva Biolabs. Cells were seeded 48 hours before treatment, when they reached exponential growth. For short-term experiments (harvest 3 days after treatment), 2 × 105 cells were seeded per 5-cm or 6-well dish (in 5 or 4 mL medium, respectively), for long-term experiments (apoptosis and senescence following TMZ) 1 × 105 cells were seeded per dish. Under these conditions, cells were kept in exponential growth for the whole experimental period.
Drugs and drug treatment
TMZ obtained from Dr. Geoff Margison (University of Manchester) was dissolved in DMSO (150 mmol/L stock) and stored in batches at −80°C until use. Immediately before use, it was diluted 1:10 in sterile distilled water and added to the cell culture medium at the desired final concentration. For low final concentrations (≤10 μmol/L), the solution was further diluted with distilled water to a 1 mmol/L stock solution. Water-diluted stocks were used only once. The amount of DMSO in the medium did not exceed 0.05% and was without any cytotoxic effect (controls).
Quantification of O6 MeG adducts
O6MeG adduct quantification was performed by measuring O6-methyl-deoxyguanosine (O6MedG), which was normalized to the deoxyguanosine (dG) content (32), according to a modified method as recently described in detail (33). Briefly, exponentially growing cells were treated with TMZ and cells were harvested at the time points indicated by trypsinization. Cells were taken up in complete medium, washed with PBS before the cell pellet was stored frozen at −80°C. DNA from cell pellets with a defined cell number was isolated using the QIAamp DNA Mini Kit (Qiagen). Briefly, lysis buffer, proteinase K, and RNase A were added and cell pellets were incubated for 20 minutes at 56°C, 1,400 rpm on a ThermoShaker. Afterwards, ethanol was added and DNA was purified and extracted on spin columns applying washing buffers and MilliQ for elution according to the manufacturer's instruction. Internal labelled standards were added to the sample and DNA was enzymatically hydrolyzed, yielding free nucleosides. Following further sample clean-up by solid-phase extraction, O6MedG was quantified by reversed-phase nano-liquid chromatography electrospray ionization high-resolution tandem mass spectrometry (nanoLC-ESI-HRMS2). The results were expressed as number of O6MeG per 107 nucleotides (nt). The results were then converted to O6MeG per cell. The calculation rests on the measured DNA content of 5.11 pg DNA per cell in the exponentially growing A172 population, and 8.22 pg DNA per cell in the exponentially growing LN229 population. It further rests on the relationship of 1 pg DNA corresponding to 109 bp (2 × 109 nt), which gives rise to the following equation: O6MeG/107nt × pg DNA/cell × 2 × 109nt/pg.
Quantification of apoptosis and necrosis
The fraction of apoptotic and necrotic cells was determined using annexin V and propidium iodide (A/PI) staining of cells and measured by flow cytometry. For analysis, trypsinized cells including the supernatant were collected, transferred into 15 mL falcon tubes, washed with PBS, and stored on ice. For labeling, cells were incubated for 15 minutes at RT in 50 μL 1× annexin binding buffer containing 2.5 μL annexin V/FITC (MiltenyiBiotec GmbH). For PI staining, 10 μL PI from a 1 mg/mL stock solution (Sigma-Aldrich) and annexin binding buffer were added to each sample. Cells were incubated for additional 10 minutes on ice. Cells were kept in the dark until measurement. Data acquisition was performed using the FACS Canto II flow cytometer (Becton Dickinson GmbH) and the data were analyzed using the Flowing Software 2 program (PerttuTerho, Turku Center for Biotechnology, University of Turku, Finland). Apoptotic cells were defined as Annexin V+/ PI− cells, whereas late apoptotic/necrotic cells were defined as Annexin V+/ PI+ cells. For the sake of simplicity, this fraction is simply called “necrosis.” A representative plot showing the Annexin V+/ PI− and Annexin V−/ PI+ fractions is shown in Supplementary Fig. S1.
Quantification of cellular senescence
Induced senescence was determined by the amount of cellular SA-β-galactosidase. To inhibit endogenous β-galactosidase activity, cells were pre-incubated with 300 μmol/L chloroquine for 30 minutes in the incubator after which C12FDG was added to each sample at a final concentration of 33 μmol/L. Following a 90 minutes incubation period, cells were washed once with cold PBS and collected by trypsinization. Cell pellets were washed again with cold PBS, resuspended in cold PBS, and stored on ice. After the addition of chloroquine, cells were kept in the dark up to harvest. Data acquisition was performed using the FACS Canto II flow cytometer (Becton Dickinson GmbH) and the data were analyzed using the Flowing Software 2 program (PerttuTerho, Turku Center for Biotechnology, University of Turku). Untreated cells served as control. The cell population showing SA-ß-GAL mediated staining higher than the control was defined as senescent.
Quantification of γH2AX and 53BP1 foci
The γH2AX and 53BP1 foci assays were conducted essentially as described (34). Evaluation of γH2AX and 53BP1 stained nuclei of cells grown on coverslips occurred by LSM from a single focus plane. At least 100 cells were assessed per experiment and all experiments were performed three times independently. Antibodies used were γH2AX (1:500, rabbit; Cell Signaling Technology, mAb #9718S) combined with Cy3 goat-anti-rabbit (1:500; Abcam, ab97075) and 53BP1 (1:500, mouse, Sigma-Aldrich, MAB 3802) combined with Alexa Fluor 488 goat-anti-mouse (1:500, Thermo Fisher Scientific Invitrogen, A11017). The foci were counted with the software ImageJ (Wayne Rasband, NIH).
Statistical analysis and mathematic assessments
Data are presented as mean of at least three independent experiments ±SEM and compared by unpaired t test with Welch's correction. The calculation of O6MeG levels per cell was described above.
Results
Amount of O6MeG induced by TMZ
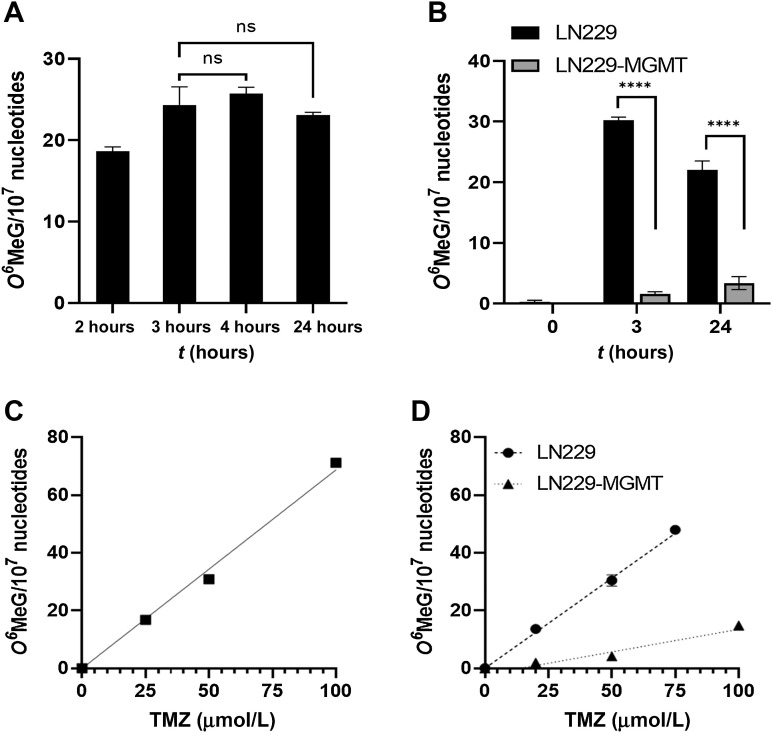
To relate phenotypic effects of TMZ exposure with primary adduct formation, first we determined the amount of O6MeG formed in glioblastoma cells following treatment with TMZ. The drug has a half-life of 1.9 hours in serum (35) and, therefore, even chronic exposure can be considered as pulse-treatment. To establish the optimal time of treatment for determining the alkylation level, we exposed cells to TMZ for different durations. As shown in Fig. 1A, O6MeG was induced in LN229 cells, which are MGMT repair incompetent, with a maximum level 3 to 4 hours after addition of 50 μmol/L TMZ to the medium and did not further increase due to the short half-life of the drug in the medium. In a separate series of experiments, we compared LN229 with LN229 stably transfected with human MGMT (LN229-MGMT) and observed that the amount of O6MeG was significantly lower in MGMT-expressing cells even 3 hours after the onset of treatment (Fig. 1B), which was expected as MGMT performs a fast repair reaction (36).
Figure 1.
Amount of O6MeG induced in glioblastoma cells (LN229) upon treatment with TMZ. A, Adducts induced after 2, 3, 4, and 24 hours exposure of LN229 cells with 50 μmol/L TMZ. There is no significant difference between the levels. Amount of adducts induced in LN229 and LN229-MGMT cells exposed for 3 and 24 hours to 50 μmol/L TMZ. Data are the mean of three independent determinations ±SEM (P < 0.0001). O6MeG in glioblastoma cells exposed to TMZ for 3 hours in A172 cells (C) and in LN229 and LN229-MGMT cells (D). Data are the mean of three independent determinations ±SEM. A172: y = 0.6873 × x; LN229: y = 0.6239 × x; LN229-MGMT: y = 0.1569 × x − 2.151. ns, not significant.
After having established the optimal treatment time for adduct quantification to be 3 hours, we determined the dose-response relationship for the induction of O6MeG. The amount of O6MeG in the DNA of A172 cells increased as a linear function of TMZ dose (Fig. 1C). Similar data were obtained for LN229 (Fig. 1D). Thus, a dose of 50 μmol/L TMZ induced 34 and 31 O6MeG/107 nucleotides in A172 and LN229 cells, respectively (for a direct comparison of A172 and LN229 see Supplementary Fig. S2). A172 and LN229 do not express MGMT due to MGMT promoter hypermethylation and are unable to repair O6MeG (37). Therefore, we used MGMT transfected cells and found that O6MeG could not be detected if cells were treated with doses below 50 μmol/L (Fig. 1D), indicating efficient and complete repair of the damage in this low-dose range. At high dose levels, however, a small increase in O6MeG was recorded (Fig. 1D), indicating increasing saturation of repair activity.
DSBs induced by TMZ
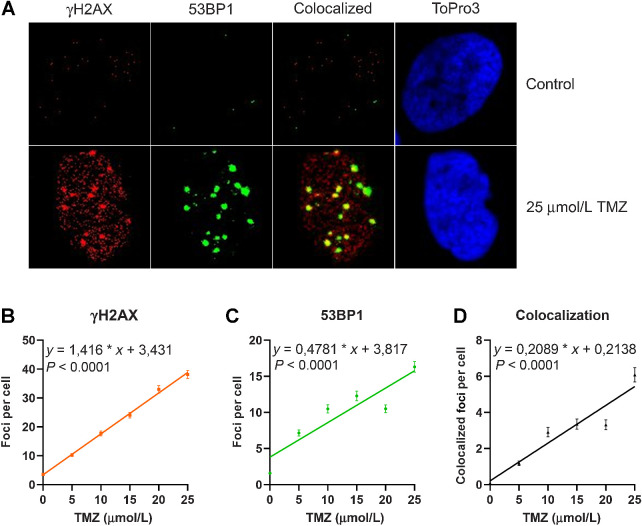
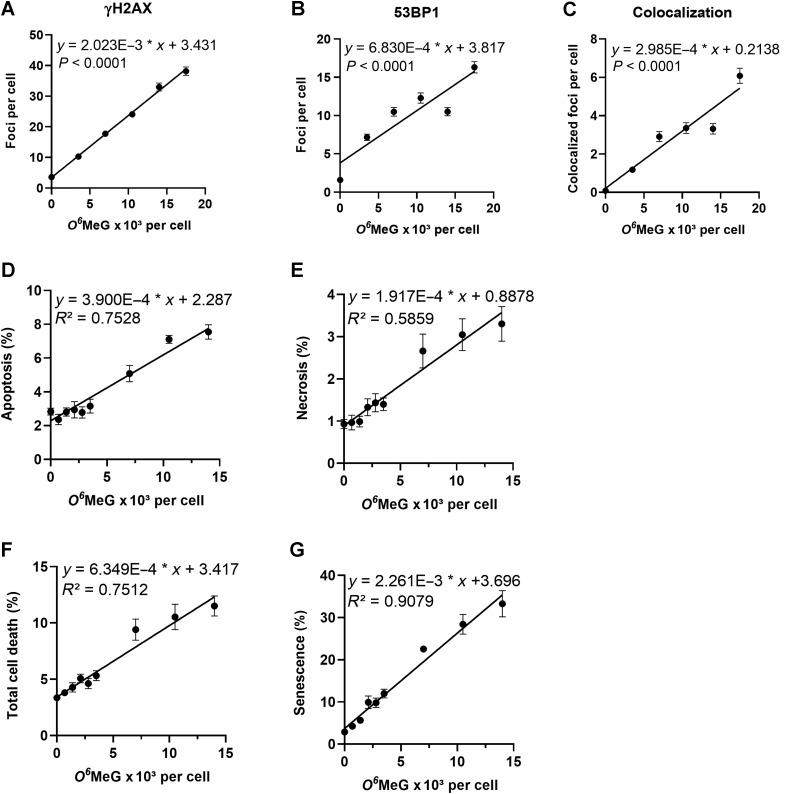
A highly sensitive and generally accepted method of detecting DSBs is based on the phosphorylation of histone 2AX (H2AX) around the DNA break, which is catalyzed by damage-activated ATR and ATM proteins, resulting in immuno-detectable nuclear foci (γH2AX). At the same time, 53BP1, an accessory protein supporting the repair of DSBs, becomes phosphorylated and recruited to the DSB sites. Thus, γH2AX foci and colocalized 53BP1 foci are considered to be reliable marker of DSBs. Therefore, we quantified γH2AX and 53BP1 foci (for representative images see Fig. 2A) in cells exposed to TMZ. The dose–response curves shown in Fig. 2B–D demonstrate a linear increase of foci without a threshold. Thus, already a low dose of 5 μmol/L TMZ gave rise to a significant increase in the DSBs, as measured 72 hours after TMZ treatment in A172 cells.
Figure 2.
Amount of γH2AX and 53BP1 foci induced in A172 glioblastoma cells upon treatment with increasing doses of TMZ. Cells were fixed and immunostained 72 hours after the addition of TMZ to the medium. A, Images of representative examples. Dose response for γH2AX (B), 53BP1 (C), and colocalized foci (D). Data are the mean of four independent determinations ± SEM. The following equations describe the curves for γH2AX y = 1.416 × x + 3.431, for 53BP1 y = 0.4781 × x + 3.817, for colocalized foci y = 0.2089 × x + 0.2138.
Induction of the DNA damage response: p53 and p21
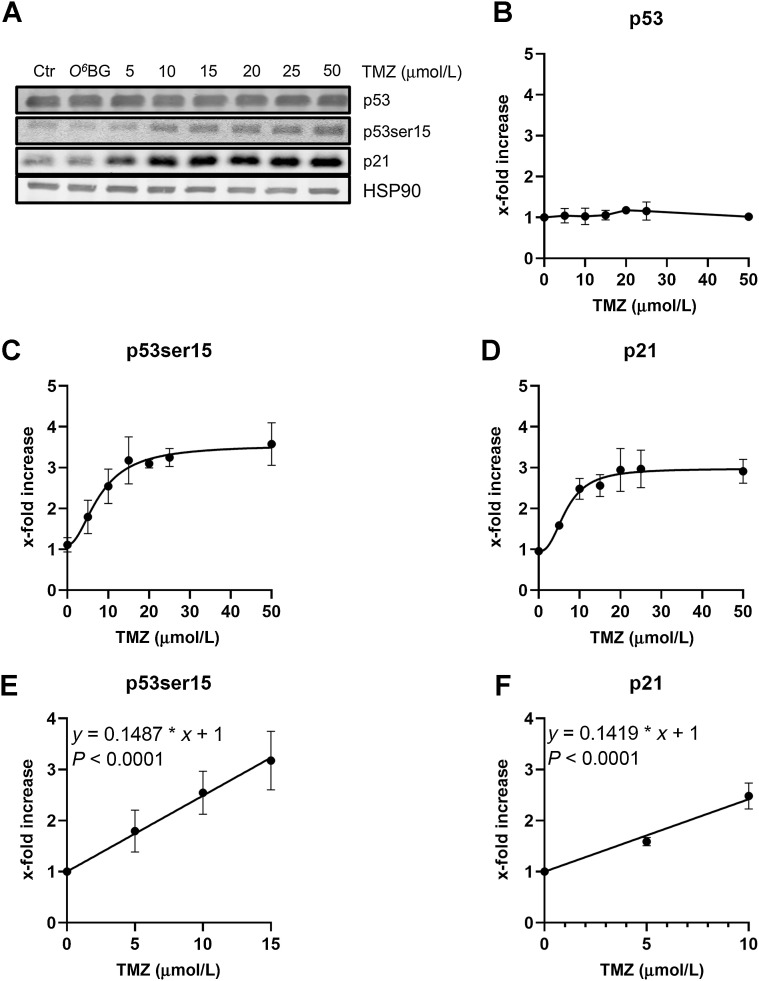
It is well established that DSBs trigger the DNA damage response (DDR), which targets downstream p53, phosphorylating p53 at serine 15. This phosphorylation leads to nuclear translocation, trimerization, and activation of p53 as transcription factor. One of the p53 target genes is p21, which is a key player in cell-cycle regulation. As shown in Fig. 3, p53ser15 and concomitantly p21 are upregulated dose-dependently until reaching a saturation level, which is achieved with a dose of about 10 to 15 μmol/L TMZ (Fig. 3A for a representative Western blot analysis, Fig. 3B for total p53 and Fig. 3C and D for the dose–response curves of p53ser15 and p21).
Figure 3.
Amount of p53, p53ser15, and p21 in A172 cells treated with various doses of temozolomide. A, Exponentially growing cells were harvested 120 hours after treatment with TMZ. HSP90 was used as loading control. A representative Western blot analysis is shown. Quantification of p53 (B), p53ser15 (C), and p21 (D) in A172 cells treated with TMZ. Data are the mean of three independent experiments ± SEM. Dose response for p53ser15 (E) and p21 (F) in A172 cells at the low TMZ dose range (up to 15 μmol/L). The curves do not indicate a threshold dose.
Mathematical analysis of the linear dose range (0–15 μmol/L) did not reveal a no-effect threshold dose (Fig. 3E and F). It should be noted that total p53 protein was not enhanced (Fig. 3B), which is due to the fact that p53 is mutated in A172 cells, leading to its stabilization without having impact on its trans-activating activity.
Induction of apoptosis and necrosis by TMZ
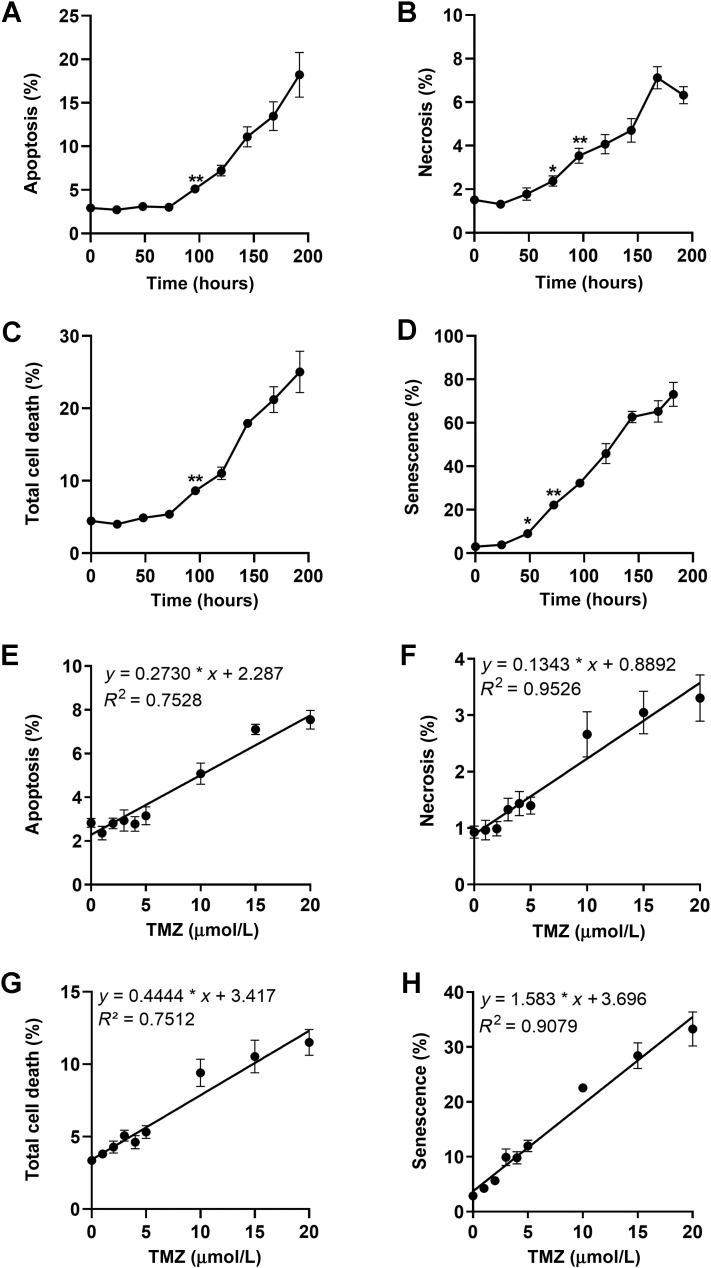
As previously shown, the induction of DSBs triggered by O6MeG precedes the activation of cell death pathways, which is a late response following DNA damage (23). The time course of apoptosis, necrosis, total cell death, and induced cellular senescence of A172 cells is shown in Fig. 4, demonstrating that cell death by apoptosis (Fig. 4A) and necrosis (Fig. 4B) starts at 96 hours significantly above the control level and increases thereafter continuously. The onset of cellular senescence was observed earlier (Fig. 4D; significant increase 48 hours after treatment P < 0.05). On the basis of these data, we decided to measure for the influence of TMZ dose on these endpoints 120 hours after addition of TMZ to the medium.
Figure 4.
Time-course of apoptosis (A), necrosis (B), total cell death (C), and senescence (D) after treatment with 50 μmol/L TMZ of A172. Data are the mean of at least three independent experiments ± SEM (*, P < 0.05; **, P < 0.01). Dose response of apoptosis (E), necrosis (F), and total cell death (G) in A172 cells treated with TMZ and measured 120 hours after the start of treatment. Data are the mean of five independent experiments measured in duplicate ± SEM. H, Cellular senescence (C12FDG+) induced by TMZ in A172 cells. Data are the mean of four independent experiments ± SEM.
We measured the rate of apoptosis, necrosis, and total cell death upon treatment with TMZ using a dose range of up to 50 μmol/L. We used defined seeding conditions to ensure exponential growth over the whole post-exposure period. As shown in the Supplementary data (Supplementary Fig. S3), apoptosis increased up to a level of max. 8% and then reached saturation at a dose of 20 μmol/L. In contrast, necrosis increased over the whole dose range linearly up to a level of 7% at 50 μmol/L. Because necrosis was a minor trait, the total cell death level reached saturation at 20 μmol/L. Induced senescence increased up to 30% to 40% and reached a saturation level at 20 μmol/L (Supplementary Fig. S3). Having established the linear dose range, we conducted further experiments using the dose range of 0 to 20 μmol/L TMZ. As shown in Fig. 4E–F, apoptosis, necrosis, and total cell death increased as a linear function of dose. An analysis of the data on the basis of the hockey stick model (38) revealed a possible threshold at 2.5 μmol/L for the endpoint apoptosis, whereas for necrosis a significant threshold was not detected (see Supplementary Fig. S4).
Cellular senescence induced by TMZ
Similar experiments were performed with the endpoint cellular senescence, which was quantified by C12FDG-flow cytometry. The level of senescence increased again as a linear function of dose. For senescence no threshold was observed (Fig. 4H), which was confirmed by hockey stick modeling of the data (Supplementary Fig. S5).
Correlation between O6MeG and DSBs, cell death, and senescence
Next, we wished to know how many O6MeG lesions per cell give rise to a DSB and cytotoxic effects. To this end, we recalculated the amount of O6MeG per unit DNA into O6MeG adducts per cell. The DNA content per cell was determined on the basis of the amount of DNA extracted from a given number of cells in an exponentially growing population; 5.11 and 8.22 pg DNA per cell for A172 and LN229, respectively. The dependence of γH2AX, 53BP1, and colocalized foci with the level of induced O6MeG adducts per cell are shown in Fig. 5A–C, respectively, for a dose range up to 25 μmol/L TMZ. Assuming a 1:1 relation between γH2AX foci and DSBs, we estimated for A172 cells that 490 O6MeG adducts per cell are required to induce one DSB (calculated from the slope of regression). Assuming that one γH2AX/53BP1 colocalized focus corresponds to one DSB, about 3,350 adducts are required for inducing one DSB.
Figure 5.
Cellular responses as a function of O6MeG induced by TMZ in A172 glioblastoma cells. γH2AX (A), 53BP1 (B), colocalized foci (C), apoptosis (D), necrosis (E), total cell death (F), senescence (G) as a function of the O6MeG content per cell. Data are from Figs. 1 to 4.
The dose–response relationships for apoptosis, necrosis, total cell death, and senescence with O6MeG adducts per cell are presented in Fig. 5D to G for a dose range up to 20 μmol/L. From this we calculated that an increase by 1% in the population of apoptotic cells requires 2,560 O6MeG adducts per cell, of necrosis 5,220 O6MeG, and total cell death 1,580 O6MeG per cell. A 1% increase of cells undergoing senescence requires only 440 O6MeG adducts per cell. It is important to note that senescence is the major endpoint triggered by O6MeG lesions (7.5% apoptosis and 3.5% necrosis compared with 35% senescence 120 hours after TMZ treatment) and, therefore, a lower amount of O6MeG adducts is required to trigger this response.
O6MeG and responses triggered in LN229 cells
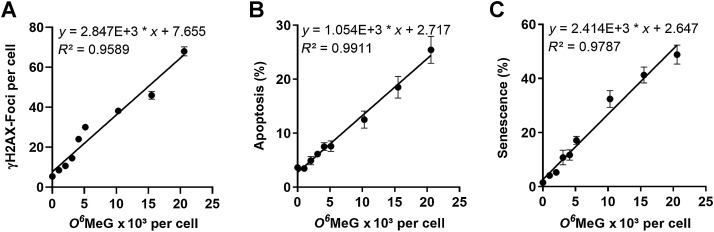
The same modeling of the relationship of various cellular responses with drug-induced adduct levels was performed for the widely used and well-characterized glioblastoma cell line LN229 (MGMT lacking, p53 functionally wt). The O6MeG adduct levels (calculated from the dose–response curve in Fig. 1) were related to DSBs, apoptosis, and senescence values from our previous report (34) and supplementary data (Supplementary Fig. S6). The data revealed that γH2AX foci (Fig. 6A), apoptosis (Fig. 6B), and senescence (Fig. 6C) increased linearly with the O6MeG adduct level per cell, and no thresholds were apparent. In this case, an increase of apoptosis by 1% in the population was calculated to require 950 O6MeG adducts per cell, and senescence 410 O6MeG adducts per cell. Assuming a 1:1 correlation between γH2AX foci and DSBs, we calculated that 350 O6MeG adducts are required for the induction of one DSB in LN229 cells. LN229 cells expressing MGMT were completely refractory to the induction of γH2AX foci, apoptosis, and senescence (Supplementary Fig. S6).
Figure 6.
Responses induced in LN229 cells by TMZ in dependence on the amount of O6MeG per cell: γH2AX (A), apoptosis (B), and senescent cells (C) in the population following treatment with increasing concentrations of TMZ. Data are from Fig. 1, ref. 34, and Supplementary Materials and Methods.
Discussion
TMZ, front-line drug in the treatment of high-grade malignant glioma, is effective if the tumor lacks MGMT or expresses the repair protein at a low level (<30 fmol/mg protein) (39). These tumors are defined as “methylated” because MGMT promoter CpG methylation correlates with silencing of the gene (40) and, as a consequence, lack of or low MGMT protein expression and enzyme activity (17). Although the curative effect is significant, the response is not dramatic and the prognosis of GBM is still bleak with a median survival of only 14.6 months (12.6 and 23.4 months in the MGMT-unmethylated and MGMT-methylated subgroups, respectively; ref. 41). Although recent phase III clinical trials revealed that the median overall survival for adult patients with newly diagnosed GBM can reached up to 20 months, indicating a trending increase in survival, the prognosis is still bad with 5-year overall survival rates of less than 10% (42). Given the mechanism of action of TMZ and the low curative response, some questions arise which are in the literature controversially discussed (27, 30) and thus need to be addressed experimentally. (i) Do GBM cells really die (by apoptosis or other forms of cell death) following TMZ treatment or does the drug just exert a cytostatic effect? (ii) Are the doses used for patient's treatment high enough to kill cancer cells in their body? This question implies that low doses are ineffective and that threshold doses do exist below which cytotoxicity does not occur. (iii) How many O6MeG adducts are required for eliciting a specific effect such as apoptosis, necrosis, and senescence? Here, we present data on the amount of O6MeG induced by TMZ in established GBM cell lines and set it in relation to the level of DSBs, apoptosis, necrosis, overall cell death, and cellular senescence.
Endpoints triggered by O6MeG
MGMT-lacking GBM cells respond to treatment with TMZ in the dose range up to 50 μmol/L with the induction of apoptosis, autophagy, and senescence. These effects were nearly completely vanished in isogenic MGMT expressing cells (Supplementary Fig. S6), supporting that O6MeG induced by TMZ is responsible for these endpoints (8). Because autophagy is a survival pathway and cellular senescence a cytostatic effect, it is pertinent to conclude that O6MeG triggers both survival and death pathways. The apoptosis pathways triggered by O6MeG are well described (25, 27); they involve activation of both the death receptor FAS/CD95/APO1 and the intrinsic mitochondrial pathway, dependent on the cellular background (43). Recently we showed, using LN229 cells, that apoptosis is a linear function of dose (34), which was substantiated in the present work with A172 cells. Using hockey-stick modeling (38), we show that there is no threshold for TMZ/O6MeG-induced apoptosis in LN229 cells, and a marginal threshold in A172 cells at a dose level of 2.5 μmol/L. In MGMT expressing cells the threshold is clearly higher, dependent on the MGMT expression level. Previous work with isogenic cell strains expressing different MGMT levels showed that the protection caused by the repair protein is a linear function of MGMT molecules per cell (4). Comparing the different endpoints triggered by O6MeG quantitatively, it is interesting to note that the yield of cell death by apoptosis (and necrosis, which represents a minor fraction) is significantly lower than senescence. Thus, we measured 120 hours after TMZ treatment in LN229 cells 25.4% apoptosis and 48.8% senescence, and in A172 cells 7.5% apoptosis and 33.3% senescence. Similar to apoptosis, senescence increased linearly with dose, without a no-effect threshold. Overall, O6MeG elicits both a cytotoxic and cytostatic effect by activating, in the same dose range, the apoptosis and the senescence pathway; the latter is the predominant trait. This occurs already at low dose levels (<50 μmol/L), which can be achieved in vivo in a therapeutic setting (44).
Is TMZ effective in killing cancer cells in vivo?
Treatment of patients with TMZ occurs daily by different schedules (2, 29, 45, 46). The serum half-life of TMZ is about 2 hours and the serum concentration of TMZ has been determined to be in the range of 20 to 70 μmol/L (1, 47–51). Thus, in a therapeutic setting with a single oral dose of 150 mg/m2, the peak plasma concentration was, on average, 28.4 μmol/L (5.5 μg/mL) and the brain interstitium concentration was 1.5 μmol/L (0.3 μg/mL; ref. 52). In another study, following oral 200 mg/m2 TMZ, the plasma peak level was 72 μmol/L, and the concentration in the cerebrospinal fluid was 9.9 μmol/L (53). Overall, the TMZ concentration at the target organ seems to be rather low (1–10 μmol/L), which may lead to the conclusion that the TMZ alkylation level is not high enough to exert a cytotoxic effect on GBM cells. Given the fact that p53 plays a supporting role in apoptosis and senescence triggered by O6MeG (31, 37), the supposition is nourished by the assumption that DNA damage activated p53 pathways involving p53ser15 and HIPK2/p53ser46 play a protective role at a low, and a proapoptotic role at a high genotoxin level, respectively (54). This model implicates that thresholds exist that regulate the balance between life and death (44). Therefore, we expected that cell death is not induced at low DNA damage levels. The view that TMZ is rather ineffective in glioma treatment is further fueled by the supposition that TMZ exerts only cytostatic, but not cytotoxic activity (30).
Here, using the well-characterized glioblastoma cell lines A172 and LN229, we show that the amount of O6MeG in the DNA and the number of DSBs increase linearly with dose of TMZ, and the same was observed for apoptosis and senescence. We did not observe a threshold for DSBs (as measured by γH2AX and 53BP1 foci formation), apoptosis, necrosis, and senescence (see also our previous report; ref. 34). This finding has implications regarding underlying mechanisms. It indicates that upon DDR activation by O6MeG-triggered secondary lesions (i.e., MMR-mediated gaps and DSBs resulting from them during replication) cells make a decision between survival and death in a stochastic manner and, at least in this cell system, there does not appear to be a threshold dose below which cell death pathways are not activated. Interestingly, at all dose levels we observed the induction of apoptosis and senescence, indicating that in the cell population cytotoxic and cytostatic effects are brought about at the same time by the same critical lesion O6MeG, although to a different extent. If we translate these findings to the tumor in vivo, we can posit that even at a low concentration TMZ elicits a cytotoxic (apoptosis and at very low degree necrosis) and a cytostatic (senescence) effect. It should also be considered the possibility that repeated low-dose treatment leads to an accumulation of O6MeG in MGMT-deficient tumor cells and an amelioration of the cytotoxic effects. This supposition is currently under study. Overall, the findings strongly support the view that repetitive treatments with low doses of TMZ (according to the metronomic schedule) are effective and represent a reasonable treatment strategy.
How many O6MeG adducts are required for inducing DSBs, p53ser15, apoptosis, necrosis, and senescence?
Here, we have quantified for the first time O6MeG adducts induced by TMZ in glioblastoma cells and set it in relation to the amount of DSBs, p53ser15, apoptosis, necrosis, and cellular senescence. From the corresponding dose–response curves, we calculated the amount of O6MeG adducts that are required for eliciting a specific effect. The results are compiled in Table 1. The data show that 48 and 21 O6MeG/107 nt give rise to 1 DSB (measured by a γH2AX focus) in A172 and LN229 cells, respectively. This corresponds to 490 and 350 O6MeG per cell that result in a DSB in A172 and LN229, respectively. The formation of one 53BP1 focus needs 1,460 O6MeG, and colocalized foci were observed at a level of 3,350 O6MeG in A172 cells. A twofold increase of phosphorylated p53ser15 needed 4,700 O6MeG, a twofold increase in p21 4,830 O6MeG, and an increase in apoptosis by one percent required an adduct level of 2,560 and 950 O6MeG in A172 and LN229, respectively. Necrosis was measured only in A172, and an increase by 1% was seen at 5,220 O6MeG. Overall, for inducing 1% cell death (apoptosis and necrosis) about 1,580 O6MeG adducts are needed. Senescence is triggered already by lower amounts of O6MeG, with 440 and 410 O6MeG adducts needed for increasing the senescence level by 1% in A172 and LN229 cells, respectively.
Table 1.
DSBs represented by γH2AX and 53BP1 foci, cell death, and cellular senescence induced in A172 and LN229 cells upon treatment with TMZ dependent of O6MeG adducts per cell.
| A172 | LN229 | |||
|---|---|---|---|---|
| Effect | O6MeG per cell | Effects triggered by 14,000 O6MeG per cell (20 μmol/L TMZ) | O6MeG per cell | Effects triggered by 20,600 O6MeG per cell (20 μmol/L TMZ) |
| γH2AX | 490 per focus | 32 foci | 350 per focus | 66 foci |
| 53BP1 | 1,460 per focus | 13 foci | nd | nd |
| Colocalization | 3,350 per focus | 4 foci | nd | nd |
| p53ser15 | 4,700 per 2-fold | 3-folda | nd | nd |
| p21 | 4,830 per 2-fold | 3-foldb | nd | nd |
| Apoptosis | 2,560 per 1% increase | 7.7% | 950 per 1% increase | 24.4% |
| Necrosis | 5,220 per 1% increase | 3.6% | nd | nd |
| Total cell death | 1,580 per 1% increase | 12.3% | nd | nd |
| Senescence | 440 per 1% increase | 35.4% | 410 per 1% increase | 52.4% |
In Table 1, we also show the effect level brought about by a dose of 20 μmol/L TMZ, which induces 14,000 and 20,600 adducts per cell in A172 and LN229, respectively. This dose induced 32 DSBs (γH2AX foci) per A172 cell, a 3-fold increase of p53ser15 (which was saturated already with a dose of 15 μmol/L TMZ), a 3-fold increase in p21 (saturated at a dose level of 10 μmol/L TMZ). Under these conditions, 12.3% of cells were dead and 35.4% of cells showed the senescence phenotype as measured 120 hours after the onset of treatment. Overall, this low-dose caused significant genotoxic and cytotoxic effects. It should be noted that long-term cultivation of TMZ-treated cells resulted in cell loss in the subsequent passages, which is explained by O6MeG triggered cell death in >2 posttreatment cell cycles, as demonstrated previously (10).
O6MeG is a most interesting DNA damage as it is not only cytotoxic, but also able to trigger genotoxic end points such as sister chromatid-exchanges (SCE), point mutations, chromosomal aberrations, and DNA single- and DSBs. In previous work, it was shown that O6-methylating agents require two cell cycles for SCEs to be formed, indicating secondary lesions to be involved (55). This was confirmed using MGMT-isogenic cell lines, demonstrating that critical events are happening in the posttreatment cell cycle (56). The conversion probability for O6MeG into SCEs was determined in two independent studies: in MGMT lacking CHO-9 cells it was shown that 30 O6MeG per cell give rise to 1 SCE (57) whereas for a glioblastoma cell line the relationship was 43 O6MeG/SCE (58). This is a very high conversion rate compared with other DNA lesions, for example, UV-induced pyrimidine dimers with 600 adducts/SCE (59). Regarding cytotoxicity, from the slope of survival curves (colony formation) it was estimated that 6,650 O6MeG induce one lethal hit (58). It is important to note that colony formation, which is frequently used as a highly sensitive indicator of cytotoxicity, is biased by the fact that nontoxic events such as cell-cycle inhibition and induced senescence reduce the size and number of colonies and therefore simulate cytotoxicity. For this reason, survival curves on the basis of colony formation can hardly be compared with dose–response curves based on apoptosis and necrosis, which are clear cell death indicators. Although the amount of O6MeG induced in A172 and LN229 cells was rather similar, the amount of DSBs and the levels of apoptosis and induced senescence were not identical, which is expected in view of the complex pathways involved, including the conversion of O6MeG into critical secondary lesions, triggering the DDR and activation of survival and death pathways.
Conclusions
O6MeG is a highly recombinogenic, mutagenic, genotoxic, autophagy, senescence, and apoptosis-inducing lesion (4, 5, 8, 25). It triggers with high efficiency SCE formation, which are cytogenetic events resulting from a damage tolerance process (60), followed by DSBs, senescence, and apoptosis. The linear dose–effect relationship for O6MeG and DSBs strongly indicates that in the therapeutic dose range, glioblastoma cells in vivo are damaged, causing responses that include cell death and cellular senescence, which appears to be the major trait. Because the O6MeG/MMR/DSB-triggered downstream survival and death pathways of senescence and apoptosis are activated at the same time in the cell population, complete elimination of cancer cells cannot be expected by this therapeutic strategy. The possibility of senescent cells being reactivated to go back into cycle and start to proliferate must also be considered. Furthermore, TMZ-induced senescent glioblastoma cells secrete proinflammatory cytokines (26), which may drive tumorigenesis. Therefore, the elimination of senescent cells by senolytic drugs should be a further goal to improve therapeutic efficacy, as opposed to purely increasing the dose of TMZ in target tumor tissue. Finally, it is reasonable to postulate that quantitative adduct formation could serve as a potential biomarker of response and/or a determinant of adequate TMZ intratumoral dose. Therefore, molecular dosimetry of O6MeG in relation to cellular effects is important for elucidating critical damage levels in cancer and normal tissues during and after therapy.
Authors' Disclosures
S.J. Sturla reports grants from Swiss National Science Foundation and Swiss Cancer Research Foundation during the conduct of the study. B. Kaina reports grants from Deutsche Forschungsgemeinschaft and Deutsche Krebshilfe during the conduct of the study. No disclosures were reported by the other authors.
Acknowledgments
The study was supported by Deutsche Forschungsgemeinschaft (DFG KA 724/31-1), German Cancer Aid, Swiss National Science Foundation (185020, 186332), and the Swiss Cancer Research Foundation (KFS-4443-02-2018-R).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is featured in Highlights of This Issue, p. 1755
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
B. Stratenwerth: Data curation, formal analysis, validation. S.M. Geisen: Conceptualization, data curation, formal analysis, validation, investigation, methodology. Y. He: Data curation. L. Beltzig: Data curation, validation, methodology. S.J. Sturla: Formal analysis, supervision, funding acquisition, validation, methodology. B. Kaina: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft.
References
- 1. Newlands E, Stevens M, Wedge S, Wheelhouse R, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev 1997;23:35–61. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 3. Villano JL, Seery TE, Bressler LR. Temozolomide in malignant gliomas: current use and future targets. Cancer Chemother Pharmacol 2009;64:647–55. [DOI] [PubMed] [Google Scholar]
- 4. Kaina B, Fritz G, Mitra S, Coquerelle T. Transfection and expression of human O6-methylguanine-DNA methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of MGMT in protection against the genotoxic effects of alkylating agents. Carcinogenesis 1991;12:1857–67. [DOI] [PubMed] [Google Scholar]
- 5. Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis 2002;17:483–7. [DOI] [PubMed] [Google Scholar]
- 6. Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutation Res 1990;231:11–30. [DOI] [PubMed] [Google Scholar]
- 7. Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair 2007;6:1079–99. [DOI] [PubMed] [Google Scholar]
- 8. Knizhnik AV, Roos WP, Nikolova T, Quiros S, Tomaszowski KH, Christmann M, et al. Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS One 2013;8:e55665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutation Res 2000;462:83–100. [DOI] [PubMed] [Google Scholar]
- 10. Quiros S, Roos WP, Kaina B. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle 2010;9:168–78. [DOI] [PubMed] [Google Scholar]
- 11. Christmann M, Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res 2013;41:8403–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christmann M, Kaina B. Epigenetic regulation of DNA repair genes and implications for tumor therapy. Mutat Res 2019;780:15–28. [DOI] [PubMed] [Google Scholar]
- 13. Margison GP, Povey AC, Kaina B, Santibanez Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis 2003;24:625–35. [DOI] [PubMed] [Google Scholar]
- 14. Goth R, Rajewsky MF. Persistence of O6-ethylguanine in rat-brain DNA: correlation with nervous system-specific carcinogenesis by ethylnitrosourea. Proc Natl Acad Sci U S A 1974;71:639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silber JR, Mueller BA, Ewers TG, Berger MS. Comparison of O6-methylguanine-DNA methyltransferase activity in brain tumors and adjacent normal brain. Cancer Res 1993;53:3416–20. [PubMed] [Google Scholar]
- 16. Preuss I, Haas S, Eichhorn U, Eberhagen I, Kaufmann M, Beck T, et al. Activity of the DNA repair protein O6-methylguanine-DNA methyltransferase in human tumor and corresponding normal tissue. Cancer Detect Prev 1996;20:130–6. [PubMed] [Google Scholar]
- 17. Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, et al. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer 2010;127:2106–18. [DOI] [PubMed] [Google Scholar]
- 18. Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350–4. [DOI] [PubMed] [Google Scholar]
- 19. Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997–1003. [DOI] [PubMed] [Google Scholar]
- 20. Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics 2016;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ensminger M, Iloff L, Ebel C, Nikolova T, Kaina B, Lbrich M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J Cell Biol 2014;206:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaina B, Christmann M. DNA repair in personalized brain cancer therapy with temozolomide and nitrosoureas. DNA Repair 2019;78:128–41. [DOI] [PubMed] [Google Scholar]
- 23. Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res 2000;60:5815–24. [PubMed] [Google Scholar]
- 24. Eich M, Roos WP, Nikolova T, Kaina B. Contribution of ATM and ATR to the resistance of glioblastoma and malignant melanoma cells to the methylating anticancer drug temozolomide. Mol Cancer Ther 2013;12:2529–40. [DOI] [PubMed] [Google Scholar]
- 25. He Y, Roos WP, Wu Q, Hofmann TG, Kaina B. The SIAH1-HIPK2-p53ser46 damage response pathway is involved in temozolomide-induced glioblastoma cell death. Mol Cancer Res 2019;17:1129–41. [DOI] [PubMed] [Google Scholar]
- 26. Aasland D, Gotzinger L, Hauck L, Berte N, Meyer J, Effenberger M, et al. Temozolomide induces senescence and repression of DNA repair pathways in glioblastoma cells via activation of ATR-CHK1, p21, and NF-kappaB. Cancer Res 2019;79:99–113. [DOI] [PubMed] [Google Scholar]
- 27. Kaina B. Temozolomide in glioblastoma therapy: role of apoptosis, senescence and autophagy. Comment on Strobel et al., temozolomide and other alkylating agents in glioblastoma therapy. Biomedicines 2019, 7, 69. Biomedicines 2019;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anton K, Baehring JM, Mayer T. Glioblastoma multiforme: overview of current treatment and future perspectives. Hematol Oncol Clin North Am 2012;26:825–53. [DOI] [PubMed] [Google Scholar]
- 29. Wick W, Winkler F. Regimen of procarbazine, lomustine, and vincristine versus temozolomide for gliomas. Cancer 2018;124:2674–6. [DOI] [PubMed] [Google Scholar]
- 30. Strobel H, Baisch T, Fitzel R, Schilberg K, Siegelin MD, Karpel-Massler G, et al. Temozolomide and other alkylating agents in glioblastoma therapy. Biomedicines 2019;7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roos WP, Batista LFZ, Naumann S, Wick W, Weller M, Menck CFM, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene 2007;26:186–97. [DOI] [PubMed] [Google Scholar]
- 32. Kraus A, McKeague M, Seiwert N, Nagel G, Geisen SM, Ziegler N, et al. Immunological and mass spectrometry-based approaches to determine thresholds of the mutagenic DNA adduct O(6)-methylguanine in vivo. Arch Toxicol 2019;93:559–72. [DOI] [PubMed] [Google Scholar]
- 33. Geisen SM, Aloisi CMN, Huber SM, Sandell ES, Escher NA, Sturla SJ. Direct alkylation of deoxyguanosine by azaserine leads to O(6)-carboxymethyldeoxyguanosine. Chem Res Toxicol 2021;34:1518–29. [DOI] [PubMed] [Google Scholar]
- 34. He Y, Kaina B. Are there thresholds in glioblastoma cell death responses triggered by temozolomide? Int J Mol Sci 2019;20:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hammond LA, Eckardt JR, Baker SD, Eckhardt SG, Dugan M, Forral K, et al. Phase I and pharmacokinetic study of temozolomide on a daily-for-5-days schedule in patients with advanced solid malignancies. J Clin Oncol 1999;17:2604–13. [DOI] [PubMed] [Google Scholar]
- 36. Pegg AE, Dolan ME, Moschel RC. Structure, function and inhibition of O6-alkylguanine-DNA-alkyltransferase. Prog Nucleic Acid Res Mol Biol 1995;51:167–223. [DOI] [PubMed] [Google Scholar]
- 37. Hermisson M, Klumpp A, Wick W, Wischhusen J, Nagel G, Roos W, et al. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem 2006;96:766–76. [DOI] [PubMed] [Google Scholar]
- 38. Lutz WK, Lutz RW. Statistical model to estimate a threshold dose and its confidence limits for the analysis of sublinear dose-response relationships, exemplified for mutagenicity data. Mutat Res 2009;678:118–22. [DOI] [PubMed] [Google Scholar]
- 39. Wiewrodt D, Nagel G, Dreimuller N, Hundsberger T, Perneczky A, Kaina B. MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome. Int J Cancer 2008;122:1391–9. [DOI] [PubMed] [Google Scholar]
- 40. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793–7. [PubMed] [Google Scholar]
- 41. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. [DOI] [PubMed] [Google Scholar]
- 42. Stepanenko AA, Chekhonin VP. Recent advances in oncolytic virotherapy and immunotherapy for glioblastoma: a glimmer of hope in the search for an effective therapy? Cancers 2018;10:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 2013;332:237–48. [DOI] [PubMed] [Google Scholar]
- 44. Oren M. Decision making by p53: life, death and cancer. Cell Death Differ 2003;10:431–42. [DOI] [PubMed] [Google Scholar]
- 45. Wick W, Steinbach JP, Kuker WM, Dichgans J, Bamberg M, Weller M. One week on/one week off: a novel active regimen of temozolomide for recurrent glioblastoma. Neurology 2004;62:2113–5. [DOI] [PubMed] [Google Scholar]
- 46. Strik HM, Marosi C, Kaina B, Neyns B. Temozolomide dosing regimens for glioma patients. Curr Neurol Neurosci Rep 2012;12:286–93. [DOI] [PubMed] [Google Scholar]
- 47. Weller M, Steinbach JP, Wick W. Temozolomide: a milestone in the pharmacotherapy of brain tumors. Future Oncol 2005;1:747–54. [DOI] [PubMed] [Google Scholar]
- 48. Batchelor T. Temozolomide for malignant brain tumours. Lancet 2000;355:1115–6. [DOI] [PubMed] [Google Scholar]
- 49. Danson SJ, Middleton MR. Temozolomide: a novel oral alkylating agent. Expert Rev Anticancer Ther 2001;1:13–9. [DOI] [PubMed] [Google Scholar]
- 50. Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res 2000;6:2585–97. [PubMed] [Google Scholar]
- 51. Marzolini C, Decosterd LA, Shen F, Gander M, Leyvraz S, Bauer J, et al. Pharmacokinetics of temozolomide in association with fotemustine in malignant melanoma and malignant glioma patients: comparison of oral, intravenous, and hepatic intra-arterial administration. Cancer Chemother Pharmacol 1998;42:433–40. [DOI] [PubMed] [Google Scholar]
- 52. Portnow J, Badie B, Chen M, Liu A, Blanchard S, Synold TW. The neuropharmacokinetics of temozolomide in patients with resectable brain tumors: potential implications for the current approach to chemoradiation. Clin Cancer Res 2009;15:7092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ostermann S, Csajka C, Buclin T, Leyvraz S, Lejeune F, Decosterd LA, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res 2004;10:3728–36. [DOI] [PubMed] [Google Scholar]
- 54. Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 2016;16:20–33. [DOI] [PubMed] [Google Scholar]
- 55. Kaina B, Aurich O. Dependency of the yield of sister-chromatid exchanges induced by alkylating agents on fixation time. Possible involvement of secondary lesions in sister-chromatid exchange induction. Mutation Res 1985;149:451–61. [DOI] [PubMed] [Google Scholar]
- 56. Kaina B, Ziouta A, Ochs K, Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutation Res 1997;381:227–41. [DOI] [PubMed] [Google Scholar]
- 57. Kaina B, Fritz G, Coquerelle T. Contribution of O6-alkylguanine and N-alkylpurines to the formation of sister chromatid exchanges, chromosomal aberrations, and gene mutations: new insights gained from studies of genetically engineered mammalian cell lines. Environ Mol Mutagen 1993;22:283–92. [DOI] [PubMed] [Google Scholar]
- 58. Rasouli-Nia A, Sibghat U, Mirzayans R, Paterson MC, Day RS. 3rd. On the quantitative relationship between O6-methylguanine residues in genomic DNA and production of sister-chromatid exchanges, mutations and lethal events in a Mer- human tumor cell line. Mutat Res 1994;314:99–113. [DOI] [PubMed] [Google Scholar]
- 59. De Weerd-Kastelein EA, Keijzer W, Rainaldi G, Bootsma D. Induction of sister chromatid exchanges in xeroderma pigmentosum cells after exposure to ultraviolet light. Mutat Res 1977;45:253–61. [DOI] [PubMed] [Google Scholar]
- 60. Kaina B. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet Genome Res 2004;104:77–86. [DOI] [PubMed] [Google Scholar]